Abstract
Recent studies have shown the p19ARF tumor suppressor to be involved in the response to oncogenic stress by regulating the activity of p53. This response is mediated by antagonizing the function of Mdm2, a negative regulator of p53, indicating a pathway for tumor suppression that involves numerous genes altered in human tumors. We previously described a transgenic mouse brain tumor model in which oncogenic stress, provided by cell-specific inactivation of the pRb pathway, triggers a p53-dependent apoptotic response. This response suppresses the growth of developing tumors and thus represents a bona fide in vivo tumor suppressor activity. We further showed that E2F1, a transcription factor known to induce p19ARF expression, was required for the response. Here, we use a genetic approach to test whether p19ARF functions to transduce the signal from E2F1 to p53 in this tumor suppression pathway. Contrary to the currently accepted hypothesis, we show that a deficiency in p19ARF has no impact on p53-mediated apoptosis or tumor suppression in this system. All measures of p53 function, including the level of apoptosis induced by pRb inactivation, the expression of p21 (a p53-responsive gene), and the rate of tumor growth, were comparable in mice with and without a functional p19ARF gene. Thus, although p19ARF is required in some cell types to transmit an oncogenic response signal to p53, it is dispensable for this function in an in vivo epithelial system. These results underscore the complexity of p53 tumor suppression and further indicate the existence of distinct cell-specific pathways that respond to similar stimuli.
p19ARF was first identified as the product of an alternative transcript within the p16INK4a gene (20). Deletions of the INK4a/ARF locus have been found in a variety of human cancers (21), and mice nullizygous at the INK4a/ARF locus develop tumors with a high frequency, usually lymphoma and sarcoma (24). Importantly, mice that specifically lack p19ARF but retain p16INK4a develop a similar spectrum of tumors (11), establishing the tumor suppressor function of p19ARF. Recent studies in fibroblasts in vitro (2, 3, 8, 16, 35) and in B cells in vivo (4, 23) suggest a role for p19ARF in p53 responses to oncogenic stress. For example, overexpression of c-Myc, E2F1, activated Ras, or E1A in primary mouse embryonic fibroblasts (MEF) induces p53-dependent growth arrest or apoptosis, and these effects are attenuated in p19ARF null cells. Furthermore, p53-deficient cells are resistant to p19ARF-induced cell cycle arrest (11), indicating that p19ARF acts upstream of p53. Biochemical analyses show that p19ARF can bind to Mdm2 and block Mdm2-induced p53 degradation, thus providing a molecular mechanism by which p19ARF can activate p53 (10, 19, 26, 32, 33).
These studies suggest a model in which p19ARF acts as a tumor suppressor by responding to oncogenic signals, possibly via direct transcriptional activation by E2F transcription factors (2, 35) and by transmitting the signal to p53 via Mdm2 regulation. Indeed, the development of B-cell lymphoma in Eμ-myc transgenic mice is significantly accelerated in Ink4a-ARF+/− (8, 23) and ARF+/− (4) backgrounds. Since a similar effect occurs in p53+/− mice (7, 23), these observations support the hypothesis that p53-mediated tumor suppression in this model is dependent on p19ARF. However, it is not known whether the same mechanism is broadly operative in diverse cell types susceptible to p53 tumor suppression.
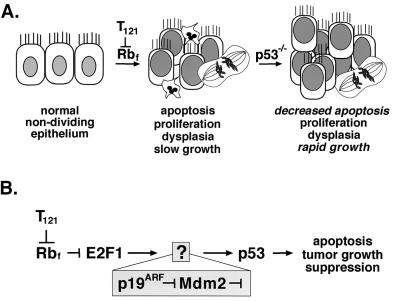
Here, we address the role of p19ARF in p53-mediated epithelial cell apoptosis and tumor suppression in vivo. Previously, we developed a transgenic mouse model (TgT121) in which epithelial brain tumors are initiated by cell-specific expression of T121, an oncoprotein derived from simian virus 40 (SV40) T antigen that specifically inactivates pRb and related proteins p107 and p130 (22, 27). Inactivation of these proteins in choroid plexus (CP) epithelial cells induces aberrant proliferation and p53-dependent apoptosis, resulting in the development of slow-growing tumors (Fig. 1A). In a p53 null background, the T121-induced apoptosis is significantly reduced, and tumor growth is accelerated sevenfold (Fig. 1A) (27). Thus, the development of tumors in this model serves as a paradigm for oncogenic stress-induced p53-mediated apoptosis and tumor suppression in epithelial cells in vivo.
FIG. 1.
p53 tumor suppression in brain epithelium. The diagram depicts previously elucidated steps in the development of choroid plexus tumors in TgT121 mice. (A) Cell-specific expression of the T121 transgene induces proliferation of normally nondividing epithelial cells by inactivating the pRb family proteins pRb, p107, and p130. p53-dependent apoptosis is then activated, as evidenced by the 85% reduction in apoptosis in TgT121;p53−/− mice. (B) Using a genetic approach, we previously showed that E2F1 acts upstream of p53 to induce apoptosis in response to T121. Previously, E2F1 was shown to activate p19ARF transcription, and p19ARF was shown to induce p53 activities by regulating Mdm2 in cultured cells (see the introduction). In the current report, we tested whether this pathway is operative to induce p53-dependent apoptosis and suppression of brain epithelial tumors in vivo.
