Abstract
Characterization of the mechanism(s) of action of trans-acting factors in higher eukaryotes requires the establishment of cellular models that test their function at endogenous target gene regulatory elements. Erythroid Krüppel-like factor (EKLF) is essential for β-globin gene transcription. To elucidate the in vivo determinants leading to transcription of the adult β-globin gene, functional domains of EKLF were examined in the context of chromatin remodeling and transcriptional activation at the endogenous locus. Human EKLF (hEKLF) sequences, linked to an estrogen-responsive domain, were studied with an erythroblast cell line lacking endogenous EKLF expression (J2eΔeklf). J2eΔeklf cells transduced with hEKLF demonstrated a dose-dependent rescue of β-globin transcription in the presence of inducing ligand. Further analysis using a series of amino-terminal truncation mutants of hEKLF identified a distinct internal domain, which is sufficient for transactivation. Interestingly, studies of the chromatin structure of the β-promoter revealed that a smaller carboxy-terminal domain generated an open promoter configuration. In vitro and in vivo binding studies demonstrated that this region interacted with BRG1, a component of the SWI/SNF chromatin remodeling complex. However, further study revealed that BRG1 interacted with an even smaller domain of EKLF, suggesting that additional protein interactions are required for chromatin remodeling at the endogenous β-promoter. Taken together, our findings support a stepwise process of chromatin remodeling and coactivator recruitment to the β-globin promoter in vivo. The J2eΔeklf inducible hEKLF system will be a valuable tool for further characterizing the temporal series of events required for endogenous β-globin gene transcription.
Appropriate control of gene transcription demands that a series of coordinated interactions occur between tissue-restricted and ubiquitously expressed transactivators, chromatin remodeling complexes, coactivators, and the general transcriptional machinery (38, 45). Although reconstitution studies of in vitro-chromatinized and nonchromatinized templates have revealed key interactions in this process, less is known about the molecular events required for gene transcription in vivo, particularly at mammalian gene loci. In this context, regulation of the five structural genes of the murine β-globin multigene cluster (5′-ɛy-βh0-βh1-βmaj-βmin-3′) provides a suitable model for studying these events given their tissue-specific and developmentally specific pattern of expression (20, 25, 51). Although a powerful upstream enhancer, the locus control region is required for high-level tissue-specific expression (28, 33), and the determinants of developmentally specific expression reside in gene-proximal promoter and enhancer sequences (18, 26, 41, 46).
Several cis-acting elements that modulate developmentally specific gene expression have been identified in β-globin-like gene promoters (14, 36, 53, 63). Of these, the proximal β-globin CACC motif is a particularly attractive candidate for study, a thalassemic state being observed in individuals with point mutations in this element (53). This alteration disrupts specifically the binding of an erythroid-specific transactivator, erythroid Krüppel-like factor (EKLF) (27, 49). The critical nonredundant role of EKLF in adult erythroid cells was further emphasized in gene disruption studies of the murine homologue. EKLF null embryos die with a lethal anemia at day 14 to 15 of gestation, definitive erythroid cells failing to produce βmaj transcripts (52, 58). Further study of these deficient embryos revealed a specific loss of a developmentally specific DNase I hypersensitivity site in the proximal β-globin promoter of day 14 erythroblasts derived from EKLF null embryos (57, 72). Since the degree of DNase I hypersensitivity of a given locus correlates with nucleosomal remodeling (56, 65, 68), these findings support the contention that EKLF is required for chromatin reorganization at the β-globin promoter in definitive erythroid cells.
The molecular mechanism by which EKLF modulates chromatin structure at the β-globin promoter has been examined in chromatin reconstitution assays (4, 37). A highly purified SWI/SNF-containing multiprotein complex, E-RC1, purified from murine erythroid cells, interacts with the Krüppel-like C2H2-type zinc finger COOH-terminal DNA binding domain of recombinant EKLF. This interaction results in remodeling and activation of a chromatinized β-globin promoter template. SWI/SNF complexes have been implicated in remodeling nucleosomes in an ATP-dependent manner in many eukaryotic organisms, permitting increased access of trans-acting factors to their cognate binding sites (40, 61, 67). Although subunit heterogeneity is a characteristic of SWI/SNF complexes purified from different cell types (69, 70), a minimum catalytic core of three components, BRG1, INI1, and BAF155 or BAF170, is sufficient to remodel nucleosomal arrays in vitro (37, 62). Interestingly, a direct protein-protein interaction between BRG1, the critical ATPase component of E-RC1, and the DNA binding domain of EKLF has been defined, this interaction being sufficient for in vitro remodeling of a chromatinized β-globin template (37).
The complexity of EKLF function is further emphasized by a review of prior structure-function studies. Initial studies of the proline-rich NH2-terminal domain of EKLF suggested that the first 102 amino acids mediate transactivation, an internal domain mediating transcriptional repression in these assays (11). However, subsequent studies utilizing transient β-promoter reporter assays have demonstrated that an internal, as yet uncharacterized domain of EKLF is sufficient for transactivation (54). Taken together, these studies raise the question of the role, necessity, and sufficiency of the domains of EKLF for transcriptional initiation at the endogenous β-globin promoter. Our ability to address these issues has been facilitated by the recent description of erythroid cell lines derived from EKLF null murine fetal liver erythroblasts, transformed with a raf/myc-containing retrovirus (15). Introduction of the murine EKLF cDNA into these cells by retrovirally mediated gene transfer resulted in rescue of human β-globin gene expression. Utilizing this resource, we have determined systematically that separate domains of EKLF subserve chromatin remodeling and transactivation functions at the endogenous murine β-globin promoter. Our studies provide evidence that EKLF-cofactor interactions, in addition to that involving E-RC1, may be required to achieve the open chromatin configuration and initiation of transcription required at the β-globin promoter in definitive erythroid cells.
MATERIALS AND METHODS
Plasmid constructions.
pSP-HA-fIEKLF was constructed by fusing a double-stranded oligonucleotide encoding consensus Kozak and influenza virus hemagglutinin (HA) tag sequences in frame with the human EKLF open reading frame utilizing the pSP-hEKLF plasmid (66). pSP-HA-Δ164 EKLF was prepared by inserting the 1,196-bp BstXI (blunt)-XhoI fragment of pSP-hEKLF into Ecl136II-XhoI sites of pSP-HA-210. pSP-HA-Δ221EKLF was made by inserting the 1,025-bp SacII (blunt)-XhoI fragment of pSP-hEKLF into the Ecl136II and XhoI sites of pSP-HA-210. pSP-HA-Δ253EKLF was made by inserting the 899-bp PstI (blunt)-SalI fragment of pSPhEKLF into the Ecl136II and XhoI sites of pSp-HA-210. pSP-HA-210 has the above-described HA oligonucleotide sequence inserted in the EcoRI site of the pSP73 polylinker (a kind gift of Derek Persons). The eukaryotic expression plasmids used in transient-transfection studies were generated by subcloning the EKLF fragment of each of the above pSP constructs into pCINeo (Promega). Glutathione S-transferase (GST) fusion constructs were generated by subcloning the EKLF fragment of the above pSP constructs into pGEX-4T-1 (Amersham Pharmacia). The retroviral vector constructs used in transduction of J2eΔeklf cells were generated by subcloning the EKLF fragment of the above pSP constructs into pMSCV-ER-irGFP vector (15). All constructs were sequenced to verify in-frame fusion with the HA, GST, and estrogen receptor (ER) domains, respectively.
The plasmids pBJ5-BRG1 and pDNBRG1 were kind gifts of G. Crabtree and R. Kingston, respectively (35, 39). The pBJ5 ΔBRG1 plasmid was made by a frameshift at amino acid 78 of BRG1. A 1,240-bp PstI kanamycin resistance cassette (pUC4K; Amersham Pharmacia Biotech) was inserted into the NsiI site of pBJ5 BRG1, resulting in complete loss of the BRG1 protein as determined by Western analysis (data not shown). The T7 polymerase expression plasmid of BRG1 was generated by inserting the 5,350-bp SgrAI (blunt)-MunI (blunt) fragment of pBJ5 BRG1 into the SmaI site of pSP73 (Promega). The MSCV-DNBRG1-irYFP vector was made by inserting the DNBRG1 coding sequence into the pMSCV-ER-irYFP vector. The pMSCV-ER-irYFP vector is a derivative vector in which the enhanced yellow fluorescent protein (irYFP; Clontech) coding sequence replaces the irGFP cDNA in the pMSCV-ER-irGFP vector.
