Abstract
RNA molecules exhibit complex structures in which a large fraction of the bases engage in non-Watson–Crick base pairing, forming motifs that mediate long-range RNA–RNA interactions and create binding sites for proteins and small molecule ligands. The rapidly growing number of three-dimensional RNA structures at atomic resolution requires that databases contain the annotation of such base pairs. An unambiguous and descriptive nomenclature was proposed recently in which RNA base pairs were classified by the base edges participating in the interaction (Watson–Crick, Hoogsteen/CH or sugar edge) and the orientation of the glycosidic bonds relative to the hydrogen bonds (cis or trans). Twelve basic geometric families were identified and all 12 have been observed in crystal structures. For each base pairing family, we present here the 4 × 4 ‘isostericity matrices’ summarizing the geometric relationships between the 16 pairwise combinations of the four standard bases, A, C, G and U. Whenever available, a representative example of each observed base pair from X-ray crystal structures (3.0 Å resolution or better) is provided or, otherwise, theoretically plausible models. This format makes apparent the recurrent geometric patterns that are observed and helps identify isosteric pairs that co-vary or interchange in sequences of homologous molecules while maintaining conserved three-dimensional motifs.
INTRODUCTION
The past 10 years have witnessed an explosion in RNA structure determination at the atomic level. An increasing number of structures of important, functionally diverse RNA molecules have been determined, including the rRNAs (5S, 16S and 23S), many tRNAs, a variety of ribozymes, part of the SRP RNA, portions of viral RNA genomes and a variety of RNA aptamers bound to their ligands (Table 1). The complexity of many of these structures challenges the ability of individual scientists to understand and visualize the diversity of interactions. Nonetheless, careful examination reveals recurrent motifs (1–4). The most fundamental motif is the edge-to-edge hydrogen bonding interaction between two bases. The prototype is the standard (canonical) Watson–Crick base pair, in which two bases interact with their Watson–Crick edges, with the glycosidic bonds oriented cis relative to the axis of the interaction (Fig. 1). Yet even the early crystallographic studies of nucleic acids revealed other modes of interaction (5). In the l980s, with only the atomic structures of tRNAs and crystal packing interactions of small oligonucleotides to work from, compilations of non-Watson–Crick pairs appeared (6). Such compilations grouped interactions according to base type (purine–purine, purine–pyrimidine and pyrimidine–pyrimidine) rather than geometry. Recently, we proposed a classification of RNA base pairs based on geometry (7). This approach is justified by the need to easily (and eventually automatically) identify recurrent structural motifs in new crystal structures and to predict the occurrences of motifs through comparative sequence analysis. This approach will lead in turn to higher quality sequence alignments of homologous RNA molecules. RNA homology modeling is based on two main assumptions (8–10). The first is that the secondary and tertiary structures are much more highly conserved than primary sequence. The second is that, just as for Watson–Crick pairs in secondary structures, those compensatory base substitutions that retain the non-Watson–Crick pairs in three-dimensional structure elements and motifs are more likely to be observed than those that cannot be accommodated. These ideas have been applied by other workers to the problem of identifying non-Watson–Crick interactions and RNA motifs using comparative sequence analysis, especially in the context of base triples (11–13).
Table 1. X-ray structures from which base pairs shown in Figures 2–14 were taken.
| NDB File | Structure description | Reference |
|---|---|---|
| AR0001 | RNA internal loop | (35) |
| AR0005 | RNA double helix | (30) |
| AR0006 | RNA 16mer duplex | (36) |
| AR0008 | 14 bp RNA duplex | (37) |
| DR0005 | Biotin-binding aptamer | (38) |
| PR0004 | EF-TU–cysteinyl tRNA complex | (39) |
| PR0005 | HDV ribozyme | (40) |
| PR0015 | 23S rRNA L11 site (E.coli) | (41) |
| PR0021 | Signal recognition particle (SRP) RNA | (42) |
| PR0022 | RNA-binding protein NOVA-2/RNA | (43) |
| PTR009 | Glu-tRNA synthetase mutant–RNA complex | (44) |
| PTR012 | EF-TU–Phe-tRNA complex | (45) |
| RR0009 | 23S rRNA L11 site (Thermotoga maritima) | (46) |
| RR0030 | 30S ribosomal subunit from T.thermophilus | (47) |
| RR0033 | 50S ribosomal subunit from H.marismortui | (48) |
| RR0051 | 50S ribosomal subunit from D.radiodurans | (49) |
| TRNA07 | Yeast tRNA (ASP) | (8) |
| TRNA09 | Yeast tRNA (PHE) | (8) |
| UR0001 | Leadzyme x-tal contact | (50) |
| UR0002 | Sarcin loop from rat 28S rRNA | (51) |
| UR0003 | Group I intron | (52) |
| UR0004 | Frameshifting pseudoknot | (53) |
| UR0008 | Cobalamin aptamer | (54) |
| URF042 | RNA hexamer | (55) |
| URL050 | RNA dodecamer | (56) |
| URL051 | Symmetric internal loop | (57) |
| URL064 | Loop E from 5S RNA | (58) |
| URX035 | Hammerhead ribozyme | (59) |
| URX053 | Group I intron | (33) |
Figure 1.
(Left) Identification of edges in the RNA bases. (Right) cis versus trans orientation of glycosidic bonds.
Here, we present matrices of observed and predicted edge-to-edge interactions based on exhaustive examination of medium to high resolution (<3.0 Å) RNA crystal structures, including the recently published structures of the ribosome. In several important cases, NMR structural work has provided the first observations of non-Watson–Crick base pairs (14–19). However, NMR geometries are not always unambiguously determined and as all base pairs have subsequently been found in X-ray crystal structures, we chose our examples from the latter. These data provide the basis for implementing algorithms to automatically identify and classify motifs mediating tertiary interactions in complex RNA structures. The data we present should also assist in the interpretation of RNA interference (20), modification (21) and instant evolution data (22), i.e. the assignment of possible geometries for a given interaction identified through these types of experiments. Finally, these data are useful and crucial to the generation of accurate structural alignments of homologous RNA sequences.
MATERIALS AND METHODS
This work relied on visual examination of high resolution X-ray crystal structures to determine hydrogen bonding patterns. Structures were obtained from the Nucleic Acid Database (http://ndbserver.rutgers.edu/NDB) and the Protein Data Bank (http://www.rcsb.org/pdb/) and were manipulated with the Swiss PDB Viewer program, available from http://www.expasy.ch/spdbv/ (23). Hydrogen bonding diagrams were prepared using the Chem3D and ChemDraw Pro programs (CambridgeSoft Corporation). Diagrams were prepared using Canvas (Deneba Software).
Figures 2–12 are available on the Internet at either http://www.bgsu.edu/departments/chem/RNA/pages or http://www-ibmc.u-strasbg.fr/upr9002/westhof/. The BGSU website also provides interactive three-dimensional views of each base pair using the CHIME plug-in.
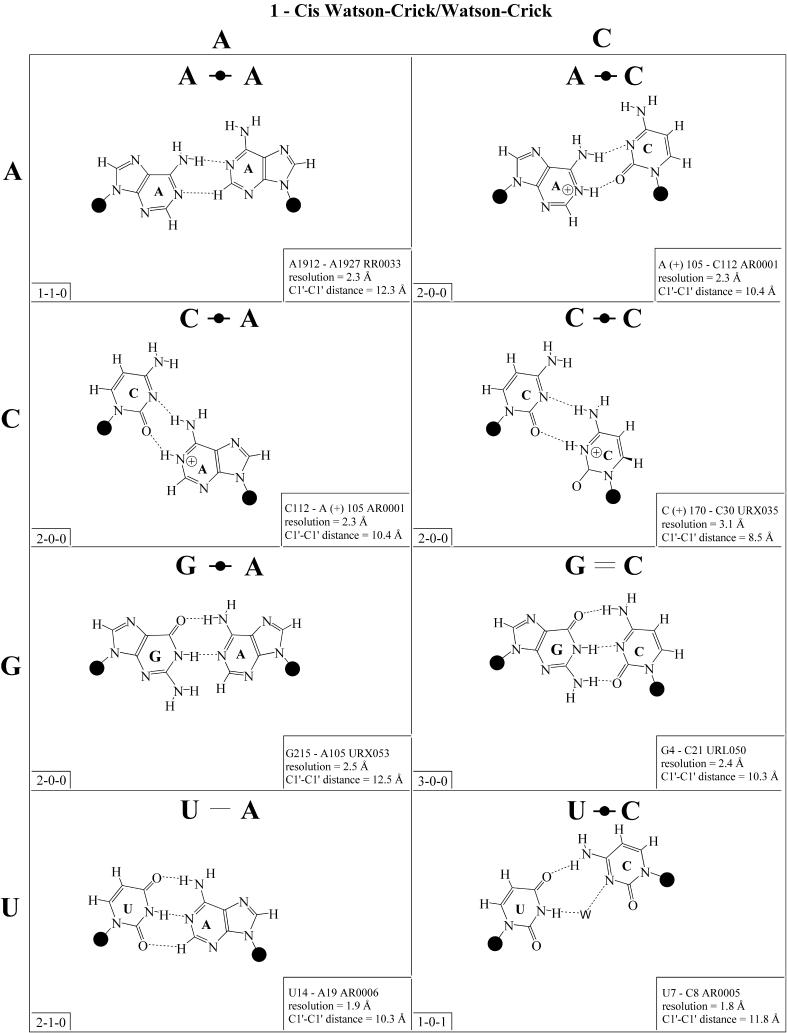
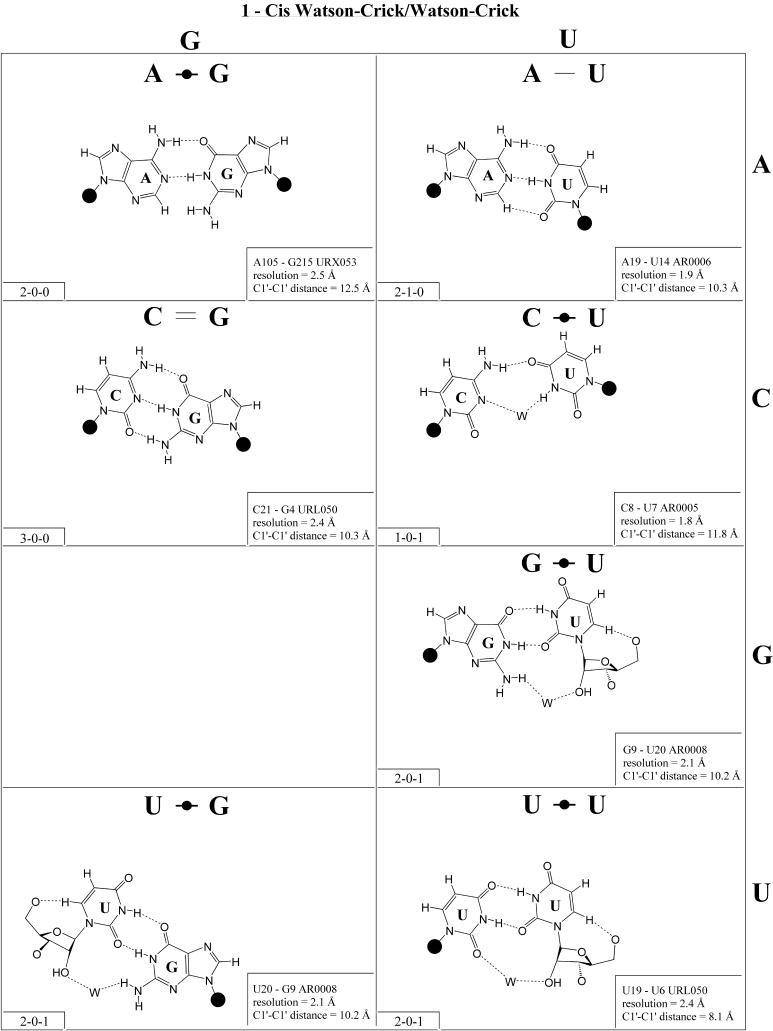
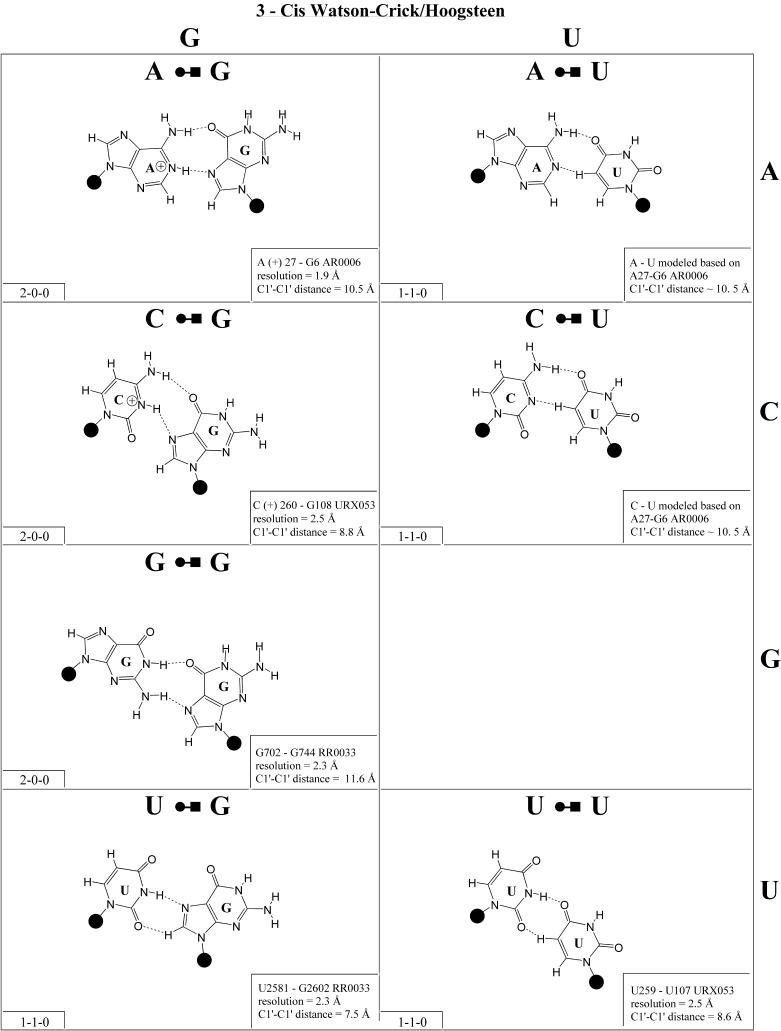
Figure 2.
