Abstract
Telomere maintenance is essential for the continuous growth of tumor cells. In most human tumors telomeres are maintained by telomerase, a specialized reverse transcriptase. Tankyrase 1, a human telomeric poly(ADP-ribose) polymerase (PARP), positively regulates telomere length through its interaction with TRF1, a telomeric DNA-binding protein. Tankyrase 1 ADP-ribosylates TRF1, inhibiting its binding to telomeric DNA. Overexpression of tankyrase 1 in the nucleus promotes telomere elongation, suggesting that tankyrase 1 regulates access of telomerase to the telomeric complex. The recent identification of a closely related homolog of tankyrase 1, tankyrase 2, opens the possibility for a second PARP at telomeres. We therefore sought to establish the role of tankyrase 1 at telomeres and to determine if tankyrase 2 might have a telomeric function. We show that endogenous tankyrase 1 is a component of the human telomeric complex. We demonstrate that telomere elongation by tankyrase 1 requires the catalytic activity of the PARP domain and does not occur in telomerase-negative primary human cells. To investigate a potential role for tankyrase 2 at telomeres, recombinant tankyrase 2 was subjected to an in vitro PARP assay. Tankyrase 2 poly(ADP-ribosyl)ated itself and TRF1. Overexpression of tankyrase 2 in the nucleus released endogenous TRF1 from telomeres. These findings establish tankyrase 2 as a bona fide PARP, with itself and TRF1 as acceptors of ADP-ribosylation, and suggest the possibility of a role for tankyrase 2 at telomeres.
Telomere integrity is essential for chromosome stability, and the maintenance of telomeric DNA is required for long-term proliferation of eukaryotic cells. Telomeres are maintained by telomerase, a reverse transcriptase that adds telomeric repeats to chromosome ends (14; reviewed in reference 31). In most normal human somatic tissue telomerase is repressed, and as a result, telomeres shorten (17, 18). Critically short telomeres lose their ability to protect chromosome ends, resulting in chromosomal degradation and fusion. In contrast to normal somatic human cells, immortalized cells (including cancer cells) and germ cells express telomerase (21, 34) and maintain their telomeres. In these cells telomere maintenance is regulated by a homeostatic mechanism (reviewed in reference 29). Thus, in the mammalian germ line telomeres show a species-specific telomere length setting which is constant over the generations (23). Regulation is also apparent in many human tumor cell lines, where despite the presence of high levels of telomerase telomeres do not grow, but rather, they are stably maintained within a given size range (9, 10).
Mammalian telomeres consist of long tandem arrays of TTAGGG repeats bound by the DNA-binding proteins, TRF1 and TRF2 (4, 5, 7; reviewed in reference 8). The TRFs are related in their primary structure; both contain carboxy-terminal Myb-type DNA-binding motifs and internal, conserved domains required for homodimerization (2, 5). The proteins do not form heterodimers (5). A distinguishing feature of the TRFs lies in their amino termini, where TRF1 is acidic and TRF2 is basic. The TRFs are ubiquitously expressed and localize predominantly to all telomeres throughout the cell cycle (5, 7). TRF1 and TRF2 remodel DNA configuration in vitro (3, 15, 16) and have been proposed to collaborate in the formation of a specific structure at telomeres, called t-loops, (16), which could protect telomere ends from DNA damage checkpoints and control access to telomerase.
Although the TRFs bear some similarities, studies indicate functional differences; TRF2 contributes to the protective function at telomeres (20, 41), whereas TRF1 functions in telomere length regulation (40). Overexpression of TRF1 in a telomerase-expressing cell line that normally maintains stable telomeres led to progressive telomere shortening. In contrast, inhibition of TRF1 binding to telomeres induced a progressive increase in telomere length. TRF1 had no effect on telomerase activity in these cell lines, suggesting that it did not act by direct modulation of telomerase. Instead it was proposed that TRF1 acts in cis to control access of telomerase at telomere termini (40). More recent studies indicate that TRF2, in addition to its protective role, can influence telomere length dynamics (39).
Two-hybrid screens using the TRFs as bait have identified a number of TRF-interacting proteins. Tankyrase 1, a 142-kDa protein, with homology to ankyrins and to the catalytic domain of PARPs, was identified as a TRF1-interacting factor (38). Tankyrase 1, through its ankyrin repeats, binds the acidic domain of TRF1 (38). This domain is unique to TRF1, and as a result, tankyrase 1 does not interact with TRF2. TIN2, a novel 40-kDa protein, binds TRF1 through the TRF1 dimerization domain (22). TIN2 shares no homology with known proteins and has few structural motifs. A two-hybrid screen with TRF2 identified hRAP1, a human ortholog of the Saccharomyces cerevisiae telomeric protein (26). All three of these TRF-interacting factors, tankyrase 1, TIN2, and Rap1, have been localized to human telomeres and, when overexpressed in human tumor cells, alter telomere length (22, 26, 37).
An unexpected and unique feature of tankyrase 1 among the TRFs and TRF-interacting factors is its homology to the poly(ADP-ribose) polymerase (PARP) family of enzymes (38). PARPs catalyze the formation of long chains of poly(ADP-ribose) onto protein acceptors using NAD (NAD+) as a substrate (reviewed in reference 1). The net effect of the negatively charged polymers is to drastically alter the properties of the protein acceptor. Over the last few years multiple members of the PARP family have been identified (reviewed in reference 35). The homology between tankyrase 1 and the PARPs is limited to the catalytic domain, with no homology outside this region (38). For example, the best characterized family member, PARP-1, promotes DNA repair in response to genotoxic stress (reviewed in reference 33). This enzyme contains a DNA-binding domain which allows its catalytic activity to be activated by DNA damage (reviewed in reference 11). In contrast, tankyrase 1 does not contain any known DNA-binding motifs, and its mechanism of activation remains to be determined.
Tankyrase 1 was found to function as a bona fide PARP. Using NAD+ as a substrate, tankyrase 1 poly(ADP-ribosyl)ated itself and TRF1 in vitro (38). Tankyrase 1-mediated poly(ADP-ribosyl)ation inhibited the ability of TRF1 to bind to telomeric repeats in vitro (38). Overexpression of tankyrase 1 in the nucleus released TRF1 from telomeres and induced telomere elongation in a telomerase-expressing tumor cell line (37). These findings led us to propose a model where tankyrase 1 functions as a positive regulator of telomere length. In this model transient poly(ADP-ribosyl)ation of TRF1 releases TRF1 from telomeres, opening up the telomeric complex and allowing access to telomerase (37).
Recently, a new member of the PARP family and a closely related homolog of tankyrase 1, termed tankyrase 2, was identified. Tankyrase 2 was found in two-hybrid screens using three distinct baits: insulin-responsive amino peptidase, a Golgi-associated protein (6); Grb14, an endosomal adapter protein (27); and the telomeric protein, TRF1 (19). In addition, two serological screens of expression libraries yielded tankyrase 2 (25, 30). Interactions with multiple binding partners from discrete subcellular locales (the Golgi complex, endosomes, and the nucleus) suggest that tankyrase 2 could be a multifunctional protein that localizes to several subcellular sites. Tankyrase 2 would then share this feature with tankyrase 1, which in addition to its telomeric location has been localized to the Golgi complex (6), nuclear pore complexes, and mitotic centrosomes (36). Moreover, the finding that tankyrases 1 and 2 have some common binding partners, TRF1 (19, 38) and insulin-responsive amino peptidase (6), suggests a possibility for redundant or related functions of these two proteins.
These observations prompted us to confirm the role of tankyrase 1 at telomeres and to investigate whether tankyrase 2 could play a role at telomeres. We demonstrate that telomere elongation by tankyrase 1 requires the catalytic activity of the PARP domain. We show that tankyrase 2 functions as a PARP in vitro with itself and TRF1 serving as acceptors of ADP-ribosylation. We further demonstrate that overexpression of tankyrase 2 in the nucleus releases endogenous TRF1 from telomeres, consistent with a potential role at telomeres for tankyrase 2.
MATERIALS AND METHODS
Plasmids.
