Abstract
Using the FDC-P1 cell line expressing the exogenous macrophage colony-stimulating factor (M-CSF) receptor, Fms, we have analyzed the role of a new mammalian DOS/Gab-related signaling protein, called Gab3, in macrophage cell development of the mouse. Gab3 contains an amino-terminal pleckstrin homology domain, multiple potential sites for tyrosine phosphorylation and SH2 domain binding, and two major polyproline motifs potentially interacting with SH3 domains. Among the growing family of Gab proteins, Gab3 exhibits a unique and overlapping pattern of expression in tissues of the mouse compared with Gab1 and Gab2. Gab3 is more restricted to the hematopoietic tissues such as spleen and thymus but is detectable at progressively lower levels within heart, kidney, uterus, and brain. Like Gab2, Gab3 is tyrosine phosphorylated after M-CSF receptor stimulation and associates transiently with the SH2 domain-containing proteins p85 and SHP2. Overexpression of exogenous Gab3 in FD-Fms cells dramatically accelerates macrophage differentiation upon M-CSF stimulation. Unlike Gab2, which shows a constant mRNA expression level after M-CSF stimulation, Gab3 expression is initially absent or low in abundance in FD cells expressing the wild-type Fms, but Gab3 mRNA levels are increased upon M-CSF stimulation. Moreover, M-CSF stimulation of FD-FmsY807F cells (which grow but do not differentiate) fails to increase Gab3 expression. These results suggest that Gab3 is important for macrophage differentiation and that differentiation requires the early phosphorylation of Gab2 followed by induction and subsequent phosphorylation of Gab3.
Growth factor receptors are important transducers of extracellular signals for regulating the growth, death, and developmental fate of individual cells of an organism. The transmembrane growth factor receptors are activated by binding an extracellular ligand and converting this event into molecular reactions mediated through the cytoplasmic portion of the receptor. These reactions constitute specific signaling pathways that ultimately determine the fate of cells and tissues through regulation of specific cellular functions or transcription of specific genes.
The macrophage colony-stimulating factor (M-CSF) receptor, Fms, is an example of this form of regulation occurring in macrophage development. The homodimeric ligand, M-CSF, interacts with the extracellular immunoglobulin domains of two Fms receptor proteins and activates their cytoplasmic tyrosine kinase domains, which transphosphorylate individual tyrosine residues of the opposing cytoplasmic domain. These tyrosine phosphorylated sites comprise motifs for high-affinity interactions with SH2-containing proteins, which transmit the molecular signals along yet incompletely understood pathways (for a review, see the work of Bourette and Rohrschneider [6]).
A primary research focus of our studies concerns the role of the Fms tyrosine kinase growth factor receptor in determining the fate of cells regarding growth versus differentiation. The signals for growth appear to involve activation of the Ras-mitogen-activated protein kinase pathway described for many growth factor systems, but the signals for differentiation and their regulation remain less well understood. Genetic studies of ommatidium development in the Drosophila compound eye have indicated that the tyrosine kinase growth factor receptor encoded by the sevenless gene is necessary for correct formation of the ommatidium R7 cells (11, 12, 52). A crucial downstream component for eye development is the DOS protein, a pleckstrin homology (PH) domain-containing scaffolding protein also required for several other receptor tyrosine kinase-regulated developmental programs throughout Drosophila (12, 34). A mammalian DOS homologue, called Gab1, has been identified and named for its ability to bind the adapter protein Grb2 (14). Gab1 interacts with the c-Met protooncogene (also known as the hepatocyte growth factor receptor), and overexpression in epithelial cells induces ligand-independent morphogenesis characteristic of c-Met activation (43). Therefore, the genetics and biochemistry of both DOS and Gab1 define a class of signaling proteins acting downstream of growth factor receptors and essential for transmitting developmental signals.
A newer member of the Gab family of proteins in mammalian cells, called Gab2, was cloned and characterized in hematopoietic cells (8, 21, 29, 51). Gab2, like the other family members, contains an N-terminal PH domain, at least two proline-rich sequence motifs for binding SH3 domain proteins, and multiple tyrosine residues within potential sites that may specify interactions with the SH2 domain portions of various signaling proteins. The Gab2 interaction with the SH2 domains of the p85 subunit of phosphatidyl inositol 3-kinase (PI3K) and SH2 domains of the tyrosine phosphatase SHP2 is inducible by growth factor stimulation, but Gab2 constitutively associates with Grb2. As with Drosophila DOS, the interaction of Gab2 with SHP2 is necessary for positive signaling (8, 13). Mutated Gab2, which lacks the two SHP2 binding sites, behaves as a dominant inhibitor of macrophage differentiation in the FD-Fms cell system (21).
In the present studies we have investigated the expression and function of a new member of the Gab family of proteins which we call Gab3. This protein is abundant in hematopoietic tissues and our results suggest a role for Gab3 in macrophage cell development.
MATERIALS AND METHODS
Cloning of Gab3.
Human Gab3 was identified out of a large expressed sequence tag database based upon sequence homology to Gab1. Clones were present in and selected from both human dendritic cell and eosinophil cDNA libraries. Murine Gab3 (mGab3) was cloned through a low-stringency screen of a mouse spleen cDNA library using a probe fragment derived from the human Gab3 cDNA clone. The mGab3 cDNA was utilized throughout our studies for analysis of signaling following expression in murine cells.
Cells and stimulation.
The BOSC 23 (33) ecotropic retroviral packaging cell line was maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS; HyClone). FDC-P1 UW cells expressing wild-type murine Fms (FD-Fms), mutant FmsY807F (FD- FmsY807F), FD-Fms (vector), and FD-Fms(Gab3V5), each from a retroviral vector, were maintained in DMEM plus 10% FBS supplemented with 0.4% conditioned medium of X63-IL-3 cells expressing recombinant interleukin-3 (IL-3) (18). Cells were starved in DMEM without FBS for 4 to 6 h and then stimulated with various concentrations of M-CSF at 37°C. For M-CSF stimulation over longer periods of time, cells were seeded at a density of 5 × 104 cells/ml and the media were replaced daily and cell density was adjusted to 5 × 104 cells/ml. Primary bone marrow (BM) cells were isolated and stimulated with M-CSF as described previously (23). The murine hematopoietic cell line BaF3 and BaF3/Flt3 were grown in RPMI 1640 plus 10% FBS and 0.3% recombinant IL-3 (conditioned medium of X63-IL-3-expressing cells), as described previously (47). Cells were starved in RPMI 1640 containing 0.8% bovine serum albumin for 6 h and BaF3 cells were stimulated with recombinant IL-3 (undiluted X63-conditioned medium), whereas BaF3/Flt3 cells were stimulated with 3 μg of Flt3 ligand (FL; a kind gift from Immunex Corporation, Seattle, Wash.) per ml at 37°C.
Antibodies and antibody production.
Polyclonal Gab3 antiserum was generated by immunizing rabbits with a glutathione S-transferase (GST) fusion protein containing amino acids 259 to 467 of murine Gab3 and was subsequently affinity purified using a Sepharose column with covalently linked GST or GST-Gab3 antigen. Polyclonal Gab2 antiserum was generated as described previously (21). Anti-V5 monoclonal antibody was purchased from Invitrogen, polyclonal anti-SHP2 antibody was from Santa Cruz Biotechnology, polyclonal anti-PI3K p85 antibody was from Upstate Biotechnology, and monoclonal anti-Grb2 antibody was from Transduction Laboratories. Polyclonal anti-Fms antibodies have been described previously (37). The 4G10 antiphosphotyrosine antibody was a kind gift from Brian Druker (Oregon Health Sciences, Portland, Oreg.).
RT-PCR.
Total RNA of various cell lines or mouse tissues was extracted as described previously (47). Poly(A)+ RNA was isolated using oligo(dT) cellulose affinity purification (New England Biolabs). Reverse transcriptase (RT)-PCR was performed as described previously (23, 46) using specific oligonucleotide primers for Gab3 (Gab3SE, 5′-GAGAGTCTCTCTCACATG; Gab3AS, 5′-GGGTGAAGCTGTGGGATA), Gab2 (Gab2SE, 5′-GGAACCTTTCTTCAGCCAG; Gab2AS, 5′-GAGCTGGCACTATTTGATC), and Gab1 (Gab1SE, 5′-CTGGCTCCTCACAAGCAC; Gab1AS, 5′-CAGCTCTTCACCCGAGAC). The specificity of oligonucleotide primers was tested using cDNA controls for murine Gab3, murine Gab2 (accession number AF104244), and murine Gab1 (accession number AJ250669). PCR conditions for Gab3, Gab2, and Gab1 were 1 cycle at 94°C for 1 min followed by 35 cycles at 94°C for 30 s, 62°C for 30 s, and 72°C for 1 min. Positive control PCR amplifications were done using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) primers (GAPDHSE, 5′-CCCATCACCATCTTCCAGGA; GAPDHAS, 5′-GGGGCCATCCACAGTCTTCT).
To estimate the relative amounts of RT-PCR products, total RNA of BM cells, FD-Fms UW cells, FD-Fms UW Y807F cells, and NFS60-Fms cells was isolated using the Qiagen RNeasy kit according to the manufacturer’s instructions. To ensure that there was no genomic DNA contamination, the RNA preparation was treated with 1 U of RNase-free DNase I (Gibco BRL) per μg of RNA for 15 min at room temperature. After termination of this reaction by the addition of EDTA to a final concentration of 2.5 mM and an incubation at 65°C for 10 min, the first-strand reaction was performed using Superscript II RT (Gibco BRL) according to the manufacturer’s instructions. Using an equivalent amount of 0.2 μg of RNA per PCR, amplification of Gab1, Gab2, Gab3, and GAPDH cDNA was performed as mentioned above. Ethidium bromide-stained gels were scanned with a Typhoon 8600 Scanner (Molecular Dynamics) at 532 nm and DNA bands were quantified using ImageQuant software. The results for Gab1, Gab2, and Gab3 were normalized to the GAPDH signals. All PCR results were not saturated, which was controlled by running the PCRs with various cycle numbers. To calibrate the quantification of the reactions, PCRs with increasing amounts of input cDNA were performed. This calibration showed that the measured signal difference was underestimated by a factor of about 3, since a twofold increase in input cDNA resulted in only a 1.3-fold increase in fluorescence signal.