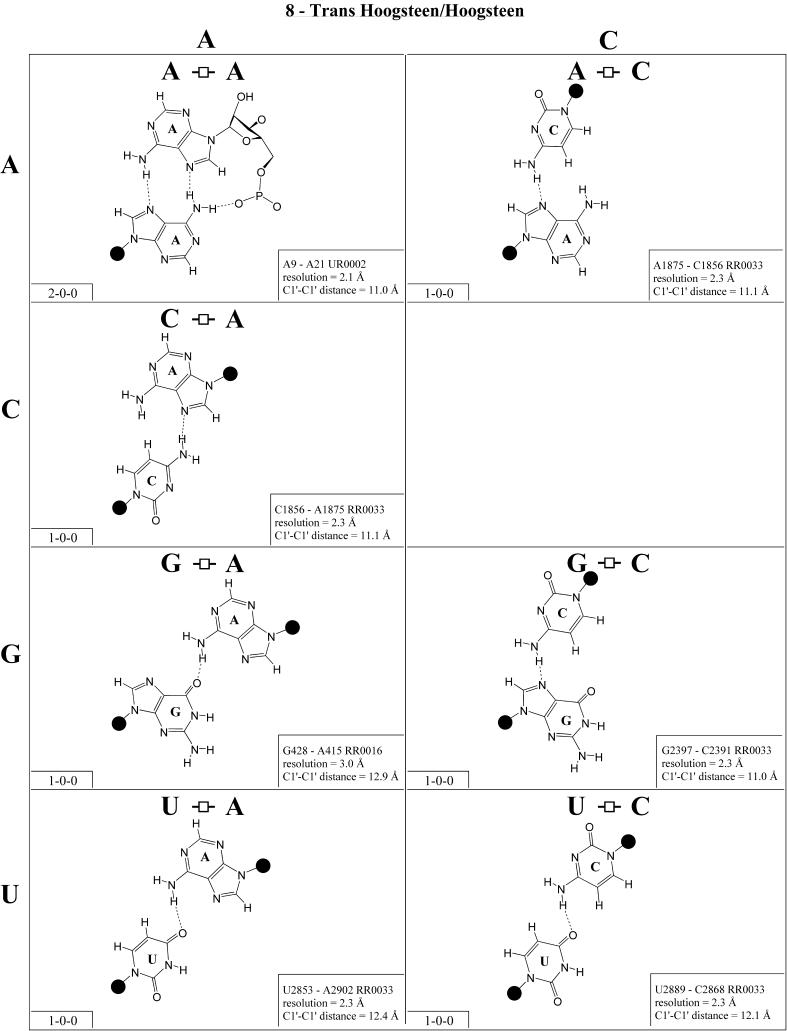
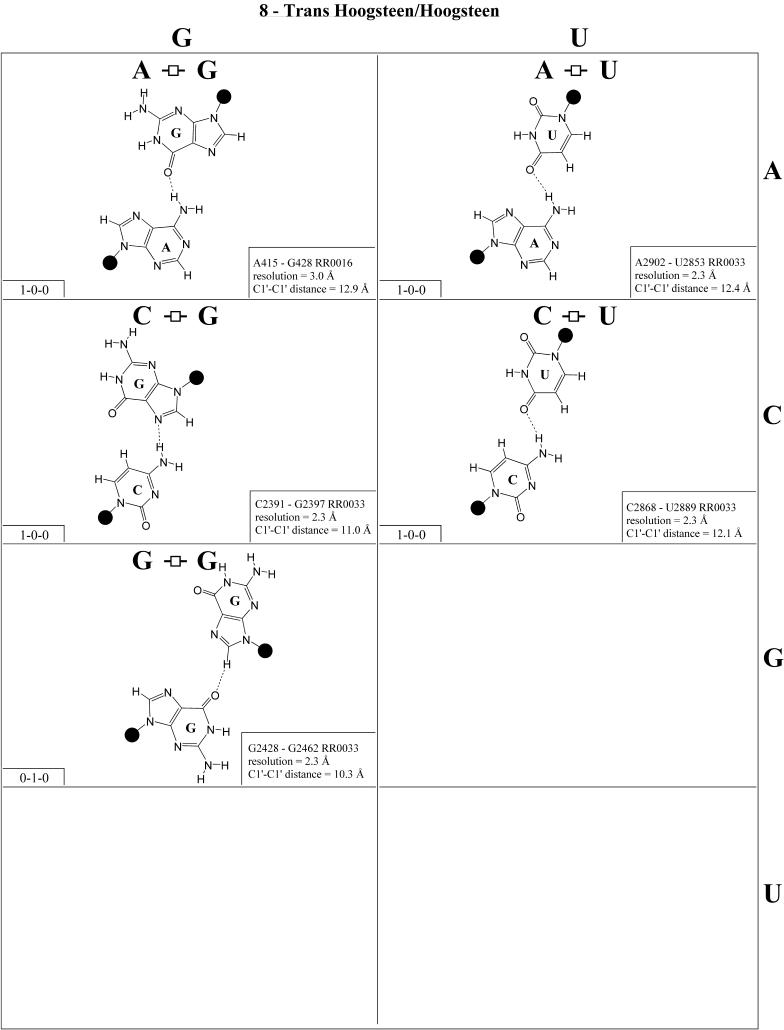
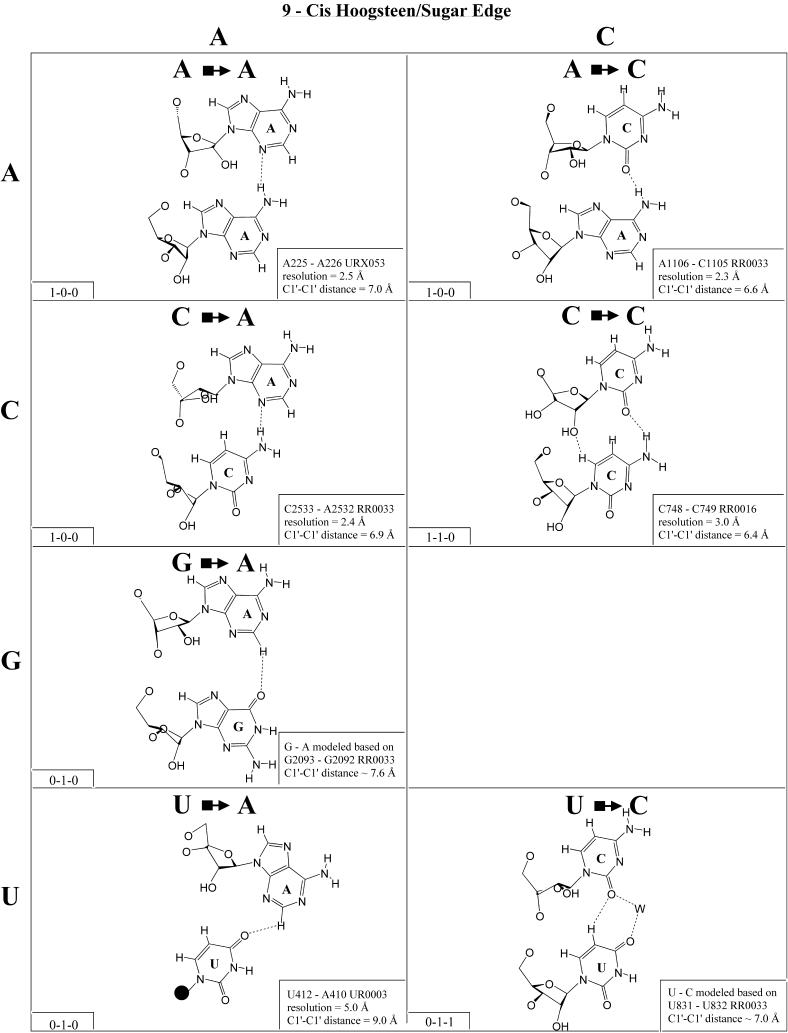
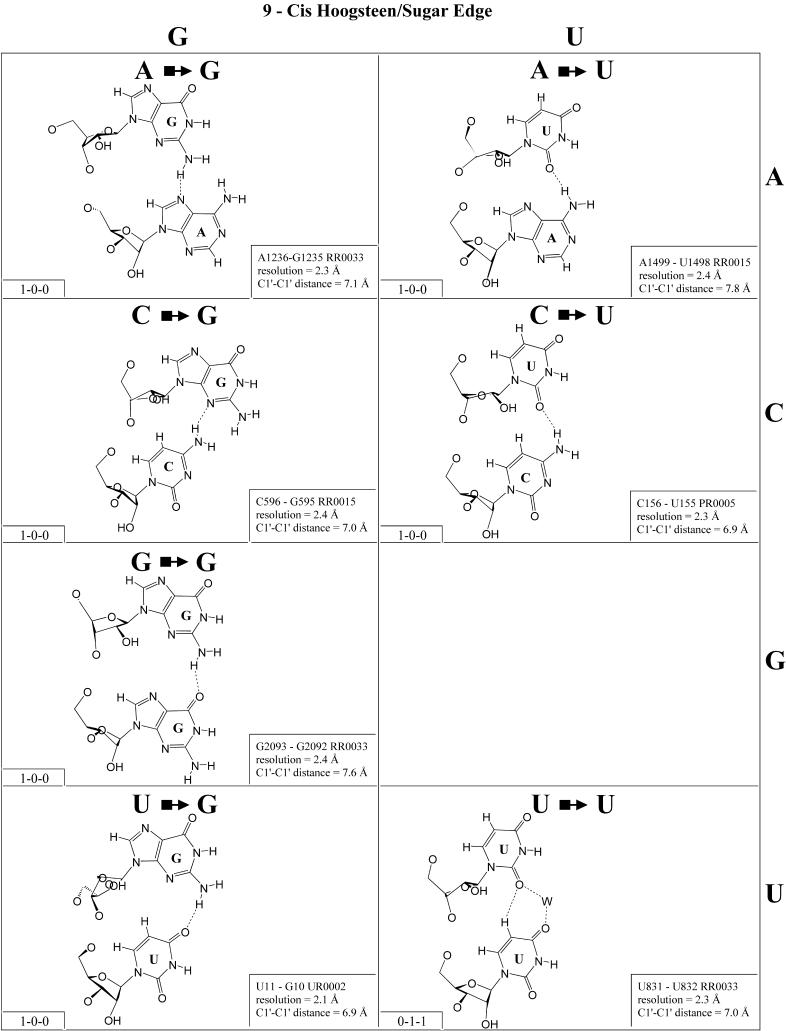
(Opposite and above) 4 × 4 matrix displaying observed base pairs belonging to the cis Watson–Crick/Watson–Crick family. The canonical Watson–Crick pairs comprise the diagonal of the matrix. Symbols used in Figures 2–14 employ circles to designate Watson–Crick edges, squares for Hoogsteen or pyrimidine CH edges, and triangles for sugar edges. Solid symbols indicate cis base pairs and open symbols trans base pairs. In the lower left-hand corner of each panel in Figures 2–14, numbers describing the hydrogen bonding are provided. The first is the number of observed or potential hydrogen bonds between two nitrogen- or oxygen-containing groups, i.e. normal hydrogen bonds. The second is the number of hydrogen bonds involving polarized C-H groups (i.e. AH2, RH8, YH5 or YH6). The third is the number of bridging water molecules.
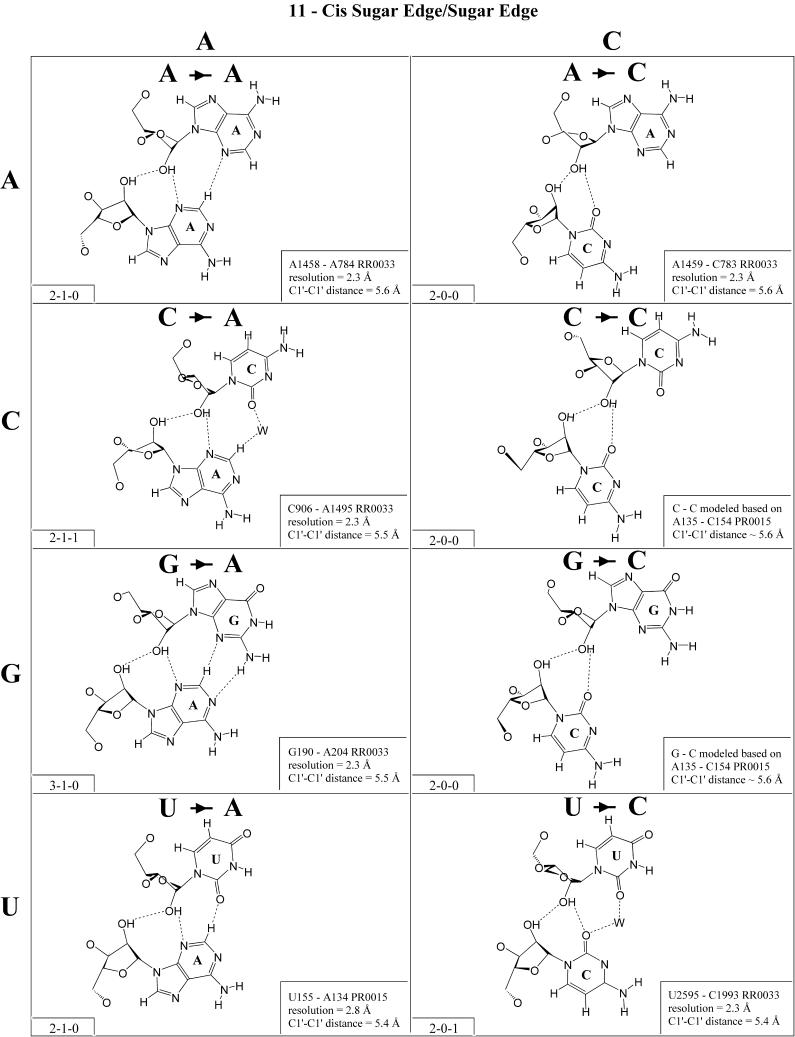
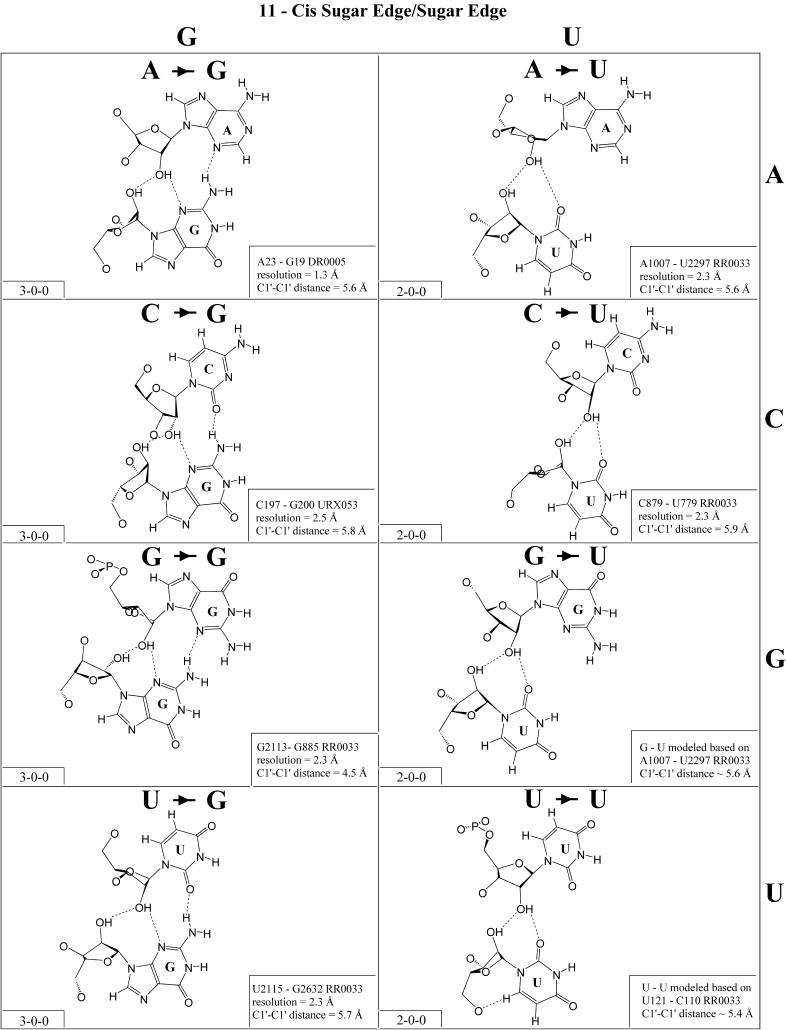
Figure 12.
