Abstract
Oxidative DNA damage may play an important role in human disease including cancer. Previously, mutational spectra have been determined using systems that include transition metal ions and hydrogen peroxide (H2O2). G→T transversions and C→T transitions were the most common mutations observed including some CC→TT tandem mutations. C→T transition mutations at methylated CpG dinucleotides are the most common mutations in human genetic diseases. It has been hypothesized that oxidative stress may increase the frequency of mutations at methylated CpG sequences. Here we have used a CpG-methylated shuttle vector to derive mutational spectra of copper/H2O2-induced DNA damage upon passage of the shuttle vector through human fibroblasts. We find that copper/H2O2 treatment produces higher numbers of CpG transition mutations when the CpGs are methylated but does not create clear C→T hotspots at these sites. More strikingly, we observed that this treatment produces a substantial frequency of mutations that were mCG→TT tandem mutations. Six of seven tandem mutations were of this type. mCG→TT mutations (6/63 = 10% of all mutations) were observed only in nucleotide excision repair-deficient (XP-A) cells but were not found in repair-proficient cells. The data suggest that this novel type of mutation may be produced by vicinal or cross-linked base damage involving 5-methylcytosine and a neighboring guanine, which is repaired by nucleotide excision repair. We suggest that the underlying oxidative lesions could be responsible for the progressive neurodegeneration seen in XP-A individuals.
INTRODUCTION
DNA damage induced by reactive oxygen species (ROS) is an important intermediate in the pathogenesis of human conditions such as cancer and aging (1–5). ROS-induced DNA damage products are both mutagenic and cytotoxic. Hydrogen peroxide (H2O2), which generates hydroxy radicals in the presence of transition metal ions, is considered an appropriate model for ROS. H2O2 is produced endogenously by several physiological processes, e.g. during oxidative phosphorylation (6) and by the inflammatory cell respiratory burst (7). Because it is freely diffusible, H2O2 can potentially reach the nucleus to interact with DNA (8). H2O2 causes strand breaks (9) and base damage (10,11) in DNA by a mechanism that requires transition metal ions, such as iron or copper (12–14). Mixtures of Cu(II) ions and H2O2, often with added ascorbic acid, produce extensive strand breaks in DNA (15–17). Strand breaks often occur near guanine residues, and it has been suggested that copper ions bind to DNA at these sites (15). Indeed, Cu(II)-dependent DNA fragmentation has been reported to be much more extensive than that produced by equimolar Fe(III) ions in comparable reaction mixtures (16,18,19). Cu(II)/ascorbate/H2O2-mediated DNA damage in aerobic aqueous solutions is believed to be induced in vitro and in vivo through formation of a DNA–Cu(I)–H2O2 complex (16,20–22). DNA damage induced by copper/H2O2 is enhanced by packaging of DNA into nucleosomes (23).
Exposure of target cells to H2O2 reproduces at least some components of the known endogenous DNA damage spectrum. More than 30 different sugar and base modifications have been identified (11). Levels of oxidative DNA damage products have been measured in tissues by a variety of techniques and, although there is some controversy about the ‘true’ level of oxidative DNA damage, the levels can be quite substantial (24,25). It is unclear which of the many different lesions produced by DNA oxidation is the one most responsible for inducing mutations. The mutations that are produced depend on the source of the ROS and the particular experimental system used to study the mutations. In general, C→T transitions (∼40–60%), G→T transversions (20–40%), as well as deletions are commonly seen (14,26,27). Candidate lesions that may have this mutational specificity include 5-hydroxycytosine for C→T (28), products of cytosine oxidation and deamination (5-hydroxyuracil and uracil glycol) for C→T (29), and 8-oxoguanine for G→T mutations (30,31). However, the mutational specificity of many of the other oxidative lesions is largely unknown and there may be as yet unidentified oxidative lesions.
Considerable attention has focused on the cause of C→T transitions at CpG sites because this is a very common mutation, detected in a range of genetic diseases as well as in many human cancers (32–35). Numerous hypotheses have been offered for the molecular events leading to this mutation, all of which emphasize the importance of methylation of cytosine residues. Methylation increases the rate of hydrolytic deamination and also increases the reactivity of neighboring guanines to electrophiles (35–37). The rate of cytosine deamination in duplex DNA is extremely slow, and hydrolysis of 5-methylcytosine is only about twice as fast (36). Since deamination of 5-methylcytosine proceeds at such a low rate and since there are multiple repair systems that operate on T/G mismatches derived from deaminated 5-methylcytosine (38,39), it has been questioned if deamination of 5-methylcytosine is the only or even the prevailing mechanism that leads to CpG transitions (35).
A possible contribution of oxidative DNA damage to mutations at methylated CpGs has never been directly investigated. The oxidation of 5-methylcytosine may contribute to the high frequency of C→T transitions at CpG sequences. Oxygen radicals can react with 5-methylcytosine to oxidize the 5,6-double bond; the intermediate product, 5-methylcytosine glycol, then deaminates to form thymine glycol (40,41). Thymine glycol, although primarily a replication blocking lesion, can base pair with adenine (42), so oxidation of 5-methylcytosine is expected to result in a C→T transition (29,40). 5-Formyl-2′-deoxycytidine can also be produced in Fenton-type reactions (43) and may produce C→T transitions and C→A transversions (44).
ROS are generated during inflammatory responses by neutrophils and phagocytes (45). Of relevance to a possible involvement of oxidative stress in CpG mutagenesis may be the fact that there is a significant excess of CpG transitions in cancers associated with an inflammatory response, such as Schistosoma-associated bladder cancers, ulcerative colitis-associated colon cancers, and esophageal cancers in certain geographic areas (46–49).
In order to assess the role of oxidative DNA damage in CpG mutagenesis, we compared mutational spectra of Cu(II)/H2O2 in CpG-methylated and unmethylated pSP189 shuttle vectors replicated in human cells which were either deficient or proficient in nucleotide excision repair.
MATERIALS AND METHODS
Cell lines and plasmids
SV-40-transformed DNA repair-deficient human XP-A fibroblasts (XP12BE) and DNA repair-proficient fibroblasts (GM00637) were obtained from the American Type Culture Collection. The cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum in a 5% CO2 humidified incubator. The pSP189 plasmid (including a randomly generated 8-bp signature sequence at the 3′ end of the supF gene) was kindly provided by Michael Seidman (50).