Cell culture.
K562 human erythroleukemia cells, SW13 human adrenal carcinoma cells, and 293T fibroblast cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Bio Whittaker) with 100 U of penicillin/ml and 0.1 mg of streptomycin (Pen/Strep; Bio Whittaker)/ml and 10% fetal bovine serum (Bio Whittaker). The EKLF-null erythroid cell line B1 (15) was maintained in Dulbecco’s modified Eagle’s medium with Pen/Strep and 15% fetal bovine serum (Bio Whittaker). Cells were cultured at 37°C in a humidified atmosphere with 5% CO2.
J2eΔeklf transductions and analysis.
Amphotropic retroviral supernatants were generated to transduce J2eΔeklf cells in a manner described previously (59). Briefly, 293T cells were cotransfected with appropriate pMSCV-flEKLF-irGFP constructs with the helper plasmid pEQPAM3 (59). Typical transfections generated titers of >105 infectious particles/ml. Viral supernatants for each construct were used repeatedly (three to six times) to transduce 5 × 104 J2eΔeklf cells. Transduced cells were allowed to recover and then expanded over 3 to 4 days in J2e growth media. The top 5 to 10% green fluorescent protein (GFP)-expressing fraction was selected by fluorescence-activated cell sorting (FACS), expanded, and re-sorted by FACS until ≥90% of the population was GFP+. Each GFP+ pool was assessed by Southern blotting to have stable, intact retroviral sequence (data not shown), and the presence of equivalent levels of EKLF-ER protein in the nucleus after tamoxifen induction was verified by Western blotting.
For nuclear localization study, nuclear extracts were prepared as described previously (2) with minor modification, using 107 J2eΔeklf cells induced with 100 nM tamoxifen. Equal amounts of total protein were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western analysis utilizing a polyclonal anti-estrogen receptor antiserum (Santa Cruz) and ECL development kit according to the manufacturer’s protocol (Amersham). Blots were stripped and reprobed with p38 antisera (Santa Cruz) as a control. The electrophoretic mobility shift assays (EMSA) were performed as described previously (19), using equal protein concentrations of each purified GST-EKLF recombinant and a CACC motif containing 32P-labeled probe.
Secondary transduction of J2eΔeklf flEKLF with DN-BRG1 was achieved by transducing J2eΔeklf flEKLF with VSV-G viral supernatants encoding dominant-negative (DN)-BRG1. Viral supernatants were generated using 293T cells as described above. The top 5 to 10% YFP-expressing fraction was selected by FACS, expanded, and re-sorted for the YFP+ GFP+ phenotype by FACS until >93% of the population was YFP+ GFP+. The YFP+ GFP+ pool was assessed to have stable, intact retroviral sequences by Southern blotting and to express DN-BRG1-ER and EKLF-ER protein after tamoxifen induction by Western blotting (data not shown).
DNase I hypersensitivity assays.
Cell cultures of approximately 0.4 × 106 cells per ml of J2eΔeklf cells were set up in duplicate. Tamoxifen (Sigma) at a final concentration of 100 nM or an equivalent diluent control was added to the induced and uninduced cultures, respectively. After 36 h of growth, nuclei from approximately 108 cells were prepared and analyzed as described previously (71). Briefly, aliquots of nuclei were digested with increasing amounts of DNase I (Pharmacia) for 10 min at 37°C. One aliquot was incubated without addition of DNase I to serve as a control for endogenous nuclease activity. The reactions were stopped with a twofold volume of 250 μg of of proteinase K (Promega)/ml, 10 mM NaCl, 0.5% SDS, 10 mM EDTA, and 10 mM Tris (pH 8.0) and incubated at 55°C for 16 h. After phenol-chloroform extraction, approximately 12 μg of DNA was digested with the restriction enzyme indicated in the text, separated on a 1% agarose gel, and transferred to a nylon membrane by Southern blotting. Analysis of DNase I hypersensitivity was performed utilizing βmaj-specific (34) or aminolevulinate synthase (ala-s)-specific probes (64).
RNase protection analysis.
Total RNA was extracted from approximately 1.5 × 107 cells, cultured in a manner similar to that described for DNase I hypersensitivity assays, utilizing the STAT 60 reagent (Ambion). RNase protection assays (RPA) were performed utilizing an RPA II kit (Ambion) and 32P-labeled riboprobes specific for murine α, βh1, and βmaj riboprobes (6, 50). Briefly, approximately 106 cpm of each riboprobe was added to 2 μg of total RNA and allowed to anneal at 42°C overnight. After RNase digestion, the protected fragments were separated on a 6% denaturing polyacrylamide gel and visualized using a Molecular Dynamics PhosphorImager (Amersham Pharmacia Biotech).
Transient-transfection studies.
K562 cells were transfected using a Bio-Rad Gene Pulser apparatus as described previously (36). Immediately following the transfection, 20 mM hemin (Sigma) was added to the growth media. SW13 cells were transfected by lipofection following the manufacturer’s protocol (Fugene6; Roche). Plasmid concentrations, optimized for the transfection studies described, were 10 μg of appropriate pCIneo construct, 5 μg of appropriate pBRG1 construct (as indicated in the text), 2 μg of HS2βluc (36), and 0.2 μg of TK-Renilla luciferase plasmid (Promega). The TK-Renilla luciferase plasmid was utilized to correct results for transfection efficiency. After 48 h posttransfection, cell lysates for each cell line were prepared and tested for luciferase activity using the Dual Luciferase Reporter Assay System (Promega). Luciferase activity was quantitated on a Monolight 2001 luminometer (Analytical Luminescence Laboratories). The results are a summary of at least three experiments in which each construct was tested in duplicate.
In vitro binding assays.
GST fusion proteins were generated for each pGST-EKLF construct as previously described with minor modifications (73). Briefly, expression of GST fusion proteins was induced by culturing logarithmically growing transformed BL21(pLysS) Escherichia coli cells (Stratagene) in the presence of 0.5 mM isopropyl-β-d-thiogalactopyranoside (Sigma) for 1.5 to 3 h. Pellets from a 500-ml culture were resuspended in 12.5 ml of TSE (50 mM Tris-HCl [pH 8], 25% sucrose, 1 mM EDTA). TSE buffer and subsequent buffers contained a cocktail of protease inhibitors (Protease Inhibitor Cocktail Tablets; Roche). The bacterial suspension was incubated at room temperature for 5 min in the presence of 5 mg of lysozyme/ml. The lysate was supplemented with 10 mM MgCl2, 10 mM MnCl2, and 10 μg of DNase I/ml and incubated for 15 min at 37°C. The lysate was further supplemented with 2 ml of 10× phosphate-buffered saline (PBS), 1 ml of 20% Tween 20, 1 ml of 20% Triton X-100, 0.2 ml of 1 M dithiothreitol, and additional protease inhibitors. Extracts were centrifuged for 20 min at 12,500 rpm in a Sorvall SS-34 rotor. Supernatants were diluted 1:1 in PBS wash (PBS plus 5 mM EDTA, 1 mM β-mercaptoethanol, and 1% Triton X-100) and were rocked with 0.5 ml of a 50% slurry of preequilibrated glutathione Sepharose (Amersham Pharmacia) at 4°C. After an overnight binding reaction, the beads were washed sequentially with 10 ml of the following: PBS wash, PBS wash with 0.5 M NaCl, PBS with 0.5% sodium deoxycholate, and PBS wash with 0.05 M NaCl.
For GST chromatography assays, 35S-labeled recombinant protein was generated in vitro using the pSP-BRG1 construct and the TNT translation system and following the manufacturer’s protocol (Promega). GST fusion protein beads were washed twice with PC-100 (20 mM HEPES [pH 7.9], 100 mM KCl, 0.2 mM EDTA, 5 mM MgCl2, 0.1% NP-40, 20% glycerol, 0.1 mg of bovine serum albumin/ml, 1 mM dithiothreitol, and a protease cocktail [Roche]) and preblocked with 30 μl of normal rabbit serum in 1 ml of PC-100 for 5 min. The beads were washed twice with PC-200 (PC-100 buffer, except with 200 mM KCl) and were typically incubated with 2.5 μl of 35S-labeled protein in 50 μl of PC-200 for 30 min. The Sepharose beads were washed three times with the PC buffer, containing the KCl salt concentration described in the text. The washed beads were boiled in 30 μl of 2× SDS-PAGE sample buffer. Eluted proteins were analyzed via SDS-PAGE and autoradiography (Molecular Dynamics PhosphorImager; Amersham Pharmacia).