Primers for subcloning the Gab3 open reading frame into pIND/V5-His TOPO vector (Invitrogen) were as follows: sense, 5′-GCCAGGATGAGCACTGGTGACACT-3′, and antisense, 5′-CACTTTGGATTGCCTCTCATCAGTC-3′ (the start codon is underlined).
V5-tagged Gab3.
The pJZen-IRES-GFP (pJIG) bicistronic retroviral expression vector (kindly provided by P. A. Algate, Fred Hutchinson Cancer Research Center, Seattle, Wash.) was constructed by inserting the internal ribosome entry site (IRES) and green fluorescent protein (GFP) (Clontech) into the pJZen-1 vector (36). The pJIG/mGab3V5 expression construct was generated by inserting the mGab3 coding region, prepared by PCR using the sense and antisense primers listed above, into the TA cloning site of the pIND/V5-His-TOPO vector (Invitrogen). The V5-tagged full-length mGab3 (mGab3V5) cDNA in pJIG/mGab3V5 was then amplified, digested with BamHI, and inserted into the pJZen-IRES-GFP vector as described previously for Gab2 (21). All PCRs were performed with HF polymerase mix (Clontech) and constructs were verified by sequencing.
Retroviral gene transfer.
Retroviral infection of murine FD-Fms UW cells was performed using transiently transfected BOSC 23 packaging cells as described previously (16). Briefly, 1.5 × 106 cells were plated onto 60-mm-diameter dishes containing 4 ml of medium 18 h prior to transfection. For transfection, 10 μg of plasmid DNA was added to each dish containing fresh medium supplemented with 25 μM chloroquine. At 7 to 10 h posttransfection, the medium was replaced with fresh medium without chloroquine, and cells were incubated for 18 h. Infections were performed by cocultivating 3 × 105 FD-Fms UW cells with transfected BOSC 23 cells for 48 h in 60-mm-diameter dishes containing 4 ml of medium supplemented with 4 μg of Polybrene per ml. Infected FD-Fms UW cells were then harvested and sorted for Fms and GFP expression on a Vantage flow cytometer (Becton-Dickinson).
IPs and IBs.
Immunoprecipitations (IPs) and immunoblots (IBs) were performed as described previously (47). Unstimulated or stimulated cells (107) were lysed in NP-40 lysis buffer (50 mM NaCl, 50 mM Tris-HCl [pH 7.3], 30 mM Na4P2O7, 50 mM NaF, 5 μM ZnCl2, 0.5% NP-40, 1 mM phenylmethylsulfonyl fluoride, 20 μg of aprotinin per ml, 2 mM orthovanadate). Lysates were cleared of cell debris by centrifugation. Supernatants (equalized for protein amount) were used for IP and incubated 3 h to overnight with 15 μl of anti-Gab3 serum, 1 μl of anti-Gab2 serum, and 20 μl of protein A-Sepharose (Amersham Pharmacia) or 1 μl of anti-V5 antibody and 20 μl of protein G-Sepharose (Amersham Pharmacia). IPs were washed four times with lysis buffer and eluted by boiling in 2× Laemmli buffer. Proteins were separated on a sodium dodecyl sulfate-6.5 to 10% polyacrylamide gel and transferred to a nitrocellulose membrane (Schleicher and Schuell) on a semidry blotting apparatus (Ellard Instrumentation Ltd., Seattle, Wash.). Membranes were blocked in TBST (300 mM NaCl, 10 mM Tris-HCl [pH 7.5], 0.5% Tween 20) containing 1% bovine serum albumin and 1% ovalbumin and incubated with primary antibodies (1:500-diluted affinity-purified anti-Gab3, 1:1,000-diluted affinity-purified anti-Gab2, and 1:2,500-diluted antiphosphotyrosine 4G10; other antibodies were used according to the manufacturer’s recommended methods), following which they were incubated with the appropriate secondary antibodies (anti-mouse or protein A) coupled to horseradish peroxidase (Bio-Rad). Immunocomplexes were visualized using enhanced chemiluminescence (Dupont-NEN).
GST constructs, protein purification, and GST pull-down assays.
For antibody production, a PCR-generated fragment using the pIND/V5-His/mGab3 construct as template, 5′-ACGTGGATCCCCAATAGAGAAATCAATGGCCCA as sense primer, and 5′-ACGTGGATCCCCTGGTTAGAGATGTGTGTT as 12antisense primer and encoding mGab3 amino acids 259 to 467 was cloned into the BamHI site of pGEX-5X-1 (Amersham Pharmacia). Constructs used for GST pull-down assays were from plasmid stocks from L. R. Rohrschneider’s laboratory and from J. Cooper’s laboratory (Fred Hutchinson Cancer Research Center). The proline-rich domain (amino acids 396 to 458) of mGab3 was PCR amplified (3-PRD-SE, 5′-TGTGTGGATCCCCGTGCCCATGAGCCCTAAAGG; 3-PRD-AS, 5′-TGTGTGAATTCGGTGGAAAGGTTTCTCGAGTC) and subcloned into the BamHI and EcoRI sites of pGEX-3X (Amersham Pharmacia). GST fusion protein expression was performed using Escherichia coli BL21(DE3) (Novagen), protein expression was induced at room temperature with 0.5 mM IPTG for 3 to 4 h, and GST fusion proteins were affinity purified using glutathione-agarose (Amersham Pharmacia). For GST pull-down assays, 3 to 5 μg of GST fusion protein coupled to beads was incubated with NP-40 lysates from unstimulated or M-CSF-stimulated FD-Fms(Gab3V5) cells. After four washes with lysis buffer, complexes were analyzed by immunoblotting.
Cell proliferation assays in liquid culture and fluorescence-activated cell sorter analysis.
(3-[4,5-Dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) proliferation assays were performed as described previously (27). In brief, FD-Fms(vector) and FD-Fms(Gab3V5) cells were plated in 96-well plates at a density of 2 × 104 cells/ml and under different growth factor conditions. The assay was performed after 3 days and proliferation was calculated relative to growth in IL-3.
For cell counts in liquid culture, FD-Fms(vector) and FD-Fms(Gab3V5) cells were plated at a final concentration of 105 cells/ml under the growth factor conditions indicated. Cells were counted every day over a 3-day period using a Coulter Counter (Coulter-Beckman). Relative proliferation was calculated based on the initial cell density. In addition, cells were plated with the same density in either IL-3 or 2,500 U of M-CSF per ml for 2 days and cell morphology was documented with a phase-contrast micrograph as well as fluorescence-activated cell sorter analysis monitoring forward and side scatter.
Soft agar assays.
These assays were performed as described previously in 35-mm-diameter petri dishes containing Iscove’s medium (Sigma), 10% FBS, and 0.31% agar (26, 37). A 1-ml top agar layer contained 700 cells, and a 1-ml bottom layer contained the same 0.31% agar medium but without cells. Growth factors were added to both top and bottom agar layers, and colony counts were made after 7 to 10 days in culture. Only colonies of 10 viable cells or greater were counted.
Nucleotide sequence accession numbers.
The amino acid and nucleotide sequences for both murine (AY057988) and human (AY057989) Gab3 have been deposited in GenBank.
RESULTS
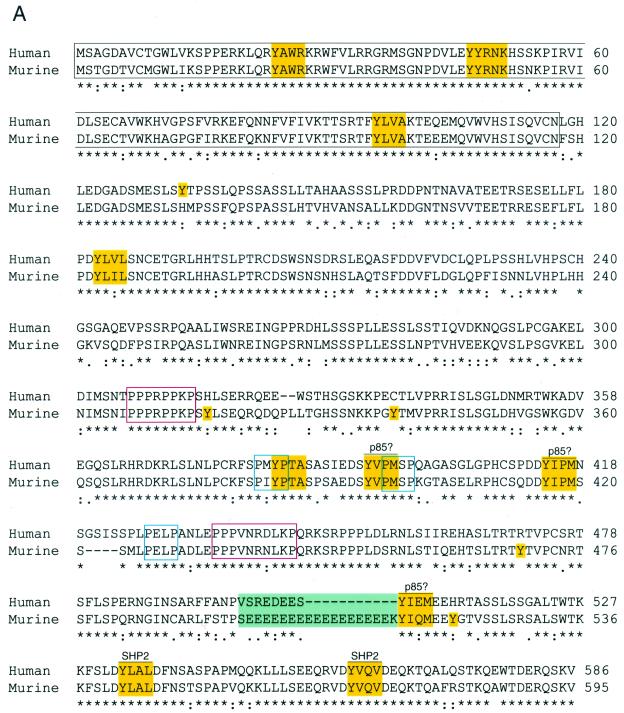
Human Gab3 was first identified from an EST database by homology with Gab1, and full-length cDNAs for both the human and murine Gab3 were isolated as described in Materials and Methods. The expected amino acid sequences for both the murine and human Gab3 proteins, based on the complete cDNA nucleotide sequence, are shown in Fig. 1A. The nucleotide sequences have been deposited in GenBank (see Materials and Methods). The 595-amino-acid murine Gab3 and 586-amino-acid human Gab3 each encode proteins with N-terminal PH domains (outlined in black) and five polyproline motifs with potential for SH3 domain interactions, of which two have close homology to known SH3 domain binding sites (outlined in red) and three do not (outlined in blue). Eleven positionally shared tyrosine residues reside adjacent to sequence motifs specifying possible SH2 domain interactions (highlighted in yellow). Three YXXM tyrosine phosphorylation motifs could each interact with an SH2 domain of the p85 subunit of PI3K (39), and all three are situated in spatially homologous regions of each respective Gab3 protein. The two tyrosine motifs, YLXL and YVXV at amino acid positions 542 and 569 of the murine Gab3 sequence, are also present in the Gab2 protein and are known to interact with the SH2 domains of SHP2 (8, 21). In addition, the murine Gab3 protein contains four tyrosine residues not matched by an equivalent tyrosine at the related position in the human sequence, and the human Gab3 protein contains one such unique tyrosine residue (all highlighted in yellow). Two short shared sequences are of interest in the Gab3 proteins. One is composed of a polyglutamic acid sequence (green background) not entirely shared between human and murine proteins. Another is the more carboxyl terminal of the polyproline sequences (outlined in red) because this sequence is known to interact with the C-terminal SH3 domain of Grb2 (7, 22).