A full-length tankyrase 2 cDNA (TNKS2.5-2-1) containing a 6,083-nucleotide insert encoding amino acids (aa) 1 to 1166 in pBluescript SK(+/−) (Stratagene) was isolated from a placental cDNA library. The human tankyrase 2 cDNA encoding aa 2 to 1166 and containing an amino-terminal epitope Myc-tag was cloned into pcDNA3 (Invitrogen), resulting in plasmid M-tankyrase 2. A simian virus 40 (SV40) nuclear localization signal (NLS) (PKKKRKVE) was inserted between the Myc epitope-tag and aa 2 of tankyrase 2 by insertion of an oligonucleotide to generate MN-tankyrase 2.
FN-tankyrase 1.HE/A, was created by replacing the histidine (H) and glutamic acid (E) residues at positions 1184 and 1291, respectively, with alanine (A) residues by site-directed mutagenesis of FN-tankyrase 1.WT (wild type) (FN-tankyrase, [37]), using the oligonucleotides 5′-CAATGAGCGCATGTTGTTTGCTGGTTCTCCTTTCATTAATGCC-3′ for H1184 and 5′-GGGCTGGCATATGCTGCATATGTCATCTACAGAGG-3′ for E1291. Mutagenesis was performed using the Stratagene quickchange site-directed mutagenesis kit according to the manufacturer’s instructions. Inserts containing FN-tankyrase 1.WT and HE/A were subcloned into the retroviral vector pLPC (32).
Immunoprecipitation of in vitro-translated proteins.
Tankyrase 2 (TNKS2.5-2-1), tankyrase 1 (TT20 [38]), or control plasmid (pBluescript) were subjected to in vitro transcription-translation with the Promega TNT coupled-reticulocyte lysate system. Reaction mixtures were diluted 16-fold in TNE buffer (10 mM Tris [pH 7.8], 1% Nonidet P-40, 0.15 M NaCl, 1 mM EDTA, and protease inhibitor cocktail [Sigma]). The reaction mixtures were precleared by incubation with normal rabbit serum for 1 h on ice, followed by addition of protein G-Sepharose (Pharmacia). Nonspecific antibody complexes and protein aggregates were removed by centrifugation, and the supernatant was used for immunoprecipitation analysis. Supernatants were incubated with normal rabbit serum (1:200), 9 μg of anti-tankyrase 465 per ml (38), 0.5 μg of anti-tankyrase 1 609 per ml, or 0.1 μg of anti-tankyrase 1 376 per ml for 1 h on ice. Antigen-antibody complexes were collected on protein G beads, washed three times with TNE buffer, fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and processed for fluorography.
Immunoprecipitation of HeLa cell extracts.
HelaI.2.11 (a subclone of HeLa1 containing telomeres of 15 to 40 kb [16]) cells were lysed in TNE buffer (1 ml per two 15-cm-diameter dishes) at 4°C with rocking for 1 h and pelleted at 14,000 × g for 10 min. Supernatant was precleared with rabbit immunoglobulin (IgG) and protein G-Sepharose on ice for 20 min. Nonspecific antibody complexes and protein aggregates were removed by centrifugation, and the supernatant was used for immunoprecipitation analysis. The supernatant was incubated with 0.35 μg of rabbit IgG per ml, rabbit anti-tankyrase 1 376, or rabbit anti-TRF1 415 at 4°C with rocking for 1 h. Antigen-antibody complexes were collected on protein G beads, washed three times with TNE buffer, and suspended in Laemmli buffer. Samples were either boiled or not boiled (to prevent the IgG from comigrating with TRF1), fractionated by SDS-PAGE, and processed for immunoblotting as described below.
Metaphase spreads.
Chromosome spreads were performed as described previously (38). HeLaI.2.11 cells were treated with colcemide (0.5 μg/ml, 60 min), harvested by trypsinization, hypotonically swollen in 10 mM Tris-HCl (pH 7.4)-10 mM NaCl-5 mM MgCl2, and sedimented onto coverslips for 15 s at 3,000 rpm in a Sorvall RT6000B tabletop centrifuge. Chromosomes were swollen for 15 min in 0.25× phosphate-buffered saline (PBS), fixed in 3.7% formaldehyde in 0.25× PBS for 10 min, and then permeabilized with 0.5% NP-40 in 0.25× PBS for 10 min. Samples were blocked with 1% bovine serum albumin in PBS and processed for indirect immunofluorescence as described below.
Transient transfections.
HelaI.2.11 cells were electroporated with FN-tankyrase 1 (WT or HE/A) and grown for 18 h or with MN-tankyrase 2 and grown for 6 h. Cells were fixed in 2% paraformaldehyde in PBS for 10 min followed by permeabilization with 0.5% NP-40 in PBS for 10 min. Samples were blocked with 1% bovine serum albumin in PBS and processed for indirect immunofluorescence as described below.
Indirect immunofluorescence.
Metaphase spreads were incubated with rabbit anti-tankyrase antibody 609 (1 μg/ml) and a mouse polyclonal serum directed against full-length baculovirus-derived TRF1 (1:10,000) (38). Tankyrase 1-transfected cells were incubated with mouse monoclonal anti-FLAG M2 (1 μg/ml; Sigma) and rabbit anti-poly(ADP-ribose) serum (1:1,000) (Biomol) or rabbit anti-TRF1 371 (0.4 μg/ml) (40). Tankyrase 2-transfected cells were incubated with mouse monoclonal anti-Myc 9E10 supernatant (1:20) and rabbit anti-TRF1 415 (0.1 μg/ml) or rabbit anti-Myc antibody (0.2 μg/ml) (Santa Cruz) and mouse monoclonal anti-TRF2 (2.5 μg/ml) (Imgenex).
Primary antibodies were detected with fluorescein isothiocyanate- or tetramethyl rhodamine isothiocyanate-conjugated donkey anti-rabbit or anti-mouse antibodies (1:100) (Jackson Laboratories). DNA was stained with 4,6-diamino-2-phenylindole (DAPI) (0.2 μg/ml). Images were acquired on a Zeiss Axioplan 2 microscope with a Photometrix SenSyn camera. Photographs were processed and merged using IPLab software.
Preparation of cell extracts.
Whole-cell extracts from HTC75 or WI38 cells were prepared by suspending cells in 4 volumes of buffer C (20 mM HEPES-KOH [pH 7.9], 420 mM KCl, 25% glycerol, 0.1 mM EDTA, 5 mM MgCl2, 0.2% NP-40, 1 mM dithiothreitol, and 0.5 mM phenylmethylsulfonyl fluoride), incubating them for 60 min on ice, and pelleting them for 10 min at 14,000 × g at 4°C. Fifty micrograms of supernatant proteins were fractionated by SDS-PAGE and processed for immunoblotting as described below.
Immunoblotting.
Proteins were transferred to nitrocellulose electrophoretically and blocked in 5% milk in PBS containing 0.1% Tween 20. Blots were incubated with the following primary antibodies: rabbit anti-poly(ADP-ribose) serum (1:1,000) (Alexis Biochemicals), rabbit anti-TRF1 415 (0.3 μg/ml), rabbit anti-tankyrase 376 (0.1 μg/ml), or mouse monoclonal anti-TRF2 (0.5 μg/ml) (Imgenex), followed by horseradish peroxidase-conjugated donkey anti-rabbit or anti-mouse IgG (Amersham) (1:2,500). Bound antibody was detected with the enhanced chemiluminescence kit (Amersham).
Yeast two-hybrid analysis.
Tankyrase 1 (WT and HE/A) (aa 1 to 1327), tankyrase 1(AR9-19) (aa 436 to 797), tankyrase 2(AR4-15) (aa 120 to 519) and ankyrinG(AR1-24) (aa 72 to 854) (24) were cloned into the vector BTM116. Expression of the LexA fusion proteins was verified by Western blotting with anti-LexA antibody. GADTRF1 was described previously (2). Two-hybrid experiments were performed in the yeast L40 strain as described previously (2). The average values for three individual transformants for each set of plasmids are reported.
Retroviruses and cell lines.
