Abstract
Nuclear factor 90 (NF90) was originally isolated in a complex that binds to the antigen recognition response element (ARRE-2) present in the interleukin-2 promoter. To characterize the transcriptional properties of NF90 in mammalian cells, we examined its ability to modulate promoter function in cellular transfection assays. NF90-Gal4 fusion proteins inhibited transcription from the adenovirus major late promoter in a fashion that was dependent on Gal4 targeting. Conversely, NF90 activated the cytomegalovirus immediate-early promoter, to which it was not targeted. These effects required distinct but overlapping domains in the C terminus of NF90, which contains a functional nuclear localization signal and two double-stranded-RNA binding motifs. NF90 is present in cellular complexes together with the NF45 protein. Transfection assays showed that NF45 binds NF90 strongly and stimulates its ability to activate but not to inhibit gene expression. This report characterizes NF90 as both a positive and negative regulator of gene expression, depending on the promoter context, and suggests a role for NF45 as a regulator of NF90.
Double-stranded-RNA (dsRNA) binding proteins play critical roles in several aspects of cellular metabolism, including transcriptional activation, translational control, and mRNA processing and localization. Most of these proteins contain a sequence referred to as a dsRNA binding motif (dsRBM) (10). This motif is well conserved through evolution and interacts with dsRNAs as well as structured RNAs such as the adenovirus 2VA RNAs (24). dsRBMs also mediate protein-protein interactions in the absence of dsRNA (39). Despite extensive characterization of the dsRBM, the functions of several of the dsRBM-containing proteins still remain to be elucidated.
Nuclear factor 90 (NF90) is a member of an expanding family of dsRNA binding proteins that have significant homology in their N terminus but differ significantly in their C-terminal regions. These homologues include DRBP76, TCP80, ILF3, and MPP4 (Homo sapiens); SPNR and ILF3 (mouse); p74 and ILF3 (rat); and 4F.1 and 4F.2 (Xenopus laevis) (2, 4, 7, 23, 31, 36, 37, 41). Recently, it was determined that several of the human proteins are derived from a single locus on chromosome 19 and are generated by alternate splicing (9). NF90 was also isolated in a Northwestern blotting assay for proteins that preferentially bound VA RNAII (19).
NF90 was originally purified as a component of a complex with nuclear factor 45 (NF45), which binds to the antigen response recognition element (ARRE-2) present in the promoter region of the interleukin-2 (IL-2) gene. Immunodepletion experiments with NF90- and NF45-specific antibodies suggested that these proteins are required for activated transcription from the IL-2 promoter in an in vitro transcription system (8, 14). Examination of the protein sequence of NF90 revealed no sequence-specific double-stranded DNA binding motifs characteristic of many transcription factors. However, two dsRBMs were found in the C-terminal region of the protein. Further characterization of these motifs showed that they preferentially bound dsRNA (17). The role of the NF90/NF45 complex in regulating transcription from genes under control of the ARRE, a key regulator of genes involved in the activation of T cells and immune response, has not been defined (33).
NF90 interacts with several additional proteins either directly or in larger complexes. NF90 has been found in a complex containing NF45, the double-stranded DNA-dependent protein kinase, the translation initiation complex eIF2, and the DNA-binding proteins Ku 70 and 80 (40). NF90, DRBP76, and p74 have all been shown to interact with and act as substrates for the dsRNA-activated protein kinase PKR (7, 17, 30, 31). The rat ILF3 was identified in a yeast two-hybrid screen as an interacting protein using protein arginine methyl transferase 1 (PRMT1) as the bait (37). In these experiments, ILF3 was shown to act as a substrate for PRMT1 and to enhance the activity of PRMT in an in vitro methylation assay. The Xenopus homologues, 4F.1/CBTF122 (CAATT box transcription factor, 122 kDa) and 4F.2/CBTF98, were recently identified as components of the CAATT-box transcription factor complex that binds to the CAATT regulatory element present in the GATA-2 promoter after the mid-blastula transition. In Xenopus, this protein complex was sequestered in the cytoplasm and bound to the large group of untranslated maternal mRNAs. Following degradation of RNA in the cytoplasm, this protein complex was translocated to the nucleus. Mutations in the dsRBM of CBTF122 that abolished dsRNA-binding activity resulted in premature nuclear localization (3, 29).
Although several of these observations implicate NF90 in transcriptional roles, its activities remain poorly defined. In this paper, we examine the ability of NF90 to regulate transcription from several reporter genes transfected into mammalian cells. We found that NF90 inhibited transcription when targeted to the adenovirus major late 1 promoter using Gal4 binding domain (BD) fusions. Conversely, NF90 activated transcription from a cytomegalovirus (CMV) reporter independent of promoter targeting. Both of these activities were enhanced by removal of the protein’s N terminus and require, to various extents, NF90’s functional nuclear localization signal and dsRBMs. The activation but not the inhibitory function of NF90 was enhanced by its heteromeric partner NF45. This work establishes a novel role for the NF90/NF45 complex in the regulation of transcription and leads to a model in which NF90’s functional domains are revealed by a conformational change induced by its association with its regulatory factor NF45.
MATERIALS AND METHODS
Plasmid constructs.
pcDNA3.1-NF90 (pNF90) was generated by digesting pGEX4T-2-NF90 (31) with EcoRI and NotI. The resulting insert was then ligated into pcDNA3.1(+) (Invitrogen Corp., Carlsbad, Calif.) cut with the same restriction enzymes. To create NF90Δ336N, pNF90 was digested with EcoRI and EcoRV, blunt-ended with DNA polymerase I Klenow fragment, and then religated to remove the intervening sequences. pcDNA3.1-NF90Δ403N was generated by PCR using primers F (5′-GCGCGAATTCCAGGCTATGAATGCCCTGAT-3′) and R (5′-GCGCGGATCCCGGCAAGCCCATGTCGTGTA-3′). The resulting product was digested with EcoRI and BamHI and ligated into pcDNA3.1(−). pcDNA3-NF45 (pNF45) was created by digesting pBSK-NF45 with EcoRI and then ligating into pcDNA3. All plasmids were sequenced for verification.
To create plasmids for immunofluorescence studies, two different enhanced green fluorescent protein (EGFP) inserts were generated by PCR. EGFP was created using the primers EGFP (F) (5′-GCGCGCTAGCATGGGATCCGTGAGCAAG-3′) and EGFP (R) (5′-GCGCGGTACCGCGGCCGCGAATTCCTTGTACAGCTC-3′). EGFP(+2), which contained a 2-base frameshift after the EGFP coding sequence, was created using primers EGFP (F) and EGFP (R + 2) (5′-GCGCGGTACCGCGGCCGCATGAATTCCTTGTACAGCTC-3′). Both PCRs used pCRUZ-GFP (Santa Cruz Biotechnology Inc., Santa Cruz, Calif.) as a template.
For pEGFP, EGFP was cut with NheI and NotI and ligated into pcDNA3.1(−) digested with the same enzymes. For the remaining vectors, EGFP and EGFP(+2) were digested with NheI and KpnI and ligated into pcDNA3.1-NF90 or pcDNA3.1-NF90Δ336N and pcDNA3.1-NF90Δ403N, respectively. All plasmids were sequenced for verification.
pGL3-G5-ML1-F.luc was created by removing the insert from pTOG5ML1 (containing the adenovirus major late promoter −51 to +62 with five Gal4 binding sites upstream; gift from C.-G. Lee) by digestion with HindIII, blunt-ending with Klenow fragment, and digestion with BamHI. This insert was ligated into pGL3-Basic (Promega Corp., Madison, Wis.) which had been digested with SmaI and BglII. The resulting plasmid was sequenced for verification. pCMV-RL (pCMV-R.luc), pSP-luc+, pSP-rluc, and pRSV-βgal were purchased from Promega. p5′VA and pSG were described previously (12, 13).
To create the NF90-BD fusion protein expression plasmid (pNF90-BD), the Gal4 BD was PCR amplified from the pGBT9 vector (Clontech Laboratories Inc., Palo Alto, Calif.) using oligonucleotides F (5′GCGCGAATTCGCCCCATGCACAACGAAGTGC-3′) and R (5′-GCGCGGATCCTCACGGCGATACAGTCAACTGTCTTTGACC-3′). The PCR product was then digested with NcoI and BamHI. NF90 was then excised from the pGEX4T-3-NF90 vector using EcoRI and NcoI. The two fragments were then ligated into pcDNA3.1(−) cut with EcoRI and BamHI. To create pcDNA3.1-NF90Δ336N-BD (pNF90Δ336N), pNF90-BD was cut with EcoRV and religated to remove the intervening sequence. To create all the other mutants, NF90-BD was removed from pNF90 using EcoRI and BamHI and ligated into the pRSETA vector in the reverse orientation to make pRS-NF90-BD(as).
