Abstract
The human Werner syndrome protein, WRN, is a member of the RecQ helicase family and contains 3′→5′ helicase and 3′→5′ exonuclease activities. Recently, we showed that the exonuclease activity of WRN is greatly stimulated by the human Ku heterodimer protein. We have now mapped this interaction physically and functionally. The Ku70 subunit specifically interacts with the N-terminus (amino acids 1–368) of WRN, while the Ku80 subunit interacts with its C-terminus (amino acids 940– 1432). Binding between Ku70 and the N-terminus of WRN (amino acids 1–368) is sufficient for stimulation of WRN exonuclease activity. A mutant Ku heterodimer of full-length Ku80 and truncated Ku70 (amino acids 430–542) interacts with C-WRN but not with N-WRN and cannot stimulate WRN exonuclease activity. This emphasizes the functional significance of the interaction between the N-terminus of WRN and Ku70. The interaction between Ku80 and the C-terminus of WRN may modulate some other, as yet unknown, function. The strong interaction between Ku and WRN suggests that these two proteins function together in one or more pathways of DNA metabolism.
INTRODUCTION
Human Werner syndrome (WS) is characterized by early onset of age-associated diseases such as arteriosclerosis, osteoporosis and diabetes mellitus type II (1). Moreover, the patients display high levels of genomic instability and are prone to cancer (2). In culture, WS cells exhibit replicative senescence, extended S phase and a variety of chromosomal aberrations, including translocations, insertions, deletions, etc. (1). The Werner gene (wrn) encodes a protein of 1432 amino acids (WRN) (3). It is a member of the RecQ family of helicases. Seven conserved helicase motifs in the central region of the WRN polypeptide share homology with bacterial RecQ, yeast sgs1 and human Bloom (BLM) and Rothmund Thomson (RecQL) proteins. The striking difference between WRN and other members of the RecQ helicase family is the N-terminal exonuclease domain of WRN, which shares homology with RNA polymerase I and RNase D of Escherichia coli (4).
Biochemical and genetic evidence suggests that WRN plays an important role in DNA metabolism, possibly by participating in DNA replication, transcription, repair and/or recombination. Purified recombinant WRN displays both 3′→5′ helicase and 3′→5′ exonuclease activity on a variety of DNA substrates (5,6). WS cells are hypersensitive to the DNA-damaging agent 4-nitroquinoline-1-oxide, topoisomerase inhibitors and DNA interstrand cross-linking agents (4). Thus, WRN is likely to have a role in the DNA damage response pathway. This notion is further strengthened by the observation that a number of important cellular proteins that are also involved in DNA damage response pathways interact with WRN and modulate its catalytic activities. This includes human replication protein A (7), p53 (8) and flap endonuclease 1 (FEN1) (9).
We have reported that a factor required for the end joining pathway for double-strand break (DSB) repair, the Ku heterodimer, interacts with WRN (10,11). WRN exonuclease is generally active on the 3′-recessed strand of a partial DNA duplex. Ku not only stimulates this function, but also relaxes substrate preference, making WRN exonuclease active on substrates like blunt end DNA duplex, 3′-protruding DNA, single-stranded DNA (12) and DNA containing oxidative DNA base lesions (13).
In eukaryotic cells, Ku has been implicated as a key molecule in DNA DSB repair by the non-homologous end joining (NHEJ) pathway (14). Ku binds to the broken DNA ends and recruits several other factors to DNA ends that are required for efficient NHEJ, including DNA-PKcs (the catalytic subunit of DNA-activated protein kinase) and the XRCC4–ligase IV complex (15,16).
NHEJ often involves significant processing of broken ends before joining can occur, but the identity of the processing factors are still only partly known. The strong physical and functional interaction between WRN and Ku suggests that the exonuclease activity of WRN might participate in the processing of DNA ends during NHEJ. Cells deficient in WRN, Ku70 or Ku80 all show genomic instability and undergo premature replicative senescence (17), consistent with the suggestion that WRN and Ku act in a common pathway in DNA metabolism (13).
Recently, another laboratory reported that the N-terminus of WRN interacts with amino acids 215–276 of Ku80 (11,12). However, that study utilized in vitro translated Ku and included no analysis of the WRN–Ku functional interaction to substantiate the physical interaction. We undertook the current studies to map the region(s) of interaction between WRN and Ku. We report here, using several approaches, that both the N- and C-termini of WRN can interact independently with Ku. The C-terminus of WRN interacts with the Ku80 subunit, while the N-terminus of WRN interacts with the Ku70 subunit. We further show that the interaction between WRN and Ku80 is not required for stimulation of exonuclease activity.