Amphotropic retroviruses were generated by transfecting pLPC, pLPC-FN-tankyrase 1.WT, or pLPC-FN-tankyrase 1.HE/A into phoenix amphotropic cells using calcium phosphate precipitation. HTC75 cells (an HT1080-derived clonal cell line [40]) or WI38 cells (human primary fibroblasts at population doubling [PD] 30; American Type Culture Collection) were infected essentially as described (32). On day 1, 10-cm-diameter dishes containing 2 × 106 cells were retrovirally infected. On day 2, infected cells were selected with 2 μg of puromycin per ml. On day 3, cells were subcultured 1:2, and upon confluence (day 5 to 6 for HTC75 and day 7 to 8 for WI38) they were designated PD 0. Cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 20% fetal calf serum (WI38) or 10% calf serum (HTC75) (HyClone) and were continuously selected in puromycin.
Genomic blotting.
Genomic DNA was isolated as described (13) and cleaved with HinfI and RsaI. Approximately 1 μg of DNA was fractionated on 0.7% agarose and transferred to a nylon membrane, and telomeric restriction fragments were detected with a TTAGGG probe as described previously (12). The mean length of telomeric restriction fragments was determined by TELO, a macro for NIH Image written by the Research Computing Department at Fox Chase Cancer Center (http://www.fccc.edu), using scanned images of autoradiograms.
In vitro PARP assays.
PARP assays were performed as described previously (38). Samples containing baculovirus-derived tankyrase 1.WT (0 to 4 μg), tankyrase 1.HE/A (0 or 4 μg), tankyrase 2 (0 or 2 μg), TRF1 (0 or 4 μg) or PARP-1 (0.2 μg) (Biomol) were incubated for 30 min at 25°C in an assay buffer (0.1 ml) containing 50 mM Tris-HCl (pH 8.0), 4 mM MgCl2, 0.2 mM dithiothreitol, 1.3 μM [32P]NAD+ (4 μCi), various concentrations of NAD+ (0 to 1 mM), and DNA (0 or 0.8 μg; untreated or DNase-treated plasmid, pSPStyll, a pSP73 vector containing an 800-bp insert of TTAGGG repeats [12]). Reactions were stopped by addition of 20% trichloroacetic acid. Acid-insoluble proteins were collected by centrifugation, rinsed in 5% trichloroacetic acid, suspended in Laemmli loading buffer, and fractionated by SDS-PAGE. Proteins were visualized by Coomassie blue staining and autoradiography.
Baculovirus-derived proteins.
To make baculovirus-derived protein, an N-terminally His6-tagged version of human tankyrase 2 or human tankyrase 1.HE/A was generated in the expression vector pFastBac HT (Gibco BRL) and used to generate a recombinant plasmid in DH10Bac Escherichia coli. The recombinant DNA was used to transfect SF21 insect cells, and recombinant virus was isolated and amplified. His-tagged full-length baculovirus derived proteins, tankyrase 1.WT, tankyrase 1.HE/A, TRF1, TRF2, and tankyrase 2 were purified as described previously (2).
Generation of antibodies.
Anti-tankyrase 1 antibody 376 was raised and affinity purified against E. coli-derived fusion protein containing tankyrase 1 aa 3 to 167 (corresponding to the HPS domain). Anti-tankyrase 1 antibody 609 (S. Smith and T. de Lange, unpublished) was raised and affinity purified against a peptide containing tankyrase 1 aa 65 to 89 (within the HPS domain). Anti-TRF1 antibody 415 was generated and affinity purified against full-length baculovirus-derived TRF1.
Northern blots.
Northern blots containing polyadenylated RNAs from adult human tissues (Clontech), total RNA from fetal tissues (Clontech), or total RNA from HeLaI.2.11 or WI38 cells at the indicated PDs were probed with DNA probes corresponding to aa 183 to 302 for tankyrase 1 or aa 36 to 152 for tankyrase 2, or β-actin (Clontech).
RESULTS
Tankyrase 1 is complexed with TRF1 at telomeres in vivo.
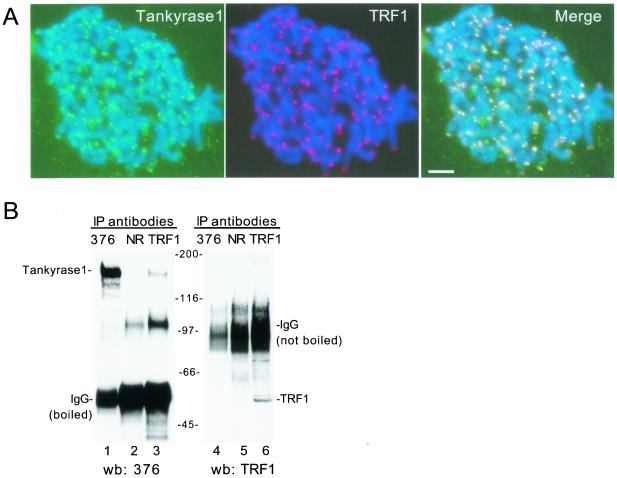
It was shown previously that endogenous tankyrase 1 localized to telomeres in metaphase spreads (38). The antibody used, 465, was raised against a subdomain of tankyrase 1 that turns out to be highly homologous to tankyrase 2 (Fig. 1A), raising the possibility that it might cross-react with tankyrase 2. Indeed, as shown in Fig. 1B, anti-tankyrase antibody 465 immunoprecipitated in vitro-translated tankyrase 2, in addition to tankyrase 1. To determine if the telomeric localization observed previously was specific for tankyrase 1 and not due to cross-reactivity of the antibody with tankyrase 2, tankyrase 1-specific antibodies were generated. We took advantage of the unique HPS domain in tankyrase 1, which has no counterpart in tankyrase 2. Two antibodies were generated, 609, an antipeptide antibody, and 376, raised against E. coli-derived recombinant HPS domain (Fig. 1A). As shown in Fig. 1B, anti-tankyrase 1 antibodies 609 and 376 specifically immunoprecipitated tankyrase 1, but not tankyrase 2.
FIG. 1.
Generation of tankyrase 1-specific antibodies. (A) Schematic representation of tankyrase 1 and 2. Percent identities to tankyrase 2 are indicated. Lines show the domains against which the indicated antibodies (Ab) were raised. HPS, homopolymeric tracts of histidine, proline, and serine. SAM, sterile alpha module. (B) Immunoprecipitation analysis of in vitro-translated proteins. Plasmids encoding tankyrase 1 (T1), tankyrase 2 (T2) or vector control (C) were subjected to in vitro transcription-translation. Reaction products were applied directly to the gel (Input; 40% of total) or immunoprecipitated (IP) with normal rabbit serum (NRS) or anti-tankyrase 1 antibodies 465, 609, or 376. Products were analyzed by SDS-PAGE and fluorography.
The tankyrase 1-specific antibodies were used to establish a telomeric association for tankyrase 1. Indirect immunofluorescence analysis of metaphase spreads probed with the tankyrase 1-specific antibody 609 showed that tankyrase 1 colocalized with TRF1 to chromosome ends (Fig. 2A). To determine if tankyrase 1 and TRF1 were complexed in vivo, immunoprecipitation analysis was performed. As shown in Fig. 2B, anti-TRF1 antibodies specifically coimmunoprecipitated tankyrase 1 from HeLa cell extracts (Fig. 2B, lane 3). Note that we were unable to coimmunoprecipitate TRF1 with anti-tankyrase 1 antibodies (Fig. 2B, lane 4), probably because the majority of tankyrase 1 resides in nontelomeric locations (6, 36) and is therefore not complexed to TRF1. Nonetheless, these colocalization and coimmunoprecipitation data confirm that tankyrase 1 is located at human telomeres and is complexed to TRF1 in vivo.
FIG. 2.
Tankyrase 1 is localized to telomeres and complexed to TRF1 in vivo. (A) Colocalization of endogenous tankyrase 1 and TRF1 at telomeres. Indirect immunofluorescence analysis of swollen-formaldehyde fixed metaphase spreads from HeLaI.2.11 cells stained with anti-tankyrase 1 antibody 609 (green) and anti-TRF1 (red). Merge represents superimposition of the red and green images. DAPI staining of DNA is shown in blue. Scale bar, 5 μm. (B) Coimmunoprecipitation of an endogenous tankyrase 1-TRF1 complex from HelaI.2.11 cells. Cell extracts were immunoprecipitated (IP) with anti-tankyrase 1 antibody 376 (lanes 1 and 4), normal rabbit IgG (NR) (lanes 2 and 5), or anti-TRF1 antibody 415 (lanes 3 and 6). Samples were processed as described in Materials and Methods, suspended in Laemmli buffer, and boiled (left panel) or not boiled (right panel). Reaction products were fractionated by SDS-PAGE and subjected to Western blot (wb) analysis with anti-tankyrase 1 antibody 376 (left panel) or anti-TRF1 antibody 415 (right panel).