Methylation and Cu(II)/ascorbate/H2O2 treatment of plasmid
The pSP189 plasmid was methylated in vitro using the CpG-specific DNA methylase SssI (New England Biolabs, Beverly, MA) according to the manufacturer’s instructions. Control DNA was mock-methylated in the absence of S-adenosylmethionine. Completion of methylation was confirmed by digesting an aliquot of the reaction mixture with the methylation-sensitive restriction endonuclease HpaII. Methylated and unmethylated pSP189 were incubated at room temperature for 30 min with 5 µM CuCl2. Chelex-treated potassium phosphate (pH 7.5), ascorbate and H2O2 were added to final concentrations of 1 mM, 100 µM, and various concentrations, respectively. After 30 min at room temperature with gentle rocking, the reaction was quenched by the addition of EDTA to a concentration of 2 mM, followed by precipitation of DNA in 0.3 M sodium acetate (pH 7.0) and 2 vol of cold ethanol. DNA was rinsed with 70% ethanol and air-dried.
Mutagenesis assay
The pSP189 shuttle vectors were transfected into cultures of repair-proficient and nucleotide excision repair-deficient human fibroblasts. Briefly, 5 × 105 cells were plated into 10 cm tissue-culture dishes in DMEM. Following a 16 h incubation of cells, a mixture of plasmid and FuGene 6 transfection agent (Roche, Indianapolis, IN) was added. After a 72-h incubation of the cells, the plasmid was rescued from the human cells by alkaline lysis. Cells were trypsinized, washed, and resuspended in suspension buffer (50 mM Tris–HCl, pH 8.0, 10 mM EDTA, 100 µg/ml RNase A), mixed with lysis buffer [0.2 M NaOH, 1% (w/v) SDS] and incubated on ice for 3–5 min, followed by the addition of neutralization buffer (3 M potassium acetate, pH 5.5). After a 15-min incubation at room temperature the mixture was centrifuged for 10 min at 16 000 g and the supernatant was extracted once with phenol–chloroform. Following ethanol precipitation, the DNA was re-suspended in DpnI reaction buffer and unreplicated plasmid was removed by digestion with DpnI which recognizes the bacterial adenine methylation pattern. Then the plasmid was electroporated into MB7070 bacteria, which carry a lacZ gene with an amber mutation. The transformed bacteria were diluted in 1 ml of SOC medium (2% tryptone, 0.5% yeast extract, 9 mM NaCl, 20 mM glucose) and plated on agar plates containing ampicillin, IPTG and X-Gal. After an overnight incubation at 37°C, wild-type (blue) and mutant (white) colonies were counted to determine the mutant frequency. Colonies containing a plasmid with a mutated supF gene were identified and the plasmids were purified and sequenced.
RESULTS
To determine a mutational spectrum by copper/H2O2 in a CpG-methylated target gene, we used the shuttle vector pSP189, which contains the supF gene as a mutational target. The vector was methylated in vitro at all CpG sequences, using the CpG-specific DNA methyltransferase SssI. In parallel experiments, pSP189 was mock-methylated in the absence of S-adenosylmethionine to produce an unmethylated counterpart. These methylated and unmethylated shuttle vectors were treated with a Fenton-type reaction including 5 µM CuCl2 and H2O2 at various concentrations. Using an aliquot of the treated plasmids, DNA damage (including damaged bases and abasic sites) was determined using the DNA glycosylase activity of the Nth or Fpg proteins. These DNA glycosylases recognize oxidized pyrimidines and purines, respectively (51). There was no substantial difference in DNA damage induction between the methylated and unmethylated plasmids. At the highest concentration of H2O2 used (100 µM), there was approximately one base lesion or strand break per 0.5 kb (data not shown). These DNA damage levels are necessary to get a sufficient increase in the mutation frequency in the small (100 bp) supF gene. In experiments using UVC irradiation, a dose of 1000 J/m2 is generally used (52–54) which produces approximately one cyclobutane pyrimidine dimer every 0.2–0.3 kb.
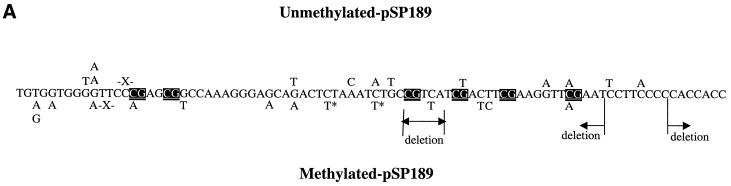
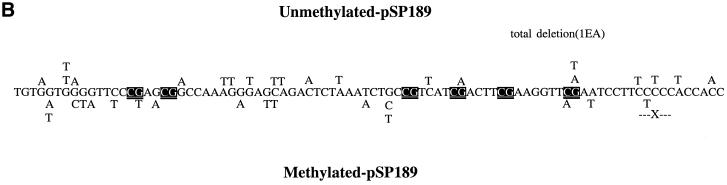
At 72 h after transfection into two different human fibroblast cell lines, the plasmids were rescued, and the DNA was cleaved with DpnI to remove unreplicated plasmids. Using HpaII and HhaI digestion, we found that the level of CpG methylation was largely (>80%) maintained in DpnI-treated DNA 72 h after transfection indicating that there is no active removal of methyl groups from the methylated plasmids and that the methylation pattern is at least partially conserved during DNA replication (data not shown). Conversely, the unmethylated plasmid did not undergo de novo methylation. The rescued plasmids were then electroporated into MB7070 bacteria, which carry a lacZ gene with an amber mutation. Plasmids were isolated from white colonies, and the supF gene was sequenced. The mutation frequencies in unmethylated and methylated CuCl2-treated supF vectors, in repair-proficient and nucleotide excision repair-deficient cells, were not substantially different (Table 1). Since there were too few colonies obtained in the absence of any treatment of the supF vector, we sequenced only mutations induced by 5 µM CuCl2 in the absence of H2O2 as a control (Fig. 1). The majority of such mutations were G/C→T/A transversions and C/G→T/A transitions (Fig. 1 and Table 2).