FIG. 1.
Amino acid sequence and domain structure of the human and murine Gab3 proteins. (A) The sequences of the 595-amino acid murine Gab3 protein and the 586-amino acid human Gab3 protein deduced from the nucleotide open reading frames of each cDNA are shown in the single-letter amino acid designations. The PH domain is outlined in black, and tyrosine-containing motifs with potential for interacting with SH2 domains when phosphorylated are highlighted in yellow (as are single, unmatched tyrosine [Y] residues). Several PXXP motifs with potential for SH3 domain interactions are shown outlined in blue. The red outlined sequences represent highly conserved regions contained in all three Gab proteins, and the green rectangle contains a polyglutamic acid stretch. The consensus line below the amino acid sequences utilizes an asterisk for complete identity, a colon for a conserved substitution, and a period for a semiconserved substitution. (B) Diagram showing the relative structural details of Gab1, Gab2, and Gab3. The amino-terminal PH domains are highlighted in black, the location of tyrosine amino acids are shown along the length of each protein (Y), and PXXP amino acid motifs are designated in black with a “P” below the relevant location. The position of the major conserved polyproline-rich sequences are designated by open red rectangles on each Gab protein. The scale above the C terminus of Gab1 indicates the length of 50 amino acids. (C) Amino acid sequence alignment of the Gab1, Gab2, and Gab3 PH domains with tyrosine residues highlighted (yellow).
The comparative organization of the major structural and domain features of the Gab1, Gab2, and Gab3 proteins is shown in Fig. 1B. Among these proteins, the amino acid identity is greatest in the PH domains (approximately 50% identity shared by all three) while the remainder of the proteins exhibit considerably less sequence identity (approximately 14% among the three). Each PH domain contains three identical tyrosine motifs for potential SH2 domain binding and the Gab1 protein exhibits an additional unique tyrosine (Fig. 1C). The unique tyrosine (Y) residue in the PH domain of Gab1 at position 83 is matched with a phenylalanine (F) amino acid residue in the respective Gab2 and Gab3 PH domain sequence, suggesting the single unmatched Y in Gab1 may serve merely a structural role. Despite the lower sequence identity among the three proteins outside the PH domains and the different overall amino acid lengths, sequence alignment demonstrates numerous identities among all Gab proteins, and some sequence motifs are also shared with the Drosophila DOS protein (not shown). The first polyproline-rich region (centered at amino acid 310 of murine Gab3) is situated identically in all four members of this protein family (i.e., Gab1, Gab2, Gab3, and DOS). In addition, the above-mentioned polyproline-rich sequence, which contains the exact motif for binding the C-terminal Grb2 SH3 domain (PPPVXRXLKP at amino acid positions 433 to 442), represents one of the most highly conserved sequence stretches among the four proteins of the DOS/Gab family (data not shown). These overall structural features of Gab3 indicate its close functional relationship to the Gab1 and Gab2 proteins and the DOS protein of Drosophila.
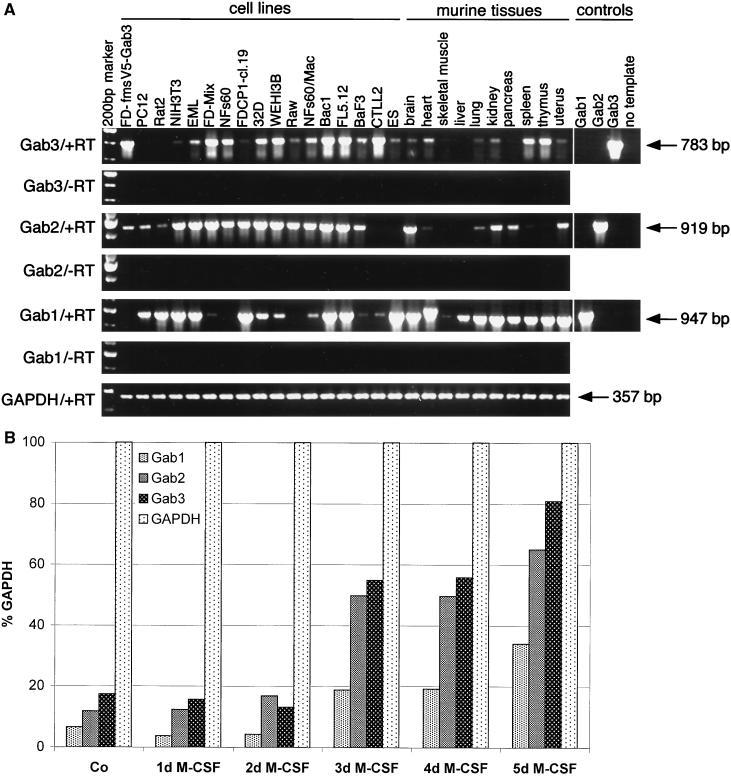
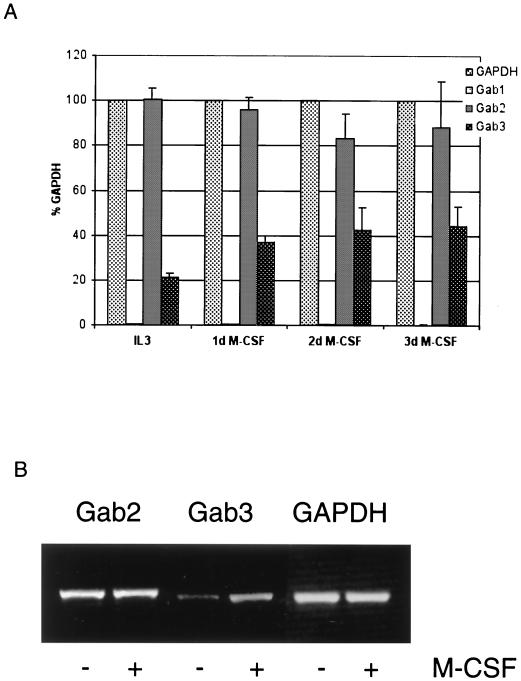
To obtain an idea about the tissue-specific functionality of Gab3, the expression of Gab3 mRNA was analyzed in several hematopoietic cell lines and tissues of the mouse by RT-PCR of mRNA (Fig. 2A). The expression of both Gab1 and Gab2 was tested concurrently. The RT-PCR primers were specific for the nucleotide sequences encoding each protein, and nonspecific priming was not evident. Gab3 mRNA was expressed in all cell lines and tissues of hematopoietic origin and was relatively abundant in spleen and thymus. Murine embryonic stem (ES) cells also expressed detectable levels of the Gab3 mRNA, as did brain, heart, lung, kidney, and uterus cells. NIH 3T3 cells showed marginally detectable expression of Gab3. Other cells or tissues not expressing Gab3 include PC12 cells, Rat2 cells, skeletal muscle, liver, and pancreas. All tested myeloid and macrophage cell lines did contain Gab3 mRNA (FDC-P1, NFS60, 32D, WEHI3B, Raw, BAC1, and NFS60/Mac), but the FDC-P1 cells showed only very weak expression.
FIG. 2.
RT-PCR analysis of Gab1, Gab2, and Gab3 mRNA expression in cell lines and tissues of the mouse (and rat). (A) The designations Gab3/+RT and Gab3/−RT indicate PCRs using primers specific for Gab3 and conducted with or without prior reverse transcription, respectively. Similar reactions were performed for detecting Gab2, Gab1, and GAPDH as positive control. Additional positive and negative RT-PCR experiments demonstrated the specificity of each primer set for its corresponding Gab protein. These reactions are shown on the right-hand end (controls), and the nucleotide length for the expected products is adjacent to each arrow. The cell lines are PC12, Rat2 (rat cell lines; all others are mouse derived); NIH 3T3 (fibroblast); EML, FD-Mix (pluripotent, hematopoietic); NFS60, FDC-P1 cl 19, 32D, WEHI 3B, Raw, NFS60/Mac, Bac1 (myeloid); FL5.12, BaF3 (B cells); CTLL2 (T cells); ES (129Sv-derived Ak7 stem cells). (B) RT-PCR analysis of Gab1, Gab2, and Gab3 in primary BM cells cultivated in the presence of M-CSF. BM cells were collected and cultured in the presence of 1,000 U of M-CSF per ml for 5 days. Total RNA was isolated before the addition of M-CSF (Co) and at the time points indicated. RT-PCRs were performed using specific primers for Gab1, Gab2, Gab3, or GAPDH. Quantification of the ethidium bromide-stained DNA bands in gels was performed with ImageQuant software. Gab1, Gab2, and Gab3 signals were normalized to the GAPDH signal and are shown as percentages of GAPDH signals. One representative example of three independent experiments is shown.