Tankyrase 1 is not activated by telomeric or damaged DNA.
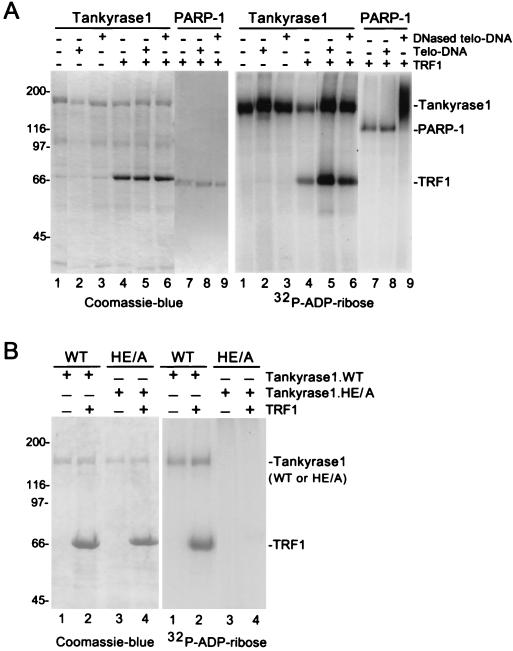
As described above, while it is known that damaged DNA activates PARP-1’s catalytic activity, the mechanism for activation of tankyrase 1’s catalytic activity is unknown. Since tankyrase 1 is complexed to TRF1 at telomeres, we asked if telomeric DNA would stimulate tankyrase 1’s PARP activity. To address this question we used an in vitro PARP assay that measures addition of radiolabeled ADP-ribose onto protein acceptors using [32P]NAD+ as a substrate. As shown in Fig. 3A (and previously [38]), incubation of recombinant tankyrase 1 with [32P]NAD+ resulted in ADP-ribosylation of tankyrase 1 (Fig. 3A, right panel, lane 1). Addition of TRF1 to the assay resulted in ADP-ribosylation of TRF1, but not in stimulation of tankyrase 1’s PARP activity (Fig. 3A, right panel, lane 4). Inclusion of telomeric DNA in the reaction had no effect on tankyrase 1’s PARP activity in the presence or absence of TRF1 (Fig. 3A, right panel, lanes 2 and 5). We occasionally saw a slight stimulation of tankyrase 1’s PARP activity by DNA; however, this effect was not reproducible. We next asked if DNase-treated telomeric DNA might stimulate tankyrase 1 since PARP-1 is activated by damaged (or DNase-treated) DNA. While DNase-treated telomeric DNA showed a dramatic stimulation of PARP-1 activity (Fig. 3A, right panel, compare lanes 7 and 9) it had no effect on tankyrase 1 in the presence or absence of TRF1 (Fig. 3A, right panel, lanes 3 and 6). Note that TRF1 is included in the PARP-1-containing reaction mixtures (Fig. 3A, left panel, lanes 7 to 9). Addition of TRF1 had no effect on PARP-1 activity, and TRF1 did not serve as an acceptor of ADP-ribosylation by PARP-1 (Fig. 3A, right panel, lanes 7 to 9). These results show that tankyrase 1 is not stimulated by intact or DNase-treated telomeric DNA in the presence or absence of TRF1.
FIG. 3.
Characterization of wild-type and mutant tankyrase 1 activity in an in vitro PARP assay. (A) Tankyrase 1 is not activated by telomeric DNA. Reaction mixtures containing 4 μg of recombinant tankyrase 1 (lanes 1 to 6) or 0.2 μg of recombinant PARP-1 (lanes 7 to 9) were subjected to an in vitro PARP assay containing 1.3 μM [32P]NAD+ substrate without (-) (lanes 1 to 3) or with (+) (lanes 4 to 9) 4 μg of TRF1 and without (-) (lanes 1, 4, and 7) or with (+) 0.8 μg of telomeric DNA (Telo-DNA) (lanes 2, 5, and 8) or 0.8 μg of DNase-treated telomeric (DNased telo-DNA) (lanes 3, 6, and 9). (B) The tankyrase 1.HE/A protein is catalytically inactive in vitro. Reaction mixtures containing 4 μg of recombinant tankyrase 1.WT (lanes 1 and 2) or 4 μg of tankyrase 1.HE/A (lanes 3 and 4) were subjected to an in vitro PARP assay containing 1.3 μM [32P]NAD+ substrate with (+) (lanes 2 and 4) or without (-) 4 μg of TRF1 (lanes 1 and 3). Reaction products in panels A and B were fractionated on SDS-PAGE gels and visualized by Coomassie blue staining (left panel) or autoradiography (right panel).
Generation of a catalytically inactive allele of tankyrase 1.
To determine if tankyrase 1’s PARP activity was required for its telomeric function, a PARP-inactive allele of tankyrase 1 was generated. The highly conserved active site residues, histidine and glutamic acid at amino acid positions 1184 and 1291, respectively, of tankyrase 1 were converted to alanine residues by site-directed mutagenesis to generate the mutant allele, tankyrase 1.HE/A. Previous studies on PARP-1 indicated that mutation to alanine of either one of the corresponding residues in PARP-1 resulted in a dramatic reduction in PARP activity (28). To determine if the HE/A mutation eliminated the PARP activity of tankyrase 1, the mutant allele was expressed in baculovirus and tested for activity in vitro. As shown in Fig. 3B, in contrast to wild-type tankyrase 1 (Fig. 3B, right panel, lanes 1 and 2), tankyrase 1.HE/A did not ADP-ribosylate itself or TRF1 (Fig. 3B, right panel, lanes 3 and 4), indicating that it was catalytically inactive. Importantly, while the mutant protein did not modify TRF1, tankyrase 1.HE/A still maintained its ability to interact with TRF1 as shown by two-hybrid analysis (see Fig. 6A) and by immunoprecipitation analysis of overexpressing cell lines (data not shown).
FIG. 6.
Two-hybrid analysis of tankyrase 1 and 2-TRF1 interactions. (A) β-Galactosidase concentrations (Miller units; average of three independent transformations) were measured for strains expressing the indicated fusion proteins. GAD, GAL4 activation domain; AR, ank repeats. (B) Northern blots of RNAs from a variety of human adult and fetal tissues and human primary and cancer cells, probed with tankyrase 1, tankyrase 2, or β-actin DNA as a control. f., fetal; PD, population doubling.
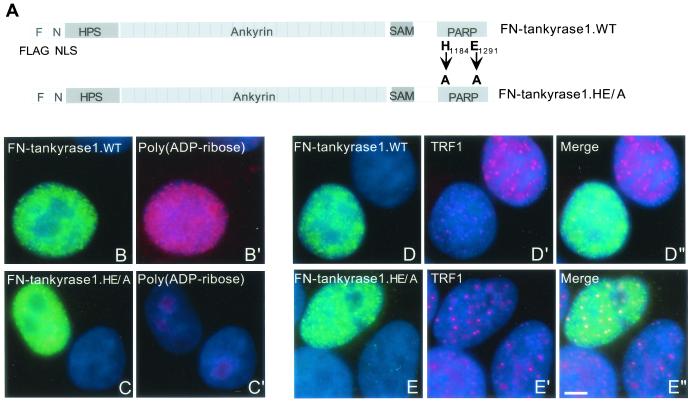
To measure the effect of catalytically inactive tankyrase 1 in vivo, the HE/A mutation was introduced into the expression construct FN-tankyrase 1.WT (Fig. 4A). This construct contains an NLS and allows localization of tankyrase 1 to the nucleus. We showed previously that overexpression of FN-tankyrase 1.WT released TRF1, but not TRF2, from telomeres and induced telomere elongation (37). Indirect immunofluorescence analysis of HeLaI.2.11 cells expressing FN-tankyrase 1.HE/A revealed a diffuse nuclear staining pattern (Fig. 4C), similar to that of FN-tankyrase 1.WT (Fig. 4B). However, upon costaining with anti-poly(ADP-ribose) antibodies, only FN-tankyrase 1.WT (Fig. 4B′), not FN-tankyrase 1.HE/A (Fig. 4C′) was detected, indicating that FN-tankyrase 1.HE/A did not synthesize ADP-ribose polymers in vivo. Moreover, unlike the wild-type protein (Fig. 4D), the HE/A mutant did not release TRF1 from telomeres. Rather, FN-tankyrase 1.HE/A remained on telomeres and colocalized with TRF1 (Fig. 4E). These results indicate that ADP-ribosylation is required for release of TRF1 from telomeres.