MATERIALS AND METHODS
Proteins
Baculovirus constructs for recombinant hexa-histidine tagged full-length WRN protein or a truncated version of WRN (N-terminal 368 amino acids, designated N-WRN) were kindly provided by Dr Matthew Gray (University of Washington, Seattle, WA). Amplified baculovirus was used to infect sf9 insect cells for overexpression of WRN protein as previously described (18). The protein was purified by passing through DEAE–Sepharose (Pharmacia), Q Sepharose (Pharmacia) and Ni–NTA (Gibco BRL) chromatography (18). Purity of the protein was routinely checked by Coomassie staining. A recombinant hexa-histidine tagged C-terminal fragment of WRN (residues 940–1432, designated C-WRN) was overexpressed in E.coli and purified as previously described (10). Human Ku heterodimer was overexpressed and purified using baculoviral constructs as previously reported (16). A Ku80 truncation mutant (1–571) was made by PCR amplifcation of the full-length human cDNA using the primers 80N (5′-acaggaattcatggtgcggtcggggaataaggca) and 80571STOP (5′-agtctagactactcagtctttaattttttagctgtaggt), followed by insertion into pFASTBAC1 (Life Technologies). Virus stocks expressing truncation mutants Ku70(330–609) and Ku70(430–542) (19) were the gift of Dr Wesley Reeves (University of Florida).
Materials
[γ-32P]ATP was purchased from New England Nuclear. ATP was purchased from Amersham Pharmacia Biotech.
Antibodies
Rabbit polyclonal anti-human WRN (NB-100) and goat polyclonal anti-human WRN (sc-9916) antibodies (both against the N-terminus of WRN) were purchased from Novus Biologicals and Santa Cruz biotechnology, respectively. Mouse monoclonal anti-WRN (W82920) and goat polyclonal anti-WRN (sc-1956) antibodies (both against the C-terminus) were obtained from Pharmingen and Santa Cruz Biotechnology, respectively. Mouse monoclonal antibodies against Ku70 (sc-5309) and Ku80 (sc-5280) were purchased from Santa Cruz Biotechnology. Mouse monoclonal anti-poly histidine antibody was obtained from Sigma Chemical Co.
Far western analysis
Far western blotting analysis was conducted essentially as described elsewhere (20). Briefly, 0.2–1.0 µg each polypeptide was electrophoresed on 8–16% acrylamide Tris–glycine gels and transferred to PVDF membranes (Amersham). All subsequent steps were performed at 4°C. Membranes were immersed twice in denaturation buffer (6 M guanidine–HCl in PBS) for 10 min followed by 6 × 10 min washes in serial dilutions (1:1) of denaturation buffer in PBS supplemented with 1 mM DTT. Membranes were blocked in TBS containing 10% powdered milk, 0.3% Tween-20 for 30 min before being incubated with purified proteins in TBS supplemented with 0.25% powdered milk, 0.3% Tween-20, 1 mM DTT and 1 mM PMSF for 60 min. Membranes were then washed for 4 × 10 min in TBS containing 0.3% Tween-20, 0.25% powdered milk. The second wash contained 0.0001% glutaraldehyde. Conventional western analysis was then performed to detect the presence of interacting proteins. Appropriate horseradish peroxidase-conjugated secondary antibody (Vector Laboratories) was used and the signal was detected using ECL (Amersham) following the manufacturer’s instructions.
Pull-down assay
Insect cells infected with baculovirus containing Ku mutant or wild-type Ku were lysed in 150 mM NaCl, 10 mM Tris pH 7.4, 1% Triton X-100, 0.2 mM PMSF, 20 µg/ml aprotinin, 10 µg/ml leupeptin, 10 U/ml DNase. The suspension was then centrifuged at 12 000 r.p.m. in a Beckman Avanti J-25I centrifuge for 20 min at 4°C. The supernatant was incubated with ∼100 ng purified proteins (either WRN or its C- or N-terminal fragments) for 1 h at 4°C followed by incubation with appropriate antibody for 4 h at 4°C. The immune complex was then precipitated with protein G plus agarose (Santa Cruz Biotechnology) and washed three times with 150 mM NaCl, 10 mM Tris pH 7.4, 1% Triton X-100, 10 U/ml DNase. Immunoprecipitated proteins were then incubated with SDS loading buffer at 90°C for 5 min and separated in an 8–16% polyacrylamide Tris–glycine gel. Conventional western blotting was performed to identify the proteins.