(Opposite and above) Observed and modeled base pairs of the cis sugar edge/sugar edge family.
RESULTS
In previous work, we showed that RNA bases (purines and pyrimidines) interact edge-to-edge using any one of three edges (Fig. 1); consequently, all base pairs involving two or more edge-to-edge hydrogen bonds belong to one of 12 geometric families (7). Each family is identified by the edges involved in the interaction and the relative orientations of the glycosidic bonds of the interacting nucleotides, cis or trans (Table 2). When the glycosidic bonds of the two bases assume the default anti configuration, the relative strand orientations are those given in the third column of Table 2 (24). In Figures 2–13, representative examples are provided of observed base pairs for each geometrical family. When the two interacting edges are different (for example Watson–Crick and Hoogsteen), a historically based priority rule is invoked (Watson–Crick > Hoogsteen > sugar edge) so the base identified with each row of a given matrix is the one interacting with the higher priority edge. Thus, in Family 3, cis Watson–Crick/Hoogsteen (Fig. 4), all the pairings in the first row involve adenine interacting with its Watson–Crick edge while all the pairings in the first column involve adenine interacting with its Hoogsteen edge. In each panel of Figures 2–13, the higher priority base appears to the left, oriented so that its Watson–Crick edge faces to the right. A list of referenced NDB files with primary references is provided in Table 1.
Table 2. The 12 families of edge-to-edge base pairs formed by nucleic acid bases, defined by the relative orientations of the glycosidic bonds of the interacting bases (column 2) and the edges used in the interaction (column 3).
Recently proposed symbols for designating each base pair family in secondary structure diagrams are given in column 4. Circles designate Watson–Crick edges, squares Hoogsteen or pyrimidine CH edges, and triangles sugar edges. Solid symbols indicate cis base pairs and open symbols trans base pairs. The local strand orientation that occurs when both bases are in the default anti conformation are in column 5; a syn orientation for one of the nucleotides would imply a reversal of orientation; for the global orientation, the stereochemistry at the phosphate groups has to be considered.
Figure 13.
(Opposite and above) Observed and modeled base pairs of the trans sugar edge/sugar edge family.
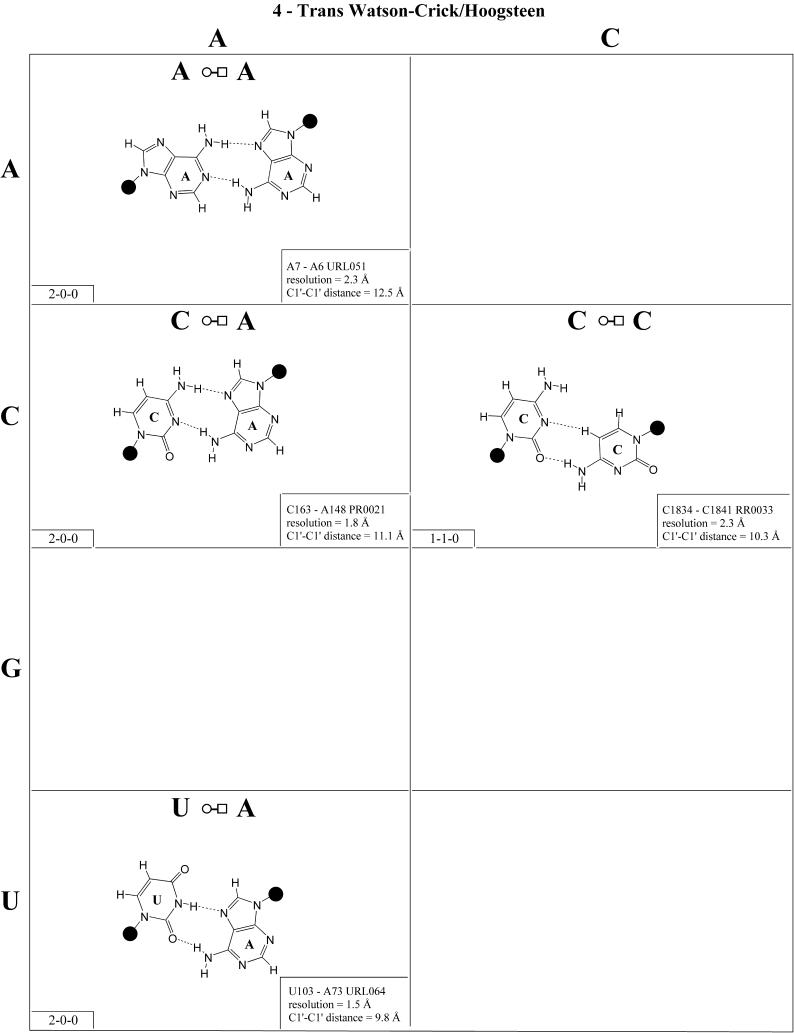
Figure 4.
(Opposite and above) Observed and modeled base pairs of the cis Watson–Crick/Hoogsteen family.
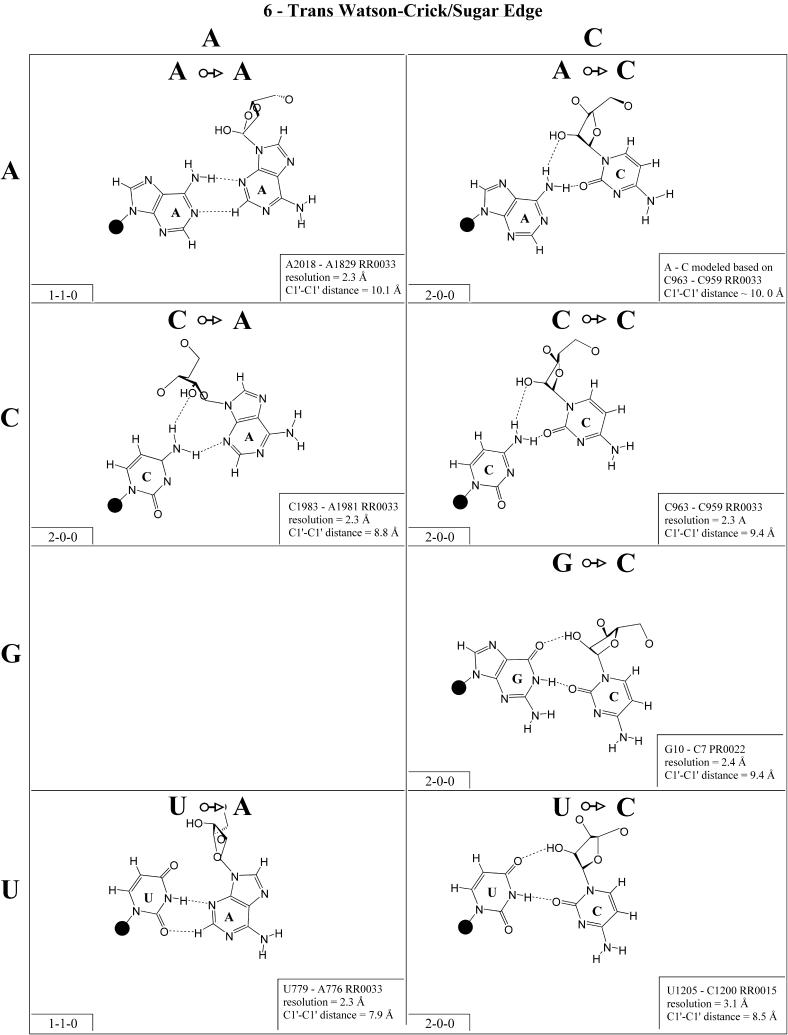
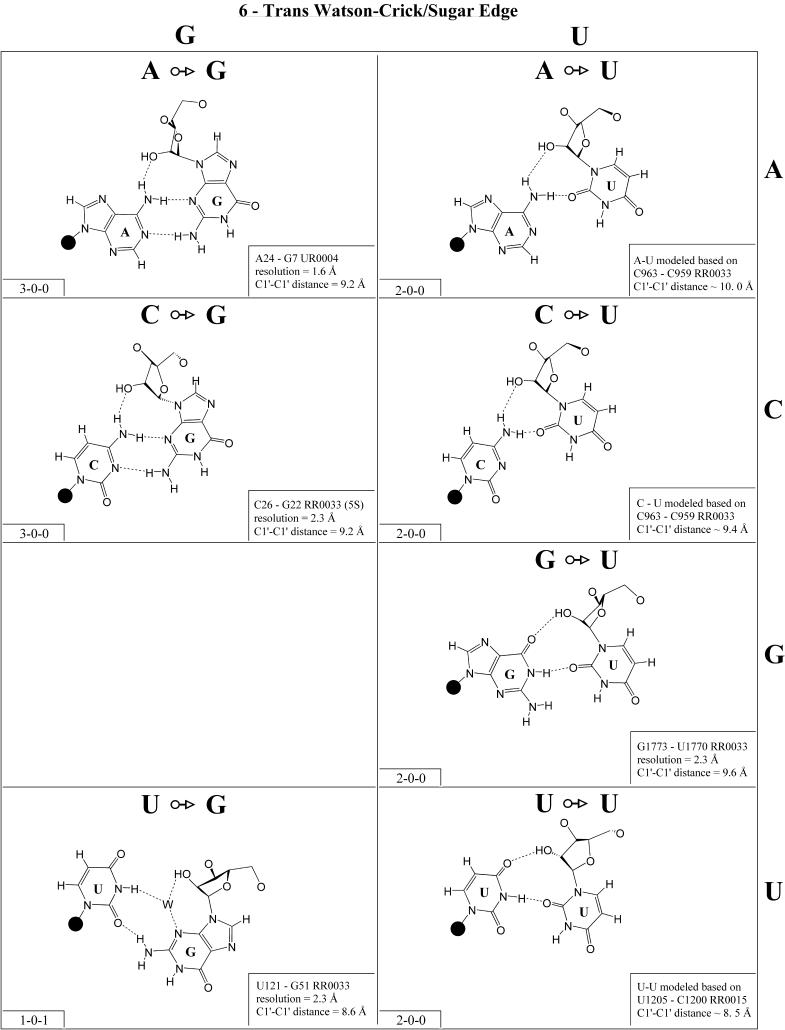
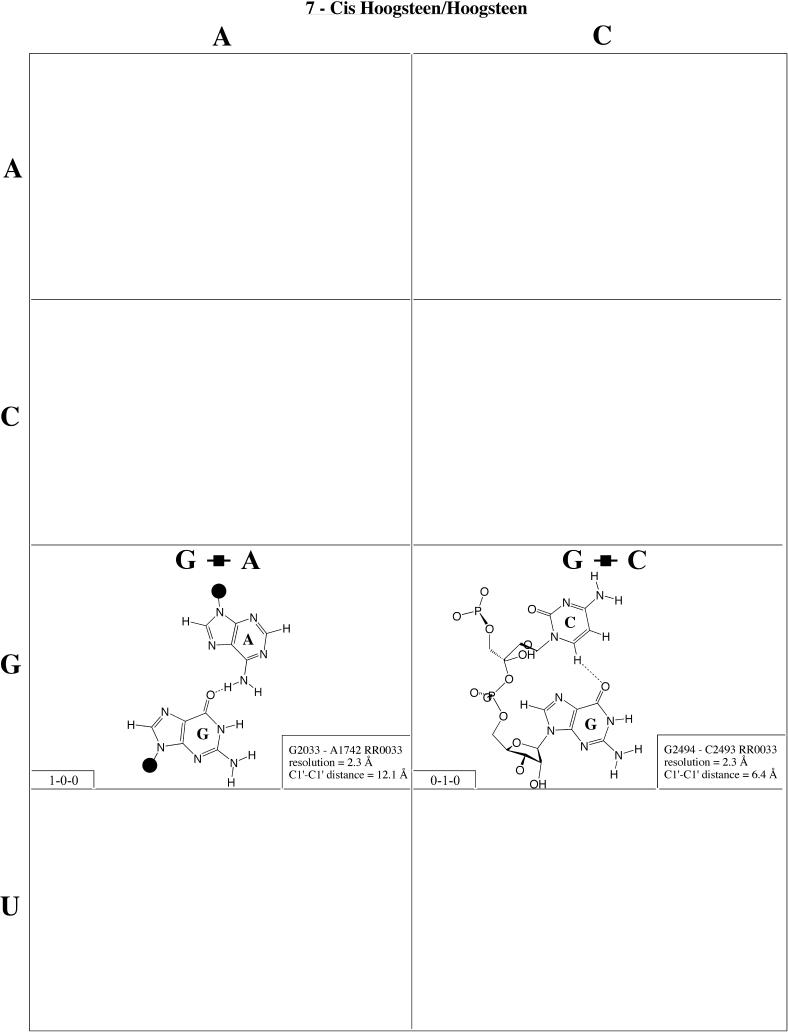
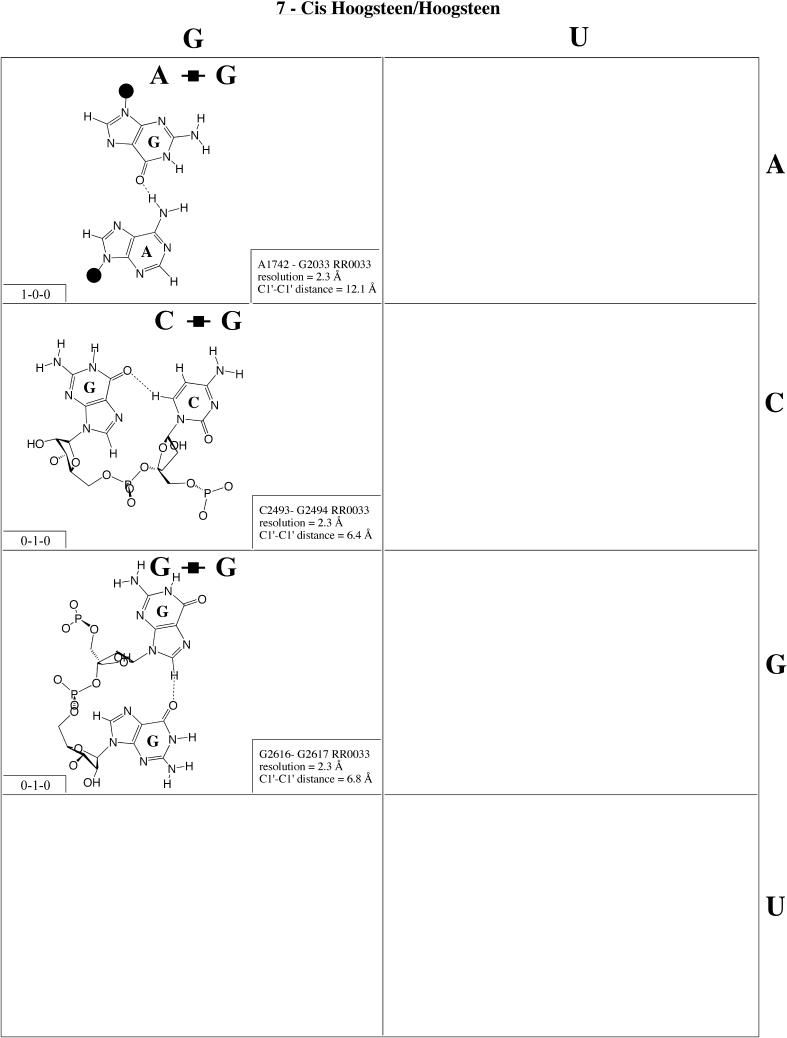
For each base pair in Figures 2–13, the source (NDB filename) and resolution of the X-ray data (in Å), as well as the C1′–C1′ distance (also in Å) are provided in the lower right corner. As higher resolution examples are obtained of each base pair, they may be conveniently substituted for the pair shown. In those cases where an example of a base pair was not found in a crystal structure, the pair was modeled using known structures as templates and basic principles of hydrogen bonding. The pairs used as templates for modeled pairs are noted in the lower right of the panel. Blank spaces in Figures 2–13 indicate base combinations for which no example has been found and for which no reasonable model could be proposed based on current knowledge. Sugar ring atoms are drawn for those cases where the O2′ participates (or could potentially participate) in hydrogen bonding to the base (or ribose O2′) of the partner nucleotide. Otherwise the entire sugar moiety is designated with a closed circle.