Similar analysis of Gab1 and Gab2 mRNA demonstrated that each of the Gab proteins exhibited nonidentical but overlapping patterns of expression within the set of cells and tissues examined (Fig. 2A). Gab2 expression was uniformly detectable throughout myeloid and B-cell lines but was absent from the CTLL2 T-cell line. Like Gab3, Gab2 was expressed in brain, heart, lung, kidney, and uterus, but the expression was weak in spleen and absent from thymus. These latter two tissues expressed Gab1 and Gab3, however. In contrast to the absence of Gab3 in pancreas, both Gab2 and Gab1 could be detected in this tissue. The ES cells did not express detectable levels of Gab2. The expression of mRNA for the Gab1 protein was uniformly positive in most mouse tissues examined and was also found in the hematopoietic cell lines, except a few myeloid cell lines (i.e., NFS60, Raw, and possibly FD-Mix). Gab1 expression was notably absent from the FD-Fms UW cells (data not shown) used to overexpress Gab3 from a retroviral vector; however, a second line of FD-Fms cells (cl 19) from our laboratory showed a strong Gab1 mRNA signal.
Expression of the three Gab mRNAs was examined in BM cells (after depletion of adherent cells by incubation on plastic) or BM cells cultured for various days in M-CSF (Fig. 2B). The semiquantitative results are plotted relative to GAPDH mRNA and indicate that during the 5-day experimental period the levels of all three Gab protein mRNAs increased. Macrophages were the primary cells present after 5-day culture of the BM cells in M-CSF.
Therefore, the overall expression analyses of Gab1, Gab2, and Gab3 demonstrate a complex regulation in which very few cell types exhibit expression of only one Gab protein (e.g., liver). More cell types express combinations of two Gab proteins (e.g., spleen, thymus, CTLL2, pancreas, NFS60), but most (at least in cell culture) may express all three Gab proteins.
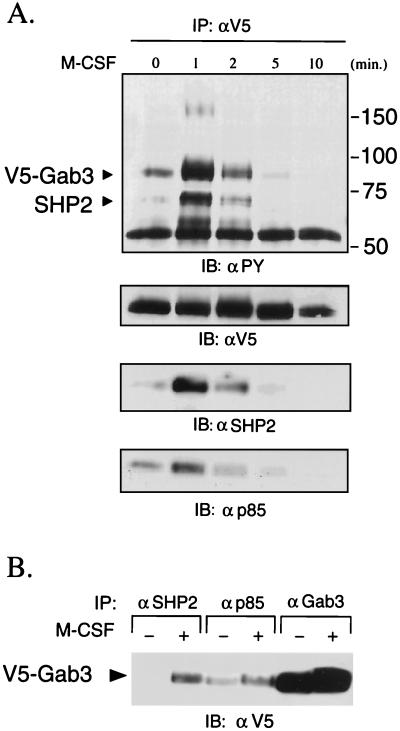
The expression of multiple Gab proteins within a single cell poses a question of whether individual Gab proteins couple to individual growth factor receptors or whether a single receptor activates all Gab proteins, perhaps in different ways. The Gab2 protein is expressed in myeloid cells as an approximately 100-kDa protein and becomes tyrosine phosphorylated in M-CSF-stimulated FD-Fms cells (21). Phosphorylated Gab2 associates with both SHP2 and the p85 subunit of the PI3K complex through their SH2 domains, which transmits signals for events such as gene activation (8, 14, 20). Therefore, FD-Fms cells expressing a V5-tagged Gab3 protein were used to determine whether Gab3 also participates in similar signaling from the activated M-CSF receptor, Fms. The FD-Fms(Gab3V5) cells were starved of growth factors and stimulated by the addition of M-CSF. The results in Fig. 3 demonstrate that Gab3 is rapidly tyrosine phosphorylated after M-CSF stimulation and, like Gab2, associates with both SHP2 and p85, as shown by co-IP with anti-V5 serum (Fig. 3A) or a reciprocal experiment where mGab3V5 was coimmunoprecipitated with anti-SHP2 or anti-p85 sera (Fig. 3B). The tyrosine-phosphorylated ≈165-kDa protein observed 1 min after M-CSF stimulation is not Fms (unpublished observation) but rather an unidentified protein. Both Gab2 and mGab3V5 participated in the signaling pathways activated by M-CSF. Therefore, in the case of Fms, a single receptor may couple to two different Gab proteins.
FIG. 3.
Gab3 is tyrosine phosphorylated by M-CSF and associates with SHP2 and p85/PI3K. (A) FD-Fms-expressing mGab3V5 cells were starved, stimulated with 2,500 U of M-CSF per ml, and lysed at the times indicated. The Gab3 proteins were immunoprecipitated (IP) with anti-V5 antibody and the immune complex was separated by polyacrylamide gel electrophoresis. After electrophoretic transfer of proteins onto nitrocellulose filters, phosphotyrosine (PY)-containing proteins were detected by immunoblotting (IB) with the 4G10 monoclonal antibody. Likewise, equal levels of V5-tagged Gab3 are shown by immunoblotting with antibody to the V5 epitope. The transient M-CSF-dependent association of mGab3V5 with SHP2 and p85 is detected by immunoblotting the anti-V5 IP with the respective antibodies shown in the bottom two panels. The migration positions of Gab3 and SHP2 are shown in the top left panel, and the positions of molecular weight marker proteins are on the right. (B) Reciprocal co-IPs showing the association of SHP2 and p85 with V5-Gab3. Cell lysates from quiescent (−) or M-CSF (2,500 U/μl)-stimulated (+) FD-Fms cells expressing mGab3V5 were immunoprecipitated with antibodies as indicated. The immunocomplexes were separated by sodium dodecyl sulfate-7.5% polyacrylamide gel electrophoresis, transferred onto nitrocellulose filters, and immunoblotted with anti-V5 antibody.
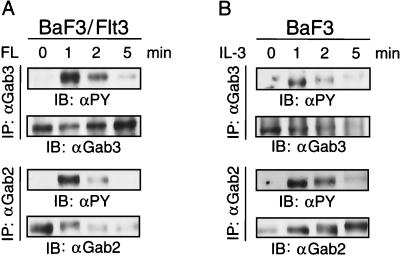
Potential roles for Gab3 and Gab2 signaling through other growth factor receptors was tested to determine whether differential utilization might exist in other systems. BaF3 cells expressing the Flt3 receptor were stimulated with FL and tyrosine phosphorylation of endogenous Gab3 and Gab2 was examined by immunoblotting (Fig. 4). Both Gab3 and Gab2 were tyrosine phosphorylated with similar kinetics following Flt3 activation (Fig. 4A). Similar results were obtained when BaF3 cells were stimulated with IL-3 (Fig. 4B). Therefore, both the Flt3 and IL-3 receptor can induce the tyrosine phosphorylation, and presumably, activation of both Gab2 and Gab3.
FIG. 4.
Tyrosine phosphorylation of Gab3 in response to activation by various ligand-receptor systems. (A) BaF3 cells expressing the Flt3 receptor were stimulated with FL for the times indicated and the endogenous Gab3 or Gab2 protein was immunoprecipitated from aliquots of the cell lysate with antibody specific to each protein. After gel electrophoresis and transfer to nitrocellulose, the filters were immunoblotted with antibody to either phosphotyrosine (IB: αPY), Gab2 (IB: αGab2), or Gab3 (IB: αGab3). (B) BaF3 cells were stimulated with IL-3 and the tyrosine phosphorylation of Gab3 and Gab2 was tested as described above.
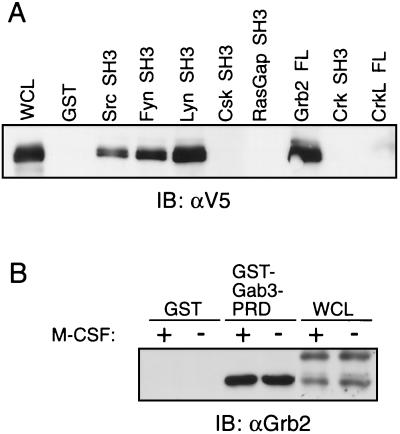
All Gab proteins encode multiple tyrosine phosphorylation motifs and polyproline regions, which could serve as potential interaction sites for SH2 domain- or SH3 domain-containing proteins, respectively. To obtain an initial idea of potential SH3-containing protein interactions with Gab3, we examined the ability of GST fused to SH3 domains derived from various signaling proteins to pull down Gab3 (Fig. 5). SH3 domains from Src, Fyn, Lyn and Grb2 all interacted avidly with the V5-tagged Gab3 in lysates from FD-Fms(Gab3V5) cells (Fig. 5A). The SH3 domains derived from Csk, RasGap, and Crk were devoid of such interactions with Gab3. The SH3 domains of Grb2 constitutively associate with polyproline regions of the Gab members Gab1 and Gab2 (8, 14, 28). A similar association was detectable between Grb2 and the proline-rich domain (PRD) encompassing amino acids 396 to 458 (see Fig. 1) corresponding to the Met-binding domain of Gab1 (43). Fused to GST, the Gab3 PRD interacts well with the 25-kDa Grb2 protein regardless of whether the FD-Fms(Gab3V5) cells are stimulated with M-CSF (Fig. 5B). This is consistent with the constitutive association of the Grb2 SH3 domains with the PRD region of Gab3.
FIG. 5.
Potential interactions of Gab3 with various SH3 domains measured by GST pull-down assays. (A) Lysates from unstimulated FD-Fms(Gab3V5) cells were reacted with GST fused to SH3 domains from the various proteins identified above the figure and the presence of the V5-tagged Gab3 protein identified by immunoblotting. The left-hand lane marked WCL (whole-cell lysate) shows the position of mGab3V5. (B) The PRD of the Gab3 protein was fused to GST and used in pull-down experiments to demonstrate an interaction with Grb2. Immunoblotting with antibody to Grb2 (IB: αGrb2) identified Grb2 in the whole-cell lysates of the FD-Fms(Gab3V5) cells and the complex with GST-Gab3-PRD but not with GST alone. Where indicated, M-CSF was present in excess for 5 min at room temperature.