Table 1. Mutation frequencies induced by CuCl2/H2O2 in unmethylated and methylated supF shuttle vectors.
| Treatment |
Repair-deficient (XP-A) |
Repair-proficient (GM0637) |
||||||
|---|---|---|---|---|---|---|---|---|
| No. of mutants/no. of total colonies | Mutant frequencies | No. of mutants/no. of total colonies | Mutant frequencies | |||||
| Unmethylated | Methylated | Unmethylated | Methylated | Unmethylated | Methylated | Unmethylated | Methylated | |
| Control (no treatment) | 3/11 660 | 2/17 830 | 0.25 × 10–3 | 0.10 × 10–3 | 13/13 274 | 7/9024 | 0.98 × 10–3 | 0.80 × 10–3 |
| CuCl2 only (5 µM) | 14/15 190 | 20/25 920 | 0.92 × 10–3 | 0.8 × 10–3 | 24/16 584 | 18/11 421 | 1.40 × 10–3 | 1.60 × 10–3 |
| CuCl2 + H2O2 (1 µM) | 19/12 925 | 11/10 956 | 1.47 × 10–3 | 1.00 × 10–3 | 40/9042 | 32/8847 | 4.40 × 10–3 | 3.60 × 10–3 |
| CuCl2 + H2O2 (10 µM) | 60/16 996 | 42/13 008 | 3.53 × 10–3 | 3.23 × 10–3 | 47/9289 | 51/6840 | 5.10 × 10–3 | 7.50 × 10–3 |
| CuCl2 + H2O2 (100 µM) | 106/5842 | 86/4153 | 18.1 × 10–3 | 20.7 × 10–3 | 69/2760 | 92/5248 | 25.0 × 10–3 | 17.5 × 10–3 |
Figure 1.
Mutation spectra induced by 5 µM CuCl2 in the absence of H2O2 in unmethylated and CpG-methylated supF shuttle vectors from repair-deficient (XP-A) cells (A) and repair-proficient fibroblasts (B). The mutations introduced into the unmethylated plasmid are shown above the sequence, and the mutations introduced into the methylated plasmid are below the sequence. X, single-base deletions (X flanked by dashes, a nucleotide was deleted within a string of two or more identical nucleotides); * indicates the occurrence of two mutations in the same plasmid. CpG sequences are emphasized by white text on a black box.
Table 2. Types of mutations induced by CuCl2/H2O2 in CpG-methylated supF shuttle vectors.
| |
Repair-deficient (XP-A) |
Repair-proficient (GM0637) |
||||||
|---|---|---|---|---|---|---|---|---|
| SupF control (CuCl2 only)a | SupF + CuCl2/H2O2 (100 µM) | supF control (CuCl2 only) | supF + CuCl2/H2O2 (100 µM) | |||||
| Unmethylated | Methylated | Unmethylated | Methylated | Unmethylated | Methylated | Unmethylated | Methylated | |
| G→T | 7 (54)b | 3 (17) | 19 (35) | 27 (43) | 7 (33) | 6 (33) | 22 (38) | 25 (38) |
| G→A | 4 (31) | 8 (44) | 18 (33) | 19 (30) | 9 (43) | 7 (39) | 18 (31) | 22 (34) |
| G→C | 0 (0) | 0 (0) | 8 (15) | 6 (10) | 0 (0) | 2 (11) | 4 (7) | 7 (11) |
| A→T | 0 (0) | 1 (5) | 1 (2) | 0 (0) | 4 (19) | 2 (11) | 0 (0) | 1 (2) |
| A→C | 1 (8) | 1 (5) | 1 (2) | 0 (0) | 0 (0) | 0 (0) | 3 (5) | 1 (2) |
| A→G | 0 (0) | 1 (5) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (2) | 0 (0) |
| Deletion | 1 (8) | 4 (22) | 2 (4) | 2 (3) | 1 (5) | 1 (6) | 9 (16) | 4 (6) |
| Insertion | 0 (0) | 0 (0) | 2 (4) | 2 (3) | 0 (0) | 0 (0) | 0 (0) | 1 (2) |
| Tandem | 0 (0) | 0 (0) | 3 (6) | 7 (11) | 0 (0) | 0 (0) | 1 (2) | 2 (3) |
| Multiple | 0 (0) | 1 (5) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 (3) |
| Total | 13 (100) | 18 (100) | 54 (100) | 63 (100) | 21 (100) | 18 (100) | 58 (100) | 65 (100) |
| Mutations at CpG | 2/13 (15) | 2/18 (11) | 10/54 (19) | 20/63 (32) | 3/21 (14) | 2/18 (11) | 6/58 (10) | 18/65 (28) |
| G→T and G→A mutations at CpG | 2/11 (18) | 2/11 (18) | 9/37 (24) | 13/46 (28) | 3/16 (19) | 2/13 (15) | 5/40 (13) | 14/47 (30) |
| Tandem mutations at CpG (CG→TT) | 0 (0) | 0 (0) | 1/3 (33%) | 6/7 (86%) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
For nucleotide excision repair-deficient cells, mutant frequencies increased between CuCl2-only- and CuCl2/H2O2-treated shuttle vectors from 0.92 × 10–3 to 18.1 × 10–3 (20-fold) for the unmethylated DNA and from 0.77 × 10–3 up to 20.7 × 10–3 (27-fold) for the methylated DNA. For repair-proficient cells, mutant frequencies were also increased between CuCl2-only- and CuCl2/H2O2-treated shuttle vectors, from 1.40 × 10–3 to 25.0 × 10–3 (18-fold) for the unmethylated DNA and from 1.60 × 10–3 to 17.5 × 10–3 (11-fold) for the methylated DNA (Table 1). The mutant frequencies increased in a dose-dependent manner with increasing concentrations of H2O2.