Exonuclease substrates
Single-stranded DNA oligomers (72mer and 53mer or 32mer and 43mer) were obtained from Gibco BRL. The 53mer or 32mer (7 pmol) was 5′-labeled with [γ-32P]ATP (60 µCi, 3000 mCi/µmol) and polynucleotide kinase (10 U) under standard conditions. For construction of a double-stranded DNA substrate with one blunt end and one 3′-recessed (5′-overhang) end, labeled oligomer was mixed with a 2-fold excess of unlabeled 72mer or 43mer, heated together at 90°C for 5 min, then cooled slowly to 25°C. Annealed double-stranded substrates were then separated from unannealed and excess single-stranded oligomers by non-denaturing polyacrylamide (12%) gel electrophoresis. Intact double-stranded DNA substrates were recovered using a Qiaex II gel extraction kit (Qiagen) and stored at 4°C.
Exonuclease assay
The assay to measure the 3′→5′ exonuclease activity of WRN was carried out in 10 µl at 37°C with 40 mM Tris pH 7.4, 5 mM MgCl2, 1 mM DTT, 0.1 mg/ml BSA, 10 mM ATP (6). Three femtomoles of exonuclease substrate was incubated with the indicated amount of proteins. The reactions were initiated by the addition of WRN and incubated at 37°C for 60 min. Reactions were stopped by addition of an equal volume of formamide loading buffer (80% formamide, 0.5× TBE, 0.1% bromophenol blue) followed by incubation at 90°C for 5 min. The digestion products of these reactions were separated on denaturing 14% polyacrylamide gels and visualized using a phosphorimager and quantitated using ImageQuant software (Molecular Dynamics).
Band shift assay
For DNA-binding activity the solution conditions were the same as used in the exonuclease assay. Approximately 3 fmol 32P-labeled exonuclease substrate was incubated on ice for 10 min with the indicated amount of proteins in buffer containing 40 mM Tris–HCl pH 7.4, 5 mM MgCl2, 1 mM DTT, 0.1 mg/ml BSA and 10 mM ATP. The DNA/protein mixture (10 µl) was analyzed by 4% non-denaturing polyacrylamide gel electrophoresis at 4°C at 200 V for 2 h in 1× Tris–acetate buffer. DNA was visualized using a phosphorimager and analyzed using ImageQuant software (Molecular Dynamics).
RESULTS
The C- and N-termini of WRN both physically interact with Ku
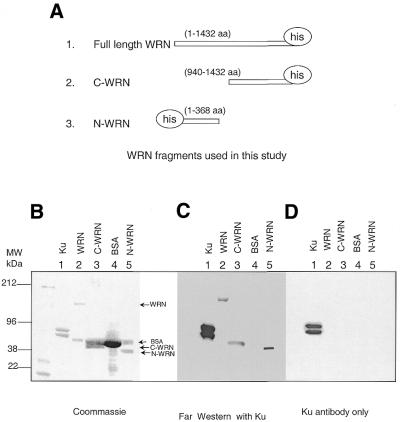
We have previously shown that the C-terminus (amino acids 940–1432) of WRN binds to Ku (10). Another research group subsequently reported that the N-terminus of WRN physically associates with a region encompassing amino acids 215–276 of Ku80 (11). We performed far western analysis with different WRN fragments (Fig. 1A) to directly determine the region of physical interaction between WRN and Ku. The gels also contained BSA and purified recombinant Ku heterodimer. In Figure 1B, Coomassie staining identified those proteins. The proteins from the other two gels were immobilized on PVDF membranes. One membrane was incubated with purified recombinant Ku70/Ku80 heterodimer to characterize the physical association between Ku and WRN fragments (Fig. 1C). The second membrane (containing the same proteins, full-length WRN, C-WRN, N-WRN, BSA and Ku) was incubated with the buffer without purified Ku (Fig. 1D). After removal of the unbound protein, the membranes were immunoblotted with antibody against Ku heterodimer. In Figure 1C, Ku antibody detects the bands corresponding to the N- and C-termini of WRN as well as full-length WRN, but not the BSA control. In the second membrane (Fig. 1D), which was incubated with buffer only, the antibody detects bands corresponding to the Ku heterodimer only, indicating that there was no cross-reaction between Werner fragments and the anti-Ku antibody (Fig. 1D).
