Abstract
Individual steps in the processing of pre-mRNA, including 5′-end cap formation, splicing, and 3′-end processing (cleavage and polyadenylation) are highly integrated and can influence one another. In addition, prior splicing can influence downstream steps in gene expression, including export of mRNA from the nucleus. However, the factors and mechanisms coordinating these steps in the maturation of pre-mRNA transcripts are not well understood. In the present study we demonstrate that SRm160 (for serine/arginine repeat-related nuclear matrix protein of 160 kDa), a coactivator of constitutive and exon enhancer-dependent splicing, participates in 3′-end formation. Increased levels of SRm160 promoted the 3′-end cleavage of transcripts both in vivo and in vitro. Remarkably, at high levels in vivo SRm160 activated the 3′-end cleavage and cytoplasmic accumulation of unspliced pre-mRNAs, thereby uncoupling the requirement for splicing to promote the 3′-end formation and nuclear release of these transcripts. Consistent with a role in 3′-end formation coupled to splicing, SRm160 was found to associate specifically with the cleavage polyadenylation specificity factor and to stimulate the 3′-end cleavage of splicing-active pre-mRNAs more efficiently than that of splicing-inactive pre-mRNAs in vitro. The results provide evidence for a role for SRm160 in mRNA 3′-end formation and suggest that the level of this splicing coactivator is important for the proper coordination of pre-mRNA processing events.
The processing of pre-mRNA to mature mRNA involves the adding of a 5′ m7GpppG cap, splicing, and 3′-end processing (cleavage and polyadenylation). Although each of these processing steps can occur independently, increasing evidence indicates they are, in fact, highly integrated and coordinated with each other as well as with transcription by RNA polymerase II (pol II) (reviewed in reference 18). Independent of transcription, formation of a 5′ cap binding complex facilitates the recognition of the adjacent, downstream, 5′ splice site, thereby promoting the definition of cap-proximal exons (21, 26). The cap binding complex can also activate the 3′-end formation of transcripts lacking introns (13). Splicing of 3′-end-most introns and 3′-end processing can stimulate each other, and interactions between splicing and polyadenylation factors are important for the definition of terminal exons in transcripts (1, 17, 27, 33–35, 43, 47).
Other studies have provided evidence that splicing and 3′-end formation are also highly coordinated with the nuclear retention and export of transcripts. Recognition of the AAUAAA polyadenylation signal by 3′-end cleavage factors is required for transcription termination as well as for 3′-end formation and therefore is necessary for the release of pol II transcripts from the nucleus. In addition, intron-containing transcripts are not normally exported because they are retained in the nucleus by interactions with splicing factors (8, 10, 24, 42). Aside from releasing transcripts from nuclear retention, it has been reported recently that splicing can promote the nuclear export of some transcripts, since the corresponding transcripts derived from intronless constructs were exported less efficiently (28, 39, 49).
Despite the numerous examples of coupling between different steps in mRNA processing and export, the factors and mechanisms involved are not well understood. Pre-mRNA splicing involves the step-wise association with transcripts of snRNPs, including U1, U2, U4/U6, and U5 snRNPs, and non-snRNP splicing factors, which include SR (serine/arginine repeat) family and SR-related proteins (reviewed in references 4, 7, 14, 15, 23, and 38). Together these factors form the spliceosome, which executes splicing catalysis. Formation of a poly(A) tail, which is specified by the highly conserved AAUAAA polyadenylation signal and a downstream G- or G/U-rich element, is catalyzed by multisubunit complexes in two steps: cleavage and then polyadenylation (reviewed in references 9 and 44). Several studies have provided evidence that different splicing factors can interact with components of the cleavage and polyadenylation machinery and either stimulate or inhibit polyadenylation (16, 17, 27, 29, 43, 47).
In previous studies we and others identified SRm160 (the SR-related nuclear matrix protein of 160 kDa), an SR-related protein which functions as a coactivator of both constitutive and exon enhancer-dependent splicing by forming cross-intron interactions with multiple splicing factors bound directly to pre-mRNA (3, 5, 12). It has been reported recently that SRm160, together with several other factors, including the acute myeloid leukemia-associated protein DEK, the splicing activator RNPS1, the hnRNP-like shuttling protein Y14, and the mRNA shuttling and export factor REF/Aly, bind to mRNAs in a splicing-dependent manner (22, 25, 31, 49). This finding has suggested that SRm160 might participate in one or more steps in mRNA metabolism influenced by prior splicing, including mRNA export.
In the present study we demonstrate that SRm160 can activate the 3′-end cleavage of transcripts both in vitro and in vivo. Consistent with a role in the coupling of splicing and 3′-end formation, SRm160 was found to interact specifically with the cleavage polyadenylation specificity factor (CPSF) and to be more active in promoting the cleavage of splicing-active substrates than of splicing-inactive substrates in vitro. Surprisingly, a consequence of overexpression of SRm160 in vivo was the uncoupling of the requirement for splicing to promote the 3′-end cleavage and transport of transcripts to the cytoplasm. The results provide evidence for a role for SRm160 in 3′-end processing and demonstrate that the level of this splicing coactivator is critical for maintaining the coordination of pre-mRNA processing events.
MATERIALS AND METHODS
Plasmids.
Details of reporter and RNase protection-probe plasmids can be found at http://www.utoronto.ca/intron/supp_info. The predicted sizes for the various RNase protection products, generated by using the probes shown in Fig. 1A, 2A, and 4B, can also be found at the above website. The pol III-adenovirus-associated (VA) reporter (pSPVA) and the corresponding RNase protection probe plasmid have been described previously (48).
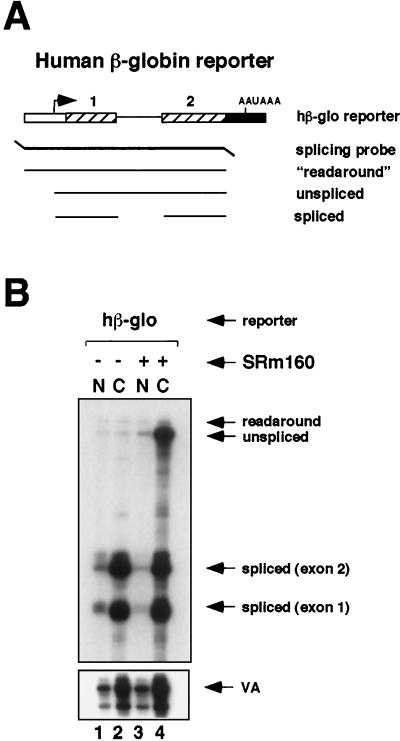
FIG. 1.
Expression of high levels of SRm160 in vivo results in the cytoplasmic accumulation of unspliced RNA. (A) Schematic representation of the RNase protection probe used to analyze splicing of transcripts from a reporter consisting of hβ-glo exons 1 and 2 with the intervening intron. The predicted RNase protection products are shown below the probe (for sizes refer to Materials and Methods and supplementary information located at http://www.utoronto.ca/intron/supp_info). (B) Human 293 cells were transiently transfected with the hβ-glo pre-mRNA reporter and a pol III reporter (pSPVA) as an internal control (lanes 1 to 4) together with a control, empty expression vector (pcDNA3-Flag) (lanes 1 and 2) or an expression vector for Flag epitope-tagged SRm160 (pcDNA3-fSRm160) (lanes 3 and 4). Proportional amounts of RNA isolated from the nuclear (N) and cytoplasmic (C) fractions were analyzed by RNase protection using the probe illustrated in panel A.
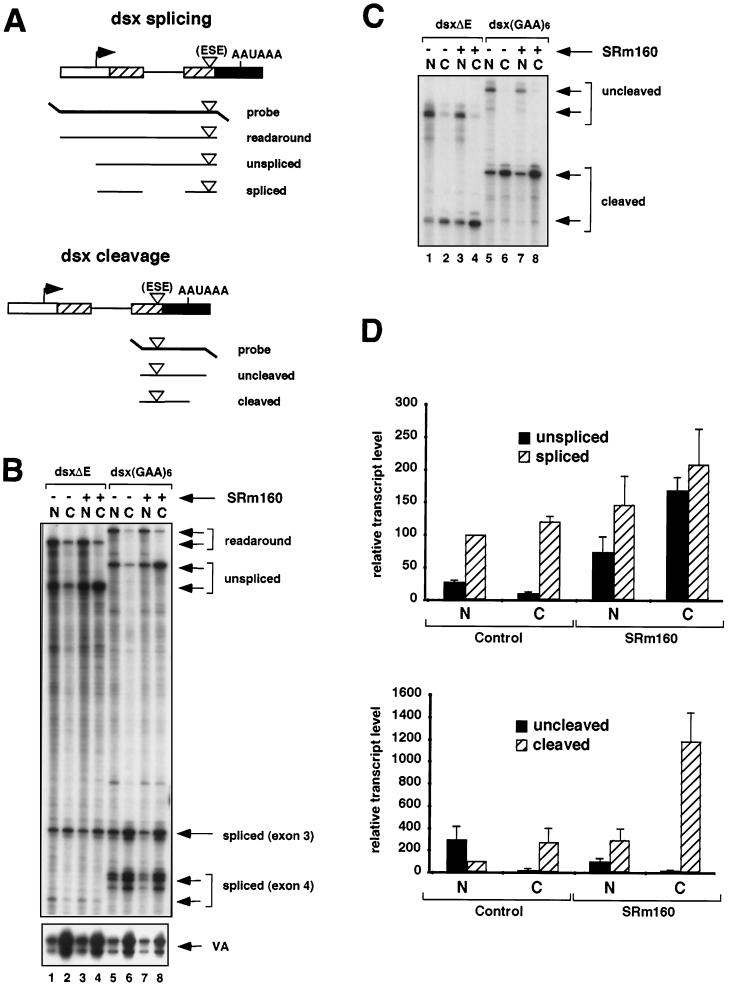
FIG. 2.