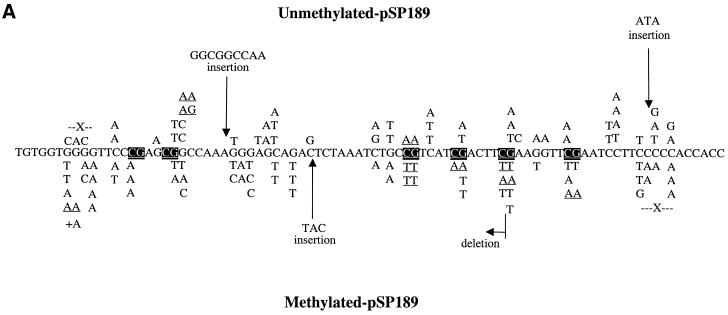
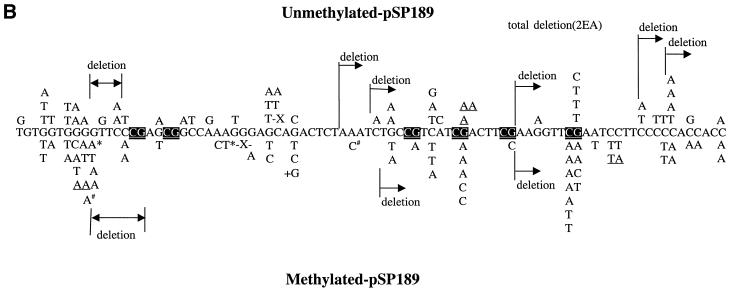
Since there was a sufficient increase in mutant frequency over background, we sequenced the plasmids obtained from the 100 µM H2O2 treatment. Figure 2 shows the mutational spectra obtained with unmethylated and methylated shuttle vectors from nucleotide excision repair-deficient and repair-proficient cells, respectively. In all cases, G→T transversions and G→A transitions were the predominant type of mutation (68–73%). For repair-deficient cells, 32% of all mutations were at CpGs in the methylated plasmid, whereas 19% were at these sites in the unmethylated plasmid. After methylation, 28% of all G→T transversions and G→A transitions were at CpG sites compared with 24% in unmethylated DNA (Table 2). Thus, there was a small increase in mutations at CpG sites after methylation of the shuttle vector. However, this did not reach statistical significance (P = 0.2; χ2 test).
Figure 2.
(A) Mutation spectra induced by Cu(II)/H2O2 in unmethylated and CpG-methylated supF shuttle vectors from nucleotide excision repair-deficient cells. The mutations introduced into the unmethylated plasmid are shown above the sequence, and the mutations introduced into the methylated plasmid are below the sequence. X, single-base deletions; +A, addition of 1 nt. Tandem mutations are underlined. CpG sequences are shown by white text on a black background. (B) Mutation spectra induced by Cu(II)/H2O2 in unmethylated and CpG-methylated supF shuttle vectors from repair-proficient cells. X, single-base deletions; +G, addition of 1 nt. Tandem mutations are underlined. * and # indicate the occurrence of two mutations in the same plasmid.
For repair-proficient cells, 28% of all mutations were at CpG sites in the methylated plasmid, whereas 10% of all mutations were at CpG sites in the unmethylated plasmid (P = 0.025; χ2 test). After methylation, 30% of all G→T transversions and G→A transitions were at CpG sites compared with 13% in unmethylated DNA (Table 2; P = 0.1; χ2 test). Thus, there was a trend linking an increased number of mutations at CpG sites and methylation, in particular for G→A transitions (Fig. 2B). Deletion mutations (11/123) from repair-proficient cells were more common than those (4/117) in repair-deficient cells.
However, the most striking difference between methylated and unmethylated shuttle vectors was that in the methylated plasmids 86% (6/7) of all tandem mutations were at CpG sites (Fig. 2A). All tandem mutations (6/6) at these CpG sites were CG→TT, whereas only 33% (one in three) of all tandem mutations were at CpG sites in the unmethylated plasmid. CG→TT mutations occurred only in nucleotide excision repair-deficient (XP-A) cells (Fig. 2A). An example of these tandem mutations is shown in Figure 3. In contrast, there was no tandem mutation at CpG sites in repair-proficient cells (Table 2 and Fig. 2B).
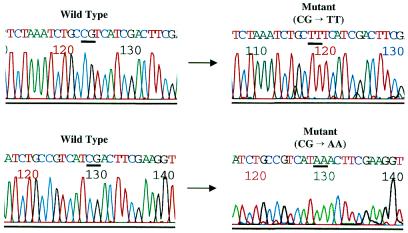
Figure 3.
CG→TT and CG→AA tandem mutations in CpG-methylated supF shuttle vectors treated with copper/H2O2. Sequence scans of two wild-type and mutant plasmids are shown.
DISCUSSION
Most of the work on the mutational specificity of oxidative DNA damage has been carried out with shuttle vectors. Invariably, the DNA used for these studies has been unmethy lated. Apparently, it is difficult to derive a mutational spectrum for oxidative DNA damage using an endogenous genomic locus. In the HPRT gene, there was less than a 3-fold induction above background and there often is a high proportion of large deletions (55). Studies with the lacI transgenic mouse cells have not been reported, presumably because the spontaneous mutation frequency in this system is too high.
We have analyzed the mutational spectra produced by treatment of unmethylated and CpG-methylated pSP189 shuttle vectors with Fenton reaction-type chemistry using Cu(II)/ascorbate/H2O2 and passage of the vectors through human fibroblasts. We find that, in accordance with previous studies, the majority of the mutations produced were G→A transitions and G→T transversions. Transition mutations at methylated CpG sequences are very common in the p53 gene in cancers related to inflammatory responses. Examples are colon cancers associated with ulcerative colitis (47) and bladder cancers associated with Schistosoma infection (46). Besides the currently accepted model that invokes deamination of 5-methylcytosine in methylated CpG sequences as the main mechanism of CpG transitions (33,35), other theories have suggested that oxidative DNA damage, perhaps via deamination of 5-methylcytosine glycol, might contribute to CpG mutagenesis (40). Our data indicate that oxidative stress, in the form of copper/H2O2-induced DNA damage, can increase the frequency of CpG transitions, albeit only in repair-proficient cells, but does not induce clear transition mutation hotspots at methylated CpG sequences. It remains to be determined if nitric oxide, also produced in inflammatory responses, may produce such mutations more efficiently. Earlier studies have not yet provided a uniform answer to this question (56–58).
Tandem base substitution mutations are generally very rare events. They have most commonly been observed in experiments with UV irradiation (59,60). CC→TT tandem mutations have long been recognized as a UV signature mutation. However, CC→TT mutations also have been described in experiments with ROS (61). In previous studies on the mutational specificity of oxidative DNA damage, CC→TT mutations were the only tandem events that were commonly observed. These earlier studies did not use mutational reporter genes that contained methylated CpG sequences. We found that methylated CpGs frequently undergo tandem CG→TT mutations after treatment of CpG-methylated DNA with copper and H2O2. This particular type of mutation has rarely been reported previously. It may be asked if the observed mCG→TT tandem mutations are specific for copper/H2O2-induced DNA damage. We cannot address this point with the presently available data. However, metal ion-dependent H2O2-induced DNA damage was more sequence specific than metal specific (62).