Figure 1.
Both the C- and N-termini of WRN interact physically with Ku. (A) Recombinant WRN protein fragments used in this study. Schematic map of the WRN fragments used. Far western analysis with purified proteins. Approximately 0.5 µg Ku, C-WRN, N-WRN and whole WRN proteins were immobilized on a PVDF membrane and incubated with purified Ku followed by immunoblotting with mouse monoclonal antibody against Ku. (B) Coomassie staining of the gel. Note that BSA was used to stabilize WRN, C-WRN and N-WRN during purification. Therefore, additional bands corresponding to BSA are seen with WRN, C-WRN and N-WRN. (C) Far western analysis with purified Ku heterodimer followed by immunoblotting with Ku antibody. (D) Immunoblot with Ku antibody only, showing no cross-reaction of the antibody with full-length WRN or its fragments.
Both Ku subunits interact with WRN
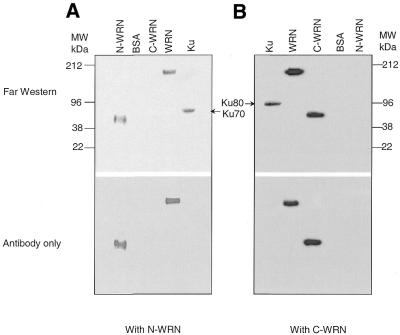
We next investigated which Ku subunit binds to the WRN fragments by far western analysis. In Figure 2A we show that the antibody against N-WRN detects a band corresponding to the Ku70 subunit. We also detect binding of the C-terminal fragment of WRN to the 80 kDa subunit of Ku (Fig. 2B). The slight difference in mobility is indicated by the arrows and is easily detetectable. In the bottom panels the membranes were immunoblotted with antibodies against N-WRN and C-WRN, respectively. None of these antibodies cross-reacted with the Ku subunits. Thus, the two different regions of WRN bind independently to the two different Ku subunits.
Figure 2.
Both subunits of Ku physically interact with WRN. Immobilized purified proteins (0.5–1 µg) on PVDF membranes were incubated with either the N- or C-terminus of WRN followed by immunoblotting with antibody against the C- and N-termini of WRN. (A) The membranes were incubated either with (top) or without (bottom) N-WRN and immunoblotted with antibody against the N-terminus of WRN. (B) Membranes were incubated either with (top) or without (bottom) C-WRN followed by incubation with antibody against the C-terminus of WRN.
To further substantiate these observations, we immunoprecipitated WRN–Ku complexes from cell extracts. Purified full-length WRN was used to pull down Ku from insect cell extracts infected with baculovirus containing either Ku80 alone, Ku70 alone or both. As a control, we used insect cell lysate that had not been infected with virus. Both Ku subunits were pulled down by WRN from the cells infected with baculovirus containing wild-type Ku (Fig. 3A, lane 2). WRN also interacted separately with Ku80 (Fig. 3A, lane 3) and Ku70 (Fig. 3A, lane 4) from insect cells infected with baculovirus containing only Ku80 and Ku70, respectively. When we incubated the cell lysate containing wild-type Ku heterodimer with anti-WRN antibody only (without prior incubation with purified WRN) we observed no interaction, thus demonstrating that WRN antibody does not react with Ku (Fig. 3A, lane 5). The same blot was reprobed with anti-polyhistidine to show that equal amounts of purified WRN were used for each pull-down assay (Fig. 3B). Also, since Ku70 is tagged with histidine, the band corresponding to Ku70 is also seen in this figure.
Figure 3.