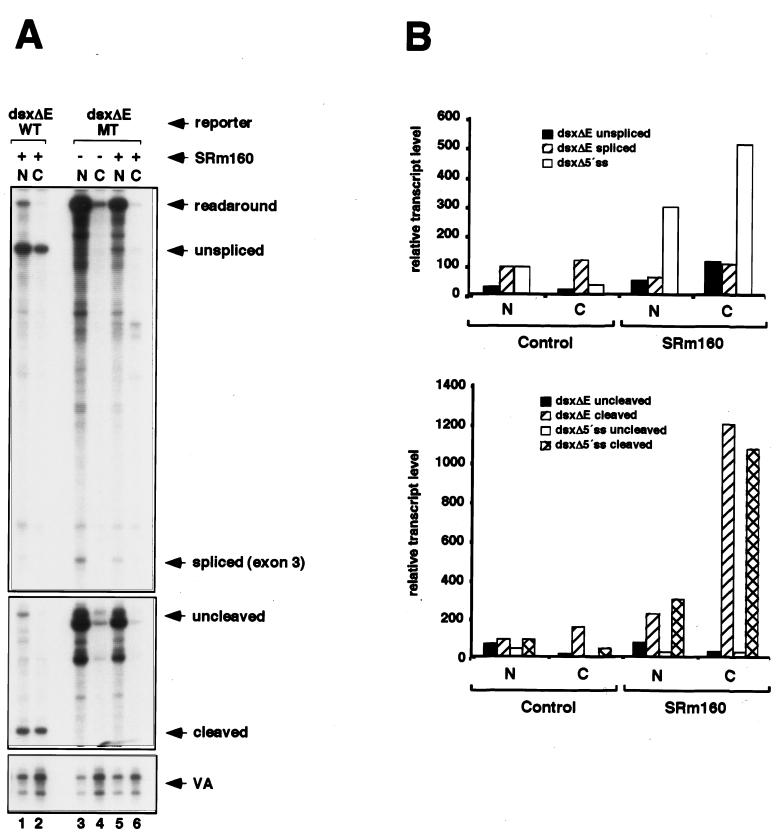
Increased expression of SRm160 promotes 3′-end cleavage in vivo. (A) Schematic representation of the RNase protection probes used to analyze splicing and 3′-end cleavage in pre-mRNAs transcribed from reporters derived from exons 3 and 4 of the Drosophila doublesex gene (dsx). The dsx reporters contained either no ESE (dsxΔE) or a six-GAA repeat ESE [dsx(GAA)6]. The predicted RNase protection products are shown below each probe (for sizes refer to Materials and Methods and supplementary information available at http://www.utoronto.ca/intron/supp_info). (B and C) Human 293 cells were transiently transfected with the dsxΔE (lanes 1 to 4) or dsx(GAA)6 reporter (lanes 5 to 8) together with a control expression vector containing no insert (pcDNA3-Flag) (lanes 1 and 2 and 5 and 6) or an expression vector for Flag epitope-tagged SRm160 (pcDNA3-fSRm160) (lanes 3 and 4 and 7 and 8); the pol III reporter (pSPVA) was cotransfected in each case as an internal control. Proportional amounts of RNA isolated from the nuclear (N) and cytoplasmic (C) fractions of the transfected cells were analyzed by RNase protection using either the splicing protection probe (B) or the 3′-end protection probe (C). The identity of each RNA species is indicated. Note that exon 4 of the dsxΔE pre-mRNA, migrating near the bottom of the gel in panel B, is less strongly detected by the [32P]UTP-labeled RNase protection probe due to the low A content of this exon. It was, however, readily detected following a longer exposure of the gel (data not shown). (D) Quantification of the effect of SRm160 expression on the yields of different RNA species transcribed from the dsxΔE reporter. RNA isolated from transfected cells was analyzed by RNase protection as described above, and the amounts of unspliced, spliced, uncleaved, and cleaved RNAs from three independent experiments were quantified. The values were normalized for both VA signal and U content. For the purpose of averaging different experiment values, the nuclear spliced RNA and the nuclear cleaved RNA were normalized to a value of 100.
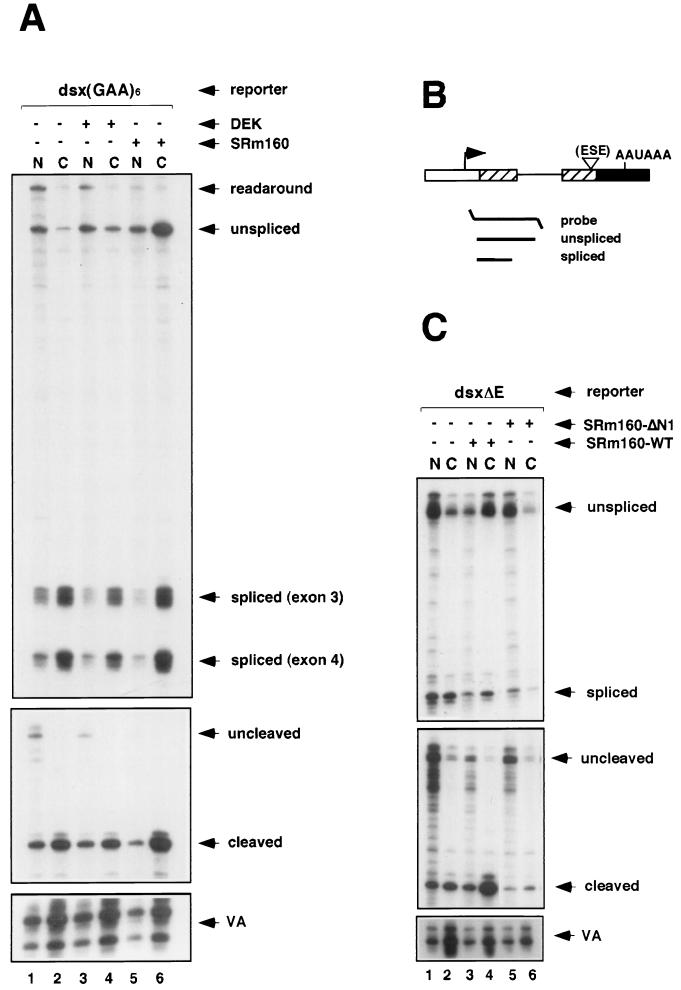
FIG. 4.
Specificity of the cleavage-stimulatory and transcript export activities of SRm160. (A) Human 293 cells were transiently transfected with the dsx(GAA)6 reporter together with a control expression vector containing no insert (pcDNA3-Flag) (lanes 1 and 2), an expression vector for HA epitope-tagged DEK (pcDNA3-DEK) (lanes 3 and 4), or an expression vector for Flag epitope-tagged SRm160 (pcDNA3-fSRm160) (lanes 5 and 6); the pol III reporter (pSPVA) was cotransfected in each case as an internal control. Proportional amounts of RNA isolated from the nuclear (N) and cytoplasmic (C) fractions of the transfected cells were analyzed by RNase protection using either a splicing protection probe or the 3′-end protection probe (refer to the legend to Fig. 2). The identity of each RNA species is indicated. (B) Schematic representation of the short splicing RNase protection probe used to analyze splicing of transcripts from the dsxΔE reporter. The predicted RNase protection products are shown below each probe (for sizes refer to Materials and Methods and supplementary information available at http://www.utoronto.ca/intron/supp_info). (C) Human 293 cells were transiently transfected with the dsxΔE reporter together with a control expression vector containing no insert (pcDNA3-Flag) (lanes 1 and 2), an expression vector for Flag epitope-tagged SRm160 (pcDNA3-fSRm160) (lanes 3 and 4), or an expression vector for Flag epitope-tagged SRm160 deleted from amino acids 1 to 155 (pcDNA3-fSRm160ΔN1) (lanes 5 and 6); the pol III reporter (pSPVA) was cotransfected in each case as an internal control. Proportional amounts of RNA isolated from the nuclear (N) and cytoplasmic (C) fractions of the transfected cells were analyzed by RNase protection using either the short splicing protection probe or the 3′-end protection probe. The identity of each RNA species is indicated.