FIG. 4.
Tankyrase 1.HE/A is inactive in vivo. (A) Schematic representation of tankyrase 1 expression constructs. F, FLAG epitope tag; N, NLS, nuclear localization signal from SV40; WT, wild type; HE/A, double point mutation converting the histidine (H) at position 1184 and the glutamic acid (E) at position 1291 to alanine (A) residues. (B to E) FN-tankyrase 1.HE/A does not synthesize ADP-ribose polymers in vivo or release TRF1 from telomeres, as shown by indirect immunofluorescence analysis of HeLaI.2.11 cells transiently transfected with FN-tankyrase 1.WT (B and D) or FN-tankyrase 1.HE/A (C and E), formaldehyde fixed, and stained with anti-FLAG antibody (green) (B, C, D, and E) and anti-poly(ADP-ribose) antibody (red) (B′ and C′), or anti-TRF1 antibody (red) (D′ and E′). Merge represents superimposition of the red and green images. DAPI staining of DNA is shown in blue. Scale bar, 5 μm.
Tankyrase 1.HE/A does not induce telomere elongation.
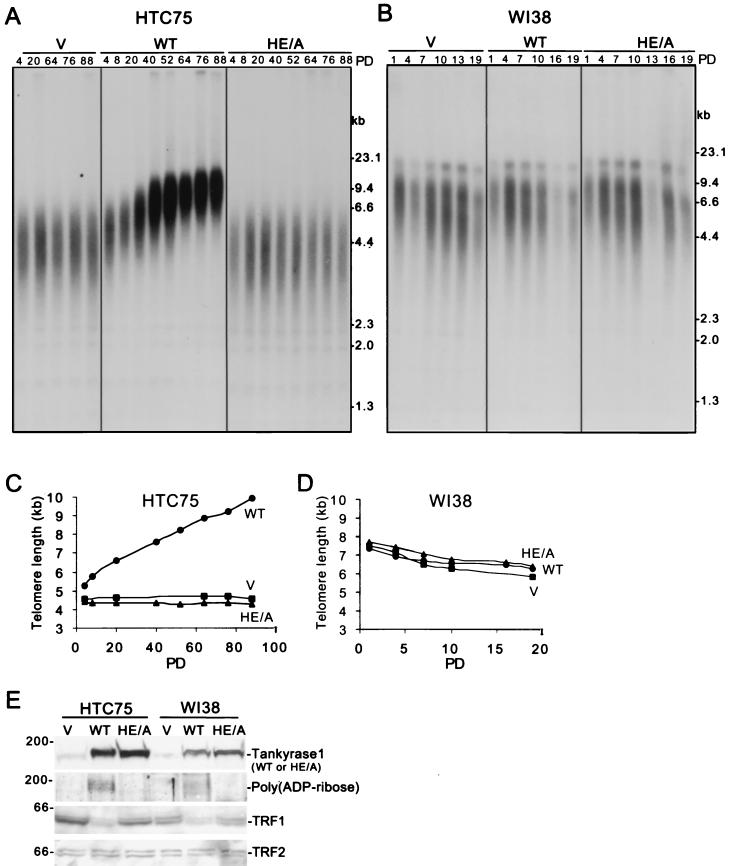
We next asked if the catalytically inactive tankyrase 1.HE/A mutant could mimic telomere elongation induced by FN-tankyrase 1.WT overexpression. Stable HTC75 tumor cell lines expressing FN-tankyrase 1.WT, HE/A, or vector were generated by retroviral infection and subjected to telomere length analysis. As shown in Fig. 5A and C, overexpression of tankyrase 1.WT induced telomere elongation. Telomeres showed progressive elongation at a rate of approximately 50 bp per PD, similar to previous results (37). In contrast, overexpression of the HE/A mutant had no effect on telomere length and was similar to the vector control (Fig. 5A and C). Immunoblot analysis indicated that the WT and HE/A alleles were overexpressed to similar extents, but only the WT allele was detected by anti-poly(ADP-ribose) antibodies (Fig. 5E). Moreover, while the WT allele induced loss of TRF1, the HE/A mutant had no effect on TRF1 levels (Fig. 5E). As expected, TRF2, which does not interact with tankyrase 1, was unaffected by overexpression of FN-tankyrase 1.WT. Together, these results demonstrate that the catalytic activity of tankyrase 1 is essential to induce loss of TRF1 and telomere elongation.
FIG. 5.
Analysis of stable cell lines expressing FN-tankyrase 1.WT or HE/A. (A and B) Southern blot analysis of HinfI/RsaI-digested genomic DNA from telomerase-positive HTC75 cell lines (A) or telomerase-negative WI38 cell lines (B) expressing vector control (V), FN-tankyrase 1.WT (WT), or FN-tankyrase 1.HE/A (HE/A). Cell lines were grown for 88 (HTC75) or 19 (WI38) population doublings (PD), and DNA samples were analyzed at the indicated PDs. Blots were probed with TTAGGG-repeat probe to detect telomeric restriction fragments. (C and D) Graphical representations of telomere length changes in HTC75 (C) or WI38 (D) cell lines expressing FN-tankyrase 1.WT, HE/A, or vector. Plots represent the mean telomere length values derived from the Southern blots analyzed in panels A and B. (E) Western blot analysis of whole-cell extracts from HTC75 (PD 52) or WI38 (PD 4) cells expressing vector control (V), FN-tankyrase 1.WT (WT), or FN-tankyrase 1.HE/A (HE/A). Blots were probed with the following antibodies: anti-tankyrase 1 376, anti-poly(ADP-ribose), anti-TRF1 415, and anti-TRF2.
Tankyrase 1-induced elongation requires telomerase-expressing cells.
Our findings are consistent with a model in which tankyrase 1 poly(ADP-ribosyl)ates TRF1, releasing it from telomeres and allowing access to telomerase. The HTC75 cells used in this study are telomerase-positive human tumor cells. The gradual and progressive increase in telomere length observed upon overexpression of FN-tankyrase 1.WT in these cells is consistent with a telomerase-based mechanism of telomere elongation. We thus asked if overexpression of tankyrase 1 would induce a similar telomere elongation in human primary, WI38 cells. These cells, like most human somatic cells, lack telomerase activity and (unlike tumor cells) show progressive telomere shortening. Stable WI38 cell lines overexpressing FN-tankyrase 1.WT, HE/A, or vector were generated by retroviral infection and subjected to telomere blot analysis. As shown in Fig. 5B and D, expression of FN-tankyrase 1.WT or HE/A did not induce telomere elongation in human primary cells. Immunoblot analysis indicated that both tankyrase 1 alleles were highly expressed and that the wild-type allele induced loss of TRF1 (Fig. 5E), as in HTC75 cells. Nonetheless, telomere length was unaffected, indicating that loss of TRF1 is not sufficient for telomere elongation and suggesting that telomere elongation by tankyrase 1 is telomerase dependent.
We have shown thus far that tankyrase 1 is a telomeric protein. Using tankyrase 1-specific antibodies (that do not react with tankyrase 2) we demonstrate that endogenous tankyrase 1 localizes to telomeres in metaphase spreads and complexes with endogenous TRF1 in vivo. Furthermore, tankyrase 1-induced loss of TRF1 and telomere elongation requires the catalytic activity of the PARP domain and does not occur in telomerase-negative cells.
Tankyrase 2 is ubiquitously expressed like tankyrase 1.