The sugar edge of purine and pyrimidine nucleotides includes the 2′-OH, when the glycosidic bond of the nucleotide is in the usual anti domain. Thus, when one or both bases interact with the sugar edge, hydrogen bonds can form with the 2′-OH group(s) acting either as donor(s) or acceptor(s). In fact, in some of the cis sugar edge/sugar edge pairs, no direct base–base hydrogen bonds occur at all. Since the position of the 2′-OH hydrogen cannot be inferred from X-ray structures of nucleic acids, the 2′-OH is drawn as a single unit in Figures 2–13. The C-H–O hydrogen bond is well established in structural chemistry on the basis of detailed analyses of small molecule crystallography (25). Thus, we also mark interactions involving adenine H2, purine (R) H8 and pyrimidine (Y) H5 or H6 as hydrogen bonds in Figures 2–13. For hydrogen bonds not involving a C-H the maximum distance between heavy atoms is 3.4 Å and for hydrogen bonds involving C-H bonds the maximum distance should be <3.9 Å. Bridging water molecules are integral elements of a number of non-Watson–Crick base pairs (26,27). Water acts as both hydrogen bond donor and acceptor in these structures but, again, the actual positions of the hydrogen atoms cannot be inferred from available crystal structures so water molecules are simply designated by W in Figures 2–13. Information regarding the hydrogen bonding is provided in the lower left-hand corner of each panel in Figures 2–13. Three numbers are given: (i) the number of observed or potential hydrogen bonds between two nitrogen or oxygen containing groups (i.e. normal hydrogen bonds); (ii) the number of hydrogen bonds involving polarized C-H groups (i.e. AH2, RH8, YH5 or YH6); (iii) the number of bridging water molecules.
It is well known that adenine can be protonated at the N1 position and cytidine at the N3 position (6). The proton cannot be directly observed in nucleic acid crystal structures, but a number of interactions cannot be readily rationalized without assuming protonation. In some rare instances, experimental or theoretical support has been obtained for protonation (28). Therefore, wherever it makes chemical sense, we have indicated protonated adenine and cytidine in Figures 2–13.
Isostericity
The three-dimensional structures of homologous RNA molecules change much more slowly than their sequences in the course of evolution (as is also true for homologous proteins). By definition, homologous molecules share a common biological origin and a conserved function. Random point mutations in structurally crucial parts of RNA molecules are accommodated by natural selection when they affect the three-dimensional structure little or when they are compensated by further mutations. Such co-variations, when they occur at positions that are cis Watson–Crick paired, have been applied with great success to predict the occurrence of conserved double helices in homologous RNA molecules. The isostericity of the standard base pairs, A-U, G=C, C=G and U-A in Figure 2, is the fundamental property. The C1′–C1′ distance in each of these pairs is identical (Fig. 2, lower right of each panel), as are the relative orientations of the glycosidic bonds, considered as vectors in three-dimensional space. When two base pairs display nearly the same C1′–C1′ distance and have their glycosidic bonds oriented in the same way, they can replace each other without drastically changing the three-dimensional path and relative geometric orientations of the phosphate–sugar backbones. We denote such base pairs as ‘isosteric’, although this does not necessarily imply that the two base pairs occupy the same total volume of space, and in many cases this, in fact, does not hold.
Isostericity matrices
Generally, base pairs belonging to the same geometric family exhibit very similar relative orientations of their glycosidic bonds, implying the maintenance of the local orientations of the strands and thus of the three-dimensional organization. However, in the general case, all possible base pairs belonging to a single geometric family are not isosteric to each other because the C1′–C1′ distances may be quite different. Thus, the C1′–C1′ distance can be used to group the base pairs within each geometric family into isosteric subsets or subfamilies. The recognition of subsets of isosteric base pairs within a family serves the purpose of identifying pairs that can substitute for each other while preserving the three-dimensional structure, crucial information for three-dimensional modeling of tertiary interactions, prediction of motifs, and the generation and refinement of accurate structural alignments. In the following, each geometric family is considered in turn and the isosteric subsets of base pairs identified from Figures 2–13 are summarized in the form of isostericity matrices in Tables 3–5.
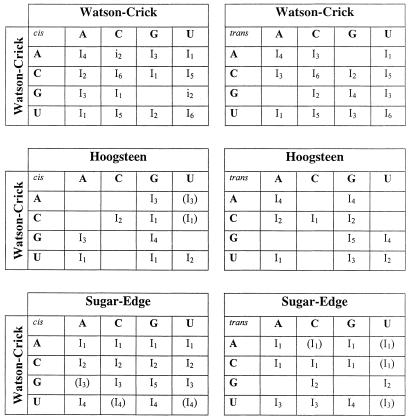
Table 3. Isostericity matrices for base pairing Families 1–6.
First row, cis and trans Watson–Crick/Watson–Crick; second row, cis and trans Watson–Crick/Hoogsteen; third row, cis and trans Watson–Crick/sugar edge. Parentheses indicate modeled interactions, not yet observed in high resolution X-ray structures. In the cis Watson–Crick/Watson–Crick table i2 is used to designate the wobble pairs G/U and A(+)/C as they are not isosteric to U/G and C/A(+); unlike the standard Watson–Crick pairs, the wobble pairs are not self-isosteric.
Table 5. Isostericity matrix for the cis bifurcated geometry.
Cis Watson–Crick/Watson–Crick (Family 1). We begin with the base pairs belonging to the cis Watson–Crick/Watson–Crick geometric family, shown in Figure 2. The (canonical) Watson–Crick pairs, A-U, U-A, G=C and C=G, form an isosteric subfamily, which we designate I1 in the isostericity matrix for this family, shown in Table 3 (first row, left). Likewise the wobble pairs G/U and A(+)/C form an isosteric subgroup I2. However, unlike I1, the wobble pairs are not self-isosteric and, thus, the wobble pairs U/G and C/A(+) comprise a third isosteric subset, which, however, is related to I2 and is therefore designated i2. In certain contexts the wobble pairs can substitute for canonical cis Watson–Crick/Watson–Crick pairs within a helix. We can say that they are compatible with the canonical pairs. However, substitution of a G/U or A(+)/C pair for a U/G or C/A(+) results in a larger structural perturbation in a helical context (29) and thus R/Y are usually not compatible with Y/R wobble pairs.
The pairs A/G and G/A constitute a fourth subfamily, designated I3. Like the canonical pairs (I1) they are self-isosteric. I4 consists solely of the A/A pair, since the G/G combination cannot occur in this geometry. C/U and U/C are self-isosteric and comprise subset I5. Interestingly, in high resolution structures this pair is consistently observed with an inserted water molecule, bridging between the imino positions of the bases, perhaps because of repulsion between the O2 atoms of the interacting pyrimidines (30). Consequently the C1′–C1′ distance for the water-inserted C/U pair is significantly larger than expected for a pyrimidine–pyrimidine pair, and close to that of cis Watson–Crick/Watson–Crick A/G. Interestingly, U/C is observed to co-vary with A/G in the anticodon stem of tRNAs (27). Thus, in certain contexts C/U and A/G are compatible.
The isosteric wobble pairs C(+)/C and U/U, both of which have been observed, comprise the final isosteric subgroup of the cis Watson–Crick/Watson–Crick geometric family, designated I6. The C1′–C1′ distance in this subfamily is significantly smaller than that of any of the others, including the water-inserted U/C.
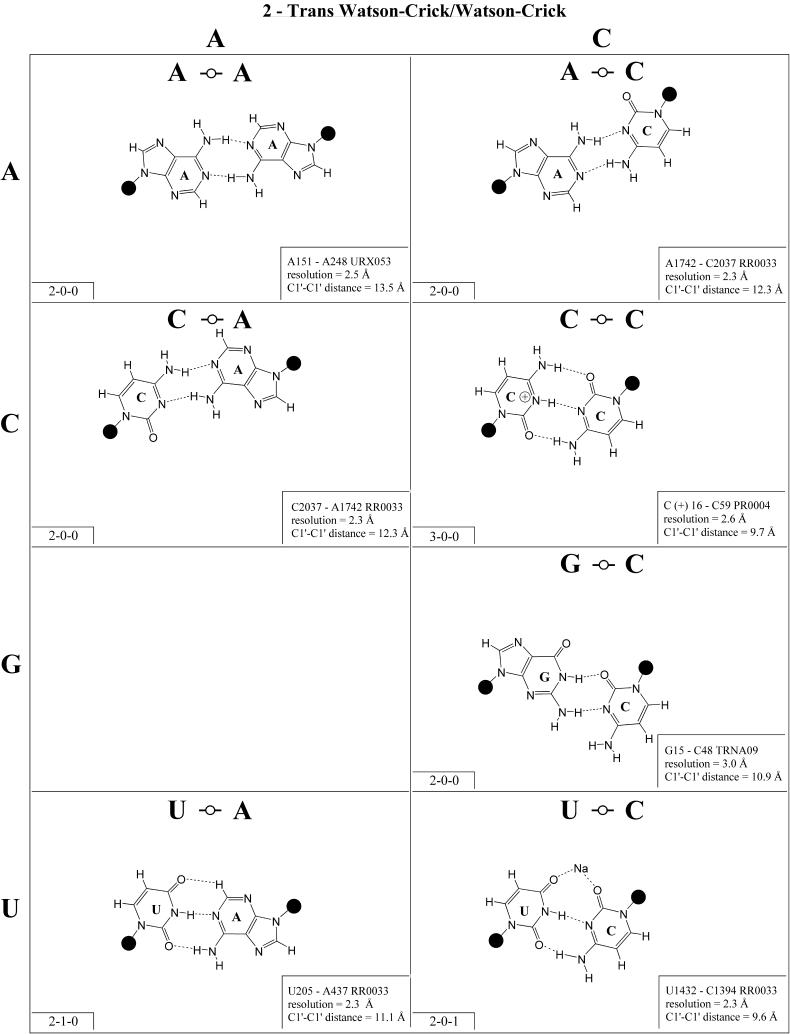
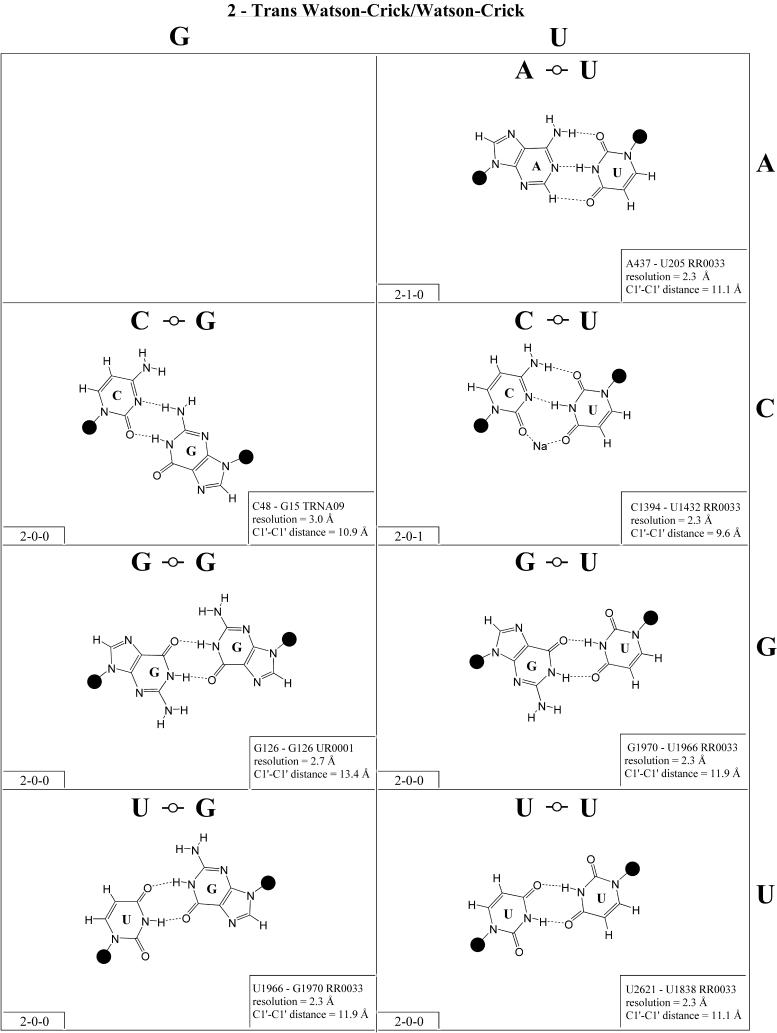
Trans Watson–Crick/Watson–Crick (Family 2). Representa tive base pairs belonging to the trans Watson–Crick/Watson–Crick geometric family are shown in Figure 3 and the isostericity matrix is shown in the right panel of the first row of Table 3. The trans orientation of the glycosidic bonds allows for a possible 2-fold axis perpendicular to and passing through the middle of the base pair. Unlike the corresponding cis pairs, the A/U (designated I1) and G/C (designated I2) pairs are not isosteric. However, these and all trans Watson–Crick/Watson–Crick pairs are self-isosteric and thus Table 3 is symmetric with respect to the main diagonal. The pairs A/C and G/U are isosteric, but not isosteric with A/U or G/C, and thus form a third group, I3. The homopurine pairs A/A and G/G are isosteric (I4) but A/G cannot form with two hydrogen bonds. As for the cis Watson–Crick/Watson–Crick family, all possible trans Watson–Crick/Watson–Crick pairs have been observed in crystal structures.