At present, the origin of the tandem mCG→TT tandem mutations is unknown. They occurred only in cells deficient in nucleotide excision repair (XP-A cells). Such tandem mutations may be generated via several pathways. These may include, but are not limited to, (i) a single-base lesion that promotes a double misincorporation event during DNA replication, (ii) cross-linked adjacent DNA bases, or (iii) adjacent double-base lesions (tandem lesions) that would promote the incorporation of two consecutive adenines opposite to the two lesions. For pathway (i) there is in fact evidence that 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxo-dG) can cause base substitution errors at neighboring template bases when copied by DNA polymerase β (63). These base substitution errors were sequence dependent. Such events might occur when a MutY class of enzyme removes adenine paired opposite 8-oxo-dG and if the resulting gap is >1 nt long and is filled by DNA polymerase β. For pathway (ii), intrastrand cross-links have been observed upon exposure of salmon sperm DNA to Fenton-type oxygen radical-generating systems (64). The copper Fenton system generated the highest total yield of these DNA lesions. After 32P post-labeling, two spots, common to all these Fenton systems, were the major oxidation products. Similar Fenton-type treatment of the purine dinucleotides dApdG and dApdA resulted in products that were chromatographically identical to the two major products generated in DNA. However, methylated CpG dinucleotides were not specifically investigated. Other cross-linked lesions have been observed (65,66). Pathway (iii), polymerase bypass of adjacent double-base lesions, has not yet been studied extensively. However, tandem lesions, in which two adjacent bases are modified but not cross-linked have been reported after treatment of DNA with ionizing radiation or with metal-catalyzed H2O2 reactions (67,68). These double-base lesions included 8-oxo-dG adjacent to a pyrimidine nucleoside which was degraded to a formamido remnant (68,69). In fact, a very high percentage of adenine insertion was observed in shuttle vectors when a formamylamine residue was part of a vicinal lesion containing 8-oxo-dG (70). Such vicinal lesions may be at the origin of mCG→TT tandem mutations. Finally, the possibility exists that 8-oxo-dG could promote deamination of a 5′-neighboring 5-methylcytosine base.
The occurrence of CG→TT mutations at methylated CpG sites predominantly in nucleotide excision repair-deficient cells would suggest that the underlying lesion is normally repaired quite efficiently by the nucleotide excision repair process. Nucleotide excision repair can operate on base damage produced by DNA oxidation, for instance on 8-oxo-dG and thymine glycol residues (71). If a cross-linked lesion were involved in generating the mCG→TT mutations, the repair of such a lesion by nucleotide excision repair would become even more plausible, since such lesions are probably unrecognizable by base excision repair enzymes and nucleotide excision repair has the specificity to recognize cross-linked DNA bases. Also, tandem base lesions may not be good substrates for base excision repair and may be funneled into the nucleotide excision repair pathway. For instance, a tandem dihydrouracil lesion was not repairable by the Escherichia coli and human endonuclease III N-glycosylases (72,73). On the other hand, both E.coli Fpg and endonuclease III proteins were able to cleave oligonucleotide chains that contained the vicinal formylamine–8-oxo-dG lesion (74).
Xeroderma pigmentosum patients belonging to complementation group XP-A are unable to perform nucleotide excision repair. In severe cases, they suffer from progressive neurodegeneration, possibly because of slow accumulation of unrepaired DNA damage in the brain (75). The brain has a high level of oxygen metabolism which may lead to accumulation of oxidative DNA damage in non-regenerating neuronal cells. The DNA excision-repair defect of xeroderma pigmentosum prevents removal of a class of oxygen free radical-induced base lesions (76). One such lesion was later shown to be the 5′,8-purine cyclodeoxynucleoside adduct (77). We suggest that the underlying oxidative lesions which cause mCG→TT tandem mutations selectively in XP-A cells, but not in NER-proficient cells, may also be candidates for a form of DNA damage that might explain the progressive neurodegeneration seen in XP-A individuals. Continued accumulation of this lesion might ultimately lead to transcriptional interference and cytotoxicity.
In summary, we describe a novel type of mutation, CG→TT tandem mutations at methylated CpG sequences, which can be produced by oxidative DNA damage via a H2O2 and transition metal ion pathway. The lesions that produce such peculiar mutations are apparently subject to nucleotide excision repair. In future experiments, the exact chemical nature of these lesions, possibly consisting of tandem or cross-linked base damage, will need to be determined.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Steven Bates for assistance with cell culture and John Termini for discussions. This work was supported by a grant from the National Institutes of Health (CA84469) to G.P.P.