Using a genetic approach, we further showed that the p53-mediated apoptosis in CP is dependent on E2F1 (Fig. 1B) (18). Hence, based on the observations described above, p19ARF was a likely candidate for transmitting the signal from E2F1 to p53 (Fig. 1B). Here, we test this hypothesis by assessing the effect of p19ARF deficiency on T121-induced p53-dependent apoptosis and tumor suppression in vivo.
MATERIALS AND METHODS
Generation of p19ARF-deficient TgT121 mice.
Characterization of TgT121 transgenic mice (C57BL6/J;DBA2) was described previously (22, 27). Mice harboring a homozygous deletion of p19ARF exon 1 (C57BL6/J; Sv129) (11) were kindly provided by C. J. Sherr and M. F. Roussel (St. Jude Children’s Hospital). TgT121;p19ARF+/− and TgT121;p19ARF−/− mice were generated by crossing hemizygous TgT121 mice with p19ARF−/− mice through two generations. In all experiments, TgT121;p19ARF+/− and TgT121;p19ARF−/− littermates were compared to control for any variability in the genetic background. TgT121;p19ARF+/+ mice were generated in a single cross between TgT121 mice and p19ARF+/− mice. TgT121 and p19ARF genotypes were identified by PCR analysis of tail DNA (11, 27).
Histology, proliferation, and apoptosis assays.
Brain tissues were fixed, embedded, and sectioned as described (18). Apoptotic cells were detected in sections using the terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assay (27). Cell proliferation was determined by in situ immunodetection of bromodeoxyuridine (BrdU) incorporation as described (18). The significance of differences in apoptosis levels and cell proliferation levels between mice with different genotypes was evaluated by t test. P < 0.05 was considered significant.
In situ RNA hybridization.
Sections were treated and hybridized as previously described (18). The p21 antisense probe was generated by T7 transcription of an EcoRI-linearized pBS-KSp21 template. The p19ARF antisense probe was generated by T3 transcription of a BamHI-linearized pBS-KSp19ARF (a kind gift of Yue Xiong). Probes were labeled with [α-35S]UTP (5 × 104 cpm/μl) and hybridized to slides at 50°C overnight. For the p19ARF-specific probe, slides were exposed for 60 days; for the p21-specific probe, slides were exposed for 3 days. The sense probes did not show any signal above background in both p21 and p19ARF in situ hybridizations.
RESULTS
p19ARF expression is induced in T121-expressing cells.
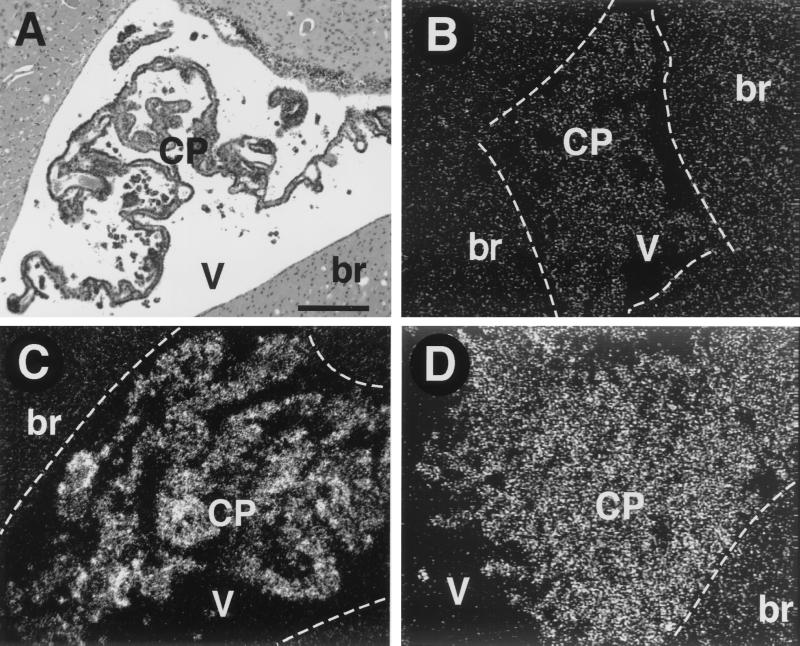
Since oncogenic stress in cultured cells results in transcriptional induction of p19ARF (2, 3, 16, 35), we first determined whether p19ARF transcripts were induced upon T121 expression coincident with p53 activation in CP. In situ RNA hybridization with a mouse p19ARF-specific probe detected a clearly positive signal in CP of TgT121 mice (Fig. 2C), whereas no signal above background was detected in normal nontransgenic CP (Fig. 2B). As predicted from previous studies, the p19ARF signal was not dependent on p53, based on the positive signal in TgT121;p53−/− CP (Fig. 2D). Although p19ARF transcripts were clearly induced in CP by T121, levels were quite low, based on the requirement for an exposure time of 8 weeks. By comparison, p21 transcripts induced by the activation of p53 were detected in 3 days using similar procedures (18) (see Fig. 5). Nonetheless, induction of p19ARF transcripts specifically by T121 expression supported the possibility that p19ARF transmits a signal to p53.