Transfections.
Human 293 cells were grown in Dulbecco’s modified Eagle medium containing 10% fetal bovine serum to a density of 2 × 106 cells per 150-mm-diameter plate prior to transient transfection using calcium phosphate precipitation. Transfections were typically performed with 5 μg of reporter plasmid, 5 to 10 μg of pcDNA3-Flag-SRm160, or the corresponding empty vector. Each transfection contained 0.5 μg of the pol III-VA RNA reporter as an internal control for transfection efficiency as well as for recovery of RNA in the nuclear and cytoplasmic fractions. Following transfection the culture medium was changed after 16 h and the cells were harvested after 40 h.
RNA preparation and analysis.
Nuclear and cytoplasmic RNA fractions from the transfected cells were prepared as previously described (19, 30). In each experiment 10% (2 to 5 μg) of the total amount of RNA recovered from each fraction was analyzed. Proportional amounts of RNA from the nuclear and cytoplasmic fractions were analyzed. RNase protections were performed using gel-purified probes, as described by Yankulov et al. (48), except that incubations with RNase were performed for 1 h. RNase protection products were quantified by using a Molecular Dynamics PhosphorImager and ImageQuant software. Each RNA species was quantified, following background subtraction and normalization for VA signal and U content of the protected probe fragment. In order to compare the relative amounts of RNAs from different experiments, the corrected signals were further normalized by being adjusted to the level of spliced or cleaved nuclear RNA (value set to 100). For quantification of splicing of the dsx pre-mRNA reporters exon 3 was measured, since it has a higher A content and was easier to visualize than exon 4. Experimental values were averaged from multiple experiments (n = 3 or 4), and standard deviations were calculated by using Microsoft Excel.
Antibodies and immunoprecipitation.
Immunoprecipitation of SRm160-containing complexes was performed as described by Eldridge et al. (12). Immunoprecipitation was performed by using a murine monoclonal antibody (MAb) specific for SRm160 (MAb-B1C8) (6, 45) and a control rabbit anti-mouse antibody. Immunoprecipitation with antibodies to CPSF-73, poly(A) polymerase (PAP), and the 77-kDa subunit of human cleavage stimulation factor (CstF-77) was performed using 10 μg of affinity-purified antibody (kind gift of D. Bentley) cross-linked to protein A-Sepharose. Prior to immunoprecipitation, nuclear extract was preincubated for 15 min at 30°C under splicing conditions, with or without the addition of an RNase cocktail (7 ng/μl; Boehringer), and in the presence of phosphatase inhibitors (potassium fluoride, sodium pyrophosphate, and sodium β-glycerophosphate). RNA isolated (by phenol-chloroform extraction and precipitation) from aliquots of the nuclear extracts incubated with or without RNase was separated on a 10% denaturing polyacrylamide gel and detected by ethidium bromide staining.
In vitro transcription and in vitro cleavage reactions.
Transcription of RNase protection probes and substrates for in vitro splicing assays was performed essentially as described previously (5). Details of the substrates used for analyzing 3′-end cleavage in vitro can be found at http://www.utoronto.ca/intron/supp_info. The MXSVL derivatives were transcribed by using SP6 RNA polymerase from DraI-linearized pMXSVL-WT and pMXSVL-Δ5′ templates, respectively (35). The cleavage reactions were performed essentially as described previously by Niwa et al. (35), except that the reaction mixtures were preincubated on ice for 10 min with or without baculovirus-expressed SRm160 (bSRm160) and then were preincubated for 2 min at 30°C. Substrates were then added and the reaction mixtures were incubated for 1 h at 30°C. Each reaction mixture contained 2.2 μl of nuclear extract and 1.5 mM MgCl2 (final concentration) and was incubated with or without 5′-cordycepin triphosphate. Purified bSRm160 was added to the reaction mixtures in the amounts described in the legend to Fig. 5.
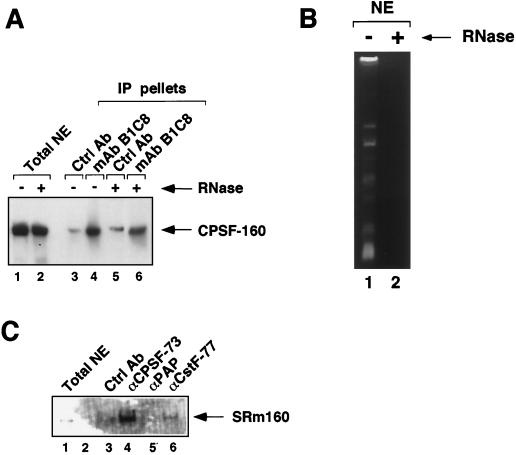
FIG. 5.
SRm160 associates with the 3′-end cleavage machinery. (A) Immunoprecipitates (IP) were collected from HeLa nuclear extract (NE) by using the SRm160-specific MAb (MAb-B1C8) (lanes 4 and 6) and excess levels of a control antibody (rabbit anti-mouse immunoglobulin; Ctrl Ab) (lanes 3 and 5). The immunoprecipitates were separated on a 7.5% sodium dodecyl sulfate-polyacrylamide gel and immunoblotted with an affinity-purified antibody specific for the 160-kDa subunit of the human CPSF (CPSF-160). Total nuclear extract, separated in lanes 1 and 2, represents 7% of the amount of extract used in each immunoprecipitation. Nuclear extract was preincubated in the presence (lanes 2, 5, and 6) or absence (lanes 1, 3, and 4) of RNase prior to immunoprecipitation. (B) Analysis of the RNA content of the nuclear extract used for immunoprecipitation shown in panel A. RNA isolated from nuclear extract pretreated with (lane 2) or without (lane 1) RNase was analyzed on a 10% denaturing acrylamide gel stained with ethidium bromide. (C) Immunoprecipitates were collected from RNase-pretreated HeLa nuclear extract by using rabbit polyclonal antibodies to the 73-kDa subunit of human CPSF (CPSF-73) (lane 4), PAP (lane 5), CstF-77 (lane 6), and a control antibody (rabbit anti-glutathione S-transferase) (lane 3). The immunoprecipitates were separated on a 7.5% sodium dodecyl sulfate- polyacrylamide gel and immunoblotted with MAb-B1C8. Total nuclear extract, separated in lane 1, represents ∼0.5% of the amount of extract used in each immunoprecipitation.
RESULTS
In order to investigate whether SRm160 influences steps in mRNA maturation besides splicing, human 293 cells were transfected with plasmids containing different pre-mRNA reporters, with or without an expression plasmid encoding SRm160 fused to an N-terminal FLAG epitope (pcDNA3-fSRm160). Each pre-mRNA reporter contained an upstream promoter derived from the adenovirus major-late region and a 3′-late poly(A) signal from simian virus 40 (SV40). Included in all transfections was an RNA pol III-VA RNA reporter plasmid (pSPVA), which serves as an internal control for monitoring the relative transfection efficiency and recovery of RNA between samples. RNA was isolated from both the nuclear and cytoplasmic fractions of the transfected cells and was analyzed by RNase protection in order to determine whether increased levels of SRm160 influence the nuclear-cytoplasmic distribution of transcripts, as well their processing.
Elevated levels of SRm160 in vivo result in the cytoplasmic accumulation of unspliced pre-mRNA transcripts.
The effect of increased expression of SRm160 was first tested on the processing of a model substrate derived from exons 1 and 2 of human β-globin (hβ-glo) pre-mRNA (Fig. 1A). Immunoblotting experiments indicated that, in a typical transfection experiment, the level of fSRm160 was approximately 10- to 20-fold higher than the level of endogenous SRm160 (data not shown). RNA in the nuclear and cytoplasmic fractions was analyzed by using the RNase protection probe illustrated in Fig. 1A, which spans from 99 bases upstream of the start site of transcription to the 3′ end of exon 2. In the control transfection the hβ-glo transcripts were processed and exported efficiently, resulting in the accumulation of spliced mRNA almost exclusively in the cytoplasmic fraction (Fig. 1B, lanes 1 and 2). Surprisingly, however, when fSRm160 was expressed a high level of unspliced transcripts accumulated in the cytoplasmic fraction in addition to spliced mRNA (lanes 3 and 4). The size of the protected product indicates that these unspliced transcripts are correctly initiated. In several independent experiments the level of the VA RNA did not change significantly following fSRm160 expression (the increase observed between lanes 2 and 4 in Fig. 1B is less than twofold and most likely is due to variation in transfection efficiency; refer also to data in Fig. 2). Therefore, the effect of increased fSRm160 expression on promoting the cytoplasmic accumulation of unspliced hβ-glo transcripts appears to be specific for pol II transcripts. Thus, increased SRm160 expression appears to prevent or bypass splicing in addition to uncoupling interactions that normally would retain unspliced transcripts in the nucleus.