Figure 3.
(Opposite and above) Observed base pairs of the trans Watson–Crick/Watson–Crick family. The pairing displays a 2-fold rotational symmetry. Thus, the matrix is symmetric.
The trans Watson–Crick/Watson–Crick C/C pair shown in Figure 3 has three hydrogen bonds and requires protonation of one cytosine at N1. It is from a crystal structure of cysteinyl tRNA at 2.6 Å resolution (PR0004). An alternative hydrogen bonding pattern can be proposed that does not require protonation but involves only two hydrogen bonds (CN1– CN4 and CN4–CN1), which would make C/C isosteric with U/U rather than U/C. This geometry is observed at lower resolution (3.5 Å) for the tertiary base pair (C1773/C2565) in the structure of the 23S rRNA of Deinococcus radiodurans (RR0051). This pair corresponds to the tertiary interaction U1838 /U2621 in the 23S rRNA of Haloarcula marismortui (U1782/U2586 in the Escherichia coli sequence) and was first identified by sequence analysis based on the co-variation of U/U and C/C for these positions (31). Thus we favor grouping U/C and C/U in one isosteric subgroup (I5) and C/C with U/U in another (I6). The observed U1432/C1394 pair (RR0033) has a sodium ion bridging UO4–CO2 (compare with cis Watson–Crick/Watson–Crick).
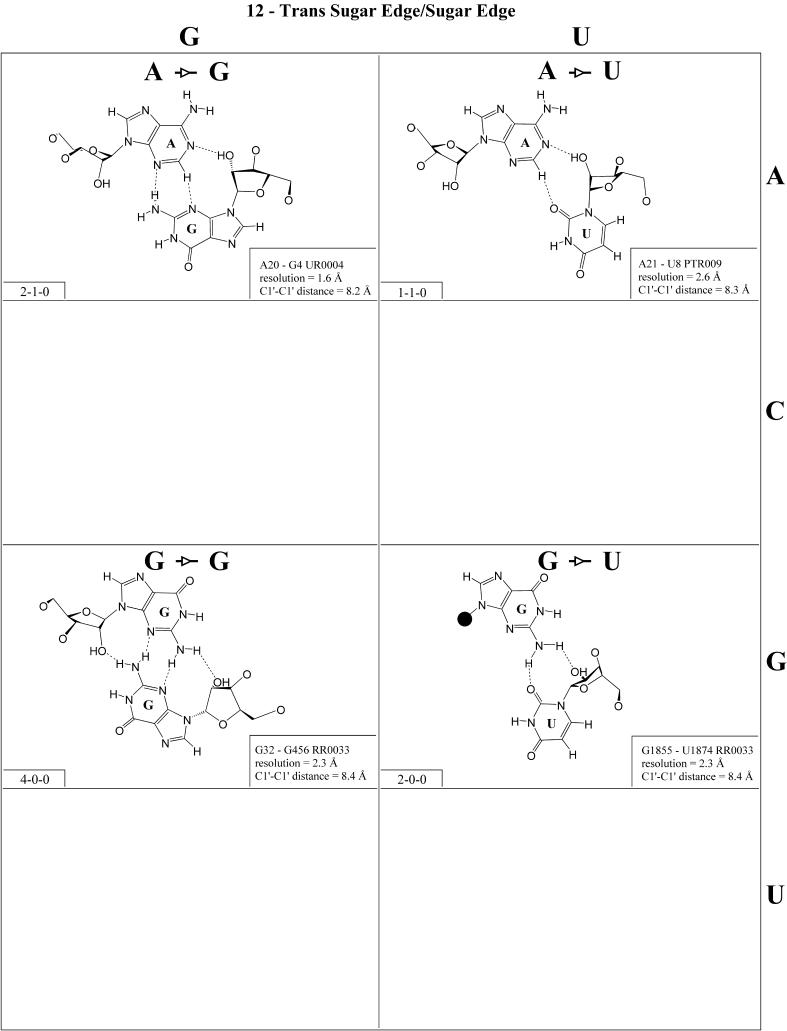
Cis Watson–Crick/Hoogsteen (Family 3). Representative pairs in this family are shown in Figure 4 and the corresponding isostericity matrix in Table 3. U/A, U/G and C(+)/G have been observed and together with C/U (modeled on C/G and A/G) are grouped into the isosteric subfamily I1. Modeled base pairs are indicated in Tables 3 and 4 in parentheses. Cytosine requires protonation at N3 to form C(+)/G. C/C and U/U have both been observed and are grouped into subfamily I2, which is related to I1 by a lateral shift in the hydrogen bonds. A(+)/G has been observed at high resolution (1.9 Å) and requires protonation of AN1 to form. A(+)/G is grouped with G/A (observed) and A/U (modeled) in subfamily I3. G/G is related to A(+)/G and G/A by a lateral shift in the hydrogen bonding positions, and thus G/G is grouped separately (I4).
Table 4. Isostericity matrices for base pairing Families 7–12.
First row, cis and trans Hoogsteen/Hoogsteen; second row, cis and trans Hoogsteen/sugar edge; third row, cis and trans sugar edge/sugar edge. Parentheses indicate modeled interactions, not yet observed in high resolution X-ray structures. For the cis Hoogsteen/sugar edge geometry, I1 indicates observed pairs in which the interacting bases are adjacent in the polynucleotide chain, while I2 indicates observed pairs in which one or two nucleotides separate the interacting bases.
The cis Watson–Crick/Hoogsteen interaction often occurs as part of a base triple. The base that interacts with its Hoogsteen edge uses its Watson–Crick edge to pair with the third base. For example, the isosteric U/U and C/C pairs comprise tertiary interactions in the conserved L11-binding site of 23S rRNA as part of such a triple. C1072· C1092=G1099 (E.coli numbering) co-varies with U·U-A in the 23S rRNAs of all phylogenetic groups. This provides another example of sequence co-variation reflecting isosteric subgroups of the isostericity matrix.
In summary, eight of the 10 pairs expected in this family have been observed. The R/R and R/Y pairs exhibit significantly longer C1′–C1′ distances than the Y/R and Y/Y pairs. In addition, isolated examples involving single hydrogen bonds and non-planar interactions have been observed (e.g. A2812/A2814 and A378/C271 in RR0033).
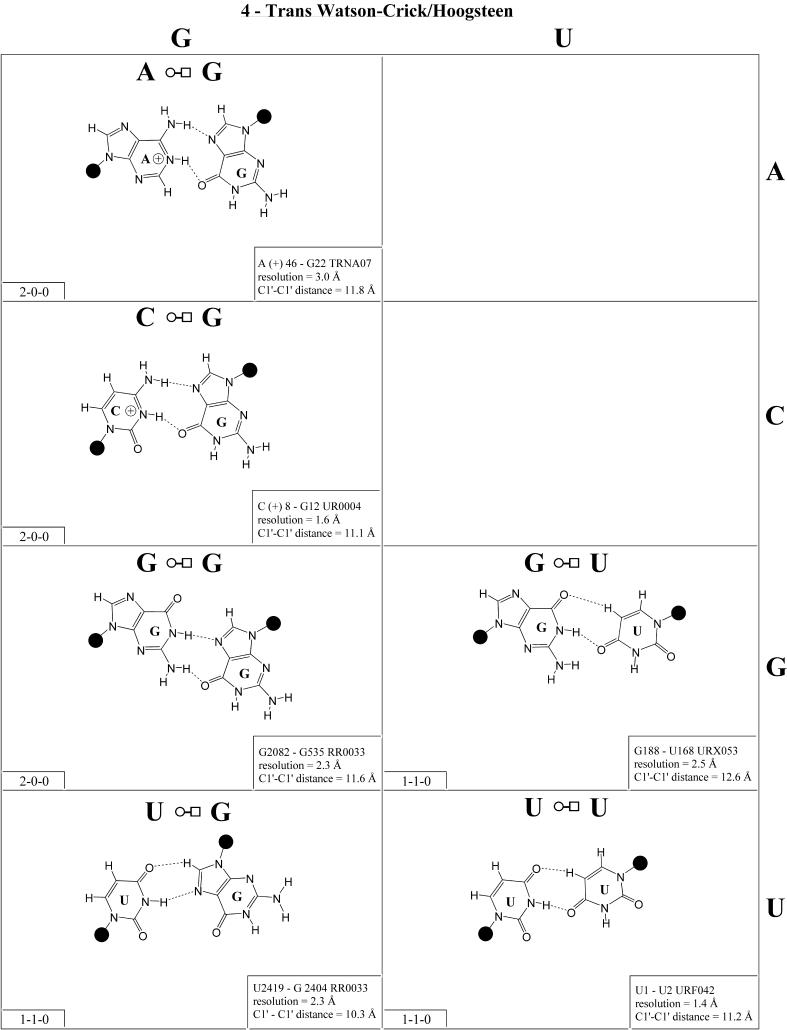
Trans Watson–Crick/Hoogsteen (Family 4). As for the corresponding cis geometry, the R/R and R/Y pairs of the trans Watson–Crick/Hoogsteen geometry exhibit significantly longer C1′–C1′ distances than the Y/R and Y/Y pairs (Fig. 5 and Table 3, second row, right). U/A and U/C are isosteric (subfamily I1) and are related by a lateral shift to C/A, C(+)/G and U/U (subfamily I2). In fact, I1 and I2 are mutually compatible, thus U/A and C/A are observed to co-vary in the loop E motifs of 5S rRNA and SRP (2,32). U/G is placed in its own group (I3) because it is rarely observed and does not co-vary with U/A or C/A, perhaps because of the repulsion between UO2 and GO6, which may destabilize pairing in the standard geometry and favor hydrogen bonding between UO4 and GC8.
Figure 5.
(Opposite and above) Observed base pairs of the trans Watson–Crick/Hoogsteen family.
Three of the four R/R combinations form base pairs. A/A and A(+)/G are isosteric and with G/U comprise subfamily I4. G/G is related by a lateral shift to A/A and A(+)/G and is thus not exactly isosteric and so is grouped separately (I5). A(+)/G requires protonation of AN1 and has been observed in tRNA (e.g. TRNA07).
In summary, all 10 pairs expected for this family have been observed. As for the cis Watson–Crick/Hoogsteen family, isolated examples involving single hydrogen bonds and non-planar interactions also occur (e.g. A2577/C2555 and G345/A305 in RR0033).
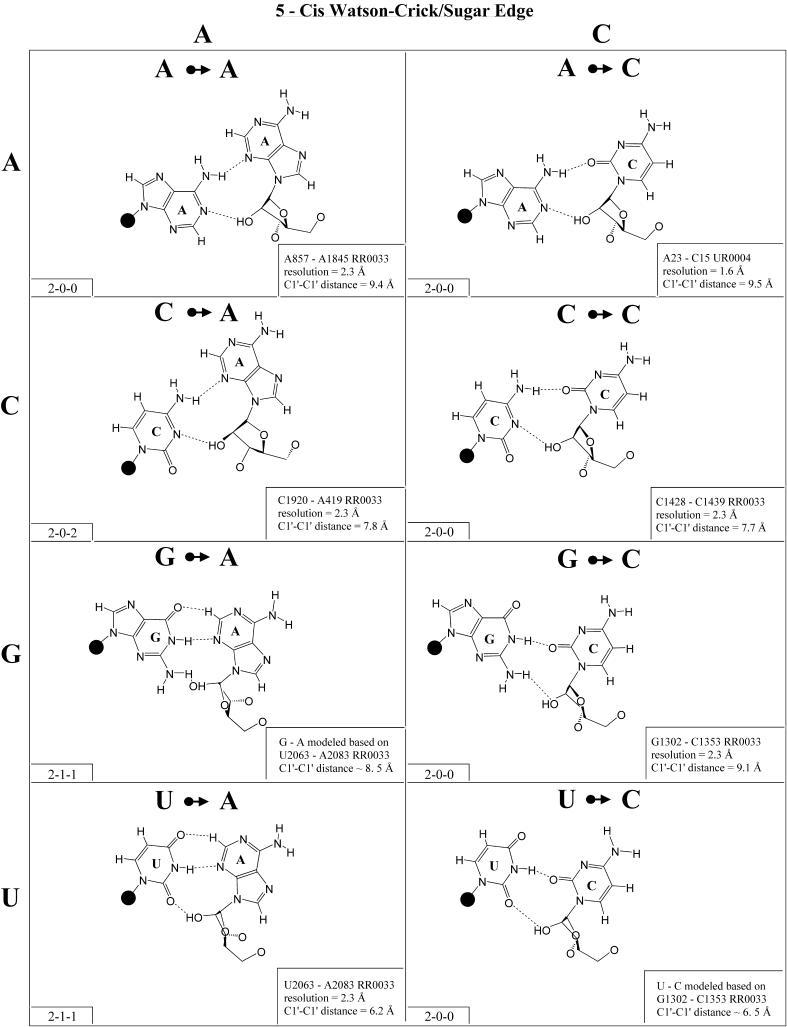
Cis Watson–Crick/sugar edge (Family 5). The cis Watson– Crick/sugar edge family (Fig. 6 and Table 3, third row, left) comprises four main isosteric subfamilies that are defined by the base that pairs using its Watson–Crick edge. Thus, all four A/N pairs are isosteric and all have been observed (subfamily I1). All four C/N pairs have been observed and comprise a second group, I2. Three of the G/N pairs have been observed (I3). G/A was modeled using U/A as a template. It should be isosteric to G/C and G/U. G/G displays a significantly longer C1′–C1′ distance and is therefore placed in its own subgroup (I5). U/A and U/G have been observed, whereas U/C and U/U were modeled based on G/C and G/U. The four U/N pairs are also expected to form a single isosteric group (I4).