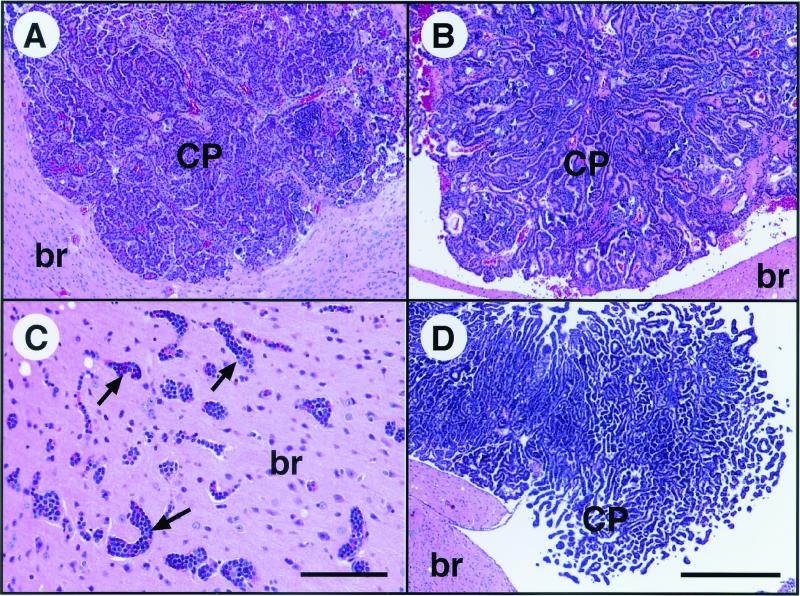
FIG. 2.
p19ARF transcripts are induced in aberrantly proliferating CP epithelium. RNA in situ hybridization was performed with an antisense p19ARF RNA probe. (A) Hematoxylin and eosin (H & E)-stained section from the same brain as in panel C shows the morphology of CP in the lateral ventricle and is in an orientation similar to that of dark-field panels B to D. The borders between brain parenchyma (br) and the ventricles (V) are depicted with white dashed lines in panels B to D. No p19ARF expression above background is detected in the CP of nontransgenic mice (B). (Compare the signal in brain with that in CP.) p19ARF transcripts are induced in TgT121 CP (C). T121-induced p19ARF expression is p53 independent; the level of p19ARF transcripts is similar in TgT121;p53−/− CP (D) and TgT121 CP (C). (A) Bar, 200 μm (magnification is the same in all panels). Sense probe hybridization showed no signal above background (not shown).
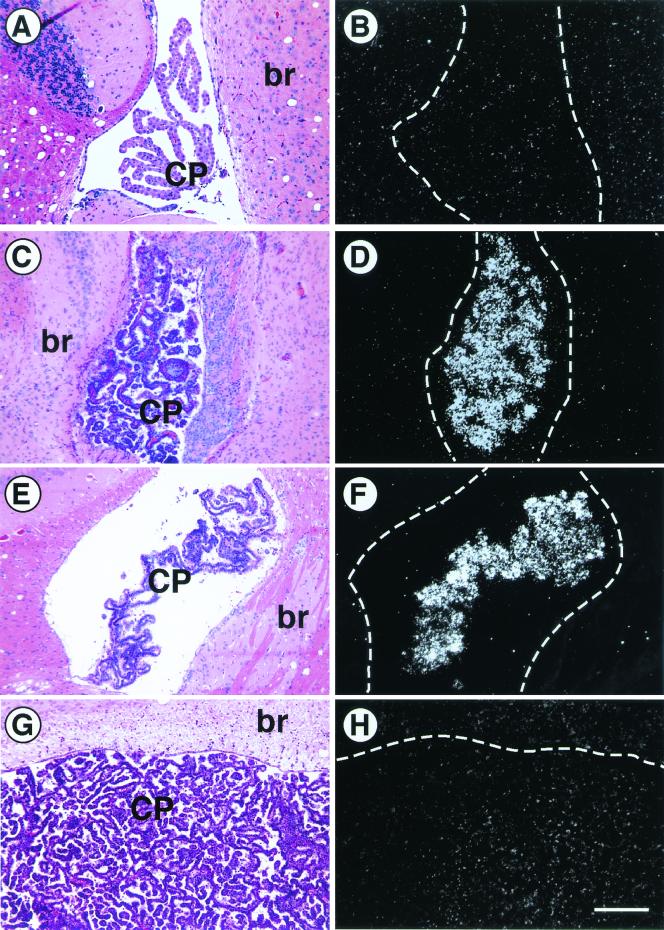
FIG. 5.
p53-mediated transactivation of p21 is unaffected by p19ARF deficiency. Brains were examined from nontransgenic (A and B), TgT121;p19ARF+/+ (C and D), TgT121;p19ARF−/− (E and F), and TgT121;p53−/− (G and H) mice. RNA in situ hybridization was performed on brain sections using an antisense p21 RNA probe and viewed by dark-field microscopy (B, D, F, and H). Adjacent sections were stained with H & E and viewed by bright-field microscopy to show the location and morphology of the CP (A, C, E, and G). No p21 expression is detected in normal CP (B). p21 expression is induced by expression of T121 (D) and remains unchanged in the absence of p19ARF (F). T121-induced p21 expression in CP requires functional p53 (H) (18). The bar in panel H is equal to 200 μm; all panels are of the same magnification. br, brain tissue.
p19ARF is not required for p53-dependent apoptosis.