We next determined whether expression of fSRm160 influences the processing and nuclear-cytoplasmic distribution of another model transcript, derived from exons 3 and 4 of the doublesex gene of Drosophila (dsx pre-mRNA) (Fig. 2). The dsx pre-mRNA contains a suboptimal 3′-splice site-polypyrimidine tract and requires an exonic splicing enhancer (ESE) in exon 4 for efficient splicing. Previously it was shown that SRm160 is required for a mammalian ESE, consisting of 6xGAA repeats, to promote the splicing of this substrate in vitro (12). In order to investigate whether SRm160 influences the processing and/or nucleocytoplasmic distribution of this substrate in vivo, we next compared the effect of its increased expression on dsx reporters containing or lacking a 6xGAA repeat ESE in exon 4, designated dsx(GAA)6 and dsxΔE, respectively (Fig. 2A).
RNA isolated from the nuclear and cytoplasmic fractions of cells transfected with the dsxΔE and dsx(GAA)6 reporters, with or without pcDNA3-fSRm160, was analyzed by RNase protection using the probes illustrated in Fig. 2A. The dsx splicing probe spans from −99 bases upstream of the start site of transcription to the 3′ end of exon 4, allowing the detection of unspliced and spliced transcripts as well as readaround transcripts, which can arise by inefficient termination of transcription or incorrect initiation at cryptic promoters. In the absence of fSRm160 expression, insertion of the 6xGAA ESE in exon 4 resulted in an increase in the efficiency of splicing, similar to its activity observed in vitro (for an example, see reference 12) (Fig. 2B, compare lanes 1 and 2 with lanes 5 and 6). Consistent with numerous previous reports demonstrating the nuclear retention of unprocessed transcripts, the unspliced pre-mRNA and readaround transcripts from both reporter plasmids were detected primarily in the nuclear fractions (compare lanes 1 and 5 with lanes 2 and 6), whereas the majority of spliced transcripts from each reporter were detected in the cytoplasmic fractions (compare lanes 2, 4, 6, and 8 with lanes 1, 3, 5, and 7).
Similar to the results obtained with the hβ-glo reporter transcripts shown in Fig. 1, fSRm160 expression resulted in a pronounced increase in the level of unspliced pre-mRNA in the cytoplasmic fraction for both the dsxΔE and dsx(GAA)6 transcripts (compare lanes 2 and 4 and lanes 6 and 8). The ratio of cytoplasmic to nuclear unspliced dsxΔE and dsx(GAA)6 pre-mRNA transcripts increased eightfold and sevenfold, respectively (average values from four independent experiments), indicating that the effect is not dependent on the presence of the ESE (see Fig. 2D; data not shown). As observed for the hβ-glo reporter transcripts, the increase in cytoplasmic unspliced pre-mRNA did not coincide with a significant decrease in the level of pre-mRNA in the nuclear fractions, again indicating that fSRm160 expression results predominantly in the cytoplasmic accumulation of a population of unspliced transcripts. Also similar to the results with the hβ-glo transcripts, increased expression of fSRm160 did not significantly affect the levels of the spliced dsxΔE or dsx(GAA)6 transcripts in the nuclear or cytoplasmic fractions (Fig. 2B, compare lanes 3 and 4 with 1 and 2 and lanes 7 and 8 with 5 and 6). In both cases it is possible that a population of pre-mRNA transcribed from these reporters is processed and exported prior to fSRm160 reaching levels which result in the cytoplasmic accumulation of unspliced transcripts (see Discussion). Thus, the results demonstrate that elevated levels of SRm160 can promote the cytoplasmic accumulation of distinct pre-mRNA transcripts.
SRm160 promotes the 3′-end cleavage of transcripts in vivo.
A prerequisite for the nuclear export of unspliced pre-mRNA is that it is released from nuclear retention factors, which can include transcribing RNA pol II as well as factors involved in the formation of splicing complexes (8, 10, 24, 42). Release of transcripts from pol II involves recognition of the AAUAAA poly(A) site by 3′-end cleavage factors, which is important for efficient transcription termination as well as 3′-end cleavage (2, 37). One possibility is that increased SRm160 expression allows the release of unspliced transcripts from the nucleus by facilitating 3′-end formation without the requirement for concomitant splicing. To investigate this we asked whether fSRm160 expression influences the 3′-end cleavage of the dsx transcripts. Accordingly, the same samples shown in Fig. 2B were analyzed with RNase protection probes designed to monitor 3′-end cleavage (Fig. 2C; refer to Fig. 2A).
Consistent with an important role for splicing in the promotion of 3′-end processing, in the absence of fSRm160 expression a significant increase in the ratio of cleaved to uncleaved transcripts was detected as a result of insertion of the 6xGAA ESE in exon 4 (Fig. 2C, compare lanes 1 and 2 with lanes 5 and 6). This increase was 9-fold in the nuclear fraction and 14-fold in the cytoplasmic fraction (data not shown). Moreover, in agreement with an important role for 3′-end processing in facilitating the release of transcripts from the nucleus, essentially all of the transcripts detected in the cytoplasmic fractions were 3′-end cleaved, whereas the uncleaved transcripts were almost entirely detected in the nuclear fractions (Fig. 2C, compare lanes 1 and 2 and lanes 5 and 6). Significantly, expression of fSRm160 resulted in a further increase [fourfold for dsxΔE and twofold for dsx(GAA)6, as determined from averaging values from three independent experiments] in the level of cleaved transcripts in the cytoplasmic fractions, concurring with the increased levels of cytoplasmic unspliced transcripts in these fractions (compare lanes 4 and 8 with 2 and 6 in Fig. 2B and C; see also Fig. 2D) (data not shown). The consistently higher level of cleavage-stimulatory activity of fSRm160 observed for the dsxΔE transcripts compared to that of the dsx(GAA)6 transcripts suggests that the level of cleavage promoted by the 6xGAA repeat ESE may already be near saturation. In agreement with the results obtained with the dsx pre-mRNA reporters and with the results in Fig. 1B, elevated levels of SRm160 also resulted in the presence of cleaved, unspliced, hβ-glo pre-mRNA transcripts in the cytoplasmic fractions (data not shown). Thus, elevated levels of SRm160 in vivo appear to facilitate the nuclear release of different unspliced transcripts by stimulating 3′-end cleavage.
Promotion of transcript release from the nucleus by SRm160 requires a wild-type polyadenylation signal.
In order to confirm whether SRm160 acts to promote the nuclear release of unspliced transcripts by activating 3′-end formation, we compared its activity on dsxΔE transcripts containing either a wild-type (AAUAAA, dsxΔE-WT) or a mutant, inactive (AAGAAA, dsxΔE-MT), SV40 late poly(A) signal (Fig. 3A). In agreement with previous experiments indicating that the AAUAAA signal is required for 3′-end cleavage and for RNA pol II to terminate (2, 37), mutation of the poly(A) signal resulted in the accumulation to high levels in the nuclear fraction of uncleaved, readaround transcripts (Fig. 3A, lanes 3 and 5). Although these readaround transcripts probably arise as a consequence of the loss of efficient termination of transcription, we cannot exclude the alternative possibility that they also arise through incorrect initiation of transcription. However, in either case the results demonstrate that fSRm160 expression did not result in the 3′-end cleavage or cytoplasmic accumulation of the dsxΔE-MT transcripts. It is noteworthy that coexpression of fSRm160 did not result in a significant change in the level of readaround transcripts in the nuclear fraction; the slight decrease observed in Fig. 3A is probably due to experimental variation, since it was not observed in repeat experiments (lanes 3 and 5) (data not shown). This indicates that the cytoplasmic accumulation of unspliced transcripts following fSRm160 expression is not a consequence of increased levels of transcription from the reporter plasmids. Results similar to those described above were observed between wild-type and mutant poly(A) signal derivatives of the dsx(GAA)6 and hβ-glo reporter transcripts (data not shown). These data confirm that activation of 3′-end cleavage by fSRm160 requires the presence of the AAUAAA poly(A) signal and also demonstrate that increased levels of SRm160 do not promote the nuclear release of transcripts without 3′-end cleavage.
FIG. 3.
Mutation of the polyadenylation signal, but not deletion of the 5′ splice site, prevents cleavage and cytoplasmic accumulation of unspliced RNA by SRm160. (A) 293 cells were transiently transfected with the dsxΔE reporter containing either a wild-type (dsxΔE-WT) (lanes 1 and 2) or mutant (dsxΔE-MT) (lanes 3 to 6) polyadenylation signal (see the text). The cells were cotransfected with a control, empty expression vector (pcDNA3-Flag) (lanes 3 and 4) or an expression vector for Flag epitope-tagged SRm160 (pcDNA3-fSRm160) (lanes 1 and 2 and 5 and 6) and with pSPVA as an internal control. Proportional amounts of RNA isolated from the nuclear (N) and cytoplasmic (C) fractions were analyzed by RNase protection using the splicing and cleavage probes shown in Fig. 2A. (B) Quantification of the effect of SRm160 expression on the nuclear and cytoplasmic levels of RNAs transcribed from a splicing-inactive dsx reporter, which lacks a functional 5′ splice site (dsxΔ5′ss), and the dsxΔE reporter. RNA isolated from transfected cells was analyzed by RNase protection as described in the legend to Fig. 2A, and the amounts of the unspliced, spliced, uncleaved, and cleaved RNAs were quantified as described in the legend to Fig. 2 (refer to supplementary information at the web site cited above for details of the dsxΔ5′ss reporter and protection probe).