In vivo binding studies.
Immunoprecipitation assays were done essentially as described previously (24). SW13 cells were transfected (Fugene6; Roche) with 3 μg of EKLF, EKLF mutant, or parent HA-tagged expression vector and 5 μg of BRG1 expression vector. Cells were harvested 48 h later in lysis buffer containing 300 mM KCl. Precleared lysates were immunoprecipitated with monoclonal anti-HA antibody (Roche). Precipitates were washed three times and subjected to SDS-PAGE and immunoblotting with a BRG1-specific antiserum (BJ2; a kind gift from G. Crabtree).
RESULTS
Human EKLF activates the endogenous murine β-globin promoter.
To explore the determinants of EKLF-dependent β-globin gene activation in vivo, we utilized a J2eΔeklf erythroid cell line (clone B1), derived previously from erythroblasts of an EKLF null embryo (15). As observed in murine EKLF studies, J2eΔeklf cells transduced by a human EKLF retrovirus showed altered growth and viability (15; also data not shown). These findings, coupled with studies of EKLF activity in transgenic mice (47), suggest that the level of EKLF alters cellular homeostasis, potentially through aberrant expression of β-globin chains. To test if titration of EKLF would alleviate toxicity and correlate with β-globin transcription, we developed a ligand-inducible system to control human EKLF expression in J2eΔeklf cells. Utilizing a murine stem cell virus (MSCV)-based retroviral vector system, we prepared viral supernatants for MSCV-EKLFirGFP and MSCV-irGFP. The MSCV-EKLFirGFP retrovirus contains full-length hEKLF cDNA (flEKLF) fused in frame with the influenza virus HA epitope at the NH2-terminal and tamoxifen-binding domain of the mutant ER at the COOH terminal (Fig. 1A). This chimeric sequence is linked by an internal ribosomal entry site (ir) to sequences encoding the enhanced GFP marker. The irGFP construct that lacks EKLF sequences served as a control. Amphotropic retroviral supernatants were generated in standard fashion (see Materials and Methods) and used to transduce a clonal population of J2eΔeklf cells. After exposure and expansion in growth media, GFP+ J2eΔeklf cells were selected by FACS, and the structural integrity of the integrated retroviral genome was confirmed by Southern analysis (data not shown).
FIG. 1.
Exogenous expression of human EKLF-ER rescues β-globin expression in J2eΔeklf null cells. The coding region of hEKLF was fused in frame to the tamoxifen response element ER in a retroviral GFP+ cloning vector (see Materials and Methods). Viral supernatants were used to transduce the hEKLF-ER sequence into J2eΔeklf cells. A pool of GFP+ cells was selected by FACS and then characterized for hEKLF expression and function. (A) Schematic of the retroviral construct utilized to test hEKLF function in J2eΔeklf cells. Note that the hEKLF cDNA was fused in frame with an HA epitope tag at the NH2 terminus to facilitate immunological detection and with the ligand-binding domain of the estrogen receptor (ERTM) at the carboxy terminus to facilitate functional control. (B) EKLF expression increases with tamoxifen induction. Crude nuclear extracts were prepared from J2eΔeklf/irGFP (lane 1) and J2eΔeklf/ flEKLF cells after 48 h in growth media with varied tamoxifen concentrations (0, 10−3, 10−2, 10−1, 101, 102, and 103, lanes 2 to 8, respectively) and analyzed by immunoblotting with HA-specific antisera. To test for loading equivalency, the blot was stripped and reprobed with anti-PCNA antibody. (C) An increase in βmaj transcripts correlates with increasing EKLF expression. Total RNA was isolated from the J2eΔeklf/flEKLF cells and subjected to RPA, utilizing βmaj and α-globin antisense riboprobes.
To assess expression of the EKLF chimeric protein, nuclear lysates from the irGFP- and EKLFirGFP-transduced populations were examined by immunoblot analysis utilizing HA-specific antisera. Detectable levels of an immunoreactive band of the correct molecular weight were present only in EKLF-transduced cells (Fig. 1B, compare lane 1 to lanes 2 to 8). To determine the dose of tamoxifen for the optimal EKLF nuclear concentration, J2eΔeklf/flEKLF cells were cultured in the absence of tamoxifen for 24 h and subsequently switched into media containing increasing concentrations of tamoxifen. After an additional 36 h in culture, nuclear extracts and total RNA were isolated from each growth condition. A small amount of immunoreactive protein was observed in the nucleus in the absence of tamoxifen induction (Fig. 1B, lane 2), which probably reflects some contamination of nuclear extracts with cytosolic material. However, an incremental increase in EKLF detection occurred at 101 to 103 nM tamoxifen inductions (lanes 5 to 7). In contrast, no further increase in EKLF expression was observed in cell populations treated with ≥103 nM concentrations (lane 8).
To determine what level of EKLF was required for β-globin gene expression, RPA were used to measure βmaj transcripts. In each sample, α-globin transcripts were measured to exclude differences in RNA concentration. After induction with tamoxifen, an increase in β-globin expression, over α-globin expression, was detected between concentrations of 100 and 102 nM (Fig. 1C, lanes 2 to 7). This increase correlated with EKLF expression (compare Fig. 1B and C). Moreover, we observed a proportionate decrease in the α- and β-globin transcripts in cells treated with tamoxifen concentrations of ≥103 nM (lane 8). Based on these observations, all subsequent studies were performed at a 102 nM concentration of tamoxifen.
Human EKLF restores an open chromatin structure at the β-globin promoter in J2eΔeklf cells.
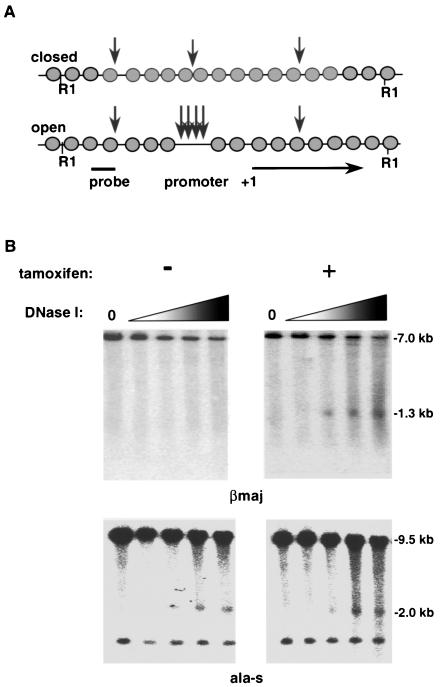
At the murine βmaj promoter, a known developmentally specific and tissue-specific hypersensitivity site (HS) has been located approximately 100 bp upstream of the transcriptional start site (16). Studies with EKLF null embryos demonstrated not only a specific loss of β-gene transcription but also a loss of the HS (72). To determine if a similar dependence occurred in J2eΔeklf cells, nuclei from tamoxifen-induced J2eΔeklf/flEKLF cells were treated with increasing amounts of DNase I. As a control, nuclei from untreated cells (no tamoxifen) were treated in the same manner. Genomic DNA was isolated from each reaction, and the HS pattern was determined by Southern blotting, using an internal probe to the 7.0-kb restriction fragment (R1) that encompasses the βmaj promoter region (Fig. 2A). As shown in Fig. 2B (top left panel), no HS was detected in the absence of EKLF expression. However, a 1.3-kb fragment was observed with EKLF expression that is identical to the wild-type HS pattern previously described (Fig. 2B, top right panel) (34). To control for the possibility that the observed DNase I sensitivity was not a result of nonspecific induction by tamoxifen, the HS patterns were determined for the erythroid-specific ala-s and non-erythroid-specific myoD (data not shown) genes. As represented by the pattern revealed using an ala-s promoter probe (Fig. 2B, bottom panels) (64), HS patterns of EKLF-independent genes were not altered by tamoxifen.
FIG. 2.
Expression of the EKLF cDNA results in restoration of DNase I hypersensitivity at the βmaj promoter. (A) Schematic representation of HS formation at the βmaj promoter. (B) Induction of DNase I hypersensitivity by tamoxifen treatment of J2eΔeklf/flEKLF cells. Cells were cultured for 48 h in the absence (−) or presence (+) of tamoxifen. Nuclei were isolated and exposed to increasing concentrations of DNase I as previously described. To determine DNase sensitivity of βmaj, DNA was harvested, digested with EcoRI, and probed with a βmaj promoter-specific probe. For DNase sensitivity of ala-s, DNA was digested with BamHI and probed with an ala-s promoter-specific probe. The DNase I concentration is 0 in the first lane of each panel and increases as shown by the shaded triangle. The autoradiographs are representative of three independent experiments.