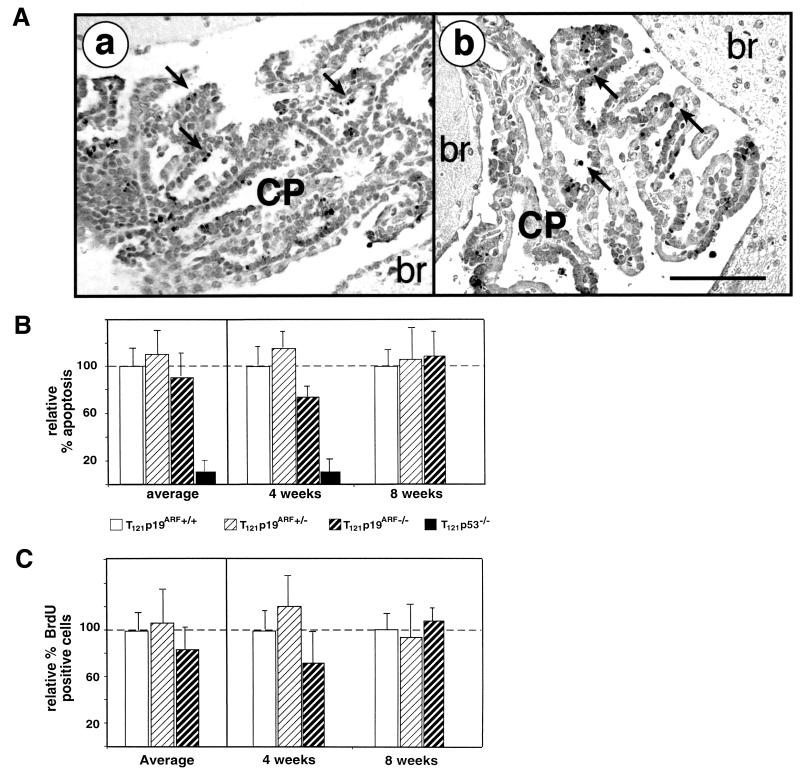
To test whether p19ARF is required for the induction of p53-dependent apoptosis by pRb protein inactivation, we generated TgT121 mice deficient in p19ARF by a series of backcrosses with p19ARF−/− mice (11). Apoptosis within the CP of several young (4 and 8 weeks) TgT121;p19ARF+/+, TgT121;p19ARF+/−, TgT121;p19ARF−/−, and p19ARF−/− mice was measured in situ using the TUNEL assay (Fig. 3A). TgT121;p53−/− mouse brain sections were assessed as a control. Although p53 deficiency dramatically reduced the level of apoptosis, p19ARF deficiency had little or no effect (Fig. 3A and B). The average apoptosis level in the CP of TgT121;p19ARF+/+ mice was 8.6% ± 1.9% (n = 6). The average relative apoptosis indices (AI) of TgT121;p19ARF−/− and TgT121;p19ARF+/− CP compared with that of TgT121;p19ARF+/+ CP (100%) were 92.9% ± 19.2% and 113.0% ± 23.1%, respectively. The small difference between the TgT121;p19ARF−/− group (n = 6) and the TgT121;p19ARF+/+ group (n = 6) is not statistically significant (P = 0.601). Moreover, as shown previously (27), the relative AI of TgT121;p53−/− CP (n = 3) was reduced to 12.4% ± 8.5%. Thus, contrary to the hypothesis, these data show that p53-dependent apoptosis in response to aberrant proliferation of epithelial cells in vivo does not require p19ARF.
FIG. 3.
T121-induced apoptosis and cell proliferation do not require p19ARF. T121 expression causes abnormal CP cell proliferation and apoptosis. About 85% of the apoptosis is p53-dependent (27) and requires E2F1 (18). The TUNEL assay was used to detect apoptosis (A). Apoptosis levels and morphology were indistinguishable between TgT121;p19ARF−/− (a) and TgT121;p19ARF+/+ (b) CP. Apoptotic nuclei are stained and appear black; representative apoptotic cells are indicated by arrows. The bar (b) is equal to 100 μm; both panels are of the same magnification. Quantitative analysis of apoptosis (B) and proliferation (C) in the CP of three mice each at 4 and 8 weeks of age was carried out. Levels are compared to that of TgT121;p19ARF+/+ tissue, which is considered to represent 100%. Average relative values for all mice of each age and genotype are shown in the left panels. For these data, error bars indicate the variation among mice. In the right panels, data for each mouse are presented, with error bars indicating the field-to-field variation in counts taken from 10 fields per brain. Different mice were used for apoptosis and proliferation assays so as to avoid any impact of BrdU incorporation on apoptosis levels. There is no significant difference in apoptotic indexes between TgT121;p19ARF+/+ and TgT121;p19ARF−/− mice (P = 0.601). Importantly, the index of TgT121;p19ARF−/− CP was significantly higher than that of TgT121;p53−/− CP (P < 0.05). The level of CP cell proliferation was determined by immunostaining of BrdU incorporated in vivo (C). The data show that p19ARF deficiency does not significantly alter cell proliferation (P = 0.288).
p19ARF deficiency does not interfere with tumor cell cycle.