SRm160 can promote the 3′-end cleavage and nuclear release of transcripts independently of splicing.
The results so far indicate that elevated levels of SRm160 in vivo can promote the 3′-end cleavage and cytoplasmic accumulation of transcripts without the requirement for concomitant splicing. To confirm whether this is the case, we next compared the ability of SRm160 to promote these activities on a splicing-inactive derivative of the dsxΔE reporter, which contains a deletion in the 5′ splice site (dsxΔ5′ss) (Fig. 3B). RNase protection analysis with a probe spanning the intron and exon sequences of this transcript confirmed that it was not spliced (data not shown). Similar to the results observed for the unspliced dsxΔE transcripts, RNase protection analysis using the dsxΔE 3′-end cleavage probe (refer to Fig. 2A) revealed that elevated levels of SRm160 resulted in a pronounced increase in the level of cleavage and cytoplasmic accumulation of the dsxΔ5′ss transcript (Fig. 3B and data not shown). This confirms that the presence of a functional intron and splicing is not required in order for SRm160 to promote 3′-end cleavage and nuclear release of transcripts. As will be expanded on below, although excess levels of SRm160 can result in the activation of 3′-end cleavage independently of splicing it may normally only promote 3′-end cleavage when coupled to splicing.
Specificity of the cleavage-stimulatory and export activities of SRm160.
In order to investigate the specificity of SRm160 in promoting 3′-end cleavage and cytoplasmic accumulation of transcripts in vivo, its activity was initially compared alongside the SRm160-interacting factor DEK, which (like SRm160) associates with splicing complexes and forms a component of a splicing-dependent exon junction complex (see the introduction). Transient transfection and RNase protection assays were performed as described above using the dsx(GAA)6 pre-mRNA reporter together with an expression plasmid encoding DEK (Fig. 4A). Although DEK was efficiently expressed, unlike SRm160 it did not result in a significant level of stimulation of 3′-end cleavage or accumulation of pre-mRNA in the cytoplasm (Fig. 4A, compare lanes 3 and 4 with lanes 1 and 2 and lanes 5 and 6) (data not shown).
Several FLAG epitope-tagged deletion derivatives of SRm160 were also tested for their ability to stimulate cleavage and cytoplasmic accumulation of transcripts. Although these deletion proteins were expressed as efficiently as wild-type fSRm160, the majority did not promote 3′-end cleavage or cytoplasmic-pre-mRNA accumulation of dsx reporter transcripts (data not shown). However, interpretation of the results was complicated, since many of the inactive deletion derivatives did not localize in the same manner as wild-type SRm160, raising the possibility that their lack of function could be a consequence of mislocalization. However, one of the deletions (fSRm160ΔN1), which lacks residues 1 to 155 corresponding to the highly conserved N-terminal domain of SRm160, localized in the same nuclear speckled pattern as wild-type SRm160 yet was not active in promoting the 3′-end cleavage and cytoplasmic accumulation of transcripts. This is demonstrated in the RNase protection analysis of dsxΔE pre-mRNA splicing and cleavage shown in Fig. 4C (compare lanes 3 and 4 with lanes 1 and 2 and lanes 5 and 6), in which cleavage was analyzed with the same probe as that described for Fig. 2A and splicing was analyzed by using the probe illustrated in Fig. 4B. This latter probe spans from the start of exon 3 to the middle of the dsx intron. It is noteworthy that, while inactive, SRm160ΔN1 retains the arginine/serine (RS) domain and other repeat motifs that are probably important for its correct localization and interactions with other pre-mRNA processing factors. This result indicates that intact SRm160 is important for stimulation of 3′-end cleavage and cytoplasmic accumulation of pre-mRNA and that both of these activities depend on the presence of the conserved N-terminal domain of the protein.
SRm160 interacts with the CPSF.
The activity of SRm160 in promoting 3′-end processing in the experiments described above could result from one or more indirect effects arising from its increased expression levels or the more interesting possibility that it participates more directly in promoting 3′-end cleavage. In order to distinguish between these possibilities we next asked whether SRm160 associates with one or more components of the 3′-end cleavage machinery.
Immunoprecipitates were collected from HeLa nuclear extract by using a MAb specific for SRm160 (MAb-B1C8) (6, 45). MAb-B1C8 has previously been shown to immunoprecipitate SRm160 in complexes containing SR family proteins, the SR-related proteins hTra2-beta and Hel117, and the oncoprotein DEK (12, 31) (our unpublished observations). The MAb-B1C8 immunoprecipitates were immunoblotted with available antisera specific for 3′-end processing factors, including CstF-77 (cleavage stimulation factor 77-kDa subunit), CPSF-160 (cleavage polyadenylation specificity factor 160-kDa subunit), and PAP [poly(A) polymerase]. Although all three of these antibodies detected proteins of the expected sizes in HeLa nuclear extract, only CPSF-160 was significantly enriched in the MAb-B1C8 immunoprecipitates (Fig. 5A, lane 4, and data not shown). Approximately 2% of CPSF-160 in the nuclear extract was immunoprecipitated with MAb-B1C8, indicating that only a low level of this cleavage factor interacts with SRm160 or else that this interaction is unstable during immunoprecipitation. Nevertheless, the interaction was specific, since CPSF-160 was not substantially coimmunoprecipitated with excess amounts of nonspecific control antibodies (lanes 3 and 5 and data not shown). Moreover, CPSF-160 was still coimmunoprecipitated by MAb-B1C8 after extensive pretreatment of the nuclear extract with RNase (Fig. 5B, compare lanes 1 and 2), indicating that it probably associates with SRm160 through protein-protein interactions (Fig. 5A, compare lanes 4 and 6).
In order to confirm whether SRm160 and CPSF associate specifically, a reciprocal immunoprecipitation experiment was performed in which immunoprecipitates were collected from RNase-pretreated HeLa nuclear extract using antisera specific for the 73-kDa subunit of CPSF (CPSF-73), PAP, and CstF-77 and then were immunoblotted with MAb-B1C8 (Fig. 5C). All of these antibodies immunoprecipitate their target proteins efficiently, whereas the anti-CPSF-160 antibody used above does not and therefore was not included in the analysis (data not shown). The anti-CPSF-73 antibody resulted in a significant level of enrichment of SRm160 (lane 4), whereas little or no coimmunoprecipitation was observed above the background level with a control antibody, the anti-PAP antibody, or the anti-CstF-77 antibody (compare lane 3 with lanes 5 and 6, respectively). Approximately 2 to 3% of SRm160 was coimmunoprecipitated with anti-CPSF-73, again indicating that a relatively low level of SRm160 and CPSF associate specifically in HeLa nuclear extract.
SRm160 stimulates 3′-end cleavage in vitro.
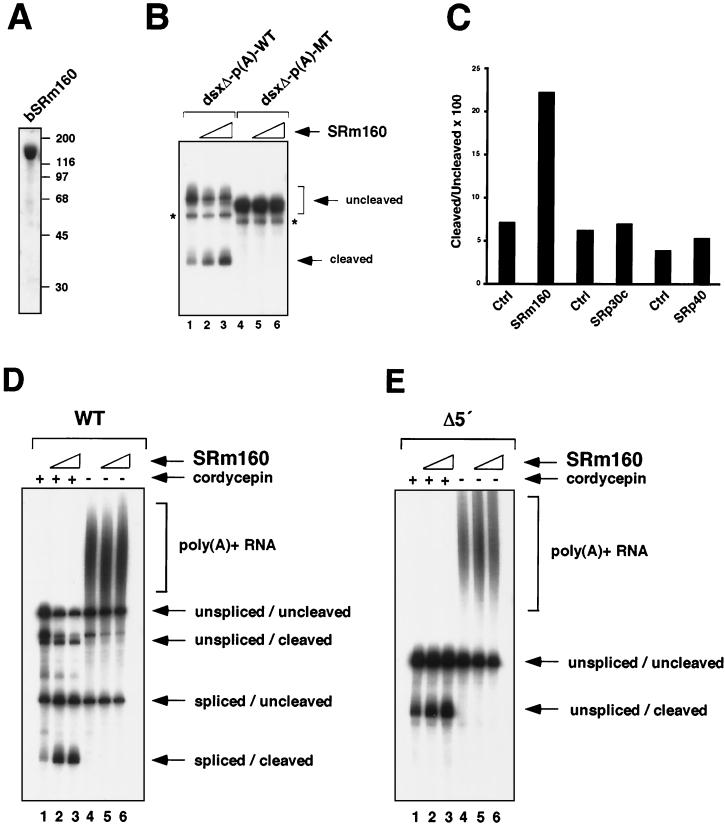
The activity of SRm160 in promoting 3′-end processing was next investigated by determining whether it can activate 3′-end cleavage of different RNA substrates with functional polyadenylation sites in vitro (Fig. 6). To first determine whether SRm160 can promote cleavage independent of splicing, highly purified baculovirus-expressed SRm160 (bSRm160) (Fig. 6A) (see reference 3) was added to 3′-end cleavage reaction mixtures containing a substrate derived from the 3′-half of exon 4 of the dsxΔE pre-mRNA, fused to either a wild-type [AAUAAA, dsxΔ-p(A)-WT] or mutant [AAGAAA, dsxΔ-p(A)-MT] late poly(A) signal from SV40 (Fig. 6B). Significantly, increasing amounts of bSRm160 stimulated, up to approximately threefold, cleavage of the dsxΔ-p(A)-WT but not the dsxΔ-p(A)-MT substrate (Fig. 6B, compare lanes 1 to 3 with lanes 4 to 6; note that although these substrates differ only by the U→G substitution in the AAUAAA sequence, they migrate differently due to a structural difference conferred by this substitution). The results demonstrate that SRm160, similar to its activity in vivo, can promote 3′-end cleavage in vitro. Moreover, its cleavage-stimulatory activity does not depend on the presence of active splice sites.