Figure 6.
(Opposite and above) Observed and modeled base pairs of the cis Watson–Crick/sugar edge family.
Trans Watson–Crick/sugar edge (Family 6). The base pairs belonging to the trans Watson–Crick/sugar edge family are shown in Figure 7 and the corresponding isostericity matrix in Table 3 (third row, right). Both A/A and A/G have been observed and are isosteric. The A/G pair is more common and probably more stable as it involves two conventional base–base hydrogen bonds and a potential A(N6)–G(O2′) hydrogen bond. This interaction can occur as part of a base triple (for example A24·G7=C14 in UR0004) or as an isolated tertiary base pair (e.g. A629·G2070 or A2018·A1829 in H.marismortui 23S rRNA, RR0033). The A/Y interactions were modeled based on C/C, but these would only involve one base–base hydrogen bond (Fig. 7) and are expected to occur in the context of base triples. All four A·N interactions should be isosteric (I1, Table 3, third row, right).
Figure 7.
(Opposite and above) Observed and modeled base pairs of the trans Watson–Crick/sugar edge family.
The C/A, C/G and C/C interactions have been observed and C/U can be modeled using C/C as a template. Like the A/R interactions, the C/R interactions can occur as part of base triples (e.g. C46·G43=C37 in H.marismortui 5S rRNA, RR0033) or as isolated tertiary interactions (e.g. C1981· A1983, RR0033). The C963/C959 pair from 23S rRNA belongs to a base triple in which C959 is Watson–Crick paired to A1005. The C/G pair is the only C/N trans Watson–Crick/sugar edge interaction to feature two conventional base–base hydrogen bonds and is the most common. All the C/N and A/N pairs are grouped in a single isosteric subfamily, designated I1.
The G/U pair occurs most commonly as the closing base pair in UUCG-type hairpin loops, with the G in the syn configuration and the strands antiparallel (see Table 2 legend). The G/C trans Watson–Crick/sugar edge pair can also occur in a hairpin loop (e.g. G10/C7 in PR0022) and is isosteric with G/U, which together form the I2 subfamily. The G/R interactions are not expected to occur and have not been observed.
Examples of U/A, U/C and U/G have been observed and U/U can be modeled based on U/C (Fig. 7). (An example of U/U exists in a low resolution structure, U106/U258 in the Group I intron, UR0003.) The U/A interaction occurs as part of a base quadruple with C879=G871 in a three-way junction in 23S rRNA (RR0033). The U/C interaction occurs as part of a base triple in 16S rRNA of Thermus thermophilus (RR0015) and U/G as a tertiary interaction in 23S rRNA (RR0033) that involves a bridging water molecule. The hydrogen bonding patterns in the U/Y and G/Y pairs are similar but the C1′–C1′ distances are greater in the G/Y pairs, so these form different isosteric subgroups. U/A can be grouped with U/Y (I3), but U·G is distinct (I4).
Cis Hoogsteen/Hoogsteen (Family 7). The only examples from this family have been observed in the ribosome (Fig. 8 and Table 4, row one, left). They are very rare. The G2494/C2493 interaction involves adjacent nucleotides. C2493 is in the rare syn conformation and thus presents its Hoogsteen edge to interact with the Hoogsteen edge of G2494, thus allowing the CH6–GO6 hydrogen bond to form in place of the unfavorable CO2–GO6 repulsive interaction. The second example, G2616/G2617, also involves adjacent nucleotides with G2616 also in the syn conformation. A1742/G2033 is a tertiary interaction with antiparallel strands. Kinks and sharp turns in the phosphodiester backbones of the antiparallel strands allow the two bases to approach each other to form the characteristic AN6–GO6 hydrogen bond.
Figure 8.
(Opposite and above) Observed base pairs of the cis Hoogsteen/Hoogsteen family.
Trans Hoogsteen/Hoogsteen (Family 8). The trans Hoogsteen/Hoogsteen pairs are shown in Figure 9 and the isostericity matrix in Table 4 (first row, right). Like the trans Watson–Crick/Watson–Crick family, these pairs are self-isosteric due to symmetry. It is interesting to notice that in this family, except for one base pair, all the pairs involve a single hydrogen bond. This pair occurs in tRNA and in sarcin/ricin motifs. The sequence variations observed for these motifs correspond closely to the observed base pairs shown in Figure 8 (2,27).
Figure 9.
(Opposite and above) Observed base pairs of the trans Hoogsteen/Hoogsteen family. The pairing displays a 2-fold rotational symmetry. Thus, the matrix is symmetric.
Cis Hoogsteen/sugar edge (Family 9). The cis Hoogsteen/sugar edge interaction can involve the bases of adjacent or more distant nucleotides in the polynucleotide chain. Generally, only a single hydrogen bond can form between the interacting bases (Fig. 10). The best known examples are the A/A ‘platform’ (33) and the U/G ‘side-by-side’ pair of the sarcin/ricin loop motif (18). In addition to these, many other pairs of this type have been observed. Eleven examples involving immediately adjacent nucleotides have been observed and are shown in Figure 10. On the basis of the U/U pair, we can propose a model for U/C, and on the basis of the G/G pair we can propose G/A. In fact, cis Hoogsteen/sugar edge G/A is observed at lower resolution (∼3.5 Å) in the 23S rRNA of D.radiodurans (G2035/A2034 in NDB file rr0051) at the position corresponding to G2093/G2092. Bases of non-adjacent nucleotides can form similar base pairs, but these are not isosteric to the adjacent pairs. All the pairs involving adjacent pairs are essentially isosteric (I1 in Table 4). Non-adjacent pairs form a second isosteric group (I2). Examples of non-adjacent cis Hoogsteen/sugar edge pairs exist for many of the adjacent pairs shown in Figure 10, but the adjacent pair is shown by preference. Examples of non-adjacent pairs include U2527/G2525, C2787/C2785 and C2575/U2473 from 23S rRNA (H.marismortui) and A56/A54 from 5S rRNA (H.marismortui).
Figure 10.
(Opposite and above) Observed and modeled base pairs of the cis Hoogsteen/sugar edge family.
One of the most remarkable cis Hoogsteen/sugar edge pairs is U832/U831, in which a water bridges between U832(O4) and U831(O2). U/U is observed to co-vary with cis Hoogsteen/sugar edge U/G in some sarcin loop motifs (N.B.Leontis and E.Westhof, manuscript in preparation).
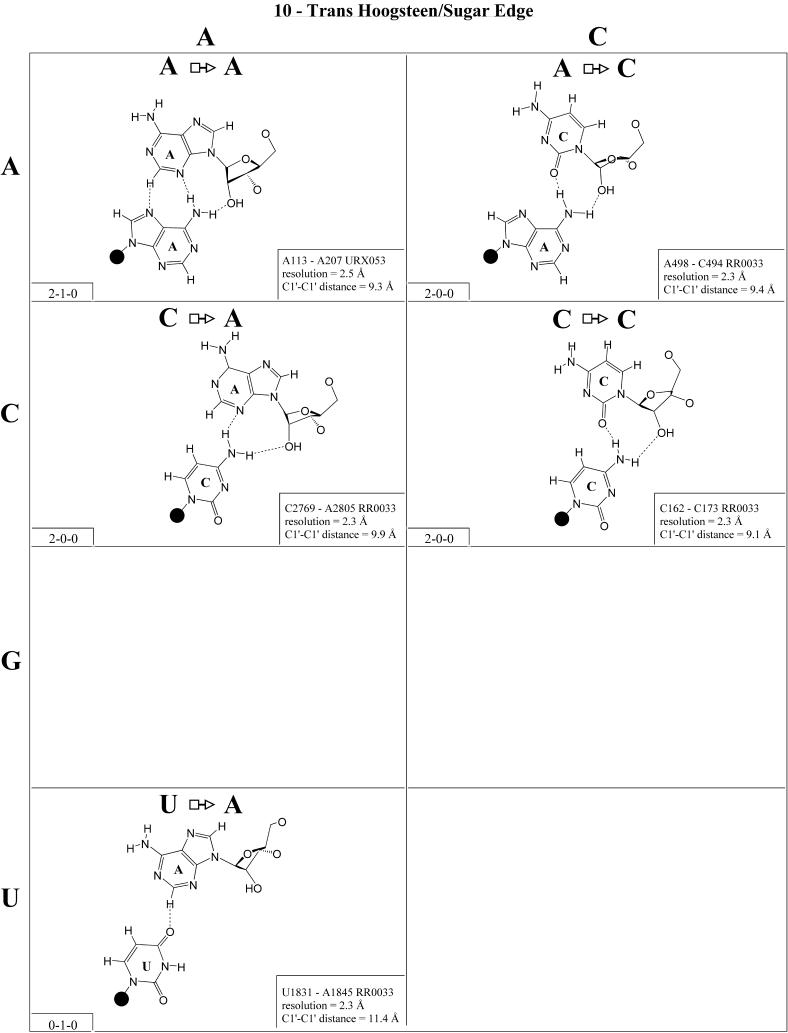
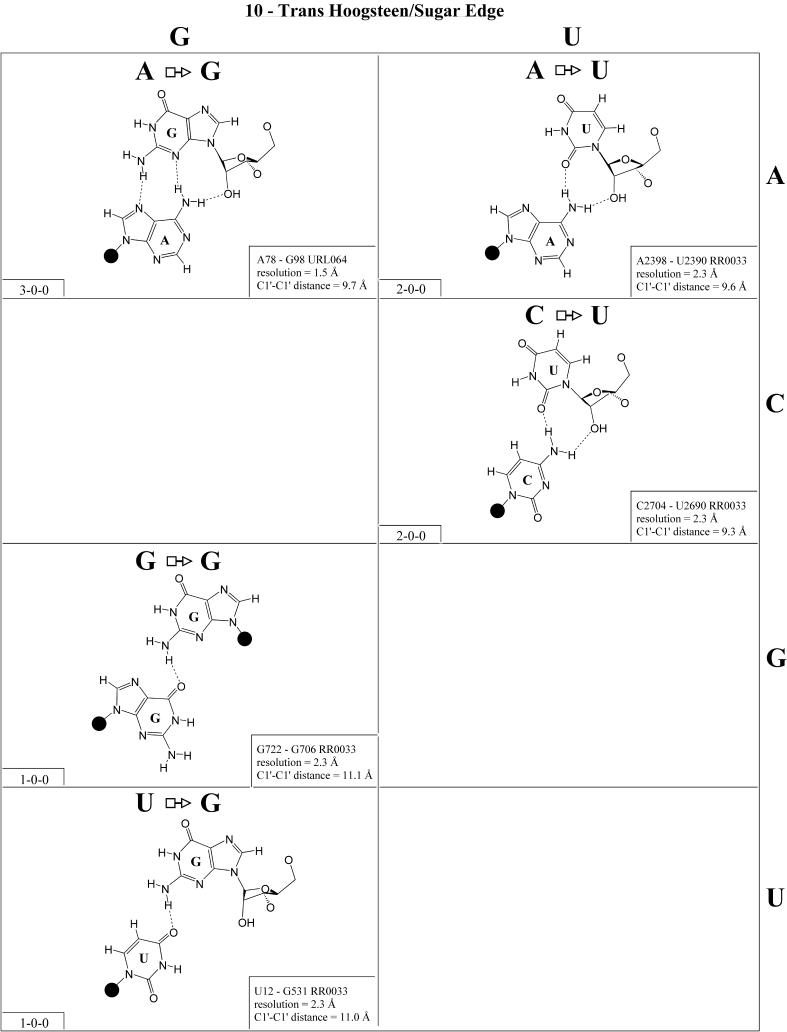
Trans Hoogsteen/sugar edge (Family 10). The most common interaction of this type is the ‘sheared’ A/G in which the Hoogsteen edge of A interacts with the sugar edge of G (Fig. 11). In fact, this is the most commonly occurring A/G base pair. This base pair occurs in loop E of 5S rRNA and in the sarcin/ricin motif of 23S rRNA. Co-variations at these positions include A/A, A/Y (Y = U or C), C/A (C Hoogsteen, A sugar edge) and C/Y. On the basis of these co-variations and the structures of the A/G and A/A pairs, models were proposed for A/Y, C/A and C/Y (32). Subsequently, all these pairs have been observed (see Fig. 11), just as modeled, and, interestingly, all are isosteric. Thus, the A/N, C/A and C/Y pairs are grouped into one isosteric subfamily, designated I1 in Table 4 (second row, right). The G/G, U/A and U/G pairs form a second isosteric subgroup (I2) that does not co-vary with the first.
Figure 11.
(Opposite and above) Observed base pairs of the trans Hoogsteen/sugar edge family.