Recent studies have shown that p19ARF could inhibit cell growth by a p53-independent pathway (29). Thus, although p19ARF was clearly dispensable for the p53-dependent apoptosis of CP tumor cells, it was possible that p19ARF affected the tumor cell cycle by unknown p53-independent mechanisms. To determine whether this was the case, the fraction of proliferating tumor cells was determined in the presence and absence of p19ARF by in situ immunodetection of BrdU incorporation. TgT121;p19ARF+/+, TgT121;p19ARF+/−, TgT121;p19ARF−/−, and p19ARF−/− mice of different ages (4 and 8 weeks) were examined. This analysis indicated that p19ARF deficiency did not affect the proliferation of CP tumor cells. The average percentage of BrdU-positive cells in the CP of TgT121;p19ARF+/+ mice was 8.5% ± 2.2% (n = 6). The relative BrdU staining levels within the CP of TgT121;p19ARF−/− and TgT121;p19ARF+/− mice were 86.5% ± 21.2% and 107.5% ± 37.6%, respectively (n = 6 in both cases) (Fig. 3C). Thus, no significant effect of p19ARF on tumor cell proliferation was detected (P = 0.288).
p19ARF deficiency does not accelerate CP tumor growth.
In previous studies of TgT121 mice, we showed that tumor growth was slow, becoming life-threatening only after a mean age of 26 weeks (22). Deficiency of p53 caused acceleration of tumor growth due to reduced apoptosis, and the animals died by 4 weeks of age (27). Given that neither apoptosis nor proliferation appeared to be altered by p19ARF deficiency, tumor growth was predicted to be unaltered as well. However, it was possible that additional unexpected parameters dependent on p19ARF could impact either the rate or morphological characteristics of tumor growth. Because TgT121;p19ARF−/− and TgT121;p19ARF+/− mice simultaneously develop multiple tumor types, including CP tumors, sarcoma, and lymphoid malignancies (summarized in Table 1), survival does not provide an appropriate assessment of CP tumor growth. However, unlike TgT121;p53−/− mice, all TgT121;p19ARF−/− mice survived beyond 4 weeks of age to a mean age of 17 weeks. This result confirms that p19ARF deficiency does not induce a phenocopy of p53 deficiency.
TABLE 1.
Tumor frequency in TgT121;p19ARF−/− terminal-stage mice
| Genotype | No. of mice | Survival (wk)a | % of mice with:
|
|||
|---|---|---|---|---|---|---|
| CP tumor | Sarcoma | Lymphoid malignancyb | Other tumorsc | |||
| TgT121;p19ARF+/+ | 12 | 45 | 100 | 0 | 0 | 0 |
| TgT121;p19ARF+/− | 10 | 25 | 100 | 40 | 70 | 20 |
| TgT121;p19ARF−/− | 10 | 17 | 100 | 80 | 70 | 40 |
| p19ARF−/− | 23 | 27 | 0 | 70 | 52 | 30 |
Mice were sacrificed when they appeared ill. All malignancies present at the time of sacrifice were scored. Most mice with altered ARF alleles harbored multiple malignancies.
Including lymphomas and leukemias.
Including pheochromocytomas and endocrine tumors.
Two mice of each genotype clearly contained infiltrating leukemia cells, as shown in Fig. 4C.
Further assessment of CP tumor histopathology showed no detectable differences between TgT121;p19ARF−/− and TgT121;p19ARF+/+ CP tumors (for example, compare panels A and B of Fig. 4). Although survival time is clearly affected by the combination of TgT121 and altered p19ARF alleles (Table 1), evidence indicates that this effect results from exacerbation by coexisting malignancies. For example, in mice with compound genotypes, the simultaneous presence of leukemia (induced by p19ARF deficiency) and CP carcinoma (caused by T121 expression) resulted in extensive infiltration of leukemia cells into the brain (Fig. 4C), a phenotype not observed in either individual background. It is likely that such effects caused the mice to die at earlier ages (Table 1). In summary, evaluation of CP tumor growth indicates that p19ARF is not required for suppression of tumorigenesis in brain epithelium.
FIG. 4.
Tumor growth and morphology are unaffected by p19ARF deficiency. H & E-stained CP tumors from 20-week-old TgT121;p19ARF+/+ (A) and TgT121;p19ARF−/− (B) mice are similar in size and morphology. In contrast, tumors become life-threatening by 4 weeks of age in TgT121;p53−/− mice (D). Leukemia caused by the p19ARF−/− mutation is frequently observed invading the brains of TgT121;p19ARF−/− mice (C), a pathology that is present only in mice with compound genotypes and is thought to further reduce survival time (Table 1). Representative leukemia-filled vessels in the brain are indicated by arrows. The bar in panel C is equal to 100 μm. The bar in panel D is equal to 200 μm; panels A, B, and D are of the same magnification. br, brain tissue.
p53 function is intact in CP of p19ARF-deficient mice.