FIG. 6.
Highly purified, recombinant SRm160 preferentially stimulates cleavage of spliced substrates in vitro. (A) Analysis of recombinant, baculovirus-expressed SRm160 (bSRm160) by sodium dodecyl sulfate gel electrophoresis and Coomassie blue staining (see Materials and Methods for purification details). bSRm160 (4.5 μg) was loaded on the gel shown. (B) bSRm160 stimulates the 3′-end cleavage of a dsx substrate lacking functional splice sites in vitro. Reactions were performed with substrates derived from the 3′ half of dsx exon 4 (no ESE present) containing either a wild-type SV40 late poly(A) site [dsxΔ-p(A)-WT] (lanes 1 to 3) or a mutant poly(A) site [dsxΔ-p(A)-MT] (lanes 4 to 6) in the presence (lanes 2 and 3 and 5 and 6) or absence (lanes 1 and 4) of bSRm160. bSRm160 (165 ng) was added to the reaction shown in lanes 2 and 5, and 330 ng of bSRm160 was added to the reaction shown in lanes 3 and 6. The reactions were performed with added 5′-cordycepin triphosphate, as described in Materials and Methods. Duplicate reactions performed in the absence of 5′-cordycepin triphosphate confirmed the identity of the 3′-end cleaved bands (data not shown). Asterisks indicate a nonspecific degradation product not related to cleavage. (C) Specificity of the 3′-end stimulatory activity of SRm160. Approximately equal amounts (176 ng) of bSRm160, bSRp30c, and bSRp40, as assessed by Bradford assay, were added to 3′-end cleavage reaction mixtures incubated with the dsxΔ-p(A)-WT substrate. The reaction products were analyzed by electrophoresis on a denaturing gel and quantified by using a Molecular Dynamics PhosphorImager and ImageQuant software. (D and E) In vitro splicing and cleavage reactions containing a wild-type (WT) (D) or a 5′-splice site-deleted (Δ5′) (E) adenovirus pre-mRNA substrate, with an SV40 late polyadenylation signal (MSXVL) (35), were performed in the presence (lanes 2 and 3 and 5 and 6) or absence (lanes 1 and 4) of bSRm160. bSRm160 (110 ng) was added to the reaction shown in lanes 2 and 5, and 220 ng of bSRm160 was added to the reaction shown in lanes 3 and 6. The reactions were performed with (lanes 1 to 3) or without (lanes 4 to 6) added 5′-cordycepin triphosphate in order to distinguish the 3′-end cleaved products (see the text).
Specificity of the cleavage-stimulatory activity of SRm160 in vitro.
In order to investigate the specificity of the 3′-end cleavage-stimulatory function of SRm160 in vitro, we compared its activity with two different baculovirus-expressed SR proteins, SRp30c and SRp40 (Fig. 6C). Both of these proteins were purified to near homogeneity and were active in splicing reconstitution assays performed in HeLa S100 reactions (data not shown). Titration of equal amounts of these proteins as SRm160 in 3′-cleavage reaction mixtures containing the dsxΔ-p(A)-WT substrate did not significantly influence the ratio of cleaved to uncleaved substrate, whereas an approximately fourfold increase was consistently observed for SRm160 (Fig. 6C and data not shown). These differences in cleavage-stimulatory activity are not a consequence of differences in the lengths of the RS domains of these proteins, since SRp40 contains a higher number of consecutive SR/RS repeats than SRm160 whereas SRp30c contains fewer repeats. Moreover, we have observed that multiple domains of SRm160 other than the RS-rich regions of the protein are important for promoting 3′-end cleavage in vivo (see Fig. 4C) (our unpublished observations). Thus, the results indicate that the activity of SRm160 in stimulating 3′-end processing in vitro is not a general feature of RS domain proteins and, moreover, does not reflect the length of the RS domains of these splicing factors.
SRm160 augments the splicing-dependent enhancement of 3′-end cleavage in vitro.
The experiments presented so far demonstrate a cleavage-stimulatory activity of SRm160 that can function independently of splicing. However, at endogenous levels SRm160 normally associates stably only with transcripts in the context of functional splicing complexes (5, 12) and may, therefore, provide an important role in 3′-end formation coupled to splicing. To investigate whether the 3′-end cleavage-stimulatory property of SRm160 is augmented by the formation of functional splicing complexes, we next compared its influence on the 3′-end cleavage of splicing-active and -inactive derivatives of an adenovirus-derived pre-mRNA substrate (MXSVL) (35), each containing the late poly(A) signal from SV40 (Fig. 6D and E). Importantly, bSRm160 was added to splicing and cleavage reaction mixtures containing these substrates at levels that were stimulatory to splicing, thus allowing its activity in 3′-end cleavage to be assessed in the context of productive splicing complex formation.
Splicing and cleavage reaction mixtures containing wild-type MXSVL (Fig. 6D) or a splicing-inactive derivative lacking a 5′ splice site MXSVL (Δ5′) (Fig. 6E) were incubated with or without the ATP analog 5′-cordycepin triphosphate in order to distinguish the 3′-end cleaved products. In the presence of 5′-cordycepin triphosphate, polyadenylation is prevented, resulting in the accumulation of 3′-end cleaved products. In the absence of 5′-cordycepin triphosphate, the 3′-end cleaved products are polyadenylated and migrate as a smear in the upper region of the gel (Fig. 6D and E, compare lanes 1 to 3 with lanes 4 to 6). The different products detected in the reactions with the MXSVL substrates correspond to those previously characterized in detail (34, 35) and were assigned accordingly.
Quantification of the reaction intermediates and products revealed that, at the highest level of bSRm160 addition (220 ng) to reaction mixtures containing the WT-MXSVL substrate, only minor (less than twofold) changes in the level of unspliced and cleaved pre-mRNA and spliced and uncleaved transcripts were observed. However, there was a sevenfold increase in the level of spliced and cleaved mRNA (Fig. 6D, compare lanes 1 to 3) (data not shown). In contrast, addition of 220 ng of bSRm160 to reaction mixtures containing the Δ5′-MXSVL substrate resulted in an approximately threefold increase in the level of cleavage of this pre-mRNA (Fig. 6E, compare lanes 1 to 3). Similarly, addition of 220 ng of bSRm160 to an MXSVL substrate lacking a functional 3′ splice site also resulted in an approximately threefold increase in 3′-end cleavage (data not shown). Thus, in agreement with the results obtained with the dsxΔ-p(A) substrate (Fig. 6B), SRm160 can promote 3′-end cleavage of the MXSVL substrate independently of splicing. However, the presence of functional splice sites and splicing of the MXSVL pre-mRNA appears to augment the activity of bSRm160 in promoting 3′-end cleavage. These results, taken together with the data shown in Fig. 5 indicating that SRm160 can associate with CPSF, provide evidence that SRm160 participates in the coupling of splicing and 3′-end processing.
DISCUSSION
The results of the present study provide new information on the coordination of splicing with 3′-end formation and the nuclear-cytoplasmic transport of transcripts. SRm160, which previously was shown to promote both constitutive and exon enhancer-dependent splicing, was found to stimulate 3′-end cleavage. At elevated levels in vivo, SRm160 increased the levels of distinct pre-mRNAs, consistent with previous evidence that its specific ratio to other splicing factors is critical for optimal splicing. Under these conditions, SRm160 activated the 3′-end cleavage of the unspliced transcripts, thereby circumventing the normal requirement for splicing to promote the cleavage of these substrates. A consequence of the activation of pre-mRNA 3′-end cleavage by SRm160 was the accumulation of the unspliced transcripts in the cytoplasm. Thus, elevated levels of SRm160 appear to bypass the normal requirement of splicing for the nuclear release of transcripts. Consistent with a more direct role in activating the 3′-end formation of transcripts, SRm160 was found to associate with CPSF and to promote 3′-end cleavage in vitro. Importantly, although SRm160 promoted 3′-end cleavage independently of splicing, its cleavage-stimulatory activity was enhanced by the concomitant splicing of a transcript in vitro. In summary, the results demonstrate a role for SRm160 in 3′-end cleavage and provide evidence that the level of this splicing coactivator is important not only for optimal splicing but also for the coordination of splicing with 3′-end formation and nuclear retention of incompletely processed transcripts.