To create the NF90Δ150C-BD mutant, pRS-NF90-BD was cut with SalI and NcoI, blunt-ended with Klenow enzyme, and religated. To create NF90Δ313C-BD, pRS-NF90-BD was cut with EcoRV and NcoI, blunt-ended with Klenow enzyme, and religated. To create NF90ΔD1-2-BD, pRS-NF90-BD was cut with SacI and religated to remove the intervening sequence. Once the mutations were constructed in the pRSETA vector, mutants were removed with EcoRI and BamHI and then religated into pcDNA3.1(−) cut with the same restriction enzymes.
To create NF90Δ403N, the PCR product generated for NF90Δ403N as described above was digested with EcoRI and NcoI and religated together with the BD (digested as described above) into pcDNA3.1(EcoRI and BamHI). NF90Δ418C was generated by PCR using primers F (5′-GCGCGGATCCGAATTCGTAAAAATGCGTCCAATG-3′) and R (5′-GCGCCCATGGACTGCAGCCCTGGCTTCAGCTG-3′). The resulting PCR product was cut with EcoRI and NcoI. This insert plus the BD digested as described above were then ligated into pcDNA3.1(−) cut with EcoRI and BamHI.
To create pNF90Δ4, pNF90-BD was cut with HindIII and BamHI. This fragment was then ligated into pcDNA3.1(+) digested with HindIII and BamHI. To create pNF90ΔM, pNF90-BD was cut with SalI, blunt-ended with Klenow fragment, and then digested with NotI. The resulting fragment was ligated into pNF90Δ403N digested with NotI and EcoRV. pNF90ΔPE was created by digesting NF90Δ418C with EcoRV and religating the plasmid to remove the intervening sequence. The insert for NF90Δ356N was created using PCR and the following oligonucleotides: NF90Δ356N (F) (5′-GCGCGAATTCATGAGCACCACCTATGCCATT-3′) and NF90 (R) (5′GCGCGGATCCTCATGTAGCCTCCATGG-3′). The resulting PCR fragment was digested with EcoRI and NotI and then ligated along with the digested BD into pcDNA3.1(−) cut with EcoRI and BamHI. NF90Δ336N(−D2) was also generated using PCR and the following primers: NF90Δ336N (F) (5′-GCGCGAATTCATGCCTCTGCCTTCC-3′) and NF90 (R2) (5′-GCGCGGATCCCCATGGTCTTGCCGTGCTTTGTCAG-3′). The resulting PCR fragment was digested with EcoRI and NotI and then ligated along with the digested BD into pcDNA3.1(−) cut with EcoRI and BamHI. All plasmids were confirmed by sequencing and tested for expression in mammalian cells by Western blot with an anti-Gal4BD monoclonal antibody (Santa Cruz Biotechnology Inc.).
Transfections and reporter assays.
Human 293-T cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Life Technologies Inc., Rockville, Md.) supplemented with 10% fetal bovine serum (FBS; Sigma Chemical Co., St. Louis, Mo.) and 1% penicillin-streptomycin (P/S; Life Technologies Inc.). Prior to transfection, cells were seeded in 12-well plates approximately 12 h prior to transfection. When the cells reached ≈80% confluency, they were transfected using the Quantum Prep Cytofectene reagent (Bio-Rad, Hercules, Calif.).
Unless otherwise stated, 300 ng of pGL3-G5-ML1-luc, 10 ng of pCMV-R.Luc, 200 ng of pBD or pNF90-BD (or mutant), and 400 ng of empty pcDNA3.1 vector were diluted to a final volume of 50 μl with serum-free medium. Then 4 μl of Cytofectene was added, and the mixture was incubated at room temperature for 20 min. Following addition of 450 μl of medium (plus FBS) to the mixture, the medium on the plate was removed and replaced with the entire transfection cocktail. After 24 h, cells were washed once with ice-cold phosphate-buffered saline (PBS) and then lysed in 150 μl of reporter lysis buffer (100 mM potassium phosphate [pH 7.8], 0.2% Triton X-100) for 30 min. Lysate (20 μl) was then analyzed for luciferase reporter activities using the dual luciferase reporter assay system (Promega Corp.) as described by the manufacturer. Luciferase activities were in the range of 27 × 103 ± 12 × 103 units/4 × 104 cells (firefly) and 9 × 103 ± 3 × 103 units/4 × 104 cells (Renilla).
For β-galactosidase activity, cell lysate was first diluted 1:20 in reporter lysis buffer, and 20 μl of the diluted lysate was then analyzed using the Galactolight reporter assay (Tropix Inc., Bedford, Mass.). Statistical analysis was carried out on data from all reporter assays using a two-tailed Student’s t test, and P values of <0.05 are indicated in the figure legends, but more significant values (<0.01) were frequently obtained.
RNase protection assay.
pSP-luc+, pSP-rluc, and p5′VA were linearized with HincII, BsaAI, and XbaI, respectively. Radioactively labeled RNA was then generated by transcription of digested plasmids with T7 RNA polymerase in the presence of [α-32P]UTP (ICN Pharmaceuticals Inc., Costa Mesa, Calif.). Each reaction was incubated for 1 h at 37°C. DNA templates were then removed by DNase I digestion, and the RNA probes were purified by gel electrophoresis. The resulting firefly, Renilla, and 5′VA probes were 390 nucleotides (nt), 245 nt, and 120 nt, respectively.
For the RNase protection assay, 293-T cells were grown in 60-mm plates and transfected as described above except that the transfection was scaled up appropriately. After 24 h, the cells were lysed using Rnazol B (Tel-Test, Inc., Friendswood, Tex.). Reporter RNA levels were then determined using the RPAIII kit (Ambion Inc., Austin, Tex.).
Immunoprecipitation and Western blot analysis.
Anti-Gal4BD antibodies (100 μg; Santa Cruz Biotechnology Inc.) were covalently attached to 100 μl of packed protein A-Sepharose beads using dimethylpimelidate (Pierce Chemical Co., Rockford, Ill.). The antibody-Sepharose conjugate was stored as a 50% slurry in 1× PBS-0.01% sodium azide at 4°C. For immunoprecipitation, 293-T cells were transfected as described above. At 24 h, cells were washed twice with ice-cold PBS and lysed in 600 μl of immunoprecipitation (IP) buffer (20 mM HEPES [pH 7.6], 150 mM NaCl, 0.2% Triton X-100, 20% glycerol, 1 mM dithiothreitol, 0.2 mM EDTA, 50 mM NaF, 15 mM β-glycerophosphate, plus 0.2 μg of aprotinin, 0.2 μg of leupeptin, and 0.1 μg of pepstatin A per ml) for 30 min at 4°C with gentle rocking. Lysates were cleared by centrifugation at 14,000 × g for 30 min. Then 5 μl of packed BD antibody-Sepharose (see above) was added to 250 μl of cell lysate and incubated for ≈3 h at 4°C with agitation. Beads were washed four times with lysis buffer and then boiled in 20 μl of 2× Laemmli buffer for 5 min. Samples were then separated in a 10% polyacrylamide-sodium dodecyl sulfate (SDS) gel and transferred to a 0.2-μm nitrocellulose membrane using a semidry transfer apparatus. Membranes were incubated with Blotto (5% milk, 1× Tris-buffered saline[TBS], 0.1% Tween 20) overnight. Membranes were then blotted with anti-NF45 antibody (1:3,000) in Blotto for 3 h, washed with TBS-T (1× TBS, 0.1% Tween 20), and incubated with anti-rabbit immunoglobulin secondary antibody (1:5,000; Santa Cruz Biotechnology Inc.) for 1 h. After extensive washing with TBS-T, blots were developed with enhanced chemiluminescence. Membranes were then stripped using the membrane regeneration kit (Chemicon International Inc., Temecula, Calif.). Membranes were then reblotted with BD antibody (1:1,000; Santa Cruz Biotechnology Inc.), using a similar procedure.
Immunofluorescence.