Although p19ARF was not required for p53-mediated apoptosis or tumor suppression in CP epithelium, we considered whether p53 activity was at all dependent on p19ARF in this cell type. Furthermore, although unlikely, it was a formal possibility that the absence of p19ARF had triggered a switch from p53-dependent apoptosis and tumor suppression to p53-independent functions, thus masking the impact on p53. Hence, we used an independent assessment of p53 function to determine whether p53 remained active in p19ARF-deficient CP.
The p21 gene is a direct transcriptional target of p53 (5). In previous work, we demonstrated that p53-dependent p21 expression was indeed induced by T121 in CP (18). Furthermore, CP tumors that underwent p53 loss of heterozygosity also lost p21 expression, providing a perfect correlation between loss of p53 function and loss of p21 expression (12). If p53 activation were mediated by p19ARF in response to the T121 oncogenic signal, then p21 induction would not be observed in TgT121;p19ARF−/− CP, as in the case of TgT121;E2F1−/− and TgT121;p53−/− mice (18). Thus, the level of p21 expression in the CP was determined for TgT121;p19ARF−/−, TgT121;p19ARF+/−, and TgT121;p19ARF+/+ mice (n > 3 in each case). In all cases, no difference in p21 induction between TgT121;p19ARF−/− and TgT121;p19ARF+/+ CP was observed (Fig. 5). These results demonstrate that the p53 pathway induced by oncogenic stress in CP epithelium does not require p19ARF function.
DISCUSSION
We previously showed that p53 suppresses the growth of brain epithelial tumors by mediating apoptosis in response to aberrant proliferation resulting from Rb pathway inactivation (27). Although p19ARF is widely believed to generally transmit oncogenic stress signals to p53 (25, 31), the studies presented here show that no defects were detected in known p53 responses in the CP of p19ARF-deficient mice. p53-dependent apoptosis, tumor suppression, and transcriptional transactivation all remained unaffected in the absence of p19ARF. Thus, these studies indicate that p19ARF does not transmit the oncogenic stress signal to p53 in this system. Furthermore, since p19ARF deficiency appeared to have no effect on CP tumor growth or morphology, p19ARF also does not appear to suppress tumors in this cell type by p53-independent mechanisms.
Previous studies by others showed that B-cell lymphoma induced in transgenic mice by c-myc overexpression is accelerated in Ink4a/ARF+/− and ARF+/− backgrounds, similar to the effect observed in p53+/− mice (see the introduction). Further experiments in primary B cells and in B-cell lymphomas indicate that deficiencies in p53, Ink4a/ARF, or ARF also reduce the level of c-myc-induced apoptosis (4, 23). Moreover, in c-myc-induced lymphomas, mutation of p53 or deletion of the Ink4a/ARF locus was frequent. These mutations were mutually exclusive (23), supporting the idea that p19ARF and p53 are on the same tumor suppression pathway in B cells. Thus, the fact that p19ARF was not required for p53-dependent apoptosis and tumor suppression in brain epithelium indicates that this pathway is not universal to all cell types and that cell-specific mechanisms exist for transmitting an oncogenic stress signal to p53 for tumor suppression. This concept is supported by a recent report that medulloblastoma induced in mice heterozygous for patched is accelerated by p53 deficiency but not by p19ARF deficiency (30). Although the mechanism for p53 tumor suppression in this model is unknown, the result clearly indicates that p19ARF is not required.
A recent study by K. Tsai and T. Jacks (personal communication) shows that p19ARF is also dispensable for p53-dependent apoptosis of embryonic central nervous system (CNS) neurons and ocular lens epithelium. pRb deficiency induces unscheduled cell proliferation in both cell types, resulting in p53-dependent apoptosis (13, 15, 17). As in the adult CP, the response in these cells requires E2F1 (28). Quantitative analysis of the apoptosis in Rb−/− p19ARF−/− embryos showed that p53-mediated apoptosis was unaffected in the CNS and was only minimally diminished in the lens. Furthermore, p53 transactivation and DNA binding remained intact in the absence of p19ARF. Previous work in the embryonic lens had shown that a deficiency in both p16Ink4a and p19ARF inhibited p53-dependent apoptosis to a greater extent, although the relative contributions of the two factors could not be resolved (19). Interestingly, in support of cell-specific mechanisms for p53 regulation, Tsai and Jacks (personal communication) show that p53-dependent proliferation suppression in embryonic peripheral neurons (13) does appear to depend on p19ARF.