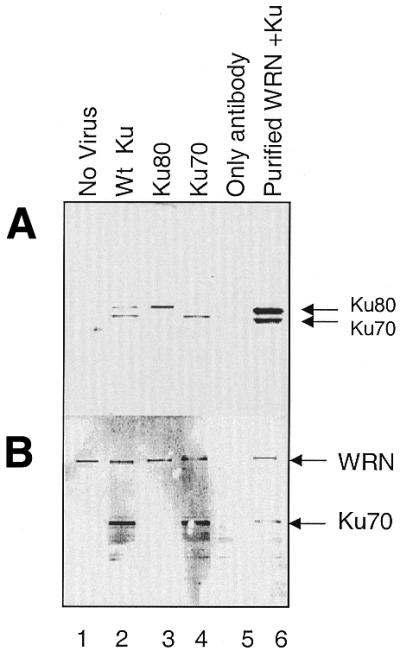
Both the Ku70 and Ku80 subunits are pulled down by WRN. (A) Insect cells infected with virus containing the designated Ku were lysed and incubated with recombinant WRN. Anti-WRN antibody was used to pull down the interacting proteins. The final immunocomplex was separated in an 8–16% acrylamide Tris–glycine gel and immunoblotted simultaneously with antibodies against Ku80 and Ku70. Insect cells without virus (no virus, lane 1) or infected with wild-type Ku and incubated only with antibody (only antibody, lane 5) were used as controls. (B) The same membrane was reprobed with antibody against polyhistidine. Bands corresponding to WRN and Ku70 were visualized, as both proteins have a polyhistidine tag.
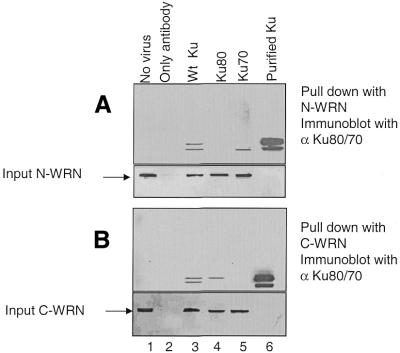
We next assayed for the ability of C- and N-WRN to differentially pull down the two subunits of Ku from insect cells infected with baculovirus containing either Ku80 alone, Ku70 alone or both. N-WRN specifically interacts with Ku70 (Fig. 4A, lane 5), while C-WRN interacts with Ku80 (Fig. 4B, lane 4). This experiment confirms our previous observations that the Ku80 subunit interacts with the C-terminal end of WRN and that the Ku70 subunit interacts with the N-terminal end of WRN. The presence of roughly equivalent amounts of WRN fragment in each pull down was verified by reprobing each blot with antibodies against N-WRN (Fig. 4A, bottom) and C-WRN (Fig. 4B, bottom).
Figure 4.
Ku70 interacts with N-WRN and Ku80 interacts with C-WRN. (A) Recombinant N-WRN was used for the pull-down assay with either wild-type Ku heterodimer-, Ku70- or Ku80-containing insect cells. (Bottom) Input N-WRN used in each pull-down reaction. (B) Recombinant C-WRN was used to pull down Ku from the same sets of insect cell lysates. (Bottom) Input C-WRN used in each pull-down reaction.
Further mapping of Ku with WRN
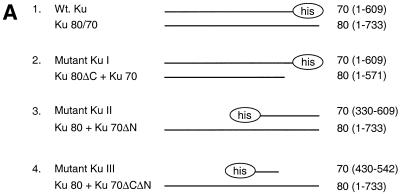
The interaction between WRN and Ku was further characterized using Ku truncation mutants (Fig. 5A). Figure 5B shows partially purified proteins from the different mutants shown in Figure 5A. The expression levels in the different mutants were then analyzed by western blot in Figure 5C. Epitope variation between the different mutant proteins required that this analysis be performed using different antibodies. Next, WRN fragments were incubated with extracts from insect cells infected with various combinations of Ku subunit-expressing constructs. Proteins were immunoprecipitated with anti-WRN antibodies and then separated in acrylamide Tris–glycine gels and transferred to PVDF membranes. The membranes were simultaneously immunoblotted with antibodies against both Ku70 and Ku80 (Fig. 6A and C). In Figure 6A, lanes 4 and 5, only one band is detected for Ku. This is because the Ku mutation removes the epitope recognized by the detecting antibody. Though expressed at lower levels than wild-type Ku, a heterodimer of full-length Ku70 paired with a fragment of Ku80 missing the C-terminal 178 amino acids still interacts with N-WRN, but not with C-WRN. The interaction with C-WRN is no higher than the background level seen in the absence of baculovirus infection (Fig. 6, lanes 2 and 4; compare panels A and C). The C-terminus of Ku80, previously shown to mediate interaction with DNA-PKcs (21), is evidently primarily required for interaction with C-WRN. In contrast, a heterodimer of full-length Ku80 paired with a fragment of Ku70 missing the N-terminal 330 amino acids interacts weakly with N-WRN, but the interaction with C-WRN is not significantly reduced (lane 5; compare panels A and C). Deletion of the C-terminal 67 amino acids of Ku70 completely abrogates interaction with N-WRN, but interaction with C-WRN is maintained (lane 6; compare panels A and C). We conclude that the Ku region required for interaction with N-WRN resides in the C-terminal 67 amino acids of Ku.