A small carboxy-terminal domain of EKLF is sufficient to remodel chromatin in vivo.
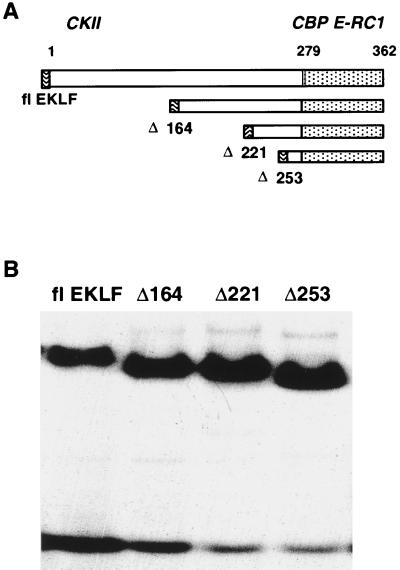
Establishment of an open promoter structure and rescue of βmaj expression by flEKLF in the J2eΔeklf cell line permitted us to explore the structure-function relationships of various domains of EKLF in the context of the endogenous locus. To delineate these functions, we prepared a series of NH2-terminal mutants of EKLF (Fig. 3A). The selection of deletions was based, in part, on domains previously defined in reporter assays and protein-protein interaction defined in vitro (4, 11, 37, 75, 76). Although each construct contained the native DNA binding domain of EKLF, additional analysis was performed to determine if the NH2-terminal truncations altered binding affinity in EMSA. As shown in a representative EMSA (Fig. 3B), recombinant forms of all mutants bound in a similar fashion to a βmaj CACC element-encoding probe compared to flEKLF.
FIG. 3.
Generation of a series of β-CACC-binding hEKLF mutants. (A) Schematic representation of EKLF mutants tested. NH2-terminal mutants were chosen based on functional properties of EKLF defined in in vitro studies. Construction of these mutants is detailed in Materials and Methods. Above the diagram are the known protein-protein interactions of EKLF. The dotted area indicates the zinc finger DNA binding domain. (B) All amino-terminal mutants of EKLF bind the cognate βmaj CACC motif. Equal amounts of bacterially expressed recombinant EKLF polypeptides were incubated with a 32P-radiolabeled β-CACC probe for 30 min at room temperature. The products of this reaction were electrophoresed through a 4% polyacrylamide gel and then gel dried and subjected to autoradiography. The representative autoradiograph shows an electrophoretic mobility shift assay for each mutant.
Subsequently a series of J2eΔeklf cell lines expressing these mutants was generated using the retrovirus-mediated expression strategy described above for flEKLF (Fig. 1A and 4A). To determine if comparable levels of EKLF in the nucleus were obtained in induced cells for each construct, Western analysis was performed on nuclear extracts collected after 48 h of growth in 100 nM tamoxifen. As shown in the representative immunoblot, an equivalent EKLF signal was found in each nuclear extract, when corrected for expression of p38, a ubiquitously expressed control (Fig. 4B). To determine the effect of the NH2-terminal deletions on formation of HS, nuclei of induced cells for each mutant were incubated with increasing amounts of DNase I (Fig. 4C). Contrary to in vitro chromatin remodeling assays (37), the analysis of the HS pattern in J2eΔeklf cells expressing Δ253EKLF demonstrated a pattern similar to that previously observed for J2eΔeklf null cells (Fig. 2B). In contrast, studies of cells expressing Δ221EKLF and Δ164EKLF showed an HS pattern similar to that observed with flEKLF. These results suggest that EKLF sequence between amino acids 221 and 253 is essential for open chromatin configuration, as measured by DNase I hypersensitivity. To confirm that nuclear localization was not altered by the NH2-terminal truncations, immunofluorescence microscopy of transfected cells was performed, and intense nuclear localization was observed for all mutant proteins (data not shown).
FIG. 4.
The DNA binding domain is required but is not sufficient for chromatin remodeling at the βmaj globin promoter. (A) Schematic diagram of flEKLF retrovirus and derivative mutants used to stably transduce J2eΔeklf cells. (B) Retrovirus-mediated transfer into J2eΔeklf cells results in comparable levels of EKLF transgene expression. Nuclear extracts were prepared from individual pools of J2eΔeklf cells transduced with EKLF mutant retroviruses after 48 h of culturing in the presence of tamoxifen. Equal amounts of protein were separated on SDS-PAGE gels, blotted, and probed with HA-specific antisera. Blots were subsequently stripped and reprobed with p38-specific antisera to confirm equivalency of gel loading. (C) Induction of DNase I hypersensitivity at the β-globin promoter requires a large carboxy-terminal domain. Cells expressing each EKLF mutant were cultured for 48 h in the presence of tamoxifen. Nuclei were isolated and exposed to increasing concentrations of DNase I as previously described. DNA was harvested, digested with EcoRI, and probed with a βmaj-specific probe. The DNase I concentration is 0 in the first lane of each panel and increases as shown by the shaded triangle. The autoradiograph is representative of several independent experiments.
The DNA binding domain of EKLF is sufficient for BRG1 interaction in vivo.
An SWI/SNF complex, E-RC1, has been demonstrated to mediate chromatin remodeling by murine EKLF in vitro (4). E-RC1 and its central catalytic component, BRG1, interact directly with the DNA binding domain of EKLF, and this interaction is sufficient to remodel a chromatinized β-promoter template (37). To extend these studies and to determine if a larger domain was required in the context of human EKLF, in vitro binding studies were performed. Recombinant forms of each mutant EKLF molecule fused in frame with a GST polypeptide were prepared in bacteria and bound to glutathione Sepharose beads. Each preparation was assayed for its ability to bind 35S-methionine-labeled BRG1. As seen in a representative autoradiograph, a BRG1 signal was present for each corresponding GST-EKLF fusion but not for GST alone (Fig. 5A). Importantly, BRG1 interacted with Δ253EKLF, which contained all the sequences required by murine EKLF to remodel in vitro-chromatinized β-globin templates (37).
FIG. 5.
BRG1, the core component of the SWI/SNF complex E-RC1, requires the zinc finger region of hEKLF for maximal interaction in vitro and in vivo. (A) The hEKLF DNA binding region directly interacts with BRG1. EKLF mutant polypeptides fused to GST or GST protein alone bound to glutathione beads was incubated with equal amounts of in vitro-translated 35S-methionine-labeled BRG1. Beads were extensively washed, and bound protein was revealed by SDS-PAGE and fluorography. (B) A similar pattern of EKLF-BRG1 interaction is observed in vivo. SW13 cells were cotransfected with BRG1- and EKLF-expressing plasmids. Nuclear extracts were prepared 48 h posttransfection and incubated with HA-epitope tag antisera. Interacting proteins were precipitated, and the products were separated by SDS-PAGE. After blotting to nylon filters was done, BRG1-specific antiserum was utilized to detect the presence of EKLF-BRG1 specific interactions.
To determine if a similar requirement occurred in vivo, we studied the abilities of flEKLF and the mutant EKLF molecules to interact with BRG1 in SW13 cells, a cell line that lacks endogenous BRG1 (24). SW13 cells were cotransfected with vectors expressing BRG1 and individual mutants and propagated for 48 h, and nuclear extracts were prepared. Immunoprecipitation of HA-tagged flEKLF with anti-HA antibody resulted in coprecipitation of BRG1 (Fig. 5B). Similar results were observed with Δ162EKLF and Δ221EKLF. Moreover, BRG1 could coprecipitate with Δ253EKLF, suggesting that the observed in vitro interaction between the DNA binding domain of EKLF and BRG1 occurs in vivo.
An internal domain of EKLF is sufficient to activate β-globin-specific expression in J2eΔeklf cells.