Hence, one or more p19ARF-independent pathways exist that trigger p53 tumor suppression in response to disrupted cell cycle regulation in vivo. What is the mechanism? One possibility is that p19ARF carries out this function when present, but a compensatory factor signals p53 in its absence, i.e., a redundancy exists at this step in the pathway. We consider this possibility unlikely since there are no known p19ARF-related proteins. Thus, any compensatory pathway, such as an alternative cell-specific pathway, would be unique and hence interesting.
p19ARF signals to p53 by regulating Mdm2 (10, 19, 26, 32, 33), a protein that specifically binds p53 and targets it for degradation via the ubiquitin pathway (1, 6). We do not yet know whether Mdm2 is also the target for p53 regulation in CP tumor suppression. Mdm2 deficiency in the mouse causes lethality in the early embryo (9, 14), so that a cell-specific deficiency will be required to test the role of Mdm2 in this or any other in vivo tumor model. We do know that E2F1, and possibly other E2Fs (34), act upstream of p53 to facilitate the apoptotic response to pRb pathway inactivation in CP (18). Thus, as with p19ARF, the signal(s) to p53 could be directly transcriptionally regulated by E2Fs. However, further experimentation will be required to uncover potential candidates. To this end, we are currently using an array-based approach to examine the genes whose transcription is induced in response to T121 in CP in an E2F1-dependent fashion.
In summary, the studies presented here indicate that, although p19ARF signaling to p53 is a critical tumor suppression mechanism in some cell types, it does not universally mediate p53 tumor suppression in response to disrupted cell cycle regulation. Importantly, additional mechanisms, likely dictated by the cell type, do exist.
Acknowledgments
The first two authors contributed equally to this work.
We thank Chuck Sherr and Martine Roussel (Saint Jude Children’s Hospital, Memphis, Tenn.) for providing ARF−/− mice. We also thank Virginia Godfrey (UNC at Chapel Hill) for histological evaluation of non-CP tumors in this study. We acknowledge the UNC Lineberger Comprehensive Cancer Center Histology Core for processing tissues used in this study and the UNC Division of Laboratory Animals for excellent animal care.
This work was supported by NCI grants 1 R01 CA 46283 and 5 U01 CA84314 to T.V.D.
REFERENCES
- 1.Ashcroft, M., and K. H. Vousden. 1999. Regulation of p53 stability. Oncogene 18:7637–7643. [DOI] [PubMed] [Google Scholar]
- 2.Bates, S., A. C. Phillips, P. A. Clark, F. Stott, G. Peters, R. L. Ludwig, and K. H. Vousden. 1998. p14ARF links the tumour suppressors RB and p53. Nature 395:124–125. [DOI] [PubMed] [Google Scholar]
- 3.de Stanchina, E., M. E. McCurrach, F. Zindy, S. Y. Shieh, G. Ferbeyre, A. V. Samuelson, C. Prives, M. F. Roussel, C. J. Sherr, and S. W. Lowe. 1998. E1A signaling to p53 involves the p19(ARF) tumor suppressor. Genes Dev. 12:2434–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eischen, C. M., J. D. Weber, M. F. Roussel, C. J. Sherr, and J. L. Cleveland. 1999. Disruption of the ARF-Mdm2–p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 13:2658–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.El-Deiry, W. S., T. Tokino, V. E. Velculescu, D. B. Levy, R. Parsons, J. M. Trent, D. Lin, W. E. Mercer, K. W. Kinzler, and B. Vogelstein. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75:817–825. [DOI] [PubMed] [Google Scholar]
- 6.Freedman, D. A., and A. J. Levine. 1999. Regulation of the p53 protein by the MDM2 oncoprotein—thirty-eighth G. H. A. Clowes Memorial Award Lecture. Cancer Res. 59:1–7. [PubMed] [Google Scholar]
- 7.Hsu, B., M. C. Marin, A. K. el-Naggar, L. C. Stephens, S. Brisbay, and T. J. McDonnell. 1995. Evidence that c-myc mediated apoptosis does not require wild-type p53 during lymphomagenesis. Oncogene 11:175–179. [PubMed] [Google Scholar]
- 8.Jacobs, J. J., B. Scheijen, J. W. Voncken, K. Kieboom, A. Berns, and M. van Lohuizen. 1999. Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes Dev. 13:2678–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones, S. N., A. E. Roe, L. A. Donehower, and A. Bradley. 1995. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature 378:206–208. [DOI] [PubMed] [Google Scholar]
- 10.Kamijo, T., J. D. Weber, G. Zambetti, F. Zindy, M. F. Roussel, and C. J. Sherr. 1998. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc. Natl. Acad. Sci. USA 95:8292–8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamijo, T., F. Zindy, M. F. Roussel, D. E. Quelle, J. R. Downing, R. A. Ashmun, G. Grosveld, and C. J. Sherr. 1997. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell 91:649–659. [DOI] [PubMed] [Google Scholar]