Figure 5.
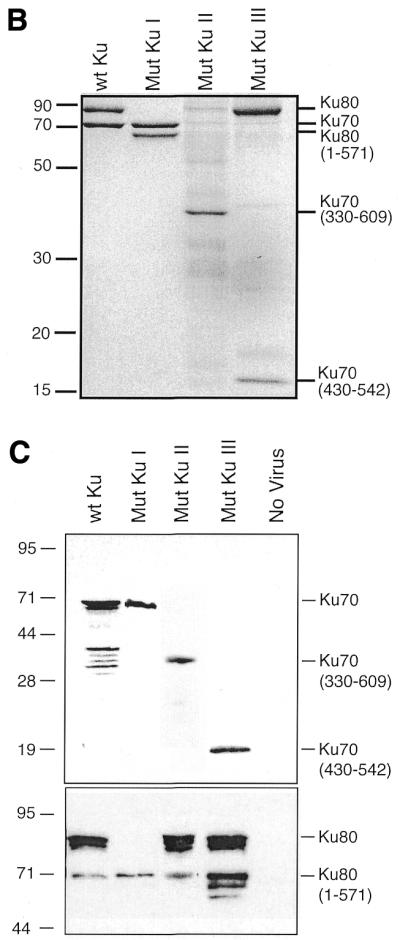
Different recombinant Ku proteins used in this study. (A) Schematic map of the mutants used. (B) Partially purified Ku variants. An aliquot of 1 µg of each preparation was run on a SDS–12% polyacrylamide gel and stained with Coomassie brilliant blue. (C) Western analysis of insect cell infections. Extract samples from ∼2 × 105 cells, infected with different Ku mutant viruses as indicated above, were subjected to western analysis. Epitope variation between the different mutant proteins required that this analysis be performed using different antibodies. These different blots were then assembled into the two panels shown above by correcting for differences in signal strength using replicate wild-type Ku samples that had been included on each different blot. (Upper) Ku70 blot probed with a polyclonal anti-Ku70 antibody, except for Mutant Ku II, probed with an anti-His tag antibody (Penta-His; Qiagen). (Lower) Ku80 blot probed with a monoclonal anti-Ku80 antibody against the Ku80 N-terminus (clone S10B1; NeoMarkers), except for Mutant Ku II, probed with a polyclonal anti-Ku80 antibody.
Figure 6.
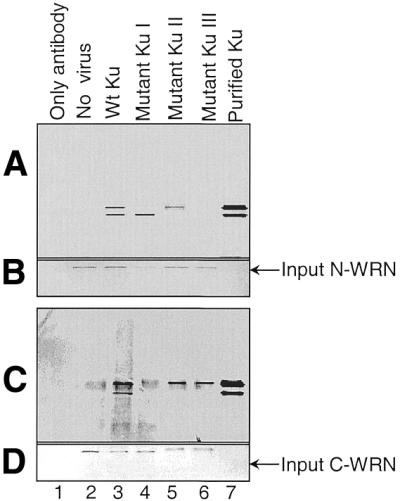
Physical interaction between WRN fragments and Ku mutants. Insect cells infected with baculovirus containing different mutant Ku were incubated with either N-WRN or C-WRN, followed by incubation with the corresponding antibody. The immunoprecipitated complex was separated by 8–16% acrylamide Tris–glycine gel electrophoresis and simultaneously immunoblotted with anti-Ku70/Ku80 antibodies. (A) Pull down with N-WRN and simultaneously immunoblotted with anti-Ku80/Ku70 antibodies. (B) The same membrane was reprobed with antibody against N-WRN. (C) Pull down with C-WRN and simultaneously immunoblotted with anti-Ku80/Ku70 antibody. (D) The same membrane was reprobed with antibody against C-WRN.