We next sought to determine if the closely related homolog tankyrase 2 displayed properties similar to those of tankyrase 1 that might implicate it in telomere length regulation. The ankyrin domain of tankyrase 2 was identified previously in a two-hybrid screen with TRF1 (19). To determine specifically which ank repeats in tankyrase 2 were required for interaction with TRF1, a construct containing an internal 12-ank repeat domain was generated for two-hybrid analysis. As shown in Fig. 6A, tankyrase 2 binds TRF1 through ank repeats 4 to 15. Since TRF1 binds to both tankyrase 1 and tankyrase 2 through their ankyrin domains, we postulated that TRF1 may recognize some structural feature of ankyrins in general. However, TRF1 did not interact with the ankyrin domain of another ankyrin family member, ankyrin G (H. Seimiya and S. Smith, unpublished data) (Fig. 6A), indicating that its interaction with tankyrase 1 and 2 may be specific for some unique feature of the tankyrase ankyrin domain.
The high homology between tankyrases 1 and 2 and the finding that they both bind TRF1 suggested that they could have redundant functions. We therefore hypothesized that the genes might be differentially expressed. Northern blot analysis was performed to determine if tankyrases 1 and 2 showed any tissue-specific differences in their patterns of gene expression. As shown in Fig. 6B, tankyrase 2 (a single transcript of approximately 7 kb) (19, 25, 27) was ubiquitously expressed in most human adult and fetal tissues, in HeLa cells, and in human primary fibroblasts. Comparison between tankyrase 1 (three transcripts of approximately 7 to 10 kb) (38) and tankyrase 2 revealed similar patterns of gene expression. Both genes were highly expressed in testis, ovary, and skeletal muscle. The only difference in expression pattern was in placenta, where tankyrase 2, but not tankyrase 1, was highly expressed.
Tankyrase 2 is a PARP that interacts with TRF1 in vitro and in vivo.
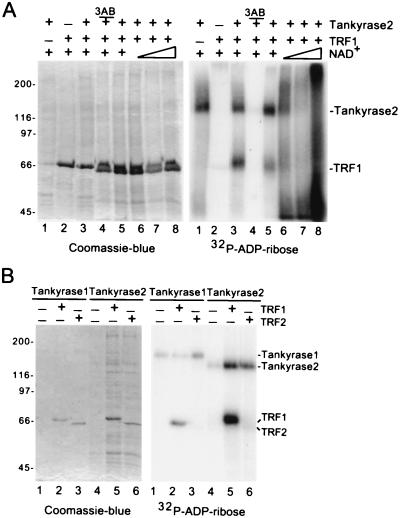
To determine if tankyrase 2 displayed enzymatic properties similar to those of tankyrase 1, recombinant baculovirus-derived tankyrase 2 was tested in the in vitro PARP assay. Incubation of tankyrase 2 with the substrate [32P]NAD+ resulted in a 32P-labeled species that comigrated with tankyrase 2 (Fig. 7A, lane 1, right panel), indicating that tankyrase 2 ADP-ribosylated itself. Inclusion of recombinant TRF1 in the assay showed a 32P-labeled species that comigrated with TRF1 (Fig. 7, lane 3, right panel). Thus, tankyrase 2 can ADP-ribosylate itself and TRF1 in vitro. Addition of increasing concentrations of the substrate NAD+ produced slower-migrating heterogeneous products characteristic of poly(ADP-ribosyl)ation (Fig. 7A, right panel, lanes 6 to 8). The reaction was inhibited by the general PARP inhibitor, 3-aminobenzamide (3AB) (Fig. 7A, right panel, lane 4).
FIG. 7.
Tankyrase 2 is a PARP that modifies itself and TRF1, but not TRF2, in vitro. (A) Tankyrase 2 ADP-ribosylates itelf and TRF1 in vitro. Reaction mixtures containing 2 μg of recombinant tankyrase 2 (lanes 1 and 3 to 8) were subjected to an in vitro PARP assay containing 1.3 μM [32P]NAD+ substrate without (-) (lane 1) or with (+) 4 μg of TRF1 (lanes 2 to 8). Three reaction mixtures were supplemented with unlabeled NAD+ (0.04, 0.2, and 1 mM, triangle) (lanes 6 to 8) and one reaction mixture contained 1 mM 3-amino benzamide (3AB) (lane 4). (B) Tankyrase 2 does not modify TRF2 in vitro. Reactions containing 0.5 μg of recombinant tankyrase 1 (lanes 1 to 3) or 2 μg of recombinant tankyrase 2 (lanes 4 to 6) were subjected to an in vitro PARP assay containing 1.3 μM [32P]NAD+ substrate with (+) 1 μg TRF1 (lanes 2 and 5) or (+) 1 μg TRF2 (lanes 3 and 6). Reaction products in panels A and B were fractionated by SDS-PAGE and visualized by Coomassie blue staining (left panel) or autoradiography (right panel).
Tankyrase 2, like tankyrase 1, interacts with the acidic, amino-terminal domain of TRF1 (19, 38). This domain is a distinguishing feature between TRF1 and TRF2; TRF2 has, instead, a basic domain at its amino terminus. To determine if tankyrase 2 interacts with TRF2 we asked if TRF2 could serve as an acceptor of ADP-ribosylation in vitro. As shown in Fig. 7B, while TRF1 served as an acceptor of ADP-ribosylation by tankyrase 1 (lane 2) or tankyrase 2 (lane 5), TRF2 was not modified by tankyrase 1 (lane 3) or by tankyrase 2 (lane 6).
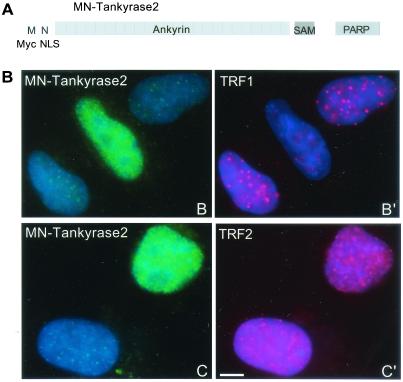
Tankyrase 2 displays properties similar to those of tankyrase 1; it binds TRF1 using an internal ank repeat domain (Fig. 6A) and it poly(ADP-ribosyl)ates itself and TRF1 in vitro (Fig. 7). Tankyrase 2 does not contain an NLS, and transfected tankyrase 2, like tankyrase 1, is excluded from the nucleus (19; B. Cook and S. Smith, unpublished data). To assess the effect of tankyrase 2 on TRF1 in vivo, an allele containing an NLS as well as a Myc-epitope tag at its amino terminus, MN-tankyrase 2, was generated (Fig. 8A). Indirect immunofluorescence analysis of HeLaI.2.11 cells expressing MN-tankyrase 2 revealed a diffuse nuclear staining pattern (Fig. 8B). As shown in Fig. 8B′, expression of MN-tankyrase 2 released TRF1 from telomeres. This effect was specific for TRF1, since overexpression of MN-tankyrase 2 had no effect on TRF2 (Fig. 8C′). These results indicate that tankyrase 2, like tankyrase 1, can modulate TRF1 at telomeres in vivo.
FIG. 8.
Overexpression of tankyrase 2 in the nucleus releases TRF1, but not TRF2, from telomeres. (A) Schematic diagram of the tankyrase 2 expression construct. M, Myc epitope tag; N, SV40 NLS. (B) Indirect immunofluorescence of HelaI.2.11 cells transiently transfected with MN-tankyrase 2, formaldehyde fixed, and stained with anti-myc antibody (green) (B and C) and anti-TRF1 antibody 415 (red) (B′) or anti-TRF2 antibody (red) (C′). DAPI staining of DNA is shown in blue. Scale bar, 5 μm.
DISCUSSION
Tankyrases 1 and 2 form a new subgroup of the expanding family of cellular PARPs. Tankyrases 1 and 2 are most closely related in their PARP catalytic domains (94% identity) but are highly conserved throughout their primary structure (83% identity overall). This structure includes a number of protein-protein interacting motifs: 24 ank repeats and a sterile alpha module (SAM) domain. The recent identification of multiple interacting (and in some cases overlapping) partners for these two proteins suggests multifunctional, and perhaps redundant roles for tankyrase 1 and 2. Here we sought to firmly establish tankyrase 1 as a telomere-specific protein and to investigate the role of tankyrase 2 at telomeres.