Based on our results so far, both Gab3 and Gab2 are tyrosine phosphorylated after stimulation of Flt3, the IL-3 receptor, or Fms. The phosphorylation of Gab3 and Gab2 by Fms could occur simultaneously or perhaps sequentially. The former mechanism would require that both proteins be expressed concurrently, whereas the latter mechanism could encompass a temporally different expression pattern. The latter case might be possible because our RT-PCR results indicated that Gab3 was only weakly expressed in the FDC-P1 cl 19 cells (Fig. 2A) and in the FD-Fms UW cells used to express exogenous V5-tagged Gab3, whereas Gab2 expression was considerably higher in both cell types (unpublished observation). However, Gab3 would need to be induced in these cells. We therefore measured the level of Gab3 mRNA by RT-PCR within the FD-Fms UW cells at times after M-CSF stimulation (Fig. 6A). The results demonstrated that Gab3 mRNA levels increased about twofold after M-CSF stimulation. However, a calibration of our RT-PCR indicated that actual differences between control cells in IL-3 and those in M-CSF were underestimated by a factor of 3 (see Materials and Methods). In the FD-Fms UW cell line used in these studies, levels for Gab1 remained uniformly low, while those for Gab2 remained high (Fig. 6A). Similar results were obtained in the NFS60-Fms cells, with Gab2 levels remaining constant but Gab3 levels increasing significantly after M-CSF stimulation (Fig. 6B). No Gab1 signals could be detected in these cells. These results indicate that Gab3 mRNA, and presumably protein, levels are increased by M-CSF stimulation of Fms-expressing cells, and suggest, at least in FD-Fms cells, that Gab2 and Gab3 may act at temporally distinct points in Fms signaling.
FIG. 6.
Expression of Gab protein mRNA in FD-Fms and NFS60-Fms cells. (A) RT-PCR analysis of Gab1, Gab2, and Gab3 in FD-Fms UW cells. FD-Fms UW cells were either cultured in IL-3 or stimulated with 2,500 U of M-CSF per ml, and total RNA was isolated at the time points indicated. Quantification of the PCR products was performed as described in the legend for Fig. 2B. Standard deviations of three independent experiments are indicated. (B) RT-PCR analysis of Gab2 and Gab3 in NFS60-Fms cells. NFS60-Fms cells were either cultured in IL-3 (−) or stimulated with 2,500 U of M-CSF per ml (+) for 3 days. Total RNA was isolated and RT-PCRs were performed using specific primers for Gab2, Gab3, or GAPDH. A representative ethidium bromide-stained gel of PCR products from three independent experiments is shown.
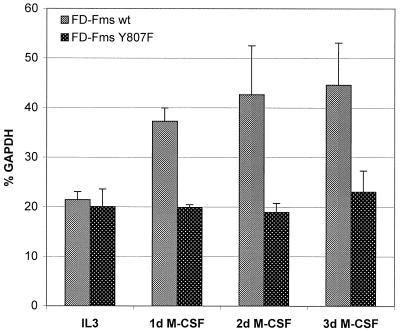
FDC-P1 cells expressing the Y807F point mutation of Fms are defective for macrophage differentiation in M-CSF, even though the in vivo tyrosine kinase activity of the receptor remains largely unaffected (4, 5). The FD-FmsY807F cells were examined for Gab3 mRNA expression at times after M-CSF stimulation, and the results were compared directly to those with FDC-P1 cells expressing the wild-type (WT) receptor (Fig. 7). Gab3 mRNA was induced in FD-Fms WT cells as early as the first day after M-CSF stimulation, whereas the levels of Gab3 mRNA in FD-FmsY807F cells remained constant throughout the 3-day experiment. The absence of both M-CSF-induced Gab3 mRNA induction and macrophage differentiation in the FD-FmsY807F cells suggests that Gab3 is important for differentiation and/or that the Gab3 promoter is activated during differentiation.
FIG. 7.
Comparison of Gab3 signals in FD-Fms WT and Y807F cells. FD-Fms WT and FD-FmsY807F cells were either cultured in IL-3 or stimulated with 2,500 U of M-CSF per ml and total RNA was isolated at the time points indicated. RT-PCRs and quantification of the PCR products were performed as described in the legend to Fig. 2B. Data represent mean values of three independent experiments and standard deviations are indicated.
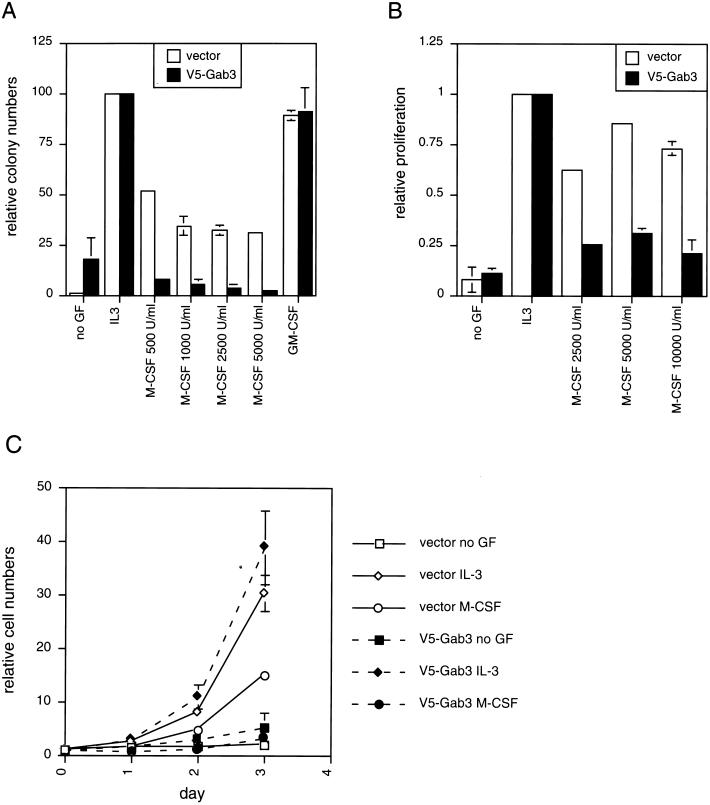
Biological effects of mGab3V5 overexpression in the FD-Fms cells were examined in colony formation assays in soft agar, growth in liquid culture, and morphological differentiation assays. The results in Fig. 8A show the colony count for FD-Fms(Gab3V5) and control FD-Fms(vector) cells grown in soft agar with no growth factor or with IL-3, GM-CSF, or M-CSF. Both IL-3 and GM-CSF stimulate growth of the FD-Fms cells without inducing morphological changes, whereas M-CSF activates a program for differentiation accompanied by a decreased rate of cell growth (4, 5). IL-3 and GM-CSF have similar effects on mGab3V5- and vector-expressing FD-Fms cells, both stimulate maximum colony numbers in soft agar, and 70 to 80% of the input FD-Fms cells consistently form colonies. Growth of the FD-Fms(Gab3V5) and control cells in increasing units of M-CSF resulted in progressively decreasing numbers of colonies and a significant reduction in growth of FD-Fms cells expressing the mGab3V5 relative to the control cells expressing the empty vector. In the absence of growth factor addition to the agar assays, FD-Fms control cells grew poorly or not at all; however, the cells expressing mGab3V5 exhibited significant and unexpected colony formation (morphologically, these colonies were composed of undifferentiated cells). These results suggest that the mGab3V5-expressing cells differentiate more rapidly than control cells in the presence of M-CSF, but they also exhibited increased factor-independent growth relative to control cells.
FIG. 8.
Biological effects of mGab3V5 overexpression on growth of FD-Fms cells. (A) Soft agar colony assays were performed on FD-Fms cells expressing either the empty vector or the vector expressing the mGab3V5. The agar assay conditions included either no growth factor, IL-3, GM-CSF, or increasing M-CSF concentrations from 500 to 5,000 U/ml. (B) An MTT proliferation assay of the vector- or mGab3V5-FD-Fms cells was performed for the IL-3- and M-CSF-stimulated cells. (C) Cell growth in liquid suspension culture was measured by cell counts at 24-h intervals after ligand addition. The vector- or Gab3-expressing FD-Fms cells were grown without ligand or with either IL-3 or M-CSF as indicated in the legend to the right of the figure.
Growth of the mGab3V5- and vector-expressing FD-Fms cells in liquid culture confirmed the effect of mGab3V5 overexpression on decreasing the overall growth in M-CSF (Fig. 8B). The liquid culture assay, however, did not show any difference in factor-independent growth between the control and mGab3V5-expressing FD-Fms cells. The kinetics of mGab3V5- and vector-expressing FD-Fms growth was monitored over a 3-day period to obtain a better idea of the relative influence of the overexpressed mGab3V5 protein (Fig. 8C). The data obtained by measuring the rate of growth as a function of cell number produced results analogous to those obtained from the soft agar assay. Expression of mGab3V5 in FD-Fms cells decreased the rate of cell growth in M-CSF relative to the control cells and resulted in increased factor-independent growth.