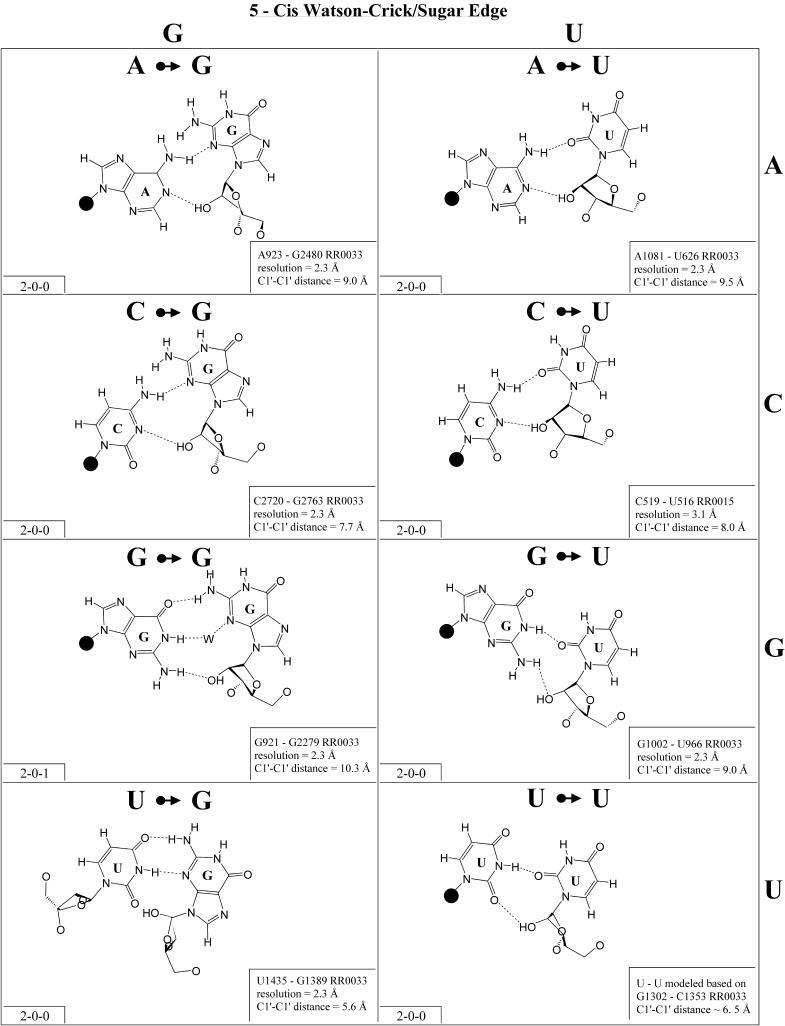
Cis sugar edge/sugar edge (Family 11). As shown in Figure 12, examples of almost all possible cis sugar edge/sugar edge pairs have been observed and all 16 combinations are expected to be isosteric (Table 4, third row, left). This interaction is not symmetric as the O2′ of one nucleotide hydrogen bonds to the base R(N3) or Y(O2) and to the hydroxyl O2′ of the other nucleotide. The former nucleotide is given priority (7). When that nucleotide is a pyrimidine (Y), there is in fact no direct base–base hydrogen bond. When it is a purine (R), there is a single base–base hydrogen bond (except for A·G, with two). This interaction occurs frequently between adjacent nucleotides belonging to two strands (with the 5′ nucleotide of one strand receiving from the hydroxyl group of the 3′ nucleotide of the other). Such a motif is referred to as the ‘ribose-zipper motif’ (33). Furthermore, the cis sugar edge/sugar edge interaction often occurs in combination with the trans sugar edge/sugar edge pair of the frequent and versatile recognition motif comprised of adjacent cis and trans sugar edge/sugar edge base pairs (3,27).
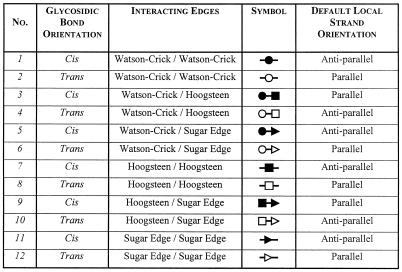
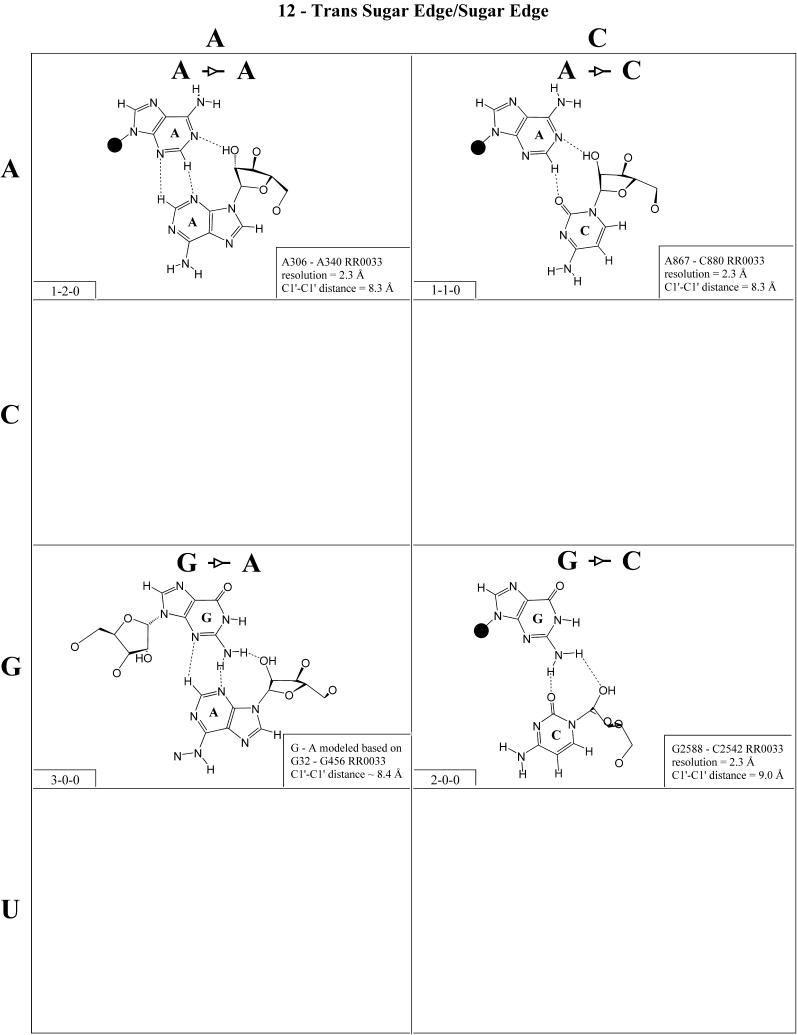
Trans sugar edge/sugar edge (Family 12). The trans sugar edge/sugar edge base pair (Fig. 13 and Table 4, third row, right) usually involves at least one adenosine. Generally, such interactions occur as part of base triples in which the adenosine (and more rarely guanosine) interacts with the sugar edge of a standard base pair. The A·A, A·G and A·C examples are of this type: A306·A340-U325 (RR0033), A867·C880=G870 (RR0033) and A20·G4=C17 (UR0004). Of these, A·G is by far the most common, since it occurs in the frequent recognition motif made of adjacent cis and trans sugar edge/sugar edge pairs. The A·U pair is found in tRNAs as part of a base triple (A21·U8·A14). The other pairs involving G are much rarer. Examples of G·G include those in which one G is canonically paired as well as isolated tertiary pairs such as G315·G336 and G2428·G2466 (RR0033). The G·U shown in Figure 13 is a tertiary pair, whereas the G·C example is part of a base triple (G2617·C2542=G2617, RR0033). The A·N pairs form one group (I1) and the G·N pairs a second group (I2).
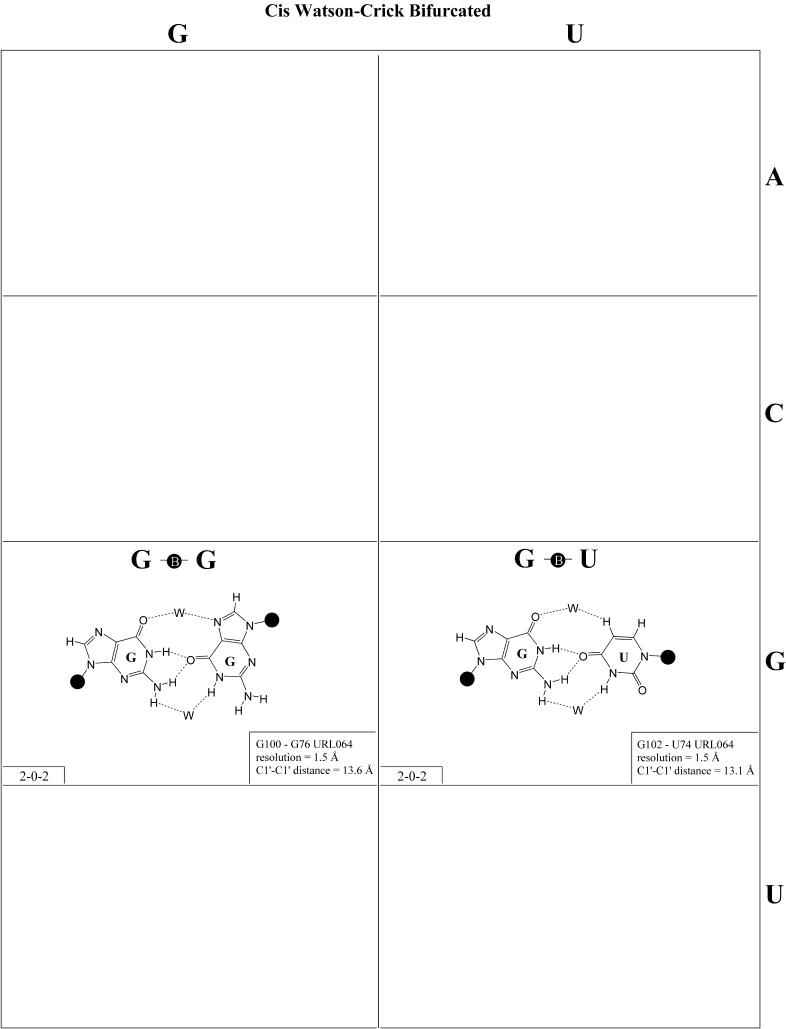
Bifurcated hydrogen bonding patterns. Bifurcated pairs are intermediate between two edge-to-edge geometries (Fig. 14 and Table 5). They involve interactions between an exocyclic functional group of one base and the edge of another. Bifurcated pairs may also show distinct patterns of co-variation and substitution. For example, the isosteric G·G and G·U cis bifurcated pairs, first observed at high resolution in the structure of loop E of bacterial 5S rRNA, were found to co-vary with each other and with A·C and A·A, both of which could be modeled in the same geometry (32). These pairs are intermediate to the cis Watson–Crick/Watson–Crick and the trans Watson–Crick/Hoogsteen families. The isostericity matrix (Table 5) was proposed for bifurcated pairs of this kind (27). Additional examples belonging to this family of bifurcated pairs have been observed in the ribosome, including C2502/C2518 (also part of a loop E type motif) and C930/A1040.
Figure 14.
(Opposite and above) Observed and modeled base pairs of the cis Watson–Crick bifurcated family.
Bifurcated pairs intermediate to the trans Watson–Crick/Hoogsteen and trans sugar edge/Hoogsteen families occur in loop E-related motifs in 16S rRNA (G581·G760, E.coli numbering), 23S rRNA (G706·G722) and the SRP (G162-G149).
Intermediate and alternative hydrogen bonding patterns. In a small number of cases, alternative hydrogen bonding patterns have been observed for particular base pair combinations. These may be due to the limited resolution of the experimental data or refinement errors or to the actual existence of distinct potential energy minima that depend on the local structural context. The symmetrical, cis Watson–Crick (wobble-like) U/U and C/C pairs provide trivial examples of the latter. For example, two uridines can pair with UO4–UN3 and UN3– UO2 hydrogen bonds or with UN3–UO4 and UO2–UN3 hydrogen bonds. Which set of hydrogen bonds occurs depends on the local context. Alternatively, U/U can open up and incorporate a bridging water molecule (34). Likewise, G and U can form a conventional wobble pair (Fig. 2) or, in certain contexts, a bifurcated pair, involving two bridging water molecules (Fig. 14). Two possible hydrogen bonding patterns for trans Watson–Crick C/C were discussed above. Higher resolution structural work complemented by computation is needed to determine which pattern is favored and whether this is context-dependent.
Another example is provided by the cis Watson–Crick (wobble) C·A pair, for which hydrogen bonding may be proposed between C(N4) and A(N1) and between C(N3) and A(C2) in place of hydrogen bonds between C(N1) and A(N6) and C(N3) and protonated A(N3), which are usually observed. An example with the alternative hydrogen bonding pattern is observed in the context of a base triple in 23S rRNA (C40·A441·A442 in RR0033). The triple consists of the A442·A441 cis Hoogsteen/sugar edge interaction and the alternative C40·A441 cis Watson–Crick/Watson–Crick pair. An additional hydrogen bond is observed between C40(O2) and A442(N6). Higher resolution is required to confirm this interaction.
In conclusion, it must be emphasized that base pairing is due to multiple weak interactions and thus a considerable degree of flexibility and deformation is expected. Thus, while one can generally classify base pairs into one of the 12 families discussed above, a particular base pair may form with a slightly different combination of hydrogen bonds or with the absence of one or more hydrogen bonds, depending on the structural context or on the resolution of the structure.
Interactions of a base with an ‘edge’ defined by two bases. A premise of the approach we have taken has been that complex interactions (base triples, quadruples, etc.) can be analyzed as combinations of base pairs. In a few cases this analysis breaks down and new patterns arise, which again reflect synergistic effects. An example is the interaction of the Watson–Crick edge of C with the Hoogsteen edge of a (standard) G=C base pair. Four interactions can be anticipated, cis or trans C·G Watson–Crick/Hoogsteen and cis or trans C·C Watson–Crick/Hoogsteen, and are in fact observed (see Tables 4 and 5). A fifth interaction, distinct from these, has also been observed. An example is provided by C113 interacting with the Hoogsteen edges of the C15=G66 base pair in 5S rRNA of H.marismortui (RR0033). This interaction is intermediate between the cis C·C Watson–Crick/Hoogsteen interaction seen in base triples such as C1072·C1092=G1099 in the L11-binding site of 23S rRNA (E.coli numbering, NDB file PR0015 or RR0009) and the trans C(N1+)·G Watson–Crick/Hoogsteen interaction seen in base triples such as C8(+)· G12=C26 in the frameshifting pseudoknot (UR0004). It can best be described as an interaction of the Watson–Crick edge of C113 with the Hoogsteen edge of the C15=G66 base pair, as it involves hydrogen bonds to both G66(O6) and C15(N4).
CONCLUSIONS
The rapidly growing database of RNA crystal structures provides examples of nearly every type of base pair. Many of the base pairs presented in Figures 2–14 were first proposed on theoretical grounds and have now been observed by X-ray crystallography at <3.0 Å resolution. Generally, the observed base pairs are as predicted (27,32). The overwhelming number of base–base interactions observed in the ribosome and the other new structures that have appeared recently can be unambiguously classified into one of the 12 families of Table 2. A small number of base pairs comprise bifurcated pairs that are intermediate between two of the 12 families (7). Furthermore, care must be taken so as not to confuse the trans sugar edge/sugar edge and trans Watson–Crick/sugar edge interactions, because frequently a Watson–Crick/2′-OH hydrogen bond can also occur in the trans sugar edge/sugar edge geometry.
Other kinds of interactions are observed in complex RNA structures which need to be analyzed and catalogued, including additional bifurcated pairs, perpendicular edge-to-edge interactions, interactions exclusively involving the ribose moiety of one or both nucleotides, and base stacking interactions.