Using tankyrase 1-specific antibodies (that do not cross-react with tankyrase 2) we show that endogenous tankyrase 1 localizes to telomeres in metaphase spreads (Fig. 2A) and complexes with TRF1 in vivo (Fig. 2B). An important question is whether endogenous tankyrase 2 is localized to telomeres. Unfortunately, since tankyrase 2 lacks a unique domain (like the HPS domain of tankyrase 1), attempts to generate tankyrase 2-specific peptide antibodies for immunofluorescence analysis have been unsuccessful. Thus, the localization of endogenous tankyrase 2 is unknown. The development of tankyrase 2-specific antibodies (such as monoclonal antibodies) will be required in order to determine if endogenous tankyrase 2 localizes to telomeres.
While tankyrase 1 localizes to telomeres in vivo, the protein does not contain an NLS and its mechanism of localization to telomeres is not known. In order to overexpress tankyrase 1 (or tankyrase 2) in the nucleus it was necessary to introduce an NLS into the construct. Analysis of stable cell lines overexpressing a wild-type tankyrase 1 allele lacking an NLS showed that tankyrase 1 had no effect on TRF1 and did not induce telomere elongation (G. Shostak and S. Smith, unpublished data), indicating that nuclear localization is required for tankyrase 1’s effect at telomeres. It will be important to determine how endogenous tankyrase 1 localizes to telomeres. It was shown previously that exogenous tankyrase 1 can be recruited to telomeres by overexpression of TRF1 (36); thus, TRF1 or another NLS-containing protein may recruit tankyrase 1 to telomeres. Alternatively, tankyrase 1 could bind to TRF1 on telomeres at mitosis when the nuclear envelope barrier breaks down.
Overexpression of tankyrase 1 in the nuclei of telomerase-positive tumor cells induced loss of TRF1 and lengthening of telomeres (Fig. 5A, C, and E) (37). In striking contrast, overexpression of tankyrase 1 in the nuclei of telomerase-negative human primary cells had no effect on telomere length (Fig. 5B and D). Indeed, even though FN-tankyrase 1.WT induced loss of TRF1 in WI38 cells (Fig. 5E), telomere elongation was not observed, indicating that loss of TRF1 may be necessary but is not sufficient to induce telomere lengthening. These findings are consistent with a model in which telomerase is required for tankyrase 1-induced telomere elongation. It is interesting that tankyrase 1 is highly expressed in telomerase-negative cells, such as WI38 cells and most human adult tissues (Fig. 6B). Tankyrase 1 could play a structural role at telomeres in telomerase-negative cells but may also have additional unrelated functions, consistent with its localization to multiple subcellular sites (6, 36).
Previous studies indicated that a carboxy-terminally deleted form of tankyrase 1 (lacking the last four ank repeats and the SAM and PARP domains) had no effect on telomere length (37). We now extend these results using a catalytically inactive, full-length allele of tankyrase 1 to show that loss of TRF1 and telomere elongation depend upon, specifically, the ADP-ribosylating activity of tankyrase 1. Thus, overexpression in the nucleus of FN-tankyrase 1.HE/A (at the same levels as FN-tankyrase 1.WT) had no effect on TRF1 and did not induce telomere elongation. These findings are consistent with a model in which tankyrase 1-mediated ADP-ribosylation of TRF1 modulates the telomeric complex to allow access of telomerase. An important question is whether telomeres will shorten in the absence of tankyrase 1. Thus, future studies will include disruption of tankyrase 1 in mouse embryonic stem cells and in mice. Of course, the possibility exists that tankyrase 1 and 2 have redundant roles at telomeres. Hence, knockout of both genes may be required to address the question.
Our studies show that when tankyrase 1 is overexpressed in an active form in the nucleus, TRF1 is released and telomeres increase in length (Fig. 4 and 5). In FN-tankyrase 1.WT-overexpressing cells, loss of TRF1 and telomere elongation occur continuously, but normally, these events are likely to be tightly regulated. Indeed, in most cells, TRF1 is observed on telomeres; and in telomerase-positive cells telomeres do not lengthen indefinitely; rather, they are maintained at a constant length setting. We therefore suggest that tankyrase 1-mediated ADP-ribosylation and release of TRF1 from telomeres is a transient reaction that may occur at every cell cycle or every few cell cycles to allow access of telomerase for telomere maintenance. According to this hypothesis, the endogenous tankyrase 1 that we observe on telomeres (Fig. 2A) is most likely inactive. It will be interesting to determine how and when tankyrase 1 is activated. We show that, unlike PARP-1, tankyrase 1 is not activated by damaged DNA or by telomeric DNA (Fig. 3A). Activation could occur by a posttranslational modification of the protein. Tankyrase 1 was found to be a target of mitogen-activated protein kinase (6). Alternatively, tankyrase 1 may be activated by interaction with another protein. Tankyrase 1 already has several known interacting factors, and its multiple protein-interacting domains suggest that there will be additional binding partners.
Our findings indicate that tankyrase 2 behaves like tankyrase 1 in a number of assays. Tankyrase 2 poly(ADP-ribosyl)ated itself and TRF1 in vitro (Fig. 7) and bound to TRF1 in a two-hybrid assay (Fig. 6A). Overexpression of tankyrase 2 in the nucleus released TRF1 from telomeres (Fig. 8), suggesting that in vivo tankyrase 2 can have an effect on TRF1 similar to that of tankyrase 1. It will be interesting to determine if long-term overexpression of tankyrase 2 in the nucleus will have an effect on telomere length similar to that of tankyrase 1. Tankyrase 1 and 2 each interact with the acidic domain of TRF1 (19, 38). Since TRF1 binds telomeric DNA as a dimer (2), it is possible that a single TRF1 dimer could bind to tankyrase 1 and 2 at the same time. Alternatively, tankyrase 1 and 2 could form separate complexes with TRF1. Although highly related, tankyrase 1 and 2 may be activated by different stimuli and have different biological properties. Indeed, recent studies indicate that overexpression of tankyrase 2, but not tankyrase 1, induced necrotic cell death (19). It remains to be determined if endogenous tankyrase 2 localizes to telomeres and if its role and that of tankyrase 1 are redundant or if it has a unique role at telomeres.
Acknowledgments
We are grateful to Hiroyuki Seimiya for two-hybrid constructs and Ben Houghtaling for help with retroviral infections. We thank Tom Meier, Hiroyuki Seimiya, and Ben Houghtaling for helpful comments on the manuscript.
This work was supported by grants from the Edward Mallinckrodt, Jr. Foundation and the New York City Council Speaker’s Fund for Biomedical Research. S.S. is a recipient of the Kimmel Scholar Award.
B. D. Cook and J. N. Dynek contributed equally to this work
REFERENCES
- 1.Ame, J., E. Jacobson, and M. Jacobson. 2001. ADP-ribose polymer metabolism, p. 1–34. In G. D. Murcia, and S. Shall (ed.), From DNA damage and stress signalling to cell death: poly(ADP-ribosyl)ation reactions. Oxford University Press, Oxford, England.