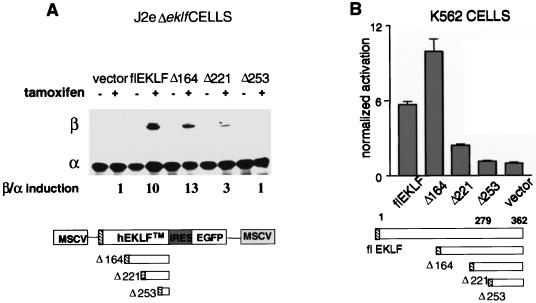
Based on prior structural analysis of EKLF, the activation domain of EKLF should reside in sequences upstream of the Δ221–253 region (11, 54). Our HS studies suggested that the in vivo transcriptional and chromatin remodeling activities of EKLF are separable. To test this hypothesis, transcriptional activity for each mutant was measured and correlated with its chromatin remodeling properties. Concurrent with HS assays, RNA of the induced samples was isolated for RPA analysis, using α and βmaj riboprobes. As anticipated, no significant level of βmaj transcript was detected in the Δ253EKLF cells (Fig. 6A). In contrast, expression of the Δ221EKLF polypeptide resulted in a small but appreciable (threefold) increase in gene transcripts. However, examination of Δ164EKLF-expressing cells revealed levels of βmaj transcripts that paralleled flEKLF expression, when compared to α-globin signal to correct for gel loading differences.
FIG. 6.
An internal domain of hEKLF is sufficient to activate β-globin gene expression to wild-type levels. (A) Addition of amino acids 164 to 221 is sufficient to confer wild-type activation potential on the DNA binding (Δ253EKLF) and chromatin remodeling (Δ221EKLF) domains of EKLF. RNA was harvested from J2eΔeklf clones expressing varying EKLF mutant moieties at 48 h post-tamoxifen induction. RPA was performed utilizing βmaj and α riboprobes. The numbers underneath the panel represent the mean fold induction of the β/α ratio for each construct assayed, using three independent experiments and the activity in cells transduced by vector alone as baseline. (B) The identical domain of hEKLF activates a β-promoter-regulated reporter gene in transient assays. EKLF mutant constructs lacking the ER element were subcloned into a mammalian expression vector. These constructs were cotransfected individually into K562 cells, along with a human β-globin promoter construct, HS2βLuc, and a Renilla luciferase control plasmid. Luciferase activity was determined by luminometry 48 h posttransfection. Fold activation of each construct is represented with respect to a control transfection of HS2βLuc and the empty expression vector. The effect on HS2β luciferase activity for each hEKLF construct was measured in six separate assays of three independent transfections. The results of the Renilla luciferase analysis were utilized to correct for variation in transfection efficiency.
Human and murine EKLF share a high degree of identity at the level of primary polypeptide structure (8, 66). Similar to the findings at the endogenous locus (Fig. 6A), a recent study demonstrated that an internal domain of murine EKLF was capable of activating a transient β-promoter reporter construct in erythroid cells (54). Although the transiently transfected constructs may acquire some degree of chromatin structure, they predominantly measure the transactivation potential of protein domains. To determine the relative abilities of hEKLF and the COOH mutants described above to induce transactivation in this context, expression plasmids encoding flEKLF or the mutants were cotransfected into K562 cells with the HS2βLuc reporter plasmid (Fig. 6B). K562 cells provide a favorable background for these studies based on their erythroid properties and lack of significant endogenous EKLF activity (23). The HS2βLuc reporter construct contained a 300-bp β-promoter fragment flanked by the hypersensitivity site 2 (HS2) enhancer region of the locus control region and a luciferase reporter gene. Cotransfection of the reporter plasmid with the DNA binding domain mutant, Δ253EKLF, failed to activate the reporter above the control. Expression of the in vivo chromatin remodeling mutant, Δ221EKLF, resulted in low-level reporter gene expression, similar to that observed at the endogenous locus. In contrast, addition of another 60 residues, Δ164EKLF, resulted in promoter activation that was comparable to that observed with flEKLF. These results suggested that these assays recapitulate the transactivation properties of hEKLF that we observed at the endogenous promoter.
In vivo activation of the β-globin gene by human EKLF is BRG1 dependent.
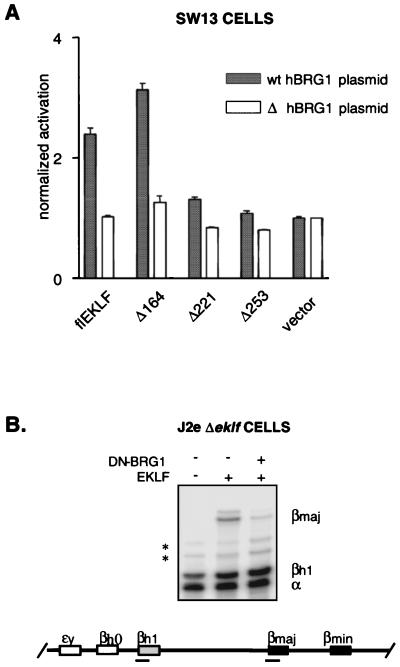
SWI/SNF complexes have been implicated not only in chromatin remodeling but also in transcriptional activation (13, 74). To determine the role of BRG1 in EKLF-specific transactivation, we established an EKLF transactivation assay in the SW13 cell line. These cells are particularly helpful in addressing this issue as they lack functional BRG1 expression (24). Activation assays were done identically to those performed with K562 cells (Fig. 6B), except that flEKLF and EKLF mutants were tested in the presence of either exogenous expressed wild-type (wt) BRG1 or a derivative BRG1 deletion mutant. Expression of flEKLF and mutants with the ΔhBRG1 plasmid failed to activate β-promoter activity (Fig. 7A, open bars). However, consistent augmentation of promoter activation occurred with coexpression of flEKLF with wt hBRG1. Similarly, coexpression of Δ164EKLF and BRG1 resulted in promoter activation. In contrast, expression of Δ221EKLF or Δ253EKLF with wt BRG1 failed to significantly activate the target promoter. These results suggested that BRG1 is required for EKLF-dependent transactivation.
FIG. 7.
BRG1 is required for EKLF function in vivo. (A) BRG1 is required for transactivation of an EKLF-dependent promoter. Plasmids expressing full-length EKLF or amino-terminal mutants of EKLF were cotransfected into SW13 cells with either wt BRG1-expressing plasmid (black bars) or the derivative pΔBRG1 alone (white bars). In addition, each transfection contained the HS2βLuc reporter construct. Luciferase activity was determined by luminometry 48 h posttransfection. Fold activation of each construct is represented with respect to a control transfection of HS2βLuc and the empty expression vector. The graph reflects HS2β luciferase activation for each hEKLF construct, measured in six separate assays of three independent transfections. (B) Dominant-negative form of BRG1 (DN-BRG1) represses specifically EKLF-dependent transcription in fetal erythroblasts. J2eΔeklf/flEKLF-irGFP/DN-BRG1-irYFP cells were generated in a fashion similar to that described for J2eΔeklf/flEKLF-irGFP cells (see Fig. 1 and Material and Methods). Total cellular RNA was harvested 48 h post-tamoxifen induction, and RPA was performed using βmaj, βhl, and α riboprobes. The autoradiograph represents the level of each transcript detected for the J2eΔeklf lines. Asterisks, nonspecific bands.
Based on these findings and those demonstrating BRG1 dependency on a reconstituted chromatinized template (4), we anticipated that BRG1-dependent, EKLF-mediated transactivation occurs at the endogenous β-globin promoter. To address this issue, we adapted the J2eΔeklf system to coexpress flEKLF with a dominant-negative mutant of BRG1 (DN-BRG1) and determined its effect on βmaj expression. The DN-BRG1 mutant carried an altered ATPase activity site, which has been shown to significantly attenuate wt BRG1 activity in vivo (21). To allow coordinate induction of DN-BRG1 and flEKLF activities, the DN-BRG1 molecule was fused to the ER domain in a fashion similar to that described for flEKLF. A J2eΔeklf line was produced that demonstrated tamoxifen-dependent nuclear localization of both molecules by immunoblotting (data not shown). As shown in a representative autoradiograph, expression of the transdominant mutant resulted in significant reduction of EKLF-dependent β-globin gene activation (Fig. 7B, compare lanes 2 and 3). In contrast, expression of DN-BRG1 did not affect α or βh1 transcripts, suggesting that the observed effect on the βmaj gene was specific.