NIH 3T3 cells cultured in DMEM-10% FBS-1% P/S were plated at a low density in a six-well dish on polylysine-treated glass cover slips prior to transfection. At 50% confluency, cells were transfected with 1 μg of plasmid DNA as described above. The Cytofectene-DNA mixture was removed 4 h after transfection and replaced with fresh medium. At 24 h posttransfection, the cells were fixed for 30 min in 3% paraformaldehyde at room temperature, permeabilized with 0.2% Triton X-100 for 5 min at room temperature, and then stained with DAPI (4′,6′-diamidino-2-phenylindole; 6.25 μg/ml; Sigma Chemical Co.) solution and a 1:200 dilution of phalloidin-rhodamine dye (300 U/ml; Molecular Probes Inc., Eugene, Oreg.) for 5 min at room temperature. Between each step, cells were washed several times with PBS. After staining, cells were mounted with 30% glycerol-PBS, and images were obtained using a Nikon TE-300 inverted fluorescence microscope.
RESULTS
NF90 has a functional NLS.
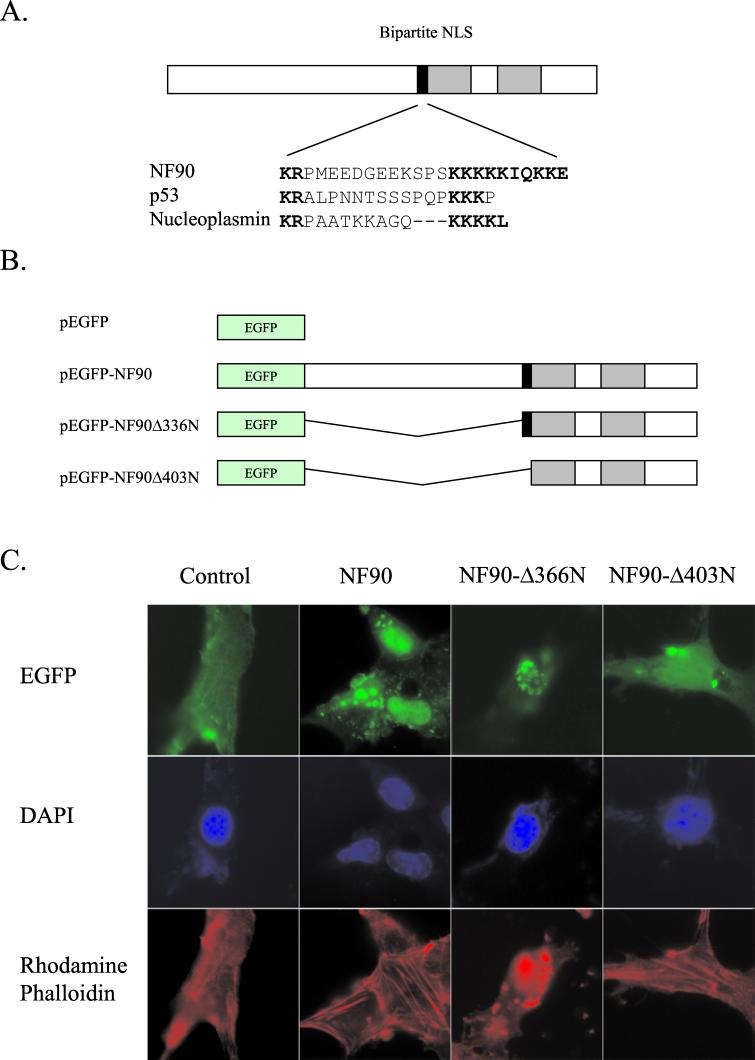
As its name implies, NF90 is detected predominantly in the nucleus by immunofluorescence of fixed cells (1, 31), although it is dispersed throughout the cell during mitosis (23) and substantial concentrations are present in cytosolic fractions after disruption (19). Inspection of the NF90 sequence revealed a potential bipartite nuclear localization signal (NLS) (18, 34) in the middle of the protein, immediately adjacent to the dsRBMs (Fig. 1A).
FIG. 1.
NF90 has an NLS. (A) Comparison of the putative bipartite NLS in NF90 with established NLS sequences in p53 and nucleoplasmin. (B) Schematic diagram of plasmid constructs used for NF90 subcellular localization studies. (C) Localization of EGFP-NF90 and EGFP-NF90 mutants in mouse NIH 3T3 cells. Cells were transfected with the indicated plasmids, fixed after 24 h, and then stained with DAPI and rhodamine-phalloidin. Slides were viewed with an inverted microscope.
To discover whether this sequence plays a role in determining the subcellular localization of NF90 in living cells, we created a set of plasmids containing fusions of the EGFP gene to intact and deletions of the NF90 cDNA (Fig. 1B) and introduced these constructs into mouse NIH 3T3 cells. Whereas the EGFP protein expressed on its own was evenly distributed throughout the cell, the NF90-EGFP fusion protein was largely restricted to the nuclei, as indicated by colocalization with the DAPI stain (Fig. 1C). The nuclear localization was not changed by removal of 336 amino acids from the N terminus of NF90 (Δ336N-EGFP), but removal of a further 67 amino acids, including the presumptive NLS (Δ403N-EGFP), caused a redistribution. This fusion protein was distributed throughout the cell, although it retained concentration within the nucleus. Similar results were obtained with 293-T cells (data not shown). These data demonstrate that there is a strong NLS just upstream of the first dsRBM and suggest that a second, weaker NLS may be present in the protein’s C-terminal half, possibly within the dsRBM region.
NF90 regulates reporter expression in a transient-expression assay.
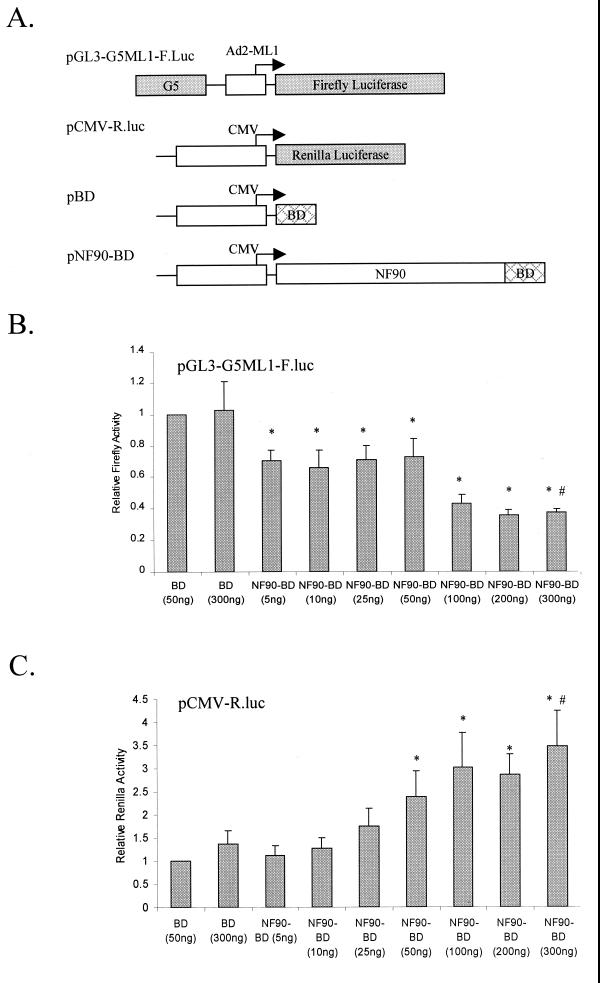
To explore the transcriptional properties of NF90 in human cells, we constructed a mammalian expression vector that generates full-length NF90 as a fusion protein carrying the S. cerevisiae Gal4 BD (amino acids 1 to 127) at its C terminus. This expression vector was transfected into human 293-T cells together with vectors encoding reporter genes (Fig. 2A). The first reporter construct, pGL3-G5-ML1-F.luc, contains five Gal4 binding sites upstream of the adenovirus major late 1 (ML1) minimal promoter, which controls the firefly luciferase gene. In transfected cells, the NF90-BD fusion protein is targeted to the ML1 minimal promoter via the Gal4 binding sites present in the plasmid construct. This approach allows us to target full-length NF90 as well as truncations of the protein to the promoter without knowledge of the protein’s DNA-binding region or the presence of a DNA-binding site for the protein in the promoter plasmid. Fusion to the Gal4 BD did not affect the ability of NF90 to interact with NF45 (see Fig. 7A) or with other known interacting proteins, such as PKR, and with dsRNA (data not shown). A second reporter construct, pCMV-R.luc, containing the Renilla luciferase gene under the control of the human CMV immediate-early promoter, was also included in the transfection assays.