Coupling of splicing and 3′-end formation.
The mechanism(s) by which the splicing and 3′-end processing machineries communicate with each other is not well understood. Several reports have provided evidence for an important role for U1 snRNP components in 3′-end formation. Antibodies to Sm and U1 snRNP proteins were shown to inhibit polyadenylation in vitro (32). Subsequently, it was shown that U1 snRNA cross-links to polyadenylation efficiency elements upstream of the poly(A) site and that the efficiency of this cross-linking correlates with the efficiency of 3′-end formation (47). It was also reported that the U1 snRNP-A protein can interact with CPSF and promote increased polyadenylation in vitro (29). Other studies indicated that the binding of U1 snRNP and the SR family protein SRp20 to an intronic splicing enhancer sequence within the alternatively spliced, calcitonin/calcitonin gene-related peptide pre-mRNA correlates with increased 3′-end processing at an adjacent poly(A) site (27). In other contexts, both U1 snRNP-A and -70k proteins inhibit polyadenylation by interacting with poly(A) polymerase, whereas an interaction between poly(A) polymerase and the U2AF-65kDa subunit has been shown to increase splicing efficiency (16, 17, 43).
The association of SRm160 with pre-mRNA splicing substrates in vitro is normally strongly dependent on U1 snRNP and is further promoted by SR family proteins and U2 snRNP. It is possible that the activity of one or more U1 snRNP components promoting 3′-end formation described above could involve interactions mediated by SRm160. Although in the present study CPSF-160 was found to associate with SRm160, this interaction could be bridged by one or more intermediary factors. For example, in previous studies it was found that SRm160 interacts with several SR family and SR-related proteins (unpublished observations and references 5 and 12). Moreover, a recent report indicates that CPSF interacts indirectly with the cleavage factor (CF) Im (11), the 68-kDa subunit of which, like SRm160, is an SR-related protein (41). Since the alternating RS domains of SR family and SR-related proteins interact and are important for the formation of protein-protein interactions, it is possible that the RS domain of CF I-68 could interact with one or more SR family and/or SR-related splicing factors, including SRm160.
A possible role for RS domain proteins (other than SRm160) in modulating 3′-end cleavage is supported indirectly by previous observations. A 22-nucleotide element from the histone H2a gene, which promotes both the 3′-end formation and export of transcripts, binds to the SR family proteins 9G8 and SRp20 (20). Although antibodies to these SR family proteins inhibited mRNA export, it was not determined whether they also interfered with cleavage. Elevated expression of the SR family protein SC35 and the SR-related, helicase-like protein HRH1(hPRP22), like that of SRm160, can result in the cytoplasmic accumulation of pre-mRNA, although the mechanism(s) underlying these effects was not investigated (36, 46). Our results suggest that these factors could facilitate the nuclear release of transcripts by stimulating cleavage, perhaps in association with SRm160, or through functionally related yet distinct interactions. However, it is important to note that the effects we have observed for SRm160 are not general properties of RS domain proteins. Expression of elevated levels of the SR family protein ASF/SF2 does not result in 3′-end cleavage or cytoplasmic accumulation of transcripts (46; S. McCracken and B. J. Blencowe, unpublished observations), and elevated expression of U1-70K inhibits splicing but prevents the export of transcripts (40). Moreover, in the present study we have shown that, unlike SRm160, the SR family proteins SRp30c and SRp40 do not significantly influence 3′-end cleavage in vitro. Thus, SRm160 may be representative of a specific subset of SR proteins that can influence 3′-end processing and the nuclear-cytoplasmic distribution of transcripts.
SRm160 and the nuclear export of transcripts.
In order for RNA pol II transcripts to be efficiently exported from the nucleus, they must first be released from nuclear retention factors. Recognition of splicing signals by factors that function early in the formation of splicing complexes is important for the retention of unspliced pre-mRNA in the nucleus (8, 10, 24, 42). Elevated expression levels of SRm160 resulted in the accumulation of unspliced transcripts in the cytoplasm, suggesting that it can either prevent, or else bypass, splicing complex formation. Moreover, the accumulation of unspliced transcripts in the cytoplasm suggests that excess SRm160 might result in the bypass of processes that normally degrade unprocessed RNA in the nucleus. In conjunction with these roles, it is also possible that excess SRm160 prematurely activates an mRNA export pathway, allowing efficient export of unprocessed transcripts (see below). However, regardless of the mechanism(s) by which increased SRm160 expression results in the accumulation of unspliced transcripts in the cytoplasm, the present results demonstrate that SRm160 can facilitate the release of transcripts from nuclear retention by stimulating 3′-end formation. In particular, increased levels of SRm160 did not relieve the nuclear retention of unspliced transcripts containing a mutant poly(A) site, indicating that its ability to promote the nuclear release of unspliced transcripts is, at least in part, a consequence of its ability to stimulate the 3′-cleavage of these transcripts.
It is important to note that, although excess levels of SRm160 could stimulate 3′-end formation independently of splicing, levels of SRm160 that were not inhibitory to splicing were more efficient in stimulating the 3′-end cleavage of splicing-active than of splicing-inactive pre-mRNAs in vitro. Taken together with previous results demonstrating a requirement for U1 snRNP and SR family proteins for the association of SRm160 with pre-mRNA (5, 12), these findings suggest that SRm160 normally promotes 3′-end formation during the formation of productive splicing complexes. Furthermore, recent work has shown that SRm160 forms part of a splicing-dependent complex, 20 to 24 nucleotides upstream of exon-exon junctions, that contains several factors, including DEK, Y14, RNPS1, and the mRNA export factor REF/Aly (25, 49). It is therefore possible that the association of SRm160 with this complex at 3′-most exon-exon junctions might facilitate mRNA export by forging interactions with one or more export factors, including REF, as well as by promoting 3′-end cleavage. Similarly, increased expression of SRm160 could result in the recruitment of export factors to unspliced transcripts, thereby facilitating their nuclear release at a step in addition to 3′-end formation.
Acknowledgments
We thank D. Bentley, S. Berget, G. Grosveld, J. Manley, J. Nickerson, J. Yeakley, and X.-D. Fu for providing plasmids and antibodies and A. Mayeda for providing baculovirus-expressed SR family proteins. We also thank D. Bentley, J. Caceres, A. Cochrane, B. Graveley, L. Harrington, E. Izaurralde, E. Rosonina, and M. Ward for critical comments on the manuscript.
This work was supported by grants to B.J.B. from the U.S. Department of Defense Breast Cancer Research Program and the Canadian Institutes of Health Research (CIHR). B.J.B. is the recipient of a Medical Research Council of Canada/CIHR Scholar Award.