DISCUSSION
Efforts to explore many of the molecular mechanisms underpinning developmentally specific transcription in vivo have been hampered by a lack of suitable models that evaluate the physiological relevance of interactions defined in vitro. Here, we report complementary biochemical and genetic analyses that define the chromatin remodeling and transactivation domains of EKLF in definitive erythroid cells. We have used the J2eΔeklf cell line to characterize the functional domains of EKLF in the context of the endogenous β-globin gene. This approach avoids deficiencies inherent to in vitro transcription and transient reporter-based assays, including the nonchromosomal nature of the DNA template and the inability to assess the effects of DNA replication. It minimizes dependency on transgenic constructs, where copy number and chromosomal position are variable. This is particularly important for studies of the β-globin locus, based on differences in the phenotypes of cis-acting mutations generated at the endogenous locus compared to single- or multiple-copy transgene constructs (10, 26, 32).
Numerous mechanisms of action have been ascribed to DNA-binding factors, including modulation of chromatin and DNA architecture, interaction with nuclear structural proteins, and recruitment and/or regulation of components of the basal transcriptional machinery (38, 45). Many factors have domains with complementary or opposing functions, their activity being dependent on promoter context (29). Indeed, studies of EKLF, utilizing reporter constructs, suggest that optimal function occurs only in the context of the β-globin promoter (5). Thus, a focused analysis of a trans activator at an established target locus provides significant advantages for dissection of the steps necessary for transcriptional activation in vivo. Complementary experiments utilizing transiently transfected reporter constructs have demonstrated that EKLF is a modular protein consisting of a carboxy-terminal DNA binding domain linked to a proline-rich transactivation domain (9, 49). Furthermore, elegant studies by Emerson and colleagues have shown that the zinc finger DNA binding domain of murine EKLF is sufficient to induce complete remodeling of a nucleosomal loaded β-promoter template, through recruitment of a multimeric SWI/SNF complex, E-RC1 (4). In the context of the native chromatin locus, we demonstrate that EKLF-dependent chromatin remodeling requires the defined DNA binding domain identified in vitro (37) as well as an adjacent region of approximately 35 amino acids (Fig. 4C). For EKLF-dependent activation, a second internal domain of hEKLF is sufficient for gene activation to levels observed with the full-length molecule (Fig. 6A). In addition to delineating this internal region, we provide evidence that the SWI/SNF factor BRG1 is required for its activity at the endogenous β-globin gene.
Several possibilities could account for the discrepancy between the in vitro and in vivo chromatin remodeling activities of EKLF. One simple explanation is that Δ253EKLF has altered nuclear localization and/or DNA binding. However, this molecule contains a functional nuclear localization signal (1) and is detected in the nucleus by cell fractionation and immunofluorescence studies (Fig. 4B and data not shown). Moreover, in electrophoretic mobility shift assays, this domain showed an affinity for the cognate β-CACC motif similar to that of the full-length polypeptide (Fig. 3B). A second possibility is that amino acid sequence variation between human and murine EKLF may result in different affinities for the BRG1 subunit of E-RC1. Although the definitive structures of these molecules are not available, the primary sequences of murine and human EKLF are highly homologous. A third possible explanation is that the additional sequence may be required for a stable interaction between hEKLF and BRG1. Indeed, recent studies of the interaction between the C/EBPβ transactivator and hBRM, a closely related homologue of hBRG1, demonstrate that sequences outside the binding domain influence the relative affinity of the interaction (42). In contrast, no appreciable difference in affinity for BRG1 was observed between Δ253EKLF and other EKLF constructs tested in vitro or in vivo (Fig. 5).
Why is the larger Δ221EKLF polypeptide required for remodeling in vivo? The most likely explanation is that a factor(s) not present in the in vitro reconstitution assays is required for the establishment and/or maintenance of the open chromatin configuration induced in vivo by EKLF. A highly attractive candidate for this additional factor would be a histone acetyl transferase, given the postulated role of histone acetylation in stabilization of SWI/SNF-induced changes in native chromatin (17, 22, 43). Indeed, previous studies have demonstrated an interaction between the carboxy-terminal zinc finger domain of murine EKLF and the coactivator CBP, a potent histone acetyl transferase (12, 75). This domain is encompassed completely in our Δ253EKLF construct. Although the possible role of CBP in modulating histone acetylation of the endogenous β-promoter has not been explored, Chen and Bieker have demonstrated that CBP interacts with, and acetylates, the carboxy-terminal zinc finger domain of murine EKLF, with a consequent increase in the ability to recruit BRG1 (12). We have shown that CBP interacts specifically with Δ253EKLF human polypeptide and does not require the amino acid 221 to 253 region for effective binding (S.P., S.M.J., and J.M.C., unpublished data). Thus, it is unlikely that CBP is the missing factor that explains the chromatin remodeling activity of the amino acid 221 to 253 domain. However, it does not exclude an indirect role of a CBP-EKLF interaction in coactivator recruitment or in the maintenance of an open chromatin configuration. Given the multisubunit composition of SWI/SNF complexes and the increasing list of chromatin remodeling complexes being described, we propose that an additional factor(s) binds the upstream domain of EKLF and facilitates chromatin remodeling in vivo. A focused search for this factor is now feasible with the tools generated by this study.
The ability to coprecipitate EKLF and BRG1 provides insight into the mechanism of gene activation, specifically the timing of the initial interaction between the SWI/SNF complex and EKLF. Previous studies of activator interactions with SWI/SNF complexes have provided evidence for two models (60). It is possible that a factor can bind its DNA element, acting as a docking site for the remodeling complex. Alternatively, the activator-SWI/SNF interaction may occur in solution, with the DNA-binding domain directing chromatin remodeling at a later step. In the case of EKLF, prior studies demonstrated that EKLF binds nucleosome-loaded templates poorly in the absence of BRG1 (4). Our experiments extend this observation by demonstrating that EKLF binds BRG1 in vivo in the absence of an available DNA binding element (Fig. 5B), supporting the second model of EKLF-mediated recruitment of E-RC1.
It is interesting that expression of the Δ221EKLF construct results in a modest increase in transcription (Fig. 6A). This observation is consistent with the concept that when the β-globin promoter becomes accessible, low-level transcription can occur. However, recruitment of other cofactors, through interactions with the EKLF upstream transactivation domain (amino acids 164 to 221), is required for high-level activation. These observations are concordant with the “poised” promoter model suggested by in vitro transcription assays and studies of stress-inducible promoters (3, 30, 31, 55). Indeed, recent studies of GAL4-mediated recruitment of the SAGA remodeling complex suggest that reorganization of the promoter chromatin structure facilitates recruitment of the RNA polymerase complex (7, 44). Thus, the dependency of β-gene expression on the BRG1-hEKLF interaction we observed both in transient assays and at the endogenous locus (Fig. 7) suggests that E-RC1 may facilitate recruitment of the basal transcriptional machinery to the open β-globin promoter in vivo. Ongoing studies utilizing chromatin immunoprecipitation and fine mapping with nucleases will allow us to determine precisely the factors required at the promoter at each step of the activation process. Moreover, the temporal relationships with GATA-1 and Sp1, β-promoter-binding factors which have been shown previously to interact with EKLF (48), can be explored.
Acknowledgments
We thank G. Crabtree, P. Curtis, D. Persons, and R. Kingston for reagents. We thank Virginia Barbour and Jin He for excellent technical support. We thank A. W. Nienhuis for his support and encouragement.
Support was provided by the NIH (S.M.J. and J.M.C., PO1HL53749 and P30CA21765; R.C.B., T32CA70089), the Assisi Foundation of Memphis (J.M.C.), the Wellcome Trust (S.M.J.), the NHMRC (S.M.J., A.P.), and the American Lebanese Syrian Associated Charities.