FIG. 2.
Dose-dependent effects of NF90 on reporter gene expression. (A) Schematic representation of plasmids used in transfection assays. Human 293-T cells in 12-well plates were transfected with increasing concentrations of NF90-BD plasmid together with 300 ng of pGL3-G5ML1-F.luc, 10 ng of CMV-R.luc, and 600 ng of total pcDNA3.1 plasmid. After 24 h, cell extracts were assayed for (B) firefly luciferase activity (from pGL3-G5ML1-F.luc) and (C) Renilla luciferase activity (from pCMV-R.luc). Luciferase expression is displayed relative to that observed with BD (50 ng) and is normalized to total protein in the extracts. The data represent results of at least three independent transfections performed in duplicate, with the error bars representing standard deviation. Asterisks (*) indicate P < 0.05 for indicated samples versus BD (50 ng) alone. The pound symbols (#) indicate P < 0.05 for NF90-BD (300 ng) versus BD (300 ng). In panel B, the Gal4-BD construct gave a luciferase expression level 1.2-fold higher than that with empty vector.
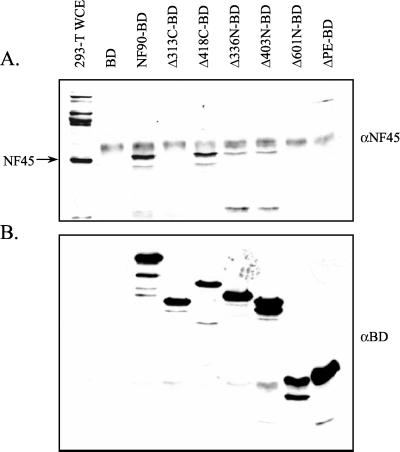
FIG. 7.
Complex formation between NF90 mutants and NF45. Human 293-T cells in 60-mm plates were transfected with 1.2 μg of pUC19, 800 ng of the indicated NF90-BD plasmid, and 1.6 μg of empty pcDNA3.1 plasmid. Following transfection, cells were lysed, and equivalent amounts of cell extracts were subjected to immunoprecipitation with anti-BD antibody-Sepharose for 3 h. Complexes were resolved in a 10% polyacrylamide-SDS gel and transferred to a nitrocellulose membrane. Western blot analysis was conducted (A) with anti-NF45 antibody (αNF45) and, after stripping, (B) with anti-BD antibody (αBD). The BD protein fragment (≈15 kDa) was too small to be resolved in the 10% polyacrylamide gel used.
The results of reporter activity assays showed that NF90 had differential effects on reporter expression. Targeting of NF90 to the adenovirus ML1 promoter resulted in a dose-dependent inhibition of reporter expression (Fig. 2B). On the other hand, NF90 stimulated the activity of the CMV reporter in a dose-dependent manner (Fig. 2C). Since both effects appeared to reach a plateau at the same plasmid concentration (200 ng/plate), this concentration was chosen for all subsequent assays.
Effector domain of NF90 maps to its C-terminal region.
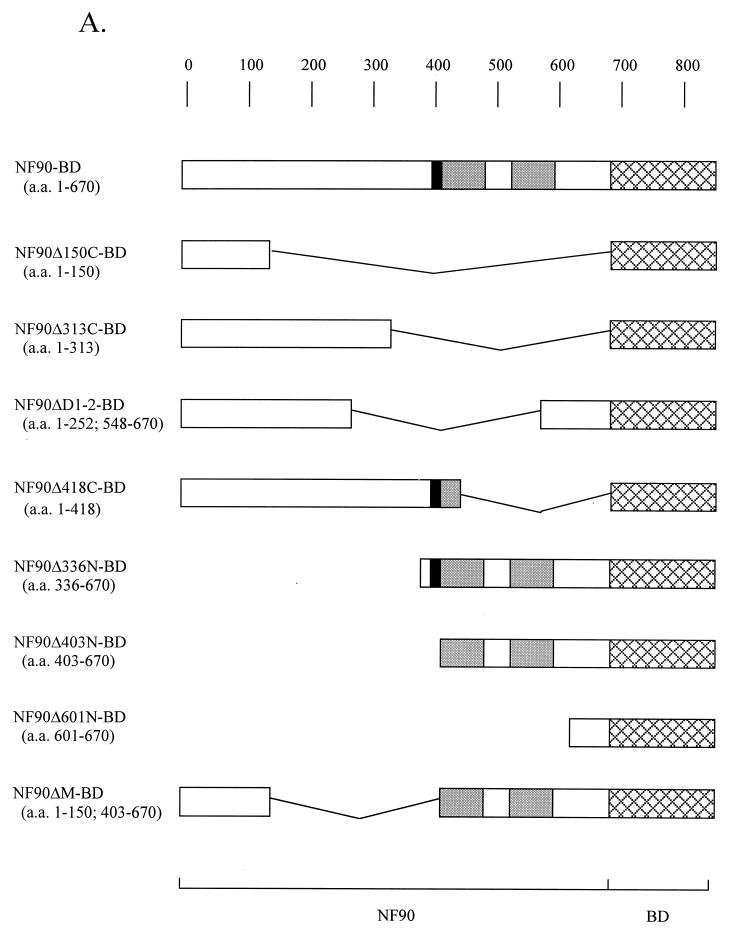
To define the regions of the NF90-BD fusion protein required for both the inhibition and stimulation of reporter expression, a series of truncation and deletion mutants were created in the form of Gal4 BD fusions (Fig. 3A). These constructs were compared to the intact fusion construct (NF90-BD) and the Gal4 BD alone (BD) in 293-T cell cotransfection assays with expression vectors driven by the G5ML1 or CMV promoter. Western blotting showed that the fusion proteins were expressed at comparable levels and that the accumulation of endogenous NF90 did not change when NF90 fusion proteins were expressed (data not shown; see Fig. 7B).
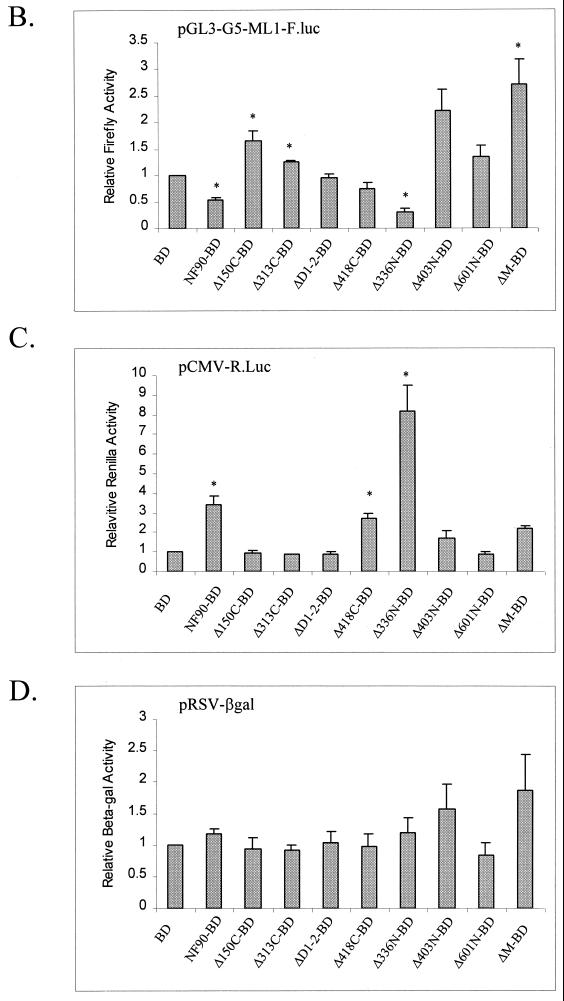
FIG. 3.
Identification of effector domain in NF90. (A) Schematic depiction of NF90-BD mutants. Shaded boxes represent the dsRBMs, black boxes represent the NLS, and cross-hatched boxes represent the BD. Human 293-T cells in 12-well plates were transfected with 300 ng of pG5-ML1-F.luc, 10 ng of CMV-R.luc, 50 ng of RSV-βgal, 200 ng of expression plasmid [pBD, pNF90-BD, or pNF90(mut)-BD], and 400 ng of pcDNA3.1 plasmid. At 24 h posttransfection, cell extracts were assayed for (B) firefly luciferase, (C) Renilla luciferase, and (D) β-galactosidase activities. Reporter gene expression is presented as in Fig. 2. Asterisks (*) indicate P < 0.05 for indicated samples versus BD alone.