REFERENCES
- 1.Bauren, G., S. Belikov, and L. Wieslander. 1998. Transcriptional termination in the Balbiani ring 1 gene is closely coupled to 3′-end formation and excision of the 3′-terminal intron. Genes Dev 12:2759–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birse, C. E., L. Minvielle-Sebastia, B. A. Lee, W. Keller, and N. J. Proudfoot. 1998. Coupling termination of transcription to messenger RNA maturation in yeast. Science 280:298–301. [DOI] [PubMed] [Google Scholar]
- 3.Blencowe, B. J., G. Bauren, A. G. Eldridge, R. Issner, J. A. Nickerson, E. Rosonina, and P. A. Sharp. 2000. The SRm160/300 splicing coactivator subunits. RNA 6:111–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blencowe, B. J., A. L. Bowman, S. McCracken, and E. Rosonina. 1999. SR-related proteins and the processing of messenger RNA precursors. Biochem. Cell Biol. 77:277–291. [PubMed] [Google Scholar]
- 5.Blencowe, B. J., R. Issner, J. A. Nickerson, and P. A. Sharp. 1998. A coactivator of pre-mRNA splicing. Genes Dev. 12:996–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blencowe, B. J., J. A. Nickerson, R. Issner, S. Penman, and P. A. Sharp. 1994. Association of nuclear matrix antigens with exon-containing splicing complexes. J. Cell Biol. 127:593–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burge, C., T. Tuschl, and P. A. Sharp. 1999. Splicing of precursors to mRNAs by the spliceosomes, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 8.Chang, D. D., and P. A. Sharp. 1989. Regulation by HIV Rev depends upon recognition of splice sites. Cell 59:789–795. [DOI] [PubMed] [Google Scholar]
- 9.Colgan, D. F., and J. L. Manley. 1997. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 11:2755–2766. [DOI] [PubMed] [Google Scholar]
- 10.Custodio, N., M. Carmo-Fonseca, F. Geraghty, H. S. Pereira, F. Grosveld, and M. Antoniou. 1999. Inefficient processing impairs release of RNA from the site of transcription. EMBO J. 18:2855–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Vries, H., U. Ruegsegger, W. Hubner, A. Friedlein, H. Langen, and W. Keller. 2000. Human pre-mRNA cleavage factor II(m) contains homologs of yeast proteins and bridges two other cleavage factors. EMBO J. 19:5895–5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eldridge, A. G., Y. Li, P. A. Sharp, and B. J. Blencowe. 1999. The SRm160/300 splicing coactivator is required for exon-enhancer function. Proc. Natl. Acad. Sci. USA 96:6125–6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flaherty, S. M., P. Fortes, E. Izaurralde, I. W. Mattaj, and G. M. Gilmartin. 1997. Participation of the nuclear cap binding complex in pre-mRNA 3′ processing. Proc. Natl. Acad. Sci. USA 94:11893–11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu, X.-D. 1995. The superfamily of arginine/serine-rich splicing factors. RNA 1:663–680. [PMC free article] [PubMed] [Google Scholar]
- 15.Graveley, B. R. 2000. Sorting out the complexity of SR protein functions. RNA 6:1197–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gunderson, S. I., M. Polycarpou-Schwarz, and I. W. Mattaj. 1998. U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol. Cell 1:255–264. [DOI] [PubMed] [Google Scholar]
- 17.Gunderson, S. I., S. Vagner, M. Polycarpou-Schwarz, and I. W. Mattaj. 1997. Involvement of the carboxyl terminus of vertebrate poly(A) polymerase in U1A autoregulation and in the coupling of splicing and polyadenylation. Genes Dev. 11:761–773. [DOI] [PubMed] [Google Scholar]
- 18.Hirose, Y., and J. L. Manley. 2000. RNA polymerase II and the integration of nuclear events. Genes Dev. 14:1415–1429. [PubMed] [Google Scholar]
- 19.Huang, Y., and G. C. Carmichael. 1996. Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol. Cell. Biol. 16:1534–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang, Y., and J. A. Steitz. 2001. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol. Cell 7:899–905. [DOI] [PubMed] [Google Scholar]
- 21.Izaurralde, E., J. Lewis, C. McGuigan, M. Jankowska, E. Darzynkiewicz, and I. W. Mattaj. 1994. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell 78:657–668. [DOI] [PubMed] [Google Scholar]
- 22.Kataoka, N., J. Yong, V. N. Kim, F. Velazquez, R. A. Perkinson, F. Wang, and G. Dreyfuss. 2000. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol. Cell 6:673–682. [DOI] [PubMed] [Google Scholar]
- 23.Kramer, A. 1996. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu. Rev. Biochem. 65:367–409. [DOI] [PubMed] [Google Scholar]
- 24.Legrain, P., and M. Rosbash. 1989. Some cis- and trans-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell 57:573–583. [DOI] [PubMed] [Google Scholar]
- 25.Le Hir, H., E. Izaurralde, L. E. Maquat, and M. J. Moore. 2000. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 19:6860–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis, J. D., E. Izaurralde, A. Jarmolowski, C. McGuigan, and I. W. Mattaj. 1996. A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5′ splice site. Genes Dev. 10:1683–1698. [DOI] [PubMed] [Google Scholar]
- 27.Lou, H., K. M. Neugebauer, R. F. Gagel, and S. M. Berget. 1998. Regulation of alternative polyadenylation by U1 snRNPs and SRp20. Mol. Cell. Biol. 18:4977–4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo, M. J., and R. Reed. 1999. Splicing is required for rapid and efficient mRNA export in metazoans. Proc. Natl. Acad. Sci. USA 96:14937–14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lutz, C. S., K. G. Murthy, N. Schek, J. P. O’Connor, J. L. Manley, and J. C. Alwine. 1996. Interaction between the U1 snRNP-A protein and the 160-kD subunit of cleavage-polyadenylation specificity factor increases polyadenylation efficiency in vitro. Genes Dev. 10:325–337. [DOI] [PubMed] [Google Scholar]
- 30.McCracken, S., E. Rosonina, N. Fong, M. Sikes, A. Beyer, K. O’Hare, S. Shuman, and D. Bentley. 1998. Role of RNA polymerase II carboxy-terminal domain in coordinating transcription with RNA processing. Cold Spring Harbor Symp. Quant. Biol. 63:301–309. [DOI] [PubMed] [Google Scholar]
- 31.McGarvey, T., E. Rosonina, S. McCracken, Q. Li, R. Arnaout, E. Mientjes, J. A. Nickerson, D. Awrey, J. Greenblatt, G. Grosveld, and B. J. Blencowe. 2000. The acute myeloid leukemia-associated protein, DEK, forms a splicing-dependent interaction with exon-product complexes. J. Cell Biol. 150:309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore, C. L., and P. A. Sharp. 1984. Site-specific polyadenylation in a cell-free reaction. Cell 36:581–591. [DOI] [PubMed] [Google Scholar]
- 33.Nesic, D., and L. E. Maquat. 1994. Upstream introns influence the efficiency of final intron removal and RNA 3′-end formation. Genes Dev. 8:363–375. [DOI] [PubMed] [Google Scholar]
- 34.Niwa, M., and S. M. Berget. 1991. Mutation of the AAUAAA polyadenylation signal depresses in vitro splicing of proximal but not distal introns. Genes Dev. 5:2086–2095. [DOI] [PubMed] [Google Scholar]
- 35.Niwa, M., S. D. Rose, and S. M. Berget. 1990. In vitro polyadenylation is stimulated by the presence of an upstream intron. Genes Dev. 4:1552–1559. [DOI] [PubMed] [Google Scholar]
- 36.Ono, Y., M. Ohno, and Y. Shimura. 1994. Identification of a putative RNA helicase (HRH1), a human homolog of yeast Prp22. Mol. Cell. Biol. 14:7611–7620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Proudfoot, N. 2000. Connecting transcription to messenger RNA processing. Trends Biochem. Sci. 25:290–293. [DOI] [PubMed] [Google Scholar]
- 38.Reed, R., and L. Palandjian. 1997. Spliceosome assembly. IRL Press, Oxford, United Kingdom.
- 39.Rodrigues, J. P., M. Rode, D. Gatfield, B. Blencowe, M. Carmo-Fonseca, and E. Izaurralde. 2001. REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc. Natl. Acad. Sci. USA 98:1030–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Romac, J. M., and J. D. Keene. 1995. Overexpression of the arginine-rich carboxy-terminal region of U1 snRNP 70K inhibits both splicing and nucleocytoplasmic transport of mRNA. Genes Dev. 9:1400–1410. [DOI] [PubMed] [Google Scholar]
- 41.Ruegsegger, U., D. Blank, and W. Keller. 1998. Human pre-mRNA cleavage factor Im is related to spliceosomal SR proteins and can be reconstituted in vitro from recombinant subunits. Mol. Cell 1:243–253. [DOI] [PubMed] [Google Scholar]
- 42.Rutz, B., and B. Seraphin. 2000. A dual role for BBP/ScSF1 in nuclear pre-mRNA retention and splicing. EMBO J. 19:1873–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vagner, S., C. Vagner, and I. W. Mattaj. 2000. The carboxyl terminus of vertebrate poly(A) polymerase interacts with U2AF 65 to couple 3′-end processing and splicing. Genes Dev. 14:403–413. [PMC free article] [PubMed] [Google Scholar]
- 44.Wahle, E., and W. Keller. 1996. The biochemistry of polyadenylation. Trends Biochem. Sci. 21:247–250. [PubMed] [Google Scholar]
- 45.Wan, K., J. A. Nickerson, G. Krockmalnic, and S. Penman. 1994. The B1C8 protein is in the dense assemblies of the nuclear matrix and relocates to the spindle and pericentriolar filaments at mitosis. Proc. Natl. Acad. Sci. USA 91:594–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, J., and J. L. Manley. 1995. Overexpression of the SR proteins ASF/SF2 and SC35 influences alternative splicing in vivo in diverse ways. RNA 1:346–355. [PMC free article] [PubMed] [Google Scholar]
- 47.Wassarman, K. M., and J. A. Steitz. 1993. Association with terminal exons in pre-mRNAs: a new role for the U1 snRNPΔ. Genes Dev. 7:647–659. [DOI] [PubMed] [Google Scholar]
- 48.Yankulov, K., J. Blau, T. Purton, S. Roberts, and D. L. Bentley. 1994. Transcriptional elongation by RNA polymerase II is stimulated by transactivators. Cell 77:749–759. [DOI] [PubMed] [Google Scholar]
- 49.Zhou, Z., M. J. Luo, K. Straesser, J. Katahira, E. Hurt, and R. Reed. 2000. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature 407:401–405. [DOI] [PubMed] [Google Scholar]