REFERENCES
- 1.Anderson, K. P., C. B. Kern, S. C. Crable, and J. B. Lingrel. 1995. Isolation of a gene encoding a functional zinc finger protein homologous to erythroid Krüppel-like factor: identification of a new multigene family. Mol. Cell. Biol. 15:5957–5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews, N. C., and D. V. Faller. 1991. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 19:2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ansari, A. Z., J. E. Bradner, and T. V. O’Halloran. 1995. DNA-bend modulation in a repressor-to-activator switching mechanism. Nature 374:371–375. [DOI] [PubMed] [Google Scholar]
- 4.Armstrong, J. A., J. J. Bieker, and B. M. Emerson. 1998. A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell 95:93–104. [DOI] [PubMed] [Google Scholar]
- 5.Asano, H., and G. Stamatoyannopoulos. 1998. Activation of beta-globin promoter by erythroid Krüppel-like factor. Mol. Cell. Biol. 18:102–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bean, T. L., and P. A. Ney. 1997. Multiple regions of p45 NF-E2 are required for beta-globin gene expression in erythroid cells. Nucleic Acids Res. 25:2509–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhaumik, S. R., and M. R. Green. 2001. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev. 15:1935–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bieker, J. J. 1996. Isolation, genomic structure, and expression of human erythroid Krüppel-like factor (EKLF). DNA Cell Biol. 15:347–352. [DOI] [PubMed] [Google Scholar]
- 9.Bieker, J. J. 1998. Erythroid-specific transcription. Curr. Opin. Hematol. 5:145–150. [DOI] [PubMed] [Google Scholar]
- 10.Bungert, J., U. Dave, K. C. Lim, K. H. Lieuw, J. A. Shavit, Q. Liu, and J. D. Engel. 1995. Synergistic regulation of human beta-globin gene switching by locus control region elements HS3 and HS4. Genes Dev. 9:3083–3096. [DOI] [PubMed] [Google Scholar]
- 11.Chen, X., and J. J. Bieker. 1996. Erythroid Krüppel-like factor (EKLF) contains a multifunctional transcriptional activation domain important for inter- and intramolecular interactions. EMBO J. 15:5888–5896. [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, X., and J. J. Bieker. 2001. Unanticipated repression function linked to erythroid Krüppel-like factor. Mol. Cell. Biol. 21:3118–3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho, H., G. Orphanides, X. Sun, X. J. Yang, V. Ogryzko, E. Lees, Y. Nakatani, and D. Reinberg. 1998. A human RNA polymerase II complex containing factors that modify chromatin structure. Mol. Cell. Biol. 18:5355–5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi, O. R., and J. D. Engel. 1988. Developmental regulation of beta-globin gene switching. Cell 55:17–26. [DOI] [PubMed] [Google Scholar]
- 15.Coghill, E., S. Eccleston, V. Fox, L. Cerruti, C. Brown, J. Cunningham, S. Jane, and A. Perkins. 2001. Erythroid Krüppel-like factor (EKLF) coordinates erythroid cell proliferation and hemoglobinization in cell lines derived from EKLF null mice. Blood 97:1861–1868. [DOI] [PubMed] [Google Scholar]
- 16.Cohen, R. B., and M. Sheffery. 1985. Nucleosome disruption precedes transcription and is largely limited to the transcribed domain of globin genes in murine erythroleukemia cells. J. Mol. Biol. 182:109–129. [DOI] [PubMed] [Google Scholar]
- 17.Cosma, M. P., T. Tanaka, and K. Nasmyth. 1999. Ordered recruitment of transcription and chromatin remodeling factors to a cell. Cell 97:299–311. [DOI] [PubMed] [Google Scholar]
- 18.Costantini, F., G. Radice, J. Magram, G. Stamatoyannopoulos, T. Papayannopoulou, and K. Chada. 1985. Developmental regulation of human globin genes in transgenic mice. Cold Spring Harbor Symp. Quant. Biol. 50:361–370. [DOI] [PubMed] [Google Scholar]
- 19.Crossley, M., M. Merika, and S. H. Orkin. 1995. Self-association of the erythroid transcription factor GATA-1 mediated by its zinc finger domains. Mol. Cell. Biol. 15:2448–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cunningham, J. M., and S. M. Jane. 1996. Hemoglobin switching and fetal hemoglobin reactivation. Semin. Hematol. 33:9–23. [PubMed] [Google Scholar]
- 21.de La Serna, I., K. A. Carlson, D. A. Hill, C. J. Guidi, R. O. Stephenson, S. Sif, R. E. Kingston, and A. N. Imbalzano. 2000. Mammalian SWI-SNF complexes contribute to activation of the hsp70 gene. Mol. Cell. Biol. 20:2839–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DiRenzo, J., Y. Shang, M. Phelan, S. Sif, M. Myers, R. Kingston, and M. Brown. 2000. BRG-1 is recruited to estrogen-responsive promoters and cooperates with factors involved in histone acetylation. Mol. Cell. Biol. 20:7541–7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donze, D., T. M. Townes, and J. J. Bieker. 1995. Role of erythroid Krüppel-like factor in human gamma- to beta-globin gene switching. J. Biol. Chem. 270:1955–1959. [DOI] [PubMed] [Google Scholar]
- 24.Dunaief, J. L., B. E. Strober, S. Guha, P. A. Khavari, K. Alin, J. Luban, M. Begemann, G. R. Crabtree, and S. P. Goff. 1994. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell 79:119–130. [DOI] [PubMed] [Google Scholar]
- 25.Engel, J. D., and K. Tanimoto. 2000. Looping, linking, and chromatin activity: new insights into beta-globin locus regulation. Cell 100:499–502. [DOI] [PubMed] [Google Scholar]
- 26.Epner, E., A. Reik, D. Cimbora, A. Telling, M. A. Bender, S. Fiering, T. Enver, D. I. Martin, M. Kennedy, G. Keller, and M. Groudine. 1998. The beta-globin LCR is not necessary for an open chromatin structure or developmentally regulated transcription of the native mouse beta-globin locus. Mol. Cell 2:447–455. [DOI] [PubMed] [Google Scholar]
- 27.Feng, W. C., C. M. Southwood, and J. J. Bieker. 1994. Analyses of beta-thalassemia mutant DNA interactions with erythroid Krüppel-like factor (EKLF), an erythroid cell-specific transcription factor. J. Biol. Chem. 269:1493–1500. [PubMed] [Google Scholar]
- 28.Forrester, W. C., C. Thompson, J. T. Elder, and M. Groudine. 1986. A developmentally stable chromatin structure in the human beta-globin gene cluster. Proc. Natl. Acad. Sci. USA 83:1359–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fry, C. J., and P. J. Farnham. 1999. Context-dependent transcriptional regulation. J. Biol. Chem. 274:29583–29586. [DOI] [PubMed] [Google Scholar]
- 30.Gilmour, D. S., and J. T. Lis. 1986. RNA polymerase II interacts with the promoter region of the noninduced hsp70 gene in Drosophila melanogaster cells. Mol. Cell. Biol. 6:3984–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goodrich, J. A., G. Cutler, and R. Tjian. 1996. Contacts in context: promoter specificity and macromolecular interactions in transcription. Cell 84:825–830. [DOI] [PubMed] [Google Scholar]
- 32.Grosveld, F. 1999. Activation by locus control regions? Curr. Opin. Genet. Dev. 9:152–157. [DOI] [PubMed] [Google Scholar]
- 33.Grosveld, F., G. B. van Assendelft, D. R. Greaves, and G. Kollias. 1987. Position-independent, high-level expression of the human beta-globin gene in transgenic mice. Cell 51:975–985. [DOI] [PubMed] [Google Scholar]
- 34.Hofer, E., R. Hofer-Warbinek, and J. E. J. Darnell. 1982. Globin RNA transcription: a possible termination site and demonstration of transcriptional control correlated with altered chromatin structure. Cell 29:887–893. [DOI] [PubMed] [Google Scholar]
- 35.Imbalzano, A. N., G. R. Schnitzler, and R. E. Kingston. 1996. Nucleosome disruption by human SWI/SNF is maintained in the absence of continued ATP hydrolysis. J. Biol. Chem. 271:20726–20733. [DOI] [PubMed] [Google Scholar]
- 36.Jane, S. M., P. A. Ney, E. F. Vanin, D. L. Gumucio, and A. W. Nienhuis. 1992. Identification of a stage selector element in the human gamma-globin gene promoter that fosters preferential interaction with the 5′ HS2 enhancer when in competition with the beta-promoter. EMBO J. 11:2961–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kadam, S., G. S. McAlpine, M. L. Phelan, R. E. Kingston, K. A. Jones, and B. M. Emerson. 2000. Functional selectivity of recombinant mammalian SWI/SNF subunits. Genes Dev. 14:2441–2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kadonaga, J. T. 1998. Eukaryotic transcription: an interlaced network of transcription factors and chromatin-modifying machines. Cell 92:307–313. [DOI] [PubMed] [Google Scholar]
- 39.Khavari, P. A., C. L. Peterson, J. W. Tamkun, D. B. Mendel, and G. R. Crabtree. 1993. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature 366:170–174. [DOI] [PubMed] [Google Scholar]
- 40.Kingston, R. E., and G. J. Narlikar. 1999. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 13:2339–2352. [DOI] [PubMed] [Google Scholar]
- 41.Kollias, G., N. Wrighton, J. Hurst, and F. Grosveld. 1986. Regulated expression of human A gamma-, beta-, and hybrid gamma beta-globin genes in transgenic mice: manipulation of the developmental expression patterns. Cell 46:89–94. [DOI] [PubMed] [Google Scholar]
- 42.Kowenz-Leutz, E., and A. Leutz. 1999. A C/EBP beta isoform recruits the SWI/SNF complex to activate myeloid genes. Mol. Cell 4:735–743. [DOI] [PubMed] [Google Scholar]
- 43.Krebs, J. E., M. H. Kuo, C. D. Allis, and C. L. Peterson. 1999. Cell cycle-regulated histone acetylation required for expression of the yeast HO gene. Genes Dev. 13:1412–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Larschan, E., and F. Winston. 2001. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 15:1946–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lemon, B., and R. Tjian. 2000. Orchestrated response: a symphony of transcription factors for gene control. Genes Dev. 14:2551–2569. [DOI] [PubMed] [Google Scholar]
- 46.Magram, J., K. Chada, and F. Costantini. 1985. Developmental regulation of a cloned adult beta-globin gene in transgenic mice. Nature 315:338–340. [DOI] [PubMed] [Google Scholar]
- 47.McMorrow, T., A. van den Wijngaard, A. Wollenschlaeger, M. van de Corput, K. Monkhorst, T. Trimborn, P. Fraser, M. van Lohuizen, T. Jenuwein, M. Djabali, S. Philipsen, F. Grosveld, and E. Milot. 2000. Activation of the beta globin locus by transcription factors and chromatin modifiers. EMBO J. 19:4986–4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Merika, M., and S. H. Orkin. 1995. Functional synergy and physical interactions of the erythroid transcription factor GATA-1 with the Krüppel family proteins Sp1 and EKLF. Mol. Cell. Biol. 15:2437–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miller, I. J., and J. J. Bieker. 1993. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Krüppel family of nuclear proteins. Mol. Cell. Biol. 13:2776–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morley, B. J., C. A. Abbott, and W. G. Wood. 1991. Regulation of human fetal and adult globin genes in mouse erythroleukemia cells. Blood 78:1355–1363. [PubMed] [Google Scholar]
- 51.Nienhuis, A. W., and G. Stamatoyannopoulos. 1994. Hemoglobin switching, p. 107–156. In G. Stamatoyannopoulos, A. W. Neinhuis, P. W. Majerus, and H. Varmus (ed.), The molecular basis of blood disease, 2nd ed. The W. B. Saunders Co., Philadelphia, Pa.