The inhibitory response of the G5ML1 promoter required the intact C-terminal region of NF90 and was seen only with Δ336N-BD (Fig. 3B). In fact, this fusion protein, which contains the NLS and dsRBMs, was reproducibly more inhibitory than the intact parent chimera NF90-BD. Conversely, constructs containing the intact dsRBM region without the NLS (Δ403N-BD and ΔM-BD) elicited a two- to threefold stimulation. This stimulation appears to be a posttranscriptional effect, since RNase protection assays failed to reveal an accompanying increase in RNA expression (data not shown). It is likely due to inhibition of PKR activation by cytoplasmically located dsRBMs, as shown previously (39). The remaining constructs had little effect in comparison with BD, although the N-terminal region was slightly stimulatory alone (Δ150C-BD) and in combination with the dsRBMs (compare Δ403N-BD with ΔM-BD). As expected, removal of the Gal 4 binding sites from the ML1 reporter abolished the repressive effect of NF90Δ336N-BD on the vector (data not shown). Similarly, the repressive effect disappeared when the BD fusion was removed from NF90Δ336N, regardless of the presence or absence of the Gal4 binding sites. Therefore, targeting of NF90 to the adenovirus ML1 promoter is essential for the protein to carry out its repressive function.
FIG. 3.
Continued.
Modulation of the CMV promoter also mapped to the C-terminal region of the NF90 protein (Fig. 3C). Stimulation of CMV-Renilla luciferase expression was elicited only by constructs that contained either the NLS (Δ418C-BD) or the dsRBM region (Δ403N-BD and ΔM-BD). When both the NLS and dsRBMs were present, in Δ336N-BD, the stimulation was strikingly amplified to over eightfold, compared with threefold for NF90-BD. NF90 also stimulates the expression of CMV-β-galactosidase (data not shown), indicating that the effect is not reporter gene specific. Furthermore, the stimulatory effect is not cell type specific, since similar responses were seen in HeLa and Cos7 cells irrespective of the presence of the Gal4 BD fusion to NF90 (data not shown). On the other hand, the NF90 fusion proteins had little effect on β-galactosidase expression from the Rous sarcoma virus (RSV) promoter (Fig. 3D), confirming that the actions of NF90 are promoter specific.
Roles of NLS and dsRBMs in gene expression.
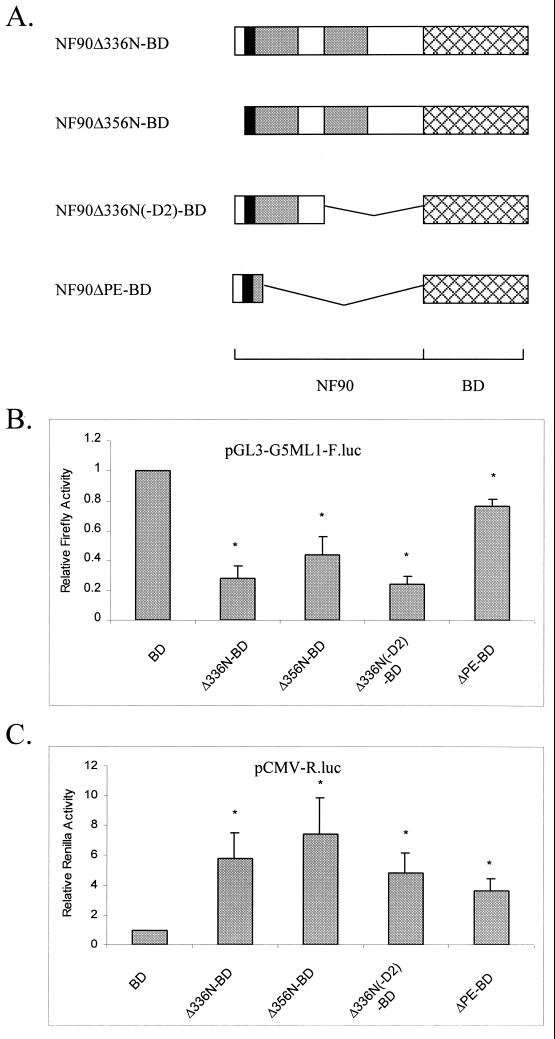
The data in Fig. 3 imply that the NLS region is sufficient to activate expression from the CMV promoter, although this stimulatory effect is enhanced by the dsRBMs. On the other hand, the inhibition of expression driven by the pG5-ML1 promoter requires both the NLS and dsRBM regions of NF90. To confirm and extend these conclusions, additional mutations were created in the NF90Δ336N-BD mutant, which showed the strongest activity in the reporter assay (Fig. 4A). These mutants were transfected into cells and assayed for their ability to activate or repress gene expression from cotransfected reporters (Fig. 4B and C).
FIG. 4.
Detailed mapping of NF90 effector domain. (A) Schematic representation of the NF90-BD mutants as in Fig. 3A. Human 293-T cells in 12-well plates were transfected as described for Fig. 3. After 24 h, cell extracts were assayed for (B) firefly luciferase and (C) Renilla luciferase activities as in Fig. 2. Asterisks (*) indicate P < 0.05 for indicated samples versus BD alone.
Consistent with the above results, the NLS region alone (ΔPE-BD) did not substantially repress G5-ML1 reporter activity. However, the mutant NF90Δ336N(−D2)-BD, which contains the NLS region in combination with only one dsRBM, was as effective as NF90Δ336N-BD (Fig. 4B). Another construct, NF90Δ356N-BD, which lacks the N-terminal 20 amino acids of NF90Δ336N-BD, leaving the NLS intact, was also an effective repressor. Thus, repression of the pG5-ML1 promoter requires the NLS region plus a single dsRBM.
The NLS region alone (ΔPE-BD) stimulated the CMV reporter to an extent similar to that seen with the C-terminal truncation mutant NF90Δ418C-BD (Fig. 3) but less than that given by NF90Δ336N-BD (Fig. 4C). Removal of the N-terminal 20 amino acids or the C-terminal dsRBM [NF90Δ356N-BD and NF90Δ336N(−D2)-BD, respectively] did not significantly diminish the stimulatory response toward NF90Δ336N-BD. Thus, activation requires the NLS and is enhanced by the presence of one or more dsRBMs. Evidently the activation and repression activities of NF90 are mediated by overlapping but distinct regions of NF90.
NF90 acts at the level of transcription.
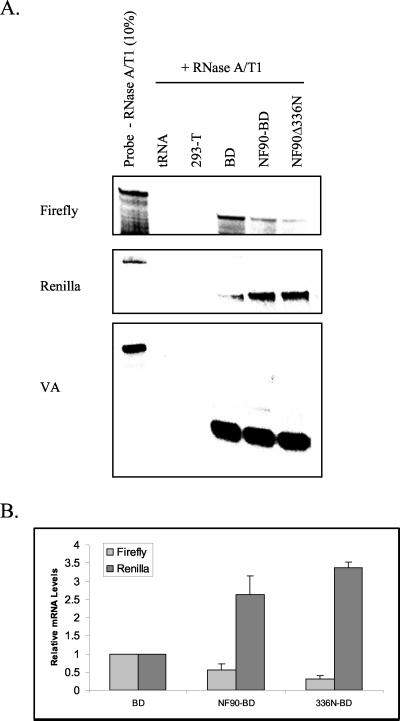
We conducted RNase protection assays to determine whether NF90 regulates gene expression at the level of transcription. RNA was isolated from 293-T cells expressing BD, NF90-BD, or NF90Δ336N-BD together with luciferase plasmids and hybridized to RNA probes for the firefly and Renilla transcripts. In addition, the plasmid pSG, encoding a nonfunctional region of the adenovirus VA RNAI gene which is transcribed by RNA polymerase III (22) and hybridizes with the 5′VA RNA probe, was included as a control for transfection efficiency. Figure 5 shows that both NF90 and the Δ336N mutant reduced accumulation of firefly RNA when targeted to the ML1 promoter by the BD. Both proteins also enhanced accumulation of Renilla RNA from the CMV promoter. These results indicate that the modulation of gene expression by NF90 is at the level of mRNA, presumably through an increase in the level of transcription. While alternative mechanisms are possible (e.g., effects on mRNA processing or stability), they are unlikely, since the effect appears to be promoter specific rather than transcript specific.