REFERENCES
- 1.Ames B.N. and Gold,L.S. (1991) Endogenous mutagens and the causes of aging and cancer. Mutat. Res., 250, 3–16. [DOI] [PubMed] [Google Scholar]
- 2.Lindahl T. (1993) Instability and decay of the primary structure of DNA. Nature, 362, 709–715. [DOI] [PubMed] [Google Scholar]
- 3.Feig D.I., Reid,T.M. and Loeb,L.A. (1994) Reactive oxygen species in tumorigenesis. Cancer Res., 54, 1890s–1894s. [PubMed] [Google Scholar]
- 4.Wang D., Kreutzer,D.A. and Essigmann,J.M. (1998) Mutagenicity and repair of oxidative DNA damage: insights from studies using defined lesions. Mutat. Res., 400, 99–115. [DOI] [PubMed] [Google Scholar]
- 5.Marnett L.J. (2000) Oxyradicals and DNA damage. Carcinogenesis, 21, 361–370. [DOI] [PubMed] [Google Scholar]
- 6.Boveris A. (1977) Mitochondrial production of superoxide radical and hydrogen peroxide. Adv. Exp. Med. Biol., 78, 67–82. [DOI] [PubMed] [Google Scholar]
- 7.Badwey A. and Kanovsky,M.L. (1980) Active oxygen species and the functions of phagocytic leukocytes. Annu. Rev. Biochem., 49, 695–726. [DOI] [PubMed] [Google Scholar]
- 8.Termini J. (2000) Hydroperoxide-induced DNA damage and mutations. Mutat. Res., 450, 107–124. [DOI] [PubMed] [Google Scholar]
- 9.Schraufstatter I.U., Hyslop,P.A., Hinshaw,D.B., Spragg,R.G., Sklar,L.A. and Cochrane,C.G. (1986) Hydrogen peroxide-induced injury of cells and its prevention by inhibitors of poly(ADP-ribose) polymerase. Proc. Natl Acad. Sci. USA, 83, 4908–4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dizdaroglu M., Nackerdien,Z., Chao,B.C., Gajewski,E. and Rao,G. (1991) Chemical nature of in vivo DNA base damage in hydrogen peroxide-treated mammalian cells. Arch. Biochem. Biophys., 285, 388–390. [DOI] [PubMed] [Google Scholar]
- 11.Dizdaroglu M. (1992) Oxidative damage to DNA in mammalian chromatin. Mutat. Res., 275, 331–342. [DOI] [PubMed] [Google Scholar]
- 12.Loeb L.A., James,E.A., Waltersdorph,A.M. and Klebanoff,S.J. (1988) Mutagenesis by the autoxidation of iron with isolated DNA. Proc. Natl Acad. Sci. USA, 85, 3918–3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blakely W.F., Fuciarelli,A.F., Wegher,B.J. and Dizdaroglu,M. (1990) Hydrogen peroxide-induced base damage in deoxyribonucleic acid. Radiat. Res., 121, 338–343. [PubMed] [Google Scholar]
- 14.Tkeshelashvili L.K., McBride,T., Spence,K. and Loeb,L.A. (1991) Mutation spectrum of copper-induced DNA damage. J. Biol. Chem., 266, 6401–6406. [PubMed] [Google Scholar]
- 15.Sagripant J.L. and Kraemer,K.H. (1989) Site-specific oxidative DNA damage at polyguanosines produced by copper plus hydrogen peroxide. J. Biol. Chem., 264, 1729–1734. [PubMed] [Google Scholar]
- 16.Stoewe R. and Prutz,W.A. (1987) Copper-catalyzed DNA damage by ascorbate and hydrogen peroxide: kinetics and yield. Free Radic. Biol. Med., 3, 97–105. [DOI] [PubMed] [Google Scholar]
- 17.Fujimoto S., Adachi,Y., Ishimitsu,S. and Ohara,A. (1986) Release of bases from deoxyribonucleic acid by ascorbic acid in the presence of Cu2+. Chem. Pharm. Bull. (Tokyo), 34, 4848–4851. [DOI] [PubMed] [Google Scholar]
- 18.Reed C.J. and Douglas,K.T. (1989) Single-strand cleavage of DNA by Cu(II) and thiols: a powerful chemical DNA-cleaving system. Biochem. Biophys. Res. Commun., 162, 1111–1117. [DOI] [PubMed] [Google Scholar]
- 19.Tachon P. (1989) Ferric and cupric ions requirement for DNA single-strand breakage by H2O2. Free Radic. Res. Commun., 7, 1–10. [DOI] [PubMed] [Google Scholar]
- 20.Goldstein S. and Czapski,G. (1986) The role and mechanism of metal ions and their complexes in enhancing damage in biological systems or in protecting these systems from the toxicity of O2. Free Radic. Biol. Med., 2, 3–11. [DOI] [PubMed] [Google Scholar]
- 21.Chevion M. (1988) A site-specific mechanism for free radical induced biological damage: the essential role of redox-active transition metals. Free Radic. Biol. Med., 5, 27–37. [DOI] [PubMed] [Google Scholar]
- 22.Drouin R., Rodriguez,H., Gao,S.W., Gebreyes,Z., O’Connor,T.R., Holmquist,G.P. and Akman,S.A. (1996) Cupric ion/ascorbate/hydrogen peroxide-induced DNA damage: DNA-bound copper ion primarily induces base modifications. Free Radic. Biol. Med., 21, 261–273. [DOI] [PubMed] [Google Scholar]
- 23.Liang Q. and Dedon,P.C. (2001) Cu(II)/H2O2-induced DNA damage is enhanced by packaging of DNA as a nucleosome. Chem. Res. Toxicol., 14, 416–422. [DOI] [PubMed] [Google Scholar]
- 24.Holmquist G.P. (1998) Endogenous lesions, S-phase-independent spontaneous mutations and evolutionary strategies for base excision repair. Mutat. Res., 400, 59–68. [DOI] [PubMed] [Google Scholar]
- 25.Burcham P.C. (1999) Internal hazards: baseline DNA damage by endogenous products of normal metabolism. Mutat. Res., 443, 11–36. [DOI] [PubMed] [Google Scholar]
- 26.Moraes E.C., Keyse,S.M. and Tyrell,R.M. (1990) Mutagenesis by hydrogen peroxide treatment of mammalian cells: a molecular analysis. Carcinogenesis, 11, 283–293. [DOI] [PubMed] [Google Scholar]
- 27.Schaaper R.M. and Dunn,R.L. (1991) Spontaneous mutation in the Escherichia coli lacI gene. Genetics, 129, 317–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feig D.I., Sowers,L.C. and Loeb,L.A. (1994) Reverse chemical mutagenesis: identification of the mutagenic lesions resulting from reactive oxygen species-mediated damage to DNA. Proc. Natl Acad. Sci. USA, 91, 6609–6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kreutzer D.A. and Essigmann,J.M. (1998) Oxidized, deaminated cytosines are a source of C→T transitions in vivo. Proc. Natl Acad. Sci. USA, 95, 3578–3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wood M.L., Dizdaroglu,M., Gajewski,E. and Essigmann,J.M. (1990) Mechanistic studies of ionizing radiation and oxidative mutagenesis: genetic effects of a single 8-hydroxyguanine (7-hydro-8-oxoguanine) residue inserted at a unique site in a viral genome. Biochemistry, 31, 7024–7032. [DOI] [PubMed] [Google Scholar]