WRN exonuclease activity with mutant Ku
We have presented evidence that Ku and WRN have two distinct and separable regions of interaction. We have previously reported that Ku functionally stimulates WRN exonuclease activity (10). Therefore, to investigate the functional significance of the interactions between Ku and WRN, different Ku truncations (Fig. 5A) were used in exonuclease stimulation assays. The tested Ku heterodimers were: (i) wild-type Ku80/Ku70; (ii) a C-terminal deletion of Ku80 (Mutant Ku I, Fig. 5A, 2), an N-terminal deletion of Ku70 (Mutant Ku II, Fig. 5A, 3) and a C- and N-truncation of Ku70 (Mutant Ku III, Fig. 5A, 4). The DNA substrate was the 5′ double-strand overhang, shown in Figure 7 (inset), and wild-type WRN was used.
Figure 7.
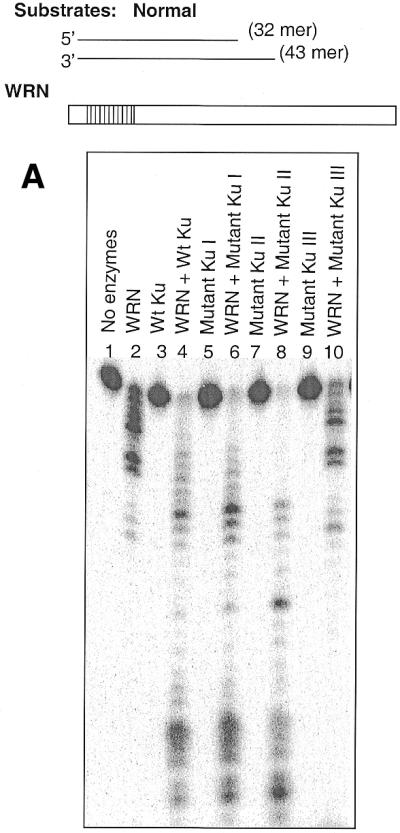
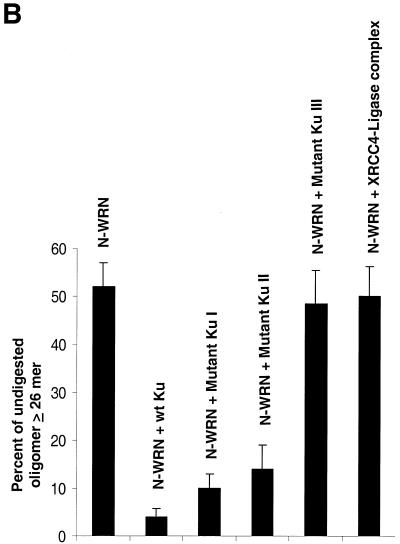
Functional interaction between WRN and Ku. (A) Different Ku mutants (15 ng each) were incubated with exonuclease substrate (32mer and 43mer) and WRN (15 ng) under standard exonuclease conditions. The DNA products were separated on a 14% denaturing polyacrylamide gel and the image was visualized with a phosphorimager. (B) Bar graph showing percent undigested oligo when N-WRN (10 ng) was used with or without different Ku mutants (10 ng) in the exonuclease assay. The representative picture is the average of two experiments.
The Ku heterodimer with full-length Ku70 and Ku80 subunits greatly stimulates the WRN exonuclease activity (Fig. 7A, compare lanes 2 and 4), while the Ku heterodimer itself has no exonuclease activity (lane 3). A Ku heterodimer with full-length Ku70 and a C-terminal truncation mutant of Ku80 (Fig. 7A, lane 6) also stimulates exonuclease activity, as does an N-terminal 330 amino acid deletion of Ku70, even though a significant portion of this Ku70 peptide is paired with co-purified Ku80 truncation products (Fig. 7A, lane 8). However, a heterodimer with a larger truncation of Ku70 (with an additional 67 amino acids deleted from the C-terminus) paired with full-length Ku80 fails to stimulate WRN exonuclease activity. Evidently, the C-terminal 67 amino acids of Ku70 are essential for the functional interaction with WRN. Similar results were also observed when a longer DNA substrate was used (53mer and 72mer; data not shown). Quantification of the lanes yielding the percent undigested oligonucleotide ≥26mer for WRN alone gave a value of 51 ± 6%, whereas in the presence of wild-type Ku, Ku80ΔC/Ku70, Ku80/Ku70ΔN and Ku80/Ku70ΔCΔN the values were 4 ± 3%, 5 ± 2.5%, 9 ± 5% and 52 ± 7%, respectively. Similar results were also observed when using 4-fold greater amounts of the mutant heterodimer (data not shown).