FIG. 5.
NF90 regulates gene expression at the level of transcription. Human 293-T cells in 60-mm plates were transfected with 1.2 μg of pG5-ML1-F.luc, 40 ng of CMV-R.luc, 40 ng of pSG, 800 ng of pBD or pNF90-BD, and 1.6 μg of empty pcDNA3.1 plasmid. After 24 h, cells were harvested for RNA and then hybridized with RNA probes for the different reporter transcripts. The pSG plasmid encodes a nonfunctional mutant of VA RNAI and served as a transfection control. (A) Autoradiogram of protected RNA fragments. (B) Quantitation of two experiments like that shown in panel A. The firefly and Renilla RNA levels were normalized to the VA RNA signal; error bars indicate standard deviations.
NF45 enhances the activity of NF90.
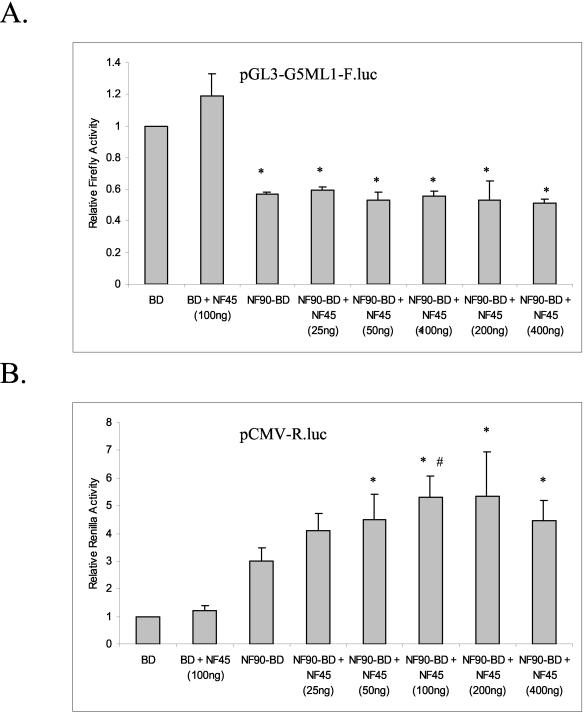
NF90 copurifies with NF45 (8, 14, 19, 40) and interacts with it in vitro (30a). To determine whether NF45 affects the activity of NF90 in the transient-expression assay, 293-T cells were cotransfected with plasmids encoding luciferase reporters and NF90-BD, together with increasing concentrations of a plasmid encoding NF45. After 24 h, cells were lysed and analyzed for both firefly and Renilla activity. NF45 had no discernible effect on the ability of NF90 to inhibit transcription from the ML-1 reporter (Fig. 6A), but it further stimulated the activator activity of NF90 on the CMV promoter (Fig. 6B). The effect was dose dependent, with a maximal enhancement of about twofold seen at 100 ng/plate. Higher concentrations of NF45 resulted in slightly decreased activation, possibly due to a squelching of other required factors by NF45.
FIG. 6.
NF45 modulates the activity of NF90. Human 293-T cells in 12-well plates were transfected with 300 ng of pG5-ML1-F.luc, 10 ng of CMV-R.luc, and 200 ng of expression plasmid (pBD or pNF90-BD), pNF45, and empty pcDNA3.1. A total amount of 400 ng of plasmid (pNF45 + pcDNA3.1 empty vector) was used. After 24 h, cell extracts were assayed for (A) firefly luciferase and (B) Renilla luciferase activities as in Fig. 2. Asterisks indicate P < 0.05 for indicated samples versus BD alone. The pound symbol indicates P < 0.05 for NF90-BD + NF45 (100 ng) versus NF90-BD alone.
To verify that NF90-BD forms complexes with NF45 in the cotransfected cells, we performed coimmunoprecipitation assays. At the same time, we mapped the interaction using a panel of NF90 deletion mutants. Cell extracts were incubated with anti-BD antibody, and the resulting immunoprecipitates were analyzed for the presence of NF45 by Western blot (Fig. 7A), and then stripped and reprobed with anti-BD antibody (Fig. 7B). Full-length NF90-BD interacted strongly with NF45, as did the mutant NF90Δ418C, implying a high-affinity binding to the N-terminal half of the NF90 protein. A shorter N-terminal fragment (NF90Δ313C) failed to coprecipitate with NF45, as did the extreme C terminus (NF90Δ610N), while NF90Δ336N and NF90Δ403N interacted weakly with NF45. These data suggest that there are two potential NF45 interaction sites in NF90 in vivo. The stronger site requires amino acids 313 to 336, and a second, weaker site lies between amino acids 403 and 601. The nature of the NF45 interaction with these two putative sites remains to be determined, but preliminary evidence indicates that the interaction between amino acids 313 and 336 is direct (Q. Li and M. B. Mathews, unpublished data).
DISCUSSION
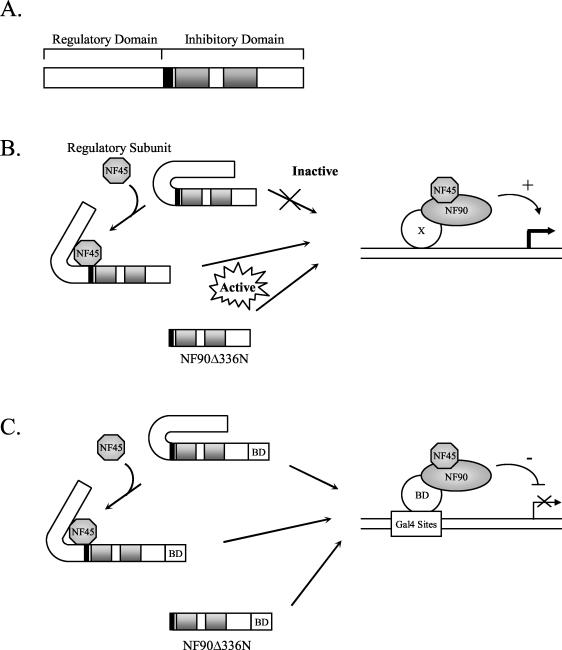
We found that NF90 can regulate gene expression both positively and negatively from different promoters and that NF45 acts as a regulatory subunit to enhance NF90’s ability to activate transcription from the CMV promoter. The enhancement of NF90 function by NF45 was almost equivalent to the effect of deleting the N terminus of the protein (compare Fig. 3C and 6B). Based on these findings, we propose that the N terminus of NF90 is a regulatory domain that inhibits the activator and inhibitory functions in the C terminus of the protein (Fig. 8A), possibly by blocking access to the NLS region and the dsRBMs of NF90.
FIG. 8.
Models for NF90 structure and function. (A) Functional domains of NF90 and proposed mechanisms of transcriptional activation (B) and inhibition (C) by NF90 and the NF90/NF45 complex. The black box indicates the NLS, and the grey boxes denote the dsRBMs. See Discussion for details.
In this model, activation of a promoter by NF90 is stimulated by its binding to NF45, which induces a conformational change, resulting in release of the C terminus of NF90 and its consequent activation (Fig. 8B). This activated NF90/NF45 complex is then capable of being recruited to a promoter region, either by interacting with a DNA-binding protein (protein X in Fig. 8B) or possibly through direct binding to DNA (Fig. 8B). The tight association of NF90 with DNA is further supported by cellular fractionation studies, in which a large percentage of NF90 copurified with chromatin from Jurkat cells (data not shown). In the absence of NF45, NF90 exists in its inactive conformation and is unable to interact with protein X or the promoter region directly and is incapable of activating transcription. Conversely, NF90Δ336N, which lacks the inhibitory N-terminal domain of NF90, is constitutively active and regulates transcription irrespective of NF45. One prediction of this model is that NF45 should not enhance the activity of the NF90Δ336N truncation. Indeed, preliminary evidence indicates that NF45 does not stimulate the activity of NF90Δ336N or any of the other mutants to which it does not bind tightly (T. W. Reichman and M. B. Mathews, unpublished data).
NF90 also acts as an inhibitor of transcription when recruited to the adenovirus ML1 promoter by the Gal4 BD. This activity of NF90 is independent of the presence of NF45 (Fig 6A). According to the model, NF90 is recruited to the promoter by the Gal4 sites and BD, so the conformational switch to a more favorable promoter-interacting form of NF90 by NF45 is not required (Fig. 8C). Therefore, increasing the amount of NF45 (by cotransfection with an NF45-expressing plasmid) has little effect on the ability of NF90 to inhibit transcription. Nevertheless, removal of the N terminus of NF90 gives a small but detectable increase in its ability to inhibit transcription (Fig. 3B), consistent with a conformational change.