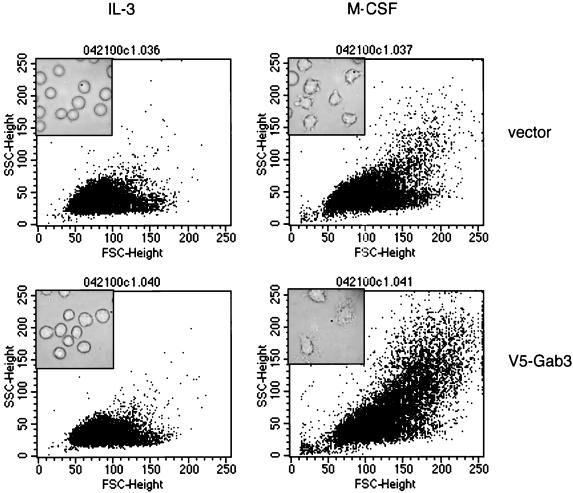
The morphology of the FD-Fms(vector) and FD-Fms(Gab3V5) cells grown in M-CSF or IL-3 was quantified by flow cytometric analysis of forward and side scatter to determine whether the M-CSF-induced decrease in growth rate of the FD-Fms(Gab3V5) cells was due to facilitated induction of macrophage differentiation (Fig. 9). The forward scatter is a function of cellular size, whereas side scatter is related to the degree of intracellular architecture and complexity. The corresponding morphology of the cells is shown by the inset phase-contrast photomicrograph. Control cells have a relatively uniform size distribution when grown in IL-3. This pattern is characteristic of undifferentiated FDC-P1 cells (4). Differentiation of these cells occurs in the presence of M-CSF and is evident from the increase in both forward and side scatter as well as in the morphological changes shown in the phase inset image. FD-Fms(Gab3V5) cells grown in IL-3 exhibit an undifferentiated morphology analogous to the control cells in IL-3, but growth in M-CSF produces a shift in forward and side scatter greater than that seen with control cells grown in M-CSF. Again, the phase-contrast photomicrograph reflects the morphological changes indirectly observed by flow cytometry. These results indicate that the overexpression of mGab3V5 in FD-Fms cells facilitates morphological differentiation.
FIG. 9.
mGab3V5 overexpression in FD-Fms cells facilitates M-CSF-induced morphological differentiation. FD-Fms(vector) and FD-Fms(mGab3V5) cells were grown in liquid culture for 2 days with the indicated growth factor (IL-3 or 2,500-U/ml M-CSF) and analyzed by flow cytometry for forward and side scatter, which reflect the size and intracellular architecture, respectively. The inset within each plot shows a phase-contrast photographic image of the corresponding cells and culture conditions.
DISCUSSION
The mammalian Gab family of proteins now includes three members, Gab1 (14), Gab2 (8), and Gab3 (presented in this publication). The relatedness of these proteins is based on the following general structural similarities: an amino-terminal PH domain, multiple motifs for tyrosine phosphorylation and SH2 domain binding, and polyproline regions potentially binding SH3 domains. The Drosophila DOS protein is the prototypical example of this larger signaling protein family (12, 34), also comprising the more distantly related mammalian Gab-type homologues like the IRS family of proteins and DOK proteins. These last protein groups are distinguished from the Gab family by the additional presence of a PTB domain (17, 42, 48, 49). Other proteins related to this group include FRS2 and LAT (19, 50), each of which exhibits distinct means of membrane attachment utilizing N-terminal myristoylation or a transmembrane domain, respectively.
The predicted amino acid sequence of murine and human Gab3 proteins defines two homologous proteins that are slightly smaller than Gab1 or Gab2 but contain all the hallmark motifs expected of Gab family members. The amino-terminal PH domain contains the most sequence identity between human and murine Gab3, as well as among all members of this family, including DOS. Among the mammalian Gab members, three tyrosine residues (two of which are adjacent) are conserved and within identical motifs. Including the DOS protein, only the twin tyrosines at amino acid 47 and 48 and the single tyrosine at amino acid 96 (see Fig. 1) are identical. Each site is predicted to have potential for an SH2 domain interaction (39); however, experimental verification is required for this possibility. The remainder of the Gab3 protein contains several additional tyrosine residues in motifs that may bind SH2 domains. From the carboxyl-terminal end of Gab3 and registering only motifs conserved between human and murine proteins, there are two sites for SHP2 binding, both of which are conserved in the mammalian Gab proteins and have been identified as SHP2-binding sites by mutagenesis of Gab2 (8, 21). Continuing upstream, there are three tyrosines in motifs with potential for binding the p85 subunit of PI3K. Further upstream, a single motif with potential for interacting with the SH2 domains of proteins such as SHIP or PLCγ (32, 39) is located at Y183. PLCγ has been reported to interact weakly with Gab1 (3), and SHIP1 interacts with both Gab1 and Gab2 (20, 21, 29).
Gab3 binds the adapter protein Grb2 constitutively. The interaction probably occurs via the two polyproline-rich motifs within the red rectangles shown in Fig. 1A. Only the more carboxyl-terminal of these motifs was contained in the GST Gab3-PRD construct used to pull down Grb2 as shown in Fig. 5B. This motif, P(X)3R(X)2KP, is identical to the motif in Gab2 known to bind the Grb2 C-terminal SH3 domain (22). The more amino-terminal polyproline-rich motif in Gab3 is a classic binding motif for Grb2 (30, 40) and may anchor Grb2 to Gab3 through the second Grb2 SH3 domain. Activated Fms binds to the Grb2 SH2 domain at the tyrosine-phosphorylated Y697 site within the kinase insert region of Fms (35, 41). Therefore, Grb2, or another adapter protein (C. Bourgin, R. P. Bourette, S. Arnoud, Y. Liu, L. R. Rohrschneider,and G. Mouchiroud, unpublished data), may tether Gab3 to the activated Fms, but because Grb2-SOS also interacts with the same FmsY697 site, some mechanism favoring exchange would be required (6). Alternatively, the adapter protein bound to Gab3 could interact with Fms at the less-well-characterized tyrosine-phosphorylated site, Y920, within the carboxyl-terminal tail region of Fms (24).
The function of the Gab proteins, defined by genetic studies of DOS in Drosophila, indicates a direct role in differentiation. The interaction of DOS with corkscrew (CSW; the SHP2 homologue) is a primary event required for development of the ommatidium R7 cell of Drosophila compound eyes (13, 38). In addition, the DOS protein participates in developmental signaling throughout other tissues of Drosophila (34). DOS interacts with signaling pathways upstream or parallel to Ras and is a substrate for the Drosophila tyrosine phosphatase CSW, which is a positive component of developmental signaling (2, 12). In contrast to Drosophila, mammalian cells express several related family members, termed Gab proteins, but each most likely follows the basic signaling mechanisms described for DOS (2, 12, 34).
The best-characterized member of the mammalian Gab family is Gab1, a component of the differentiation pathway in kidney epithelial cells (25, 43, 44). In addition, we have previously shown that Gab2 is part of the Fms signaling pathway. Gab2 is tyrosine phosphorylated early after M-CSF stimulation of FD-Fms cells. Gab2 exhibits inducible association with SHP2, p85, and SHIP, but constitutive association with Grb2. Overexpression of WT Gab2 in FD-Fms cells has a mild effect on enhancing macrophage development, but overexpression of Gab2 mutated at the two SHP2 binding sites dominantly inhibits macrophage differentiation (21). This biological behavior is consistent with the results of studies on DOS differentiation signaling in Drosophila and suggests that Gab2 may function similarly in macrophage differentiation.
The results presented in this paper indicate that the novel DOS family member, Gab3, is involved in several growth factor signaling pathways and likewise is a component of the Fms signaling pathway. Using RT-PCR analysis, Gab3 mRNA could be detected in macrophages, B cells, and T cells, in some of their progenitor cells, and in ES cells. Within tissues of the mouse, Gab3 mRNA was most abundant in spleen and thymus. M-CSF stimulation of FD-Fms cells overexpressing the V5-tagged murine Gab3 induced tyrosine phosphorylation of Gab3V5 and association with the p85 subunit of PI3K and SHP2. The same FD-Fms(Gab3V5) cells differentiated considerably better than the FD-Fms cells not expressing the exogenous Gab3V5, and concomitantly the rate of growth decreased. This suggests that Gab3 is a critical messenger for macrophage development within the FD-Fms cells; however, RT-PCR analysis of the FDC-P1 cells demonstrated that Gab3 mRNA was expressed at very low levels in these myeloid progenitor cells (Fig. 2A). A time-course analysis of Gab3 mRNA levels after M-CSF stimulation of FD-Fms cells demonstrated that Gab3 expression was increased by M-CSF stimulation. Conversely, we utilized FD-FmsY807F cells, which contain a point mutation in the Fms tyrosine kinase activation loop, to investigate any potential relationship between differentiation and Gab3 induction. The Y807F mutation in Fms has little effect on the receptor tyrosine kinase activity and endogenous substrate phosphorylation is near normal. However, FmsY807F is uniquely defective in the induction of differentiation when expressed in the FDC-P1 cells (4). Likewise, the FD-FmsY807F cells did not increase Gab3 expression following M-CSF stimulation (Fig. 7). Therefore, the expression of Gab3 in FDC-P1 cells containing either WT or Y807F mutant Fms correlates with macrophage differentiation, suggesting a role for Gab3 in this developmental process.
Both Gab2 and Gab3 have similar structural features and appear to have similar roles in macrophage development, but do they differ in their signaling functions? Are they redundant, or does each have some unique functions? One primary difference between Gab3 and both Gab1 and Gab2 is the array of adapter proteins which bind to each. All three Gab proteins have conserved motifs for binding the SH3 domains of Grb2 and the SH2 domains of SHP2 and p85 (plus two additional conserved motifs potentially binding yet uncharacterized proteins). Gab1 and Gab2 each contain five polyproline motifs, which can interact with the SH2 domains of Crk/CRKL when phosphorylated, and four of these motifs show positional conservation. However, Gab3 exhibits only one such motif in the murine protein but none in the human protein. Experiments using GST fused to full-length CRKL or the SH3 domain of Crk failed to detect an interaction with tyrosine-phosphorylated or unphosphorylated murine Gab3 (Fig. 5A and unpublished observations). Therefore, unlike Gab1 and Gab2, Gab3 may not interact with Crk or CRKL and the partner proteins coupled through these adapter proteins. In addition, Gab3 may interact with uncharacterized adapters and their partner proteins through one of the four unique YXXX motifs or additional uncharacterized motifs.