Preliminary analyses, some of which have been presented here, indicate that there is a close correspondence between the isosteric subfamilies identified on structural grounds and the patterns of co-variation and base substitution that are observed in homologous RNA, when they are properly aligned. The primary significance of this work is that it provides a basis for evaluating and refining structural alignments for homologous RNA molecules. Consideration of the isostericity matrix corresponding to each base pair is essential for producing correct alignments at positions involved in non-Watson–Crick base pairing or determining that one motif has in fact been replaced by another in a set of homologous sequences.
Here, we have emphasized the geometrical aspects of base pairing in order to aid in their classification. Clearly, depending on the edges involved, various groups or sites will be available for interactions with another RNA segment, a protein or a small molecule. For example, when the Watson–Crick sites are not engaged, they can be used for interaction with phosphate groups. Similarly, the Hoogsteen sites are used for interactions with amino acid side chains in complexes between proteins and helices. Besides conferring geometrical similarity, the isostericity matrices contain information on compensating changes that would occur between base pairs at the level of a given functional group or a set of functional groups.
Acknowledgments
ACKNOWLEDGEMENTS
The authors acknowledge fruitful discussions with Pascal Auffinger and Luc Jaeger. This work was supported by NSF REU grant CHE-9732563 and NIH grant 2R15-GM55898.
REFERENCES
- 1.Woese C.R., Winker,S. and Gutell,R.R. (1990) Architecture of ribosomal RNA: constraints on the sequence of “tetra-loops”. Proc. Natl Acad. Sci. USA, 87, 8467–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leontis N.B. and Westhof,E. (1998) A common motif organizes the structure of multi-helix loops in 16 S and 23 S ribosomal RNAs. J. Mol. Biol., 283, 571–583. [DOI] [PubMed] [Google Scholar]
- 3.Nissen P., Ippolito,J.A., Ban,N., Moore,P.B. and Steitz,T.A. (2001) RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc. Natl Acad. Sci. USA, 98, 4899–4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moore P.B. (1999) Structural motifs in RNA. Annu. Rev. Biochem., 68, 287–300. [DOI] [PubMed] [Google Scholar]
- 5.Hoogsteen K. (1963) The crystal and molecular structure of a hydrogen-bonded complex between 1-methylthymine and 9-methyladenine. Acta Crystallogr., 16, 907–916. [Google Scholar]
- 6.Saenger W. (1984) Principles of Nucleic Acid Structure. Spring-Verlag, New York, NY.
- 7.Leontis N.B. and Westhof,E. (2001) Geometric nomenclature and classification of RNA base pairs. RNA, 7, 499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Westhof E., Dumas,P. and Moras,D. (1988) Restrained refinement of two crystalline forms of yeast aspartic acid and phenylalanine transfer RNA crystals. Acta Crystallogr. A, 44, 112–123. [PubMed] [Google Scholar]
- 9.Michel F., Hanna,M., Green,R., Bartel,D.P. and Szostak,J.W. (1989) The guanosine binding site of the Tetrahymena ribozyme. Nature, 342, 391–395. [DOI] [PubMed] [Google Scholar]
- 10.Michel F. and Westhof,E. (1990) Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J. Mol. Biol., 216, 585–610. [DOI] [PubMed] [Google Scholar]
- 11.Gautheret D., Damberger,S.H. and Gutell,R.R. (1995) Identification of base-triples in RNA using comparative sequence analysis. J. Mol. Biol., 248, 27–43. [DOI] [PubMed] [Google Scholar]
- 12.Gautheret D. and Gutell,R.R. (1997) Inferring the conformation of RNA base pairs and triples from patterns of sequence variation. Nucleic Acids Res., 25, 1559–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Macke T.J., Ecker,D.J., Gutell,R.R., Gautheret,D., Case,D.A. and Sampath,R. (2001) RNAMotif, an RNA secondary structure definition and search algorithm. Nucleic Acids Res., 29, 4724–4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Varani G., Cheong,C. and Tinoco,I.,Jr (1991) Structure of an unusually stable RNA hairpin. Biochemistry, 30, 3280–3289. [DOI] [PubMed] [Google Scholar]
- 15.Heus H.A. and Pardi,A. (1991) Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science, 253, 191–194. [DOI] [PubMed] [Google Scholar]
- 16.Wimberly B., Varani,G. and Tinoco,I.,Jr (1993) The conformation of loop E of eukaryotic 5S ribosomal RNA. Biochemistry, 32, 1078–1087. [DOI] [PubMed] [Google Scholar]
- 17.SantaLucia J. Jr and Turner,D.H. (1993) Structure of (rGGCGAGCC)2 in solution from NMR and restrained molecular dynamics. Biochemistry, 32, 12612–12623. [DOI] [PubMed] [Google Scholar]
- 18.Szewczak A.A. and Moore,P.B. (1995) The sarcin/ricin loop, a modular RNA. J. Mol. Biol., 247, 81–98. [DOI] [PubMed] [Google Scholar]
- 19.Butcher S.E., Allain,F.H. and Feigon,J. (1999) Solution structure of the loop B domain from the hairpin ribozyme. Nature Struct. Biol., 6, 212–216. [DOI] [PubMed] [Google Scholar]
- 20.Ryder S.P., Ortoleva-Donnelly,L., Kosek,A.B. and Strobel,S.A. (2000) Chemical probing of RNA by nucleotide analog interference mapping. Methods Enzymol., 317, 92–109. [DOI] [PubMed] [Google Scholar]
- 21.Ehresmann C., Baudin,F., Mougel,M., Romby,P., Ebel,J.P. and Ehresmann,B. (1987) Probing the structure of RNAs in solution. Nucleic Acids Res., 15, 9109–9128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee K., Varma,S., SantaLucia,J.,Jr and Cunningham,P.R. (1997) In vivo determination of RNA structure-function relationships: analysis of the 790 loop in ribosomal RNA. J. Mol. Biol., 269, 732–743. [DOI] [PubMed] [Google Scholar]
- 23.Guex N. and Peitsch,M.C. (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis, 18, 2714–2723. [DOI] [PubMed] [Google Scholar]
- 24.Westhof E. (1992) Westhof’s rule. Nature, 358, 459–460. [DOI] [PubMed] [Google Scholar]
- 25.Desiraju G.R. (1996) The C-H–O hydrogen bond: structural implications and supramolecular design. Acc. Chem. Res., 29, 441–449. [DOI] [PubMed] [Google Scholar]
- 26.Auffinger P. and Westhof,E. (1998) Hydration of RNA base pairs. J. Biomol. Struct. Dyn., 16, 693–707. [DOI] [PubMed] [Google Scholar]
- 27.Leontis N.B. and Westhof,E. (1998) Conserved geometrical base-pairing patterns in RNA. Q. Rev. Biophys., 31, 399–455. [DOI] [PubMed] [Google Scholar]
- 28.Csaszar K., Spackova,N., Stefl,R., Sponer,J. and Leontis,N.B. (2001) Molecular dynamics of the frame-shifting pseudoknot from beet western yellows virus: the role of non-Watson-Crick base-pairing, ordered hydration, cation binding and base mutations on stability and unfolding. J. Mol. Biol., 313, 1073–1091. [DOI] [PubMed] [Google Scholar]
- 29.Masquida B. and Westhof,E. (2000) On the wobble GoU and related pairs. RNA, 6, 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanaka Y., Fujii,S., Hiroaki,H., Sakata,T., Tanaka,T., Uesugi,S., Tomita,K. and Kyogoku,Y. (1999) A′-form RNA double helix in the single crystal structure of r(UGAGCUUCGGCUC). Nucleic Acids Res., 27, 949–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gutell R.R., Power,A., Hertz,G.Z., Putz,E.J. and Stormo,G.D. (1992) Identifying constraints on the higher-order structure of RNA: continued development and application of comparative sequence analysis methods. Nucleic Acids Res., 20, 5785–5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leontis N.B. and Westhof,E. (1998) The 5S rRNA loop E: chemical probing and phylogenetic data versus crystal structure. RNA, 4, 1134–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cate J.H., Gooding,A.R., Podell,E., Zhou,K., Golden,B.L., Kundrot,C.E., Cech,T.R. and Doudna,J.A. (1996) Crystal structure of a group I ribozyme domain: principles of RNA packing. Science, 273, 1678–1685. [DOI] [PubMed] [Google Scholar]
- 34.Vicens Q. and Westhof,E. (2001) Crystal structure of paromomycin docked into the eubacterial ribosomal decoding A site. Structure (Camb.), 9, 647–658. [DOI] [PubMed] [Google Scholar]
- 35.Jang S.B., Hung,L.W., Chi,Y.I., Holbrook,E.L., Carter,R.J. and Holbrook,S.R. (1998) Structure of an RNA internal loop consisting of tandem C-A+ base pairs. Biochemistry, 37, 11726–11731. [DOI] [PubMed] [Google Scholar]
- 36.Pan B., Mitra,S.N. and Sundaralingam,M. (1999) Crystal structure of an RNA 16-mer duplex R(GCAGAGUUAAAUCUGC)2 with nonadjacent G(syn).A+(anti) mispairs. Biochemistry, 38, 2826–2831. [DOI] [PubMed] [Google Scholar]
- 37.Trikha J., Filman,D.J. and Hogle,J.M. (1999) Crystal structure of a 14 bp RNA duplex with non-symmetrical tandem GxU wobble base pairs. Nucleic Acids Res., 27, 1728–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nix J., Sussman,D. and Wilson,C. (2000) The 1.3 Å crystal structure of a biotin-binding pseudoknot and the basis for RNA molecular recognition. J. Mol. Biol., 296, 1235–1244. [DOI] [PubMed] [Google Scholar]
- 39.Nissen P., Thirup,S., Kjeldgaard,M. and Nyborg,J. (1999) The crystal structure of Cys-tRNACys-EF-Tu-GDPNP reveals general and specific features in the ternary complex and in tRNA. Struct. Fold Des., 7, 143–156. [DOI] [PubMed] [Google Scholar]
- 40.Ferre-D’Amare A.R., Zhou,K. and Doudna,J.A. (1998) Crystal structure of a hepatitis delta virus ribozyme. Nature, 395, 567–574. [DOI] [PubMed] [Google Scholar]
- 41.Conn G.L., Draper,D.E., Lattman,E.E. and Gittis,A.G. (1999) Crystal structure of a conserved ribosomal protein-RNA complex. Science, 284, 1171–1174. [DOI] [PubMed] [Google Scholar]
- 42.Batey R.T., Rambo,R.P., Lucast,L., Rha,B. and Doudna,J.A. (2000) Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science, 287, 1232–1239. [DOI] [PubMed] [Google Scholar]
- 43.Lewis H.A., Musunuru,K., Jensen,K.B., Edo,C., Chen,H., Darnell,R.B. and Burley,S.K. (2000) Sequence-specific RNA binding by a Nova KH domain: implications for paraneoplastic disease and the fragile X syndrome. Cell, 100, 323–332. [DOI] [PubMed] [Google Scholar]
- 44.Arnez J.G. and Steitz,T.A. (1996) Crystal structures of three misacylating mutants of Escherichia coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP. Biochemistry, 35, 14725–14733. [DOI] [PubMed] [Google Scholar]
- 45.Nissen P., Kjeldgaard,M., Thirup,S., Polekhina,G., Reshetnikova,L., Clark,B.F. and Nyborg,J. (1995) Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science, 270, 1464–1472. [DOI] [PubMed] [Google Scholar]
- 46.Wimberly B.T., Guymon,R., McCutcheon,J.P., White,S.W. and Ramakrishnan,V. (1999) A detailed view of a ribosomal active site: the structure of the L11-RNA complex. Cell, 97, 491–502. [DOI] [PubMed] [Google Scholar]
- 47.Carter A.P., Clemons,W.M., Brodersen,D.E., Morgan-Warren,R.J., Wimberly,B.T. and Ramakrishnan,V. (2000) Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature, 407, 340–348. [DOI] [PubMed] [Google Scholar]
- 48.Ban N., Nissen,P., Hansen,J., Moore,P.B. and Steitz,T.A. (2000) The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science, 289, 905–920. [DOI] [PubMed] [Google Scholar]
- 49.Schlunzen F., Zarivach,R., Harms,J., Bashan,A., Tocilj,A., Albrecht,R., Yonath,A. and Franceschi,F. (2001) Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature, 413, 814–821. [DOI] [PubMed] [Google Scholar]
- 50.Wedekind J.E. and McKay,D.B. (1999) Crystal structure of a lead-dependent ribozyme revealing metal binding sites relevant to catalysis. Nature Struct. Biol., 6, 261–268. [DOI] [PubMed] [Google Scholar]
- 51.Correll C.C., Munishkin,A., Chan,Y.L., Ren,Z., Wool,I.G. and Steitz,T.A. (1998) Crystal structure of the ribosomal RNA domain essential for binding elongation factors. Proc. Natl Acad. Sci. USA, 95, 13436–13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Golden B.L., Gooding,A.R., Podell,E.R. and Cech,T.R. (1998) A preorganized active site in the crystal structure of the Tetrahymena ribozyme. Science, 282, 259–264. [DOI] [PubMed] [Google Scholar]
- 53.Su L., Chen,L., Egli,M., Berger,J.M. and Rich,A. (1999) Minor groove RNA triplex in the crystal structure of a ribosomal frameshifting viral pseudoknot. Nature Struct. Biol., 6, 285–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sussman D., Nix,J.C. and Wilson,C. (2000) The structural basis for molecular recognition by the vitamin B12 RNA aptamer. Nature Struct. Biol., 7, 53–57. [DOI] [PubMed] [Google Scholar]
- 55.Wahl M.C., Rao,S.T. and Sundaralingam,M. (1996) The structure of r(UUCGCG) has a 5′-UU-overhang exhibiting Hoogsteen-like trans U.U base pairs. Nature Struct. Biol., 3, 24–31. [DOI] [PubMed] [Google Scholar]
- 56.Lietzke S.E., Barnes,C.L., Berglund,J.A. and Kundrot,C.E. (1996) The structure of an RNA dodecamer shows how tandem U-U base pairs increase the range of stable RNA structures and the diversity of recognition sites. Structure, 4, 917–930. [DOI] [PubMed] [Google Scholar]
- 57.Baeyens K.J., De Bondt,H.L., Pardi,A. and Holbrook,S.R. (1996) A curved RNA helix incorporating an internal loop with G.A and A.A non-Watson-Crick base pairing. Proc. Natl Acad. Sci. USA, 93, 12851–12855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Correll C.C., Freeborn,B., Moore,P.B. and Steitz,T.A. (1997) Metals, motifs and recognition in the crystal structure of a 5S rRNA domain. Cell, 91, 705–712. [DOI] [PubMed] [Google Scholar]
- 59.Scott W.G., Finch,J.T. and Klug,A. (1995) The crystal structure of an all-RNA hammerhead ribozyme: a proposed mechanism for RNA catalytic cleavage. Cell, 81, 991–1002. [DOI] [PubMed] [Google Scholar]