The coordinated function of NF90 and NF45 in our in vivo reporter assay correlates well with the earlier observation that both of these proteins are required for transcriptional activation of an ARRE-2 containing G-less template in vitro (8). Although the two proteins copurify (8, 19, 40), recent findings suggest that they do not invariably exist together as a complex in the cell (30a). Protein expression profiles in various cell types revealed that the levels of the two proteins and the ratios between them vary considerably among different cell lines. Furthermore, in S. cerevisiae, which lacks any known homologue of NF90 or NF45, these proteins act alone to regulate the activity of PKR. In this system, their phenotypic effects are in opposite directions: whereas NF90 functions as a PKR activator, NF45 serves to overcome the effects of PKR on yeast growth (Parker et al., submitted). The ability of these proteins to function in the absence of their heteromeric partner may help explain their differential actions on promoters (Fig. 3).
The dual function of NF90 in activating and inhibiting gene expression is shared with several transcriptional regulators, including p53, BRCA1, c-Myc, YY1, c-Myb, Wilms’ tumor protein 1 (WT1), and the retinoic acid receptor (6, 16, 21, 25–27, 38). In some cases, this property is clearly related to the physiological functions of the transcription factor and is dictated by the architecture of the promoter to which the factor is recruited. For example, when bound to their enhancer element in the absence of ligand, the retinoic acid receptors recruit repressor complexes that inhibit transcription. Once activated by ligand, the repressor is dislodged through a conformational change, and activator complexes are recruited (25). The WT1 protein has also been shown to act as both a repressor and an activator of transcription of several cellular genes. As in the case of NF90, targeting of a WT1-Gal4 BD fusion protein to a promoter results in inhibition of transcription from a transfected reporter gene (20).
Indications that the dual transcriptional activities of NF90 are physiologically relevant come from its implication as both an activator and repressor of gene expression from different cellular promoters. NF90 is required for activated transcription of an ARRE-2 reporter construct in vitro (14), and it was identified as a component of the CCAAT box transcription complex, CBTF, that is thought to regulate the GATA-2 promoter in Xenopus (3). On the other hand, NF90 has been isolated as a protein that binds to a unique palindrome sequence (DHS-22) present in the DNase I-hypersensitive site II (DHS II) in the HLA-DRα gene, and it is hypothesized to serve as a negative regulator of transcription from this gene (35). Upon tetradecanoyl phorbol acetate (TPA) stimulation of THP-1 cells, NF90 was downregulated at the mRNA level in parallel with a decrease in binding of the protein complex to the DHS II element, presumably resulting in gene expression (35). Further clarification of these properties of NF90 will require a more complete assessment of the cellular genes regulated by NF90 and the identification of promoter elements that govern its inhibitory and activator activities.
The C-terminal region of NF90 that is responsible for its transcriptional actions contains an NLS and two dsRBMs. The NLS region was shown to be required for strong localization of NF90 to the nucleus but was not the sole determinant of nuclear localization, since the dsRBMs alone (in mutant Δ403N) also appeared to concentrate a percentage of the protein in the nucleus. Therefore, the NLS region might also serve to mediate the interaction of some as yet unknown transcriptional modulator with NF90. The NLS region itself is basic and proline-rich; interestingly, it contains two of the four putative M-phase phosphorylation sites (23).
Both the inhibitory and stimulatory effects of NF90 on reporter gene expression were enhanced by the presence of at least one complete dsRBM, suggestive of a role for dsRNA binding in positively or negatively regulating one or both of these activities. Binding of dsRNA to the dsRBM of PKR leads to a conformational change resulting in the activation of the kinase from its latent state (5). During viral infection, the kinase binds dsRNA and homodimerizes through its dsRBMs, resulting in autophosphorylation and kinase activation. The activated kinase then phosphorylates the α-subunit of eIF2, which inhibits translational initiation (15).
Several viruses encode RNAs as well as proteins that block the activation of PKR by dsRNA (32). For example, adenovirus encodes VA RNAI, a short, highly structured RNA which competes with dsRNA in binding to PKR and inhibits its activation. As mentioned previously, a second adenovirus-encoded RNA, VA RNAII, specifically binds to NF90 (19). It will be interesting to determine whether dsRNA and VA RNAII have any effect on the regulation of gene expression by NF90. It is also possible that the dsRBMs of NF90 are mediating protein-protein or protein-RNA-protein interactions. NF90 has been shown to interact with PKR (7, 17, 30, 31) and with the dsRBM-containing transcriptional coactivator RNA helicase A (I. Fierro-Monti and M. B. Mathews, unpublished data). RNA helicase A can act as a coactivator of CREB-mediated transcription by bridging CBP with RNA polymerase II (28).
This report shows that NF90 acts as a positive and negative regulator of transcription, consistent with the roles imputed previously. Its interactions with PKR and viral RNAs suggest that NF90 might act to regulate certain genes involved in the viral defense pathway and inhibition of viral replication. Conversely, NF90 was upregulated in nasopharyngeal carcinomas, suggesting a role in the promotion of cell proliferation and the cell cycle, as postulated for the NF90 homologue MPP4 (11, 23). Now that specific activities for NF90 have been identified, the next challenge will be the identification of cellular targets regulated by NF90 to help further decipher its function in normal cell homeostasis and function.
Acknowledgments
We thank Peter Kao for the kind gift of the NF45 polyclonal antibody and the pBS-KS(−)-NF45 plasmid and Raymond Birge for his help with the microscopy.
This work was supported by a grant from the National Institutes of Health, R01 AI34552, to M.B.M. T.W.R. was the recipient of predoctoral fellowship 12013CCRS0 from the New Jersey State Commission on Cancer Research.
REFERENCES
- 1.Aoki, Y., G. Zhao, D. Qiu, L. Shi, and P. N. Kao. 1998. CsA-sensitive purine-box transcriptional regulator in bronchial epithelial cells contains NF45, NF90, and Ku. Am. J. Physiol. 275:L1164–L1172. [DOI] [PubMed] [Google Scholar]
- 2.Bass, B. L., S. R. Hurst, and J. D. Singer. 1994. Binding properties of newly identified Xenopus proteins containing dsRNA-binding motifs. Curr. Biol. 4:301–314. [DOI] [PubMed] [Google Scholar]
- 3.Brzostowski, J., C. Robinson, R. Orford, S. Elgar, G. Scarlett, T. Peterkin, M. Malartre, G. Kneale, M. Wormington, and M. Guille. 2000. RNA-dependent cytoplasmic anchoring of a transcription factor subunit during Xenopus development. EMBO J. 19:3683–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buaas, F. W., K. Lee, S. Edelhoff, C. Disteche, and R. E. Braun. 1999. Cloning and characterization of the mouse interleukin enhancer binding factor 3 (Ilf3) homolog in a screen for RNA binding proteins. Mamm. Genome 10:451–456. [DOI] [PubMed] [Google Scholar]
- 5.Carpick, B. W., V. Graziano, D. Schneider, R. K. Maitra, X. Lee, and B. R. Williams. 1997. Characterization of the solution complex between the interferon-induced, double-stranded RNA-activated protein kinase and HIV-I trans-activating region RNA. J. Biol. Chem. 272:9510–9516. [DOI] [PubMed] [Google Scholar]
- 6.Claassen, G. F., and S. R. Hann. 1999. Myc-mediated transformation: the repression connection. Oncogene 18:2925–2933. [DOI] [PubMed] [Google Scholar]