Another relevant question is whether Gab2 and Gab3 interact with Fms simultaneously or sequentially. In the former case, Gab2 and Gab3 may interact with Fms through the adapter proteins Grb2 or Mona (Bourgin et al., unpublished data), forming hetero-complexes [Gab2-(Fms)2-Gab2, Gab2-(Fms)2-Gab3, or Gab3-(Fms)2-Gab3], and each complex might initiate different signaling pathways or combinations of pathways. The alternative mechanism is that each Gab protein acts sequentially, and in fact, this mechanism may be favored. In the FDC-P1 cells, the initial level of Gab2 is relatively high compared to that of Gab3, but Gab3 levels increase several hours after M-CSF stimulation. Thus, Gab2 and Gab3 would be temporally separated in their expression, activation, and function. This possibility might also suggest a reason why the FD-Fms(Gab3V5) cells exhibit greater differentiation potential in M-CSF than the FD-Fms cells, which rely on the induction and increased expression of endogenous Gab3. The FD-Fms(Gab3V5) cells express Gab3V5 prior to M-CSF stimulation and therefore can differentiate earlier than the cells without exogenous Gab3. Thus, the greater differentiation seen in the FD-Fms(Gab3V5) cells than in the FD-Fms cells at 2 days after M-CSF stimulation (Fig. 9) may be due to eliminating the time lag required for endogenous Gab3 induction.
Overall, the results in this paper identify a new gene product, called Gab3, and suggest new steps involving Gab3 participation in a molecular mechanism regulating macrophage differentiation. In general, M-CSF activation of Fms in appropriate target cells initially activates proliferation signals, which ultimately change into differentiation signals upon Gab3 expression and activation. The proliferation signals follow the general activation of the Ras-MAPK pathway initiated by the interaction of Grb2-SOS with activated Fms at the tyrosine-phosphorylated Y697 site. The Grb2-Gab2 complex becomes activated in these early signaling events by still incompletely understood mechanisms (21) but probably requires prior activation of PI3K (p85/PI3K binds to activated Fms at the Y721 site) for generation of the plasma membrane PtdIns(3,4,5)P3, which associates with the Gab2 PH domain at the cytoplasmic surface. The SH2 domain of Grb2 may then link Gab2 to Fms within the complex. Fms, Gab2, or both Fms and Gab2 then transmit signals for increasing Gab3 transcription. Following accumulation of sufficient Gab3 protein, specific signals for differentiation are generated and interpreted for the formation of a mature macrophage.
The general role of Fms, Gab2, and Gab3 in this model for the differentiation process is consistent with our knowledge of the related DOS protein in Drosophila and with known functions of the Gab1 protein in epithelial cells of mammals (14, 25, 44). The mechanism also shares some similarities with fibroblast growth factor (FGF) signaling through the related scaffolding protein FRS2, which then associates with Gab1 (31). Similarly, both Gab1 and Gab2 have been reported to participate in erythropoietin signaling (45).
To test the functions of Gab proteins in vivo, knockout mice have been generated (9, 10, 15). Both Gab1 and FRS2 knockouts lead to embryonic lethality, indicating the fundamental importance of these genes in embryonic development. However, the lethality has precluded in vivo functional analysis of these proteins in differentiation signaling pathways. On the other hand, Gab2 knockout mice are viable and show defects in the allergic response, but no apparent phenotype was observed in hematopoietic lineages. It is presently not clear whether this is due to redundant functions of Gab family proteins or incomplete knockout of Gab2 functions. Therefore, in future studies it would be helpful to address the functional specificities and redundancies of the Gab family members utilizing conditional knockout experiments and combinations of Gab gene knockouts.
Acknowledgments
We thank Tami Anderson, Mario Cepeda, and Dana Bressette for significant technical help in performing several sets of these experiments.
The research was supported in part by grants from the National Cancer Institute (CA40987). M.S. was supported by the Mildred-Scheel-Stiftung. A portion of this work was conducted while L.R.R. was a fellow of the John Simon Guggenheim Memorial Foundation, on sabbatical in the laboratory of Guy Mouchiroud, Lyon, France.
REFERENCES
- 1.Allard, J. D., H. C. Chang, R. Herbst, H. McNeill, and M. A. Simon. 1996. The SH2-containing tyrosine phosphatase corkscrew is required during signaling by sevenless, Ras1 and Raf. Development 122:1137–1146. [DOI] [PubMed] [Google Scholar]
- 2.Bardelli, A., P. Longati, D. Gramaglia, M. C. Stella, and P. M. Comoglio. 1997. Gab1 coupling to the HGF/Met receptor multifunctional docking site requires binding of Grb2 and correlates with the transforming potential. Oncogene 15:3103–3111. [DOI] [PubMed] [Google Scholar]
- 3.Bausenwein, B. S., M. Schmidt, B. Mielke, and T. Raabe. 2000. In vivo functional analysis of the daughter of sevenless protein in receptor tyrosine kinase signaling. Mech. Dev. 90:205–215. [DOI] [PubMed] [Google Scholar]
- 4.Bourette, R. P., G. M. Myles, K. Carlberg, and L. R. Rohrschneider. 1995. Uncoupling of the proliferation and differentiation signals mediated by the murine macrophage colony-stimulating factor receptor expressed in myeloid FDC-P1 cells. Cell Growth Differ. 6:631–645. [PubMed] [Google Scholar]
- 5.Bourette, R. P., G. M. Myles, J.-L. Choi, and L. R. Rohrschneider. 1997. Sequential activation of phosphatidylinositol 3-kinase and phospholipase C-γ2 by the M-CSF receptor is necessary for differentiation signaling. EMBO J. 16:5880–5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bourette, R. P., and L. R. Rohrschneider. 2000. Early events in M-CSF receptor signaling. Growth Factors 17:155–166. [DOI] [PubMed] [Google Scholar]
- 7.Feng, S., J. K. Chen, H. Yu, J. A. Simon, and S. L. Schreiber. 1994. Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions. Science 266:1241–1247. [DOI] [PubMed] [Google Scholar]
- 8.Gu, H., J. C. Pratt, S. J. Burakoff, and B. G. Neel. 1998. Cloning of p97/Gab2, the major SHP2-binding protein in hematopoietic cells, reveals a novel pathway for cytokine-induced gene activation. Mol. Cell 2:729–740. [DOI] [PubMed] [Google Scholar]
- 9.Gu, H., K. Saito, L. D. Kaman, J. Shen, T. Fleming, Y.-P. Wang, J. C. Pratt, G. Lin, B. Lim, J.-P. Kinet, and B. G. Neel. 2001. Essential role for Gab2 in the allergic response. Nature 412:186–190. [DOI] [PubMed] [Google Scholar]
- 10.Hadari, Y. R., N. Gotoh, H. Kouhara, I. Lax, and J. Schlessinger. 2001. Critical role for the docking-protein FRS2α in FGF receptor-mediated signal transduction pathways. Proc. Natl. Acad. Sci. USA 98:8578–8583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hafen, E., B. Dickson, T. Raabe, D. Brunner, N. Oellers, and A. van der Straten. 1993. Genetic analysis of the sevenless signal transduction pathway of Drosophila. Dev. Suppl. 41–46. [PubMed]
- 12.Herbst, R., P. M. Carroll, J. D. Allard, J. Schilling, T. Raabe, and M. A. Simon. 1996. Daughter of sevenless is a substrate of the phosphotyrosine phosphatase corkscrew and functions during sevenless signaling. Cell 85:899–909. [DOI] [PubMed] [Google Scholar]
- 13.Herbst, R., X. Zhang, J. Qin, and M. A. Simon. 1999. Recruitment of the protein tyrosine phosphatase CSW by DOS is an essential step during signaling by the Sevenless receptor tyrosine kinase. EMBO J. 18:6950–6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holgado-Madruga, M., D. R. Emlet, D. K. Moscatello, A. K. Godwin, and A. J. Wong. 1996. A Grb2-associated docking protein in EGF- and insulin-receptor signalling. Nature 379:560–564. [DOI] [PubMed] [Google Scholar]
- 15.Itoh, I., Y. Yoshida, K. Nishida, M. Narimatsu, M. Hibi, and T. Hirano. 2000. Role of Gab1 in heart, placenta, and skin development and growth factor-and cytokine-induced extracellular signal-regulated kinase mitogen-activated protein kinase activation. Mol. Cell. Biol. 20:3695–3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jenkins, B. J., C. J. Bagley, J. Woodcock, A. F. Lopez, and T. J. Gonda. 1996. Interacting residues in the extracellular region of the common beta subunit of the human granulocyte-macrophage colony-stimulating factor, interleukin (IL)-3, and IL-5 receptors involved in constitutive activation. J. Biol. Chem. 271:29707–29714. [DOI] [PubMed] [Google Scholar]
- 17.Jones, N., and D. J. Dumont. 1998. The Tek/Tie2 receptor signals through a novel Dok-relating docking protein, Dok-R. Oncogene 17:1097–1108. [DOI] [PubMed] [Google Scholar]
- 18.Karasuyama, H., and F. Melchers. 1988. Establishment of mouse cell lines which constitutively secrete large quantities of interleukin 2, 3, 4, or 5, using modified cDNA expression vectors. Eur. J. Immunol. 18:97–104. [DOI] [PubMed] [Google Scholar]
- 19.Kouhara, H., Y. R. Hadari, T. Spivak-Kroizman, J. Schilling, D. Bar-Sagi, I. Lax, and J. Schlessinger. 1997. A lipid-anchored Grb2-binding that links FGF-receptor activation to the Ras/MAPK signaling pathway. Cell 89:693–702. [DOI] [PubMed] [Google Scholar]
- 20.Lecoq-Lafon, C., F. Verdier, S. Fichelson, S. Chrétien, S. Gisselbrecht, C. Lacombe, and P. Mayeux. 1999. Erythropoietin induces the tyrosine phosphorylation of GAB1 and its association with SHC, SHP2, SHIP, and phosphatidylinositol 3-kinase. Blood 93:2578–2585. [PubMed] [Google Scholar]
- 21.Liu, Y., B. Jenkins, J.-L. Shin, and L. R. Rohrschneider. 2000. Scaffolding protein Gab2 mediates differentiation signaling downstream of FMS receptor tyrosine kinase. Mol. Cell. Biol. 21:3047–3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lock, L. S., I. Royal, M. A. Naujokas, and M. Park. 2000. Identification of an atypical Grb2 carboxyl-terminal SH3 domain binding site in Gab2 docking proteins reveals Grb2-dependent and -independent recruitment of Gab1 to receptor tyrosine kinases. J. Biol. Chem. 275:31336–31345. [DOI] [PubMed] [Google Scholar]
- 23.Lucas, D. M., and L. R. Rohrschneider. 1999. A novel spliced form of SH2-containing inositol phosphatase SHIP is expressed during myeloid development. Blood 93:1922–1933. [PubMed] [Google Scholar]
- 24.Mancini, A., R. Niedenthal, H. Joos, A. Koch, S. Trouliaris, H. Niemann, and T. Tamura. 1997. Identification of a second Grb2 binding site in the v-Fms tyrosine kinase. Oncogene 15:1565–1572. [DOI] [PubMed] [Google Scholar]
- 25.Maroun, C. R., D. K. Moscatello, M. A. Naujokas, M. Holgado-Madruga, A. J. Wong, and M. Park. 1999. A conserved inositol phospholipid binding site within the pleckstrin homology domain of the Gab1 docking protein is required for epithelial morphogenesis. J. Biol. Chem. 274:31719–31726. [DOI] [PubMed] [Google Scholar]
- 26.Metcalf, D. 1984. The hemopoietic colony stimulating factors. Elsevier Science Publishers, New York, N.Y.