- 52.Nuez, B., D. Michalovich, A. Bygrave, R. Ploemacher, and F. Grosveld. 1995. Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature 375:316–318. [DOI] [PubMed] [Google Scholar]
- 53.Orkin, S. H., S. E. Antonarakis, and H. H. J. Kazazian. 1984. Base substitution at position −88 in a beta-thalassemic globin gene. Further evidence for the role of distal promoter element ACACCC. J. Biol. Chem. 259:8679–8681. [PubMed] [Google Scholar]
- 54.Pandya, K., D. Donze, and T. M. Townes. 2001. Novel transactivation domain in erythroid Krüppel-like factor (eklf). J. Biol. Chem. 276:8239–8243. [DOI] [PubMed] [Google Scholar]
- 55.Panov, K. I., J. K. Friedrich, and J. C. Zomerdijk. 2001. A step subsequent to preinitiation complex assembly at the ribosomal RNA gene promoter is rate limiting for human RNA polymerase I-dependent transcription. Mol. Cell. Biol. 21:2641–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pazin, M. J., P. Bhargava, E. P. Geiduschek, and J. T. Kadonaga. 1997. Nucleosome mobility and the maintenance of nucleosome positioning. Science 276:809–812. [DOI] [PubMed] [Google Scholar]
- 57.Perkins, A. C., K. M. Gaensler, and S. H. Orkin. 1996. Silencing of human fetal globin expression is impaired in the absence of the adult beta-globin gene activator protein EKLF. Proc. Natl. Acad. Sci. USA 93:12267–12271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Perkins, A. C., A. H. Sharpe, and S. H. Orkin. 1995. Lethal beta-thalassaemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature 375:318–322. [DOI] [PubMed] [Google Scholar]
- 59.Persons, D. A., M. G. Mehaffey, M. Kaleko, A. W. Nienhuis, and E. F. Vanin. 1998. An improved method for generating retroviral producer clones for vectors lacking a selectable marker gene. Blood Cells Mol. Dis. 24:167–182. [DOI] [PubMed] [Google Scholar]
- 60.Peterson, C. L., and C. Logie. 2000. Recruitment of chromatin remodeling machines. J. Cell. Biochem. 78:179–185. [DOI] [PubMed] [Google Scholar]
- 61.Peterson, C. L., and J. L. Workman. 2000. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 10:187–192. [DOI] [PubMed] [Google Scholar]
- 62.Phelan, M. L., S. Sif, G. J. Narlikar, and R. E. Kingston. 1999. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell 3:247–253. [DOI] [PubMed] [Google Scholar]
- 63.Raich, N., T. Enver, B. Nakamoto, B. Josephson, T. Papayannopoulou, and G. Stamatoyannopoulos. 1990. Autonomous developmental control of human embryonic globin gene switching in transgenic mice. Science 250:1147–1149. [DOI] [PubMed] [Google Scholar]
- 64.Schoenhaut, D. S., and P. J. Curtis. 1989. Structure of a mouse erythroid 5-aminolevulinate synthase gene and mapping of erythroid-specific DNAse I hypersensitive sites. Nucleic Acids Res. 17:7013–7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Steger, D. J., and J. L. Workman. 1996. Remodeling chromatin structures for transcription: what happens to the histones? Bioessays 18:875–884. [DOI] [PubMed] [Google Scholar]
- 66.van Ree, J. H., M. A. Roskrow, A. M. Becher, R. McNall, V. A. Valentine, S. M. Jane, and J. M. Cunningham. 1997. The human erythroid-specific transcription factor EKLF localizes to chromosome 19p13.12-p13.13. Genomics 39:393–395. [DOI] [PubMed] [Google Scholar]
- 67.Vignali, M., A. H. Hassan, K. E. Neely, and J. L. Workman. 2000. ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol. 20:1899–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wall, G., P. D. Varga-Weisz, R. Sandaltzopoulos, and P. B. Becker. 1995. Chromatin remodeling by GAGA factor and heat shock factor at the hypersensitive Drosophila hsp26 promoter in vitro. EMBO J. 14:1727–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang, W., J. Cote, Y. Xue, S. Zhou, P. A. Khavari, S. R. Biggar, C. Muchardt, G. V. Kalpana, S. P. Goff, M. Yaniv, J. L. Workman, and G. R. Crabtree. 1996. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 70.Wang, W., Y. Xue, S. Zhou, A. Kuo, B. R. Cairns, and G. R. Crabtree. 1996. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 10:2117–2130. [DOI] [PubMed] [Google Scholar]
- 71.Weintraub, H., and M. Groudine. 1976. Chromosomal subunits in active genes have an altered conformation. Science 193:848–856. [DOI] [PubMed] [Google Scholar]
- 72.Wijgerde, M., J. Gribnau, T. Trimborn, B. Nuez, S. Philipsen, F. Grosveld, and P. Fraser. 1996. The role of EKLF in human beta-globin gene competition. Genes Dev. 10:2894–2902. [DOI] [PubMed] [Google Scholar]
- 73.Yang, C., L. H. Shapiro, M. Rivera, A. Kumar, and P. K. Brindle. 1998. A role for CREB binding protein and p300 transcriptional coactivators in Ets-1 transactivation functions. Mol. Cell. Biol. 18:2218–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yudkovsky, N., C. Logie, S. Hahn, and C. L. Peterson. 1999. Recruitment of the SWI/SNF chromatin remodeling complex by transcriptional activators. Genes Dev. 13:2369–2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang, W., and J. J. Bieker. 1998. Acetylation and modulation of erythroid Krüppel-like factor (EKLF) activity by interaction with histone acetyltransferases. Proc. Natl. Acad. Sci. USA 95:9855–9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang, W., S. Kadam, B. M. Emerson, and J. J. Bieker. 2001. Site-specific acetylation by p300 or CREB binding protein regulates erythroid Krüppel-like factor transcriptional activity via its interaction with the SWI-SNF complex. Mol. Cell. Biol. 21:2413–2422. [DOI] [PMC free article] [PubMed] [Google Scholar]