- 7.Coolidge, C. J., and J. G. Patton. 2000. A new double-stranded RNA-binding protein that interacts with PKR. Nucleic Acids Res. 28:1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corthesy, B., and P. N. Kao. 1994. Purification by DNA affinity chromatography of two polypeptides that contact the NF-AT DNA binding site in the interleukin 2 promoter. J. Biol. Chem. 269:20682–20690. [PubMed] [Google Scholar]
- 9.Duchange, N., J. Pidoux, E. Camus, and D. Sauvaget. 2000. Alternative splicing in the human interleukin enhancer binding factor 3 (ILF3) gene. Gene 261:345–353. [DOI] [PubMed] [Google Scholar]
- 10.Fierro-Monti, I., and M. B. Mathews. 2000. Proteins binding to duplexed RNA: one motif, multiple functions. Trends Biochem. Sci. 25:241–246. [DOI] [PubMed] [Google Scholar]
- 11.Fung, L. F., A. K. Lo, P. W. Yuen, Y. Liu, X. H. Wang, and S. W. Tsao. 2000. Differential gene expression in nasopharyngeal carcinoma cells. Life Sci. 67:923–936. [DOI] [PubMed] [Google Scholar]
- 12.Gunnery, S., Y. Ma, and M. B. Mathews. 1999. Termination sequence requirements vary among genes transcribed by RNA polymerase III. J. Mol. Biol. 286:745–757. [DOI] [PubMed] [Google Scholar]
- 13.Herrmann, C. H., and M. B. Mathews. 1989. The adenovirus E1B 19-kilodalton protein stimulates gene expression by increasing DNA levels. Mol. Cell. Biol. 9:5412–5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kao, P. N., L. Chen, G. Brock, J. Ng, J. Kenny, A. J. Smith, and B. Corthesy. 1994. Cloning and expression of cyclosporin A- and FK506-sensitive nuclear factor of activated T-cells: NF45 and NF90. J. Biol. Chem. 269:20691–20699. [PubMed] [Google Scholar]
- 15.Kaufman, R. 2000. Double-stranded RNA activated protein kinase PKR, p. 503–528. In N. Sonenberg, J. Hershey, and M. Mathews (ed.), Translational control of gene expression, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 16.Ko, L. J., and C. Prives. 1996. p53: puzzle and paradigm. Genes Dev. 10:1054–1072. [DOI] [PubMed] [Google Scholar]
- 17.Langland, J. O., P. N. Kao, and B. L. Jacobs. 1999. Nuclear factor-90 of activated T-cells: a double-stranded RNA-binding protein and substrate for the double-stranded RNA-dependent protein kinase, PKR. Biochemistry 38:6361–6368. [DOI] [PubMed] [Google Scholar]
- 18.Liang, S. H., and M. F. Clarke. 1999. A bipartite nuclear localization signal is required for p53 nuclear import regulated by a carboxyl-terminal domain. J. Biol. Chem. 274:32699–32703. [DOI] [PubMed] [Google Scholar]
- 19.Liao, H. J., R. Kobayashi, and M. B. Mathews. 1998. Activities of adenovirus virus-associated RNAs: purification and characterization of RNA binding proteins. Proc. Natl. Acad. Sci. USA 95:8514–8519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Madden, S. L., D. M. Cook, and F. J. Rauscher III. 1993. A structure-function analysis of transcriptional repression mediated by the WT1, Wilms’ tumor suppressor protein. Oncogene 8:1713–1720. [PubMed] [Google Scholar]
- 21.Madden, S. L., and F. J. Rauscher III. 1993. Positive and negative regulation of transcription and cell growth mediated by the EGR family of zinc-finger gene products. Ann. N. Y. Acad. Sci. 684:75–84. [DOI] [PubMed] [Google Scholar]
- 22.Mathews, M. B., and T. Shenk. 1991. Adenovirus virus-associated RNA and translation control. J. Virol. 65:5657–5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsumoto-Taniura, N., F. Pirollet, R. Monroe, L. Gerace, and J. M. Westendorf. 1996. Identification of novel M phase phosphoproteins by expression cloning. Mol. Biol. Cell 7:1455–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mellits, K. H., M. Kostura, and M. B. Mathews. 1990. Interaction of adenovirus VA RNAl with the protein kinase DAI: nonequivalence of binding and function. Cell 61:843–852. [DOI] [PubMed] [Google Scholar]
- 25.Minucci, S., and P. G. Pelicci. 1999. Retinoid receptors in health and disease: co-regulators and the chromatin connection. Semin. Cell. Dev. Biol. 10:215–225. [DOI] [PubMed] [Google Scholar]
- 26.Mizuguchi, G., C. Kanei-Ishii, T. Takahashi, T. Yasukawa, T. Nagase, M. Horikoshi, T. Yamamoto, and S. Ishii. 1995. c-Myb repression of c-erbB-2 transcription by direct binding to the c-erbB-2 promoter. J. Biol. Chem. 270:9384–9389. [DOI] [PubMed] [Google Scholar]
- 27.Monteiro, A. N. 2000. BRCA1: exploring the links to transcription. Trends Biochem. Sci. 25:469–474. [DOI] [PubMed] [Google Scholar]
- 28.Nakajima, T., C. Uchida, S. F. Anderson, C. G. Lee, J. Hurwitz, J. D. Parvin, and M. Montminy. 1997. RNA helicase A mediates association of CBP with RNA polymerase II. Cell 90:1107–1112. [DOI] [PubMed] [Google Scholar]
- 29.Orford, R. L., C. Robinson, J. M. Haydon, R. K. Patient, and M. J. Guille. 1998. The maternal CCAAT box transcription factor which controls GATA-2 expression is novel and developmentally regulated and contains a double-stranded-RNA-binding subunit. Mol. Cell. Biol. 18:5557–5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parker, L. M., I. Fierro-Monti, and M. B. Mathews. 2001. Nuclear factor 90 is a substrate and regulator of the eIF2 kinase PKR. J. Biol. Chem. 276:32522–32530. [DOI] [PubMed] [Google Scholar]
- 30a.Parker, L. M., I. Fierro-Monti, T. W. Reichman, S. Gunnery, and M. B. Mathews. Double-stranded RNA binding proteins and the control of protein synthesis. Cold Spring Harbor Symp. Quant. Biol., in press. [DOI] [PubMed]
- 31.Patel, R. C., D. J. Vestal, Z. Xu, S. Bandyopadhyay, W. Guo, S. M. Erme, B. R. Williams, and G. C. Sen. 1999. DRBP76, a double-stranded RNA-binding nuclear protein, is phosphorylated by the interferon-induced protein kinase, PKR. J. Biol. Chem. 274:20432–20437. [DOI] [PubMed] [Google Scholar]
- 32.Pe’ery, T., and M. Mathews. 2000. Viral translational strategies and host defense mechanisms, p. 371–424. In N. Sonenberg, J. Hershey, and M. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Rao, A., C. Luo, and P. G. Hogan. 1997. Transcription factors of the NFAT family: regulation and function. Annu. Rev. Immunol. 15:707–747. [DOI] [PubMed] [Google Scholar]
- 34.Robbins, J., S. M. Dilworth, R. A. Laskey, and C. Dingwall. 1991. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell 64:615–623. [DOI] [PubMed] [Google Scholar]
- 35.Sakamoto, S., K. Morisawa, K. Ota, J. Nie, and T. Taniguchi. 1999. A binding protein to the DNase I hypersensitive site II in HLA-DR alpha gene was identified as NF90. Biochemistry 38:3355–3361. [DOI] [PubMed] [Google Scholar]
- 36.Schumacher, J. M., K. Lee, S. Edelhoff, and R. E. Braun. 1995. Spnr, a murine RNA-binding protein that is localized to cytoplasmic microtubules. J. Cell Biol. 129:1023–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang, J., P. N. Kao, and H. R. Herschman. 2000. Protein-arginine methyltransferase I, the predominant protein-arginine methyltransferase in cells, interacts with and is regulated by interleukin enhancer-binding factor 3. J. Biol. Chem. 275:19866–19876. [DOI] [PubMed] [Google Scholar]
- 38.Thomas, M. J., and E. Seto. 1999. Unlocking the mechanisms of transcription factor YY1: are chromatin modifying enzymes the key?. Gene 236:197–208. [DOI] [PubMed] [Google Scholar]
- 39.Tian, B., and M. B. Mathews. 2001. Functional characterization of and cooperation between the double-stranded RNA-binding motifs of the protein kinase PKR. J. Biol. Chem. 276:9936–9944. [DOI] [PubMed] [Google Scholar]
- 40.Ting, N. S., P. N. Kao, D. W. Chan, L. G. Lintott, and S. P. Lees-Miller. 1998. DNA-dependent protein kinase interacts with antigen receptor response element binding proteins NF90 and NF45. J. Biol. Chem. 273:2136–2145. [DOI] [PubMed] [Google Scholar]
- 41.Xu, Y. H., and G. A. Grabowski. 1999. Molecular cloning and characterization of a translational inhibitory protein that binds to coding sequences of human acid beta-glucosidase and other mRNAs. Mol. Genet. Metab. 68:441–454. [DOI] [PubMed] [Google Scholar]