- 12.Lu, X., G. Magrane, C. Yin, D. Louis, J. Gray, and T. Van Dyke. 2001. Selective inactivation of p53 facilitates mouse epithelial tumor progression without chromosomal instability. Mol. Cell. Biol. 21:6017–6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Macleod, K. F., Y. Hu, and T. Jacks. 1996. Loss of RB activates both p53-dependent and independent cell death pathways in the developing mouse nervous system. EMBO J. 15:6178–6188. [PMC free article] [PubMed] [Google Scholar]
- 14.Montes de Oca Luna, R., D. S. Wagner, and G. Lozano. 1995. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 378:203–206. [DOI] [PubMed] [Google Scholar]
- 15.Morgenbesser, S., B. Williams, T. Jacks, and R. DePinho. 1994. p53-dependent apoptosis produced by Rb-deficiency in the developing mouse lens. Nature 371:72–74. [DOI] [PubMed] [Google Scholar]
- 16.Palmero, I., C. Pantoja, and M. Serrano. 1998. p19ARF links the tumour suppressor p53 to Ras. Nature 395:125–126. [DOI] [PubMed] [Google Scholar]
- 17.Pan, H., and A. E. Griep. 1995. Temporally distinct patterns of p53-dependent and p53-independent apoptosis during mouse lens development. Genes Dev. 9:2157–2169. [DOI] [PubMed] [Google Scholar]
- 18.Pan, H., C. Yin, N. Dyson, E. Harlow, L. Yamasaki, and T. Van Dyke. 1998. A key role for E2F1 in p53-dependent apoptosis and cell division within developing tumors. Mol. Cell 2:283–292. [DOI] [PubMed] [Google Scholar]
- 19.Pomerantz, J., N. Schreiber-Angus, N. J. Liegeois, A. Silverman, L. Alland, L. Chin, J. Potes, K. Chen, I. Orlow, H. Lee, C. Cordon-Cardo, and R. DePinho. 1998. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM’s inhibition of p53. Cell 92:713–723. [DOI] [PubMed] [Google Scholar]
- 20.Quelle, D. E., F. Zindy, R. A. Ashmun, and C. J. Sherr. 1995. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell 83:993–1000. [DOI] [PubMed] [Google Scholar]
- 21.Ruas, M., and G. Peters. 1998. The p16INK4a/CDKN2A tumor suppressor and its relatives. Biochim. Biophys. Acta 1378:F115–F177. [DOI] [PubMed] [Google Scholar]
- 22.Saenz-Robles, M. T., H. Symonds, J. Chen, and T. Van Dyke. 1994. Induction versus progression of brain tumor development: differential functions for the pRB- and p53-targeting domains of simian virus 40 T antigen. Mol. Cell. Biol. 14:2686–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmitt, C. A., M. E. McCurrach, E. de Stanchina, R. R. Wallace-Brodeur, and S. W. Lowe. 1999. INK4a/ARF mutations accelerate lymphomagenesis and promote chemoresistance by disabling p53. Genes Dev. 13:2670–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Serrano, M., H. Lee, L. Chin, C. Cordon-Cardo, D. Beach, and R. A. DePinho. 1996. Role of the INK4a locus in tumor suppression and cell mortality. Cell 85:27–37. [DOI] [PubMed] [Google Scholar]
- 25.Sherr, C. J. 1998. Tumor surveillance via the ARF-p53 pathway. Genes Dev. 12:2984–2991. [DOI] [PubMed] [Google Scholar]
- 26.Stott, F. J., S. Bates, M. C. James, B. B. McConnell, M. Starborg, S. Brookes, I. Palmero, K. Ryan, E. Hara, K. H. Vousden, and G. Peters. 1998. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 17:5001–5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Symonds, H., L. Krall, L. Remington, M. Saenz-Robles, S. Lowe, T. Jacks, and T. Van Dyke. 1994. p53-dependent apoptosis suppresses tumor growth and progression in vivo. Cell 78:703–711. [DOI] [PubMed] [Google Scholar]
- 28.Tsai, K. Y., Y. Hu, K. F. Macleod, D. Crowley, L. Yamasaki, and T. Jacks. 1998. Mutation of E2F-1 suppresses apoptosis and inappropriate S phase entry and extends survival of Rb-deficient mouse embryos. Mol. Cell 2:293–304. [DOI] [PubMed] [Google Scholar]
- 29.Weber, J. D., J. R. Jeffers, J. E. Rehg, D. H. Randle, G. Lozano, M. F. Roussel, C. J. Sherr, and G. P. Zambetti. 2000. p53-independent functions of the p19(ARF) tumor suppressor. Genes Dev. 14:2358–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wetmore, C., D. E. Eberhart, and T. Curran. 2001. Loss of p53 but not ARF accelerates medulloblastoma in mice heterozygous for patched. Cancer Res. 61:513–516. [PubMed] [Google Scholar]
- 31.Yarbrough, W. and Y. Xiong. INK4a and ARF: crossroads of tumorigenesis. In J. R. Bertino (ed.), Encyclopedia of cancer, 2nd ed., in press. Academic Press, San Diego, Calif.
- 32.Zhang, Y., and Y. Xiong. 2001. Control of p53 ubiquitination and nuclear export by MDM2 and ARF. Cell Growth Differ. 12:175–186. [PubMed] [Google Scholar]
- 33.Zhang, Y., Y. Xiong, and W. G. Yarbrough. 1998. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell 92:725–734. [DOI] [PubMed] [Google Scholar]
- 34.Ziebold, U., T. Reza, A. Caron, and J. A. Lees. 2001. E2F3 contributes both to the inappropriate proliferation and to the apoptosis arising in Rb mutant embryos. Genes Dev. 15:386–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zindy, F., C. M. Eischen, D. H. Randle, T. Kamijo, J. L. Cleveland, C. J. Sherr, and M. F. Roussel. 1998. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 12:2424–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]