- 27.Mosmann, T. 1983. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods 65:55–63. [DOI] [PubMed] [Google Scholar]
- 28.Nguyen, L., M. Holgado-Madruga, C. Maroun, E. D. Fixman, D. Kamikura, T. Fournier, A. Charest, M. L. Tremblay, A. J. Wong, and M. Park. 1997. Association of the multisubstrate docking protein Gab1 with the hepatocyte growth factor receptor requires a functional Grb2 binding site involving tyrosine 1356. J. Biol. Chem. 272:20811–20819. [DOI] [PubMed] [Google Scholar]
- 29.Nishida, K., Y. Yoshida, M. Itoh, T. Fukada, T. Ohtani, T. Shirogane, T. Atsumi, M. Takahashi-Tezuka, K. Ishihara, M. Hibi, and T. Hirano. 1999. Gab-family adapter proteins act downstream of cytokine and growth factor receptors and T- and B-cell antigen receptors. Blood 93:1809–1816. [PubMed] [Google Scholar]
- 30.Oehrl, W., C. Kardinal, S. Ruf, K. Adermann, J. Groffen, G. S. Feng, J. Blenis, T. H. Tan, and S. M. Feller. 1998. The germinal center kinase (GCK)-related protein kinases HPK1 and KHS are candidates for highly selective signal transducers of Crk family adapter proteins. Oncogene 17:1893–1901. [DOI] [PubMed] [Google Scholar]
- 31.Ong, S. H., Y. R. Hadari, N. Gotoh, G. R. Guy, J. Schlessinger, and I. Lax. 2001. Stimulation of phosphatidylinositol 3-kinase by fibroblast growth factor receptor receptors is mediated by coordinated recruitment of multiple docking proteins. Proc. Natl. Acad. Sci. USA 98:6074–6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Osborne, M. A., G. Zenner, M. Lubinus, X. Zhang, Z. Songyang, L. C. Cantley, P. Majerus, P. Burn, and J. P. Kochen. 1996. The inositol 5′-phosphatase SHIP binds to immunoreceptor signaling motifs and responds to high affinity IgE receptor aggregation. J. Biol. Chem. 271:29271–29278. [DOI] [PubMed] [Google Scholar]
- 33.Pear, W. S., G. P. Nolan, M. L. Scott, and D. Baltimore. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA 90:8392–8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raabe, T., J. Riesgo-Escovar, X. Liu, B. S. Bausenwein, P. Deak, P. Maröy, and E. Hafen. 1996. DOS, a novel pleckstrin homology domain-containing protein required for signal transduction between sevenless and Ras1 in Drosophila. Cell 85:911–920. [DOI] [PubMed] [Google Scholar]
- 35.Reedijk, M., X. Liu, P. van der Geer, K. Letwin, M. D. Waterfield, T. Hunter, and T. Pawson. 1992. Tyr721 regulates specific binding of the CSF-1 receptor kinase insert to PI 3′-kinase SH2 domains: a model for SH2-mediated receptor-target interactions. EMBO J. 11:1365–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rohrschneider, L. R., V. M. Rothwell, and N. A. Nicola. 1989. Transformation of murine fibroblasts by a retrovirus encoding the murine c-Fms proto-oncogene. Oncogene 4:1015–1022. [PubMed] [Google Scholar]
- 37.Rohrschneider, L. R., and D. Metcalf. 1989. Induction of macrophage colony stimulating factor-dependent growth and differentiation after introduction of the murine c-Fms gene into FDC-P1 cells. Mol. Cell. Biol. 9:5081–5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simon, M. A. 1994. Signal transduction during the development of the Drosophila R7 photoreceptor. Dev. Biol. 166:431–442. [DOI] [PubMed] [Google Scholar]
- 39.Songyang, Z., and L. C. Cantley. 1995. Recognition and specificity in protein tyrosine kinase-mediated signaling. Trends Biochem. Sci. 20:470–475. [DOI] [PubMed] [Google Scholar]
- 40.Sparks, A. B., J. E. Rider, N. G. Hoffman, D. M. Fowlkes, L. A. Quillam, and B. K. Kay. 1996. Distinct ligand preferences of Src homology 3 domains from Src, Yes, Abl, Cortactin, p53bp2, PLCgamma, Crk, and Grb2. Proc. Natl. Acad. Sci. USA 93:1540–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van der Geer, P., and T. Hunter. 1991. Tyrosine 706 and 807 phosphorylation site mutants in the murine colony-stimulating factor-1 receptor are unaffected in their ability to bind or phosphorylate phosphatidylinositol-3 kinase but show differential defects in their ability to induce early response gene transcription. Mol. Cell. Biol. 11:4698–4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Voliovitch, H., D. G. Schindler, Y. R. Hadari, S. I. Taylor, D. Accili, and Y. Zick. 1995. Tyrosine phosphorylation of insulin receptor substrate-1 in vivo depends upon the presence of its pleckstrin homology region. J. Biol. Chem. 270:18083–18087. [DOI] [PubMed] [Google Scholar]
- 43.Weidner, K. M., S. Di Cesare, M. Sachs, V. Brinkmann, J. Behrens, and W. Birchmeier. 1996. Interaction between Gab1 and the c-Met receptor tyrosine kinase is responsible for epithelial morphogenesis. Nature 384:173–176. [DOI] [PubMed] [Google Scholar]
- 44.Weidner, K. M., M. Sachs, D. Riethmacher, and W. Birchmeier. 1995. Mutation of juxtamembrane tyrosine residue 1001 suppresses loss-of-function mutations of the met receptor in epithelial cells. Proc. Natl. Acad. Sci. USA 92:2597–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wickrema, A., S. Uddin, A. Sherma, F. Chen, Y. Alsayed, S. Ahmad, S. T. Sawyer, G. Krystal, T. Yi, K. Hishada, M. Hibi, T. Hirano, and L. C. Platanais. 1999. Engagement of Gab1 and Gab2 in erythropoietin signaling. J. Biol. Chem. 274:24469–24474. [DOI] [PubMed] [Google Scholar]
- 46.Wolf, I., D. M. Lucas, P. A. Algate, and L. R. Rohrschneider. 2000. Cloning of the genomic locus of mouse SH2-containing inositol 5-phosphatase SHIP and a novel 110 kDa splice isoform SHIPδ. Genomics 69:104–112. [DOI] [PubMed] [Google Scholar]
- 47.Wolf, I., and L. R. Rohrschneider. 1999. Fiz1, a novel zinc finger protein interacting with the receptor tyrosine kinase Flt3. J. Biol. Chem. 274:21478–21484. [DOI] [PubMed] [Google Scholar]
- 48.Yamanashi, Y., and D. Baltimore. 1997. Identification of the Abl- and ras-GAP-associated 62 kDa protein as a docking protein, Dok. Cell 88:205–211. [DOI] [PubMed] [Google Scholar]
- 49.Yenush, L., and M. F. White. 1997. The IRS-signalling system during insulin and cytokine action. BioEssays 19:491–500. [DOI] [PubMed] [Google Scholar]
- 50.Zhang, W., C. L. Sommers, D. N. Burshtyn, C. C. Stebbins, J. B. DeJarnette, R. P. Trible, A. Grinberg, H. C. Tsay, H. M. Jacobs, C. M. Kessler, E. O. Long, P. E. Love, and L. E. Samelson. 1999. Essential role of LAT in T cell development. Immunity 10:323–332. [DOI] [PubMed] [Google Scholar]
- 51.Zhao, C., D.-H. Yu, R. Shen, and G.-S. Feng. 1999. Gab2, a new pleckstrin homology domain-containing adapter protein, acts to uncouple signaling from ERK kinase to Elk-1. J. Biol. Chem. 274:19649–19654. [DOI] [PubMed] [Google Scholar]
- 52.Zipursky, S. L., and G. M. Rubin. 1994. Determination of neuronal cell fate: lessons from the R7 neuron of Drosophila. Annu. Rev. Neurosci. 17:373–397. [DOI] [PubMed] [Google Scholar]