- 2.Bianchi, A., S. Smith, L. Chong, P. Elias, and T. de Lange. 1997. TRF1 is a dimer and bends telomeric DNA. EMBO J. 16:1785–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bianchi, A., R. M. Stansel, L. Fairall, J. D. Griffith, D. Rhodes, and T. de Lange. 1999. TRF1 binds a bipartite telomeric site with extreme spatial flexibility. EMBO J 18:5735–5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bilaud, T., C. Brun, K. Ancelin, C. E. Koering, T. Laroche, and E. Gilson. 1997. Telomeric localization of TRF2, a novel human telobox protein. Nat. Genet. 17:236–239. [DOI] [PubMed] [Google Scholar]
- 5.Broccoli, D., A. Smogorzewska, L. Chong, and T. de Lange. 1997. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat. Genet. 17:231–235. [DOI] [PubMed] [Google Scholar]
- 6.Chi, N. W., and H. F. Lodish. 2000. Tankyrase is a Golgi-associated MAP kinase substrate that interacts with IRAP in GLUT4 vesicles. J. Biol. Chem. 275:38437–38444. [DOI] [PubMed] [Google Scholar]
- 7.Chong, L., B. van Steensel, D. Broccoli, H. Erdjument-Bromage, J. Hanish, P. Tempst, and T. de Lange. 1995. A human telomeric protein. Science 270:1663–1667. [DOI] [PubMed] [Google Scholar]
- 8.Collins, K. 2000. Mammalian telomeres and telomerase. Curr. Opin. Cell Biol. 12:378–383. [DOI] [PubMed] [Google Scholar]
- 9.Counter, C. M., A. A. Avilion, C. E. LeFeuvre, N. G. Stewart, C. W. Greider, C. B. Harley, and S. Bacchetti. 1992. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 11:1921–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Counter, C. M., F. M. Botelho, P. Wang, C. B. Harley, and S. Bacchetti. 1994. Stabilization of short telomeres and telomerase activity accompany immortalization of Epstein-Barr virus-transformed human B lymphocytes. J. Virol. 68:3410–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D’Amours, D., S. Desnoyers, I. D’Silva, and G. G. Poirier. 1999. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 342:249–268. [PMC free article] [PubMed] [Google Scholar]
- 12.de Lange, T. 1992. Human telomeres are attached to the nuclear matrix. EMBO J. 11:717–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Lange, T., L. Shiue, R. M. Myers, D. R. Cox, S. L. Naylor, A. M. Killery, and H. E. Varmus. 1990. Structure and variability of human chromosome ends. Mol. Cell. Biol. 10:518–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greider, C. W., and E. H. Blackburn. 1985. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43:405–413. [DOI] [PubMed] [Google Scholar]
- 15.Griffith, J., A. Bianchi, and T. de Lange. 1998. TRF1 promotes parallel pairing of telomeric tracts in vitro. J. Mol. Biol. 278:79–88. [DOI] [PubMed] [Google Scholar]
- 16.Griffith, J. D., L. Comeau, S. Rosenfield, R. M. Stansel, A. Bianchi, H. Moss, and T. de Lange. 1999. Mammalian telomeres end in a large duplex loop. Cell 97:503–514. [DOI] [PubMed] [Google Scholar]
- 17.Harley, C. B., A. B. Futcher, and C. W. Greider. 1990. Telomeres shorten during ageing of human fibroblasts. Nature 345:458–460. [DOI] [PubMed] [Google Scholar]
- 18.Hastie, N. D., M. Dempster, M. G. Dunlop, A. M. Thompson, D. K. Green, and R. C. Allshire. 1990. Telomere reduction in human colorectal carcinoma and with ageing. Nature 346:866–868. [DOI] [PubMed] [Google Scholar]
- 19.Kaminker, P. G., S. H. Kim, R. D. Taylor, Y. Zebarjadian, W. D. Funk, G. B. Morin, P. Yaswen, and J. Campisi. 2001. TANK2, a new TRF1-associated PARP, causes rapid induction of cell death upon overexpression. J. Biol. Chem. 276:35891–35899. [DOI] [PubMed] [Google Scholar]
- 20.Karlseder, J., D. Broccoli, Y. Dai, S. Hardy, and T. de Lange. 1999. p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science 283:1321–1325. [DOI] [PubMed] [Google Scholar]
- 21.Kim, N. W., M. A. Piatyszek, K. R. Prowse, C. B. Harley, M. D. West, P. L. Ho, G. M. Coviello, W. E. Wright, S. L. Weinrich, and J. W. Shay. 1994. Specific association of human telomerase activity with immortal cells and cancer. Science 266:2011–2015. [DOI] [PubMed] [Google Scholar]
- 22.Kim, S. H., P. Kaminker, and J. Campisi. 1999. TIN2, a new regulator of telomere length in human cells. Nat. Genet. 23:405–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kipling, D., and H. J. Cooke. 1990. Hypervariable ultra-long telomeres in mice. Nature 347:400–402. [DOI] [PubMed] [Google Scholar]
- 24.Kordeli, E., S. Lambert, and V. Bennett. 1995. AnkyrinG. A new ankyrin gene with neural-specific isoforms localized at the axonal initial segment and node of Ranvier. J. Biol. Chem. 270:2352–2359. [DOI] [PubMed] [Google Scholar]
- 25.Kuimov, A. N., D. V. Kuprash, V. N. Petrov, K. K. Vdovichenko, M. J. Scanlan, C. V. Jongeneel, M. A. Lagarkova, and S. A. Nedospasov. 2001. Cloning and characterization of TNKL, a member of tankyrase gene family. Genes Immun. 2:52–55. [DOI] [PubMed] [Google Scholar]
- 26.Li, B., S. Oestreich, and T. de Lange. 2000. Identification of human Rap1: implications for telomere evolution. Cell 101:471–483. [DOI] [PubMed] [Google Scholar]
- 27.Lyons, R. J., R. Deane, D. K. Lynch, Z. S. Ye, G. M. Sanderson, H. J. Eyre, G. R. Sutherland, and R. J. Daly. 2001. Identification of a novel human Tankyrase through its interaction with the adapter protein Grb14. J. Biol. Chem. 22:17172–17180. [DOI] [PubMed] [Google Scholar]
- 28.Marsischky, G. T., B. A. Wilson, and R. J. Collier. 1995. Role of glutamic acid 988 of human poly-ADP-ribose polymerase in polymer formation. Evidence for active site similarities to the ADP-ribosylating toxins. J. Biol. Chem. 270:3247–3254. [DOI] [PubMed] [Google Scholar]
- 29.McEachern, M. J., A. Krauskopf, and E. H. Blackburn. 2000. Telomeres and their control. Annu. Rev. Genet. 34:331–358. [DOI] [PubMed] [Google Scholar]
- 30.Monz, D., A. Munnia, N. Comtesse, U. Fischer, W. I. Steudel, W. Feiden, B. Glass, and E. U. Meese. 2001. Novel tankyrase-related gene detected with meningioma-specific sera. Clin. Cancer Res. 7:113–119. [PubMed] [Google Scholar]
- 31.Nugent, C. I., and V. Lundblad. 1998. The telomerase reverse transcriptase: components and regulation. Genes Dev. 12:1073–1085. [DOI] [PubMed] [Google Scholar]
- 32.Serrano, M., A. W. Lin, M. E. McCurrach, D. Beach, and S. W. Lowe. 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88:593–602. [DOI] [PubMed] [Google Scholar]
- 33.Shall, S., and G. de Murcia. 2000. Poly(ADP-ribose) polymerase-1: what have we learned from the deficient mouse model? Mutat. Res. 460:1–15. [DOI] [PubMed] [Google Scholar]
- 34.Shay, J. W., and S. Bacchetti. 1997. A survey of telomerase activity in human cancer. Eur. J. Cancer 33:787–791. [DOI] [PubMed] [Google Scholar]
- 35.Smith, S. 2001. The world according to PARP. Trends Biochem. Sci. 26:174–179. [DOI] [PubMed] [Google Scholar]
- 36.Smith, S., and T. de Lange. 1999. Cell cycle dependent localization of the telomeric PARP, tankyrase, to nuclear pore complexes and centrosomes. J. Cell Sci. 112:3649–3656. [DOI] [PubMed] [Google Scholar]
- 37.Smith, S., and T. de Lange. 2000. Tankyrase promotes telomere elongation in human cells. Curr. Biol. 10:1299–1302. [DOI] [PubMed] [Google Scholar]
- 38.Smith, S., I. Giriat, A. Schmitt, and T. de Lange. 1998. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science 282:1484–1487. [DOI] [PubMed] [Google Scholar]
- 39.Smogorzewska, A., B. van Steensel, A. Bianchi, S. Oelmann, M. Schaefer, G. Schnapp, and T. de Lange. 2000. Control of human telomere length by TRF1 and TRF2. Mol. Cell. Biol. 20:1659–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Steensel, B., and T. de Lange. 1997. Control of telomere length by the human telomeric protein TRF1. Nature 385:740–743. [DOI] [PubMed] [Google Scholar]
- 41.van Steensel, B., A. Smogorzewska, and T. de Lange. 1998. TRF2 protects human telomeres from end-to-end fusions. Cell 92:401–413. [DOI] [PubMed] [Google Scholar]