- 31.Shibutani S., Takeshita,M. and Grollman,A.P. (1991) Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature, 349, 431–434. [DOI] [PubMed] [Google Scholar]
- 32.Cooper D.N. and Youssoufian,H. (1988) The CpG dinucleotide and human genetic disease. Hum. Genet., 78, 151–155. [DOI] [PubMed] [Google Scholar]
- 33.Gonzalgo M.L. and Jones,P.A. (1997) Mutagenic and epigenetic effects of DNA methylation. Mutat. Res., 386, 107–118. [DOI] [PubMed] [Google Scholar]
- 34.Hainaut P. and Hollstein,M. (2000) p53 and human cancer: the first ten thousand mutations. Adv. Cancer Res., 77, 81–137. [DOI] [PubMed] [Google Scholar]
- 35.Pfeifer G.P. (2000) p53 mutational spectra and the role of methylated CpG sequences. Mutat. Res., 450, 155–166. [DOI] [PubMed] [Google Scholar]
- 36.Shen J.C., Rideout,W.M.,III and Jones,P.A. (1994) The rate of hydrolytic deamination of 5-methylcytosine in double-stranded DNA. Nucleic Acids Res., 22, 972–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Denissenko M.F., Chen,J.X., Tang,M.S. and Pfeifer,G.P. (1997) Cytosine methylation determines hot spots of DNA damage in the human P53 gene. Proc. Natl Acad. Sci. USA, 94, 3893–3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neddermann P., Gallinari,P., Lettieri,T., Schmid,D., Truong,O., Hsuan,J.J., Wiebauer,K. and Jiricny,J. (1996) Cloning and expression of human G/T mismatch-specific thymine-DNA glycosylase. J. Biol. Chem., 271, 12767–12774. [DOI] [PubMed] [Google Scholar]
- 39.Hendrich B., Hardeland,U., Ng,H.-H., Jiricny,J. and Bird,A. (1999) The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature, 401, 301–304. [DOI] [PubMed] [Google Scholar]
- 40.Zuo S., Boorstein,R.J. and Teebor,G.W. (1995) Oxidative damage to 5-methylcytosine in DNA. Nucleic Acids Res., 25, 3239–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bienvenu C. and Cadet,J. (1996) Synthesis and kinetic study of the deamination of the cis diastereomers of 5,6-dihydroxy-5,6-dihydro-5-methyl-2′-deoxycytidine. J. Org. Chem., 61, 2632–2637. [DOI] [PubMed] [Google Scholar]
- 42.Evans J., Maccabee,M., Hatahet,Z., Courcelle,J., Bockrath,R., Ide,H. and Wallace,S. (1993) Thymine ring saturation and fragmentation products: lesion bypass, misinsertion and implications for mutagenesis. Mutat. Res., 299, 147–156. [DOI] [PubMed] [Google Scholar]
- 43.Murata-Kamiya N., Kamiya,H., Karino,N., Ueno,Y., Kaji,H., Matsuda,A. and Kasai,H. (1999) Formation of 5-formyl-2′-deoxycytidine from 5-methyl-2′-deoxycytidine in duplex DNA by Fenton-type reactions and gamma-irradiation. Nucleic Acids Res., 27, 4385–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karino N., Ueno,Y. and Matsuda,A. (2001) Synthesis and properties of oligonucleotides containing 5-formyl-2′-deoxycytidine: in vitro DNA polymerase reactions on DNA templates containing 5-formyl-2′-deoxycytidine. Nucleic Acids Res., 29, 2456–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jackson A.L. and Loeb,L.A. (2001) The contribution of endogenous sources of DNA damage to the multiple mutations in cancer. Mutat. Res., 477, 7–21. [DOI] [PubMed] [Google Scholar]
- 46.Warren W., Biggs,P.J., El-Baz,M., Ghoneim,M.A., Stratton,M.R. and Venitt,S. (1995) Mutations in the p53 gene in schistosomal bladder cancer: a study of 92 tumours from Egyptian patients and a comparison between mutational spectra from schistosomal and non-schistosomal urothelial tumours. Carcinogenesis, 16, 1181–1189. [DOI] [PubMed] [Google Scholar]
- 47.Hussain S.P., Amstad,P., Raja,K., Ambs,S., Nagashima,M., Bennett,W.P., Shields,P.G., Ham,A.J., Swenberg,J.A., Marrogi,A.J. and Harris,C.C. (2000) Increased p53 mutation load in noncancerous colon tissue from ulcerative colitis: a cancer-prone chronic inflammatory disease. Cancer Res., 60, 3333–3337. [PubMed] [Google Scholar]
- 48.Biramijamal F., Allameh,A., Mirbod,P., Groene,H.J., Koomagi,R. and Hollstein,M. (2001) Unusual profile and high prevalence of p53 mutations in esophageal squamous cell carcinomas from northern Iran. Cancer Res., 61, 3119–3123. [PubMed] [Google Scholar]
- 49.Sepehr A., Taniere,P., Martel-Planche,G., Zia’ee,A.A., Rastgar-Jazii,F., Yazdanbod,M., Etemad-Moghadam,G., Kamangar,F., Saidi,F. and Hainaut,P. (2001) Distinct pattern of TP53 mutations in squamous cell carcinoma of the esophagus in Iran. Oncogene, 20, 7368–7374. [DOI] [PubMed] [Google Scholar]
- 50.Canella K.A. and Seidman,M.M. (2000) Mutation spectra in supF: approaches to elucidating sequence context effects. Mutat. Res., 450, 61–73. [DOI] [PubMed] [Google Scholar]
- 51.Cunningham R.P. (1997) DNA glycosylases. Mutat. Res., 383, 189–196. [DOI] [PubMed] [Google Scholar]
- 52.Tobi S.E., Levy,D.D., Seidman,M.M. and Kraemer,K.H. (1999) Sequence-dependent mutations in a shuttle vector plasmid replicated in a mismatch repair deficient human cell line. Carcinogenesis, 20, 1293–1301. [DOI] [PubMed] [Google Scholar]
- 53.Marionnet C., Armier,J., Sarasin,A. and Stary,A. (1998) Cyclobutane pyrimidine dimers are the main mutagenic DNA photoproducts in DNA repair-deficient trichothiodystrophy cells. Cancer Res., 58, 102–108. [PubMed] [Google Scholar]
- 54.Otoshi E., Yagi,T., Mori,T., Matsunaga,T., Nikaido,O., Kim,S.T., Hitomi,K., Ikenaga,M. and Todo,T. (2000) Respective roles of cyclobutane pyrimidine dimers, (6–4)photoproducts, and minor photoproducts in ultraviolet mutagenesis of repair-deficient xeroderma pigmentosum A cells. Cancer Res., 60, 1729–1735. [PubMed] [Google Scholar]
- 55.Oller A.R. and Thilly,W.G. (1992) Mutational spectra in human B-cells. Spontaneous, oxygen and hydrogen peroxide-induced mutations at the hprt gene. J. Mol. Biol., 228, 813–826. [DOI] [PubMed] [Google Scholar]
- 56.Schmutte C., Rideout,W.M.,III, Shen,J.C. and Jones,P.A. (1994) Mutagenicity of nitric oxide is not caused by deamination of cytosine or 5-methylcytosine in double-stranded DNA. Carcinogenesis, 15, 2899–2903. [DOI] [PubMed] [Google Scholar]
- 57.Felley-Bosco E., Mirkovitch,J., Ambs,S., Mace,K., Pfeifer,A., Keefer,L.K. and Harris,C.C. (1995) Nitric oxide and ethylnitrosourea: relative mutagenicity in the p53 tumor suppressor and hypoxanthine-phosphoribosyltransferase genes. Carcinogenesis, 16, 2069–2074. [DOI] [PubMed] [Google Scholar]
- 58.Souici A.C., Mirkovitch,J., Hausel,P., Keefer,L.K. and Felley-Bosco,E. (2000) Transition mutation in codon 248 of the p53 tumor suppressor gene induced by reactive oxygen species and a nitric oxide-releasing compound. Carcinogenesis, 21, 281–287. [DOI] [PubMed] [Google Scholar]
- 59.Sage E. (1993) Distribution and repair of photolesions in DNA: genetic consequences and the role of sequence context. Photochem. Photobiol., 57, 163–174. [DOI] [PubMed] [Google Scholar]
- 60.Pfeifer G.P. (1997) Formation and processing of UV photoproducts: effects of DNA sequence and chromatin environment. Photochem. Photobiol., 65, 270–283. [DOI] [PubMed] [Google Scholar]
- 61.Reid T.M. and Loeb,L.A. (1993) Tandem double CC→TT mutations are produced by reactive oxygen species. Proc. Natl Acad. Sci. USA, 90, 3904–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rodriguez H., Holmquist,G.P., D’Agostino,R., Keller,J. and Akman,S.A. (1997) Metal ion-dependent hydrogen peroxide-induced DNA damage is more sequence specific than metal dependent. Cancer Res., 57, 2394–2403. [PubMed] [Google Scholar]
- 63.Efrati E., Tocco,G., Eritja,R., Wilson,S.H. and Goodman,M.F. (1999) ‘Action-at-a-distance’ mutagenesis. 8-oxo-7,8-dihydro-2′-deoxyguanosine causes base substitution errors at neighboring template sites when copied by DNA polymerase beta. J. Biol. Chem., 274, 15920–15926. [DOI] [PubMed] [Google Scholar]
- 64.Lloyd D.R., Phillips,D.H. and Carmichael,P.L. (1997) Generation of putative intrastrand cross-links and strand breaks in DNA by transition metal ion-mediated oxygen radical attack. Chem. Res. Toxicol., 10, 393–400. [DOI] [PubMed] [Google Scholar]
- 65.Box H.C., Budzinski,E.E., Dawidzik,J.D., Wallace,J.C., Evans,M.S. and Gobey,J.S. (1996) Radiation-induced formation of a crosslink between base moieties of deoxyguanosine and thymidine in deoxygenated solutions of d(CpGpTpA). Radiat. Res., 145, 641–643. [PubMed] [Google Scholar]
- 66.Box H.C., Budzinski,E.E., Dawidzik,J.B., Wallace,J.C. and Iijima,H. (1998) Tandem lesions and other products in X-irradiated DNA oligomers. Radiat. Res., 149, 433–439. [PubMed] [Google Scholar]
- 67.Box H.C., Freund,H.G., Budzinski,E.E., Wallace,J.C. and Maccubbin,A.E. (1995) Free radical-induced double base lesions. Radiat. Res., 141, 91–94. [PubMed] [Google Scholar]
- 68.Box H.C. (2001) Double lesions are produced in DNA oligomer by ionizing radiation and by metal-catalyzed H2O2 reactions. Radiat. Res., 155, 634–636. [DOI] [PubMed] [Google Scholar]
- 69.Douki T., Riviere,J. and Cadet,J. (2002) DNA Tandem lesions containing 8-oxo-7,8-dihydroguanine and formamido residues arise from intramolecular addition of thymine peroxyl radical to guanine. Chem. Res. Toxicol., 15, 445–454. [DOI] [PubMed] [Google Scholar]
- 70.Gentil A., Le Page,F., Cadet,J. and Sarasin,A. (2000) Mutation spectra induced by replication of two vicinal oxidative DNA lesions in mammalian cells. Mutat. Res., 452, 51–56. [DOI] [PubMed] [Google Scholar]
- 71.Reardon J.T., Bessho,T., Kung,H.C., Bolton,P.H. and Sancar,A. (1997) In vitro repair of oxidative DNA damage by human nucleotide excision repair system: possible explanation for neurodegeneration in xeroderma pigmentosum patients. Proc. Natl Acad. Sci. USA, 94, 9463–9468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Venkhataraman R., Donald,C.D., Roy,R., You,H.J., Doetsch,P.W. and Kow,Y.W. (2001) Enzymatic processing of DNA containing tandem dihydrouracil by endonucleases III and VIII. Nucleic Acids Res., 29, 407–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weinfeld M., Rasouli-Nia,A., Chaudhry,M.A. and Britten,R.A. (2001) Response of base excision repair enzymes to complex DNA lesions. Radiat. Res., 156, 584–589. [DOI] [PubMed] [Google Scholar]
- 74.Bourdat A.G., Gasparutto,D. and Cadet,J. (1999) Synthesis and enzymatic processing of oligodeoxynucleotides containing tandem base damage. Nucleic Acids Res., 27, 1015–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bootsma D., Kraemer,K.H., Cleaver,J.E. and Hoeijmakers,J.H.J. (1998) Nucleotide excision repair syndromes: xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy. In Vogelstein,B. and Kinzler,K.W. (eds), The Genetic Basis of Human Cancer. McGraw-Hill, New York, pp. 245–274.
- 76.Satoh M.S., Jones,C.J., Wood,R.D. and Lindahl,T. (1993) DNA excision-repair defect of xeroderma pigmentosum prevents removal of a class of oxygen free radical-induced base lesions. Proc. Natl Acad. Sci. USA, 90, 6335–6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kuraoka I., Bender,C., Romieu,A., Cadet,J., Wood,R.D. and Lindahl,T. (2000) Removal of oxygen free-radical-induced 5′,8-purine cyclodeoxynucleosides from DNA by the nucleotide excision-repair pathway in human cells. Proc. Natl Acad. Sci. USA, 97, 3832–3837. [DOI] [PMC free article] [PubMed] [Google Scholar]