We have previously reported that the purified N-terminal fragment (amino acids 1–368) of WRN retains similar exonuclease activity to wild-type WRN (6). We next used the purified N-terminal fragment of WRN to further characterize the Ku interactions (Fig. 8, inset). The bar graph in Figure 7B represents quantification of the scanned lanes and the undigested oligonucleotide ≥26mer for N-WRN with or without different Ku mutant proteins. Of all Ku heterodimer mutants, only the C- and N-terminal truncated Ku70 heterodimer was unable to stimulate the N-WRN exonuclease activity significantly, as demonstrated by the percent undigested oligonucleotide remaining (Fig. 7B).
Figure 8.
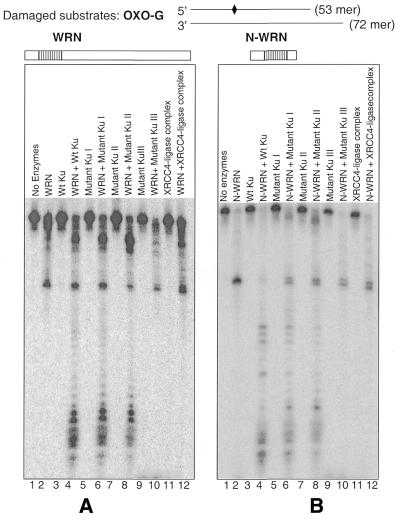
Effect of Ku mutant on WRN exonuclease DNA damage bypass. (A) The Ku mutant proteins (15 ng) were incubated with exonuclease substrate containing 8-oxoguanine and WRN (15 ng) under standard exonuclease conditions. The products were separated on a 14% denaturing polyacrylamide gel and visualized with a phosphorimager. (B) The same sets of purified Ku mutant (10 ng) were incubated with N-WRN (10 ng) and damage-containing substrate. As a negative control purified XRCC4–ligase complex was used, which has no effect on either WRN or N-WRN exonuclease activity.
We have recently shown that certain types of DNA base damage (8-oxoguanine or 8-oxoadenine) specifically block WRN exonuclease (6). The interaction with Ku greatly facilitates the ability of WRN to bypass these types of DNA damage (13). Thus, another way to map the functional interaction between WRN and Ku is to examine the ability of different mutant Ku proteins to facilitate WRN bypass of base damage in a DNA substrate. We assayed WRN exonuclease activity on a substrate containing 8-oxoguanine at a position 26 nt from the 3′ end with different Ku mutant proteins. The WRN wild-type protein (Fig. 8A) and N-WRN (Fig. 8B) were both used in these experiments. The Ku70ΔCΔN/Ku80 mutant heterodimer failed to stimulate the ability of either full-length WRN or N-WRN to bypass 8-oxoguanine (Fig. 8A and B, lane 10). As a control, we note that a similarly prepared protein (XRCC4–ligase IV complex) failed to demonstrate functional stimulation of WRN or N-WRN exonuclease activity (Fig. 8A and B, lane 12, respectively).
DNA-binding capability of WRN and Ku mutant
Binding of Ku to linear double-stranded DNA enhances the DNA end processivity of WRN. We performed electrophoretic mobility shift analyses to determine the DNA-binding ability of WRN and different mutant Ku complexes. WRN failed to bind the DNA substrate (Fig. 9, lane 2). Wild-type Ku binds with the substrate efficiently (lane 3). The formation of two complexes (Fig. 9, compare bands b and c) with wild-type Ku or Ku mutants may indicate the assembly of more than one Ku molecule per DNA molecule. Addition of WRN to the DNA–Ku complex resulted in a further shift of the band (Fig. 9, compare bands a and b), indicating that Ku mediates association between WRN and DNA (lane 4). The mutant Ku heterodimer I (Ku80ΔC/Ku70) (lanes 5 and 6) and the mutant Ku heterodimer II (Ku80/Ku70ΔN) (lanes 7 and 8) efficiently form a complex with DNA and WRN, whereas the mutant Ku heterodimer III (Ku80/Ku70ΔCΔN) fails to interact with DNA and does not form a complex on DNA with WRN (lanes 9 and 10). Mutant Ku heterodimer III did not stimulate WRN exonuclease activity, but it heterodimerizes (19) and binds with the C-terminus of WRN, as shown in Figure 6, lane 6.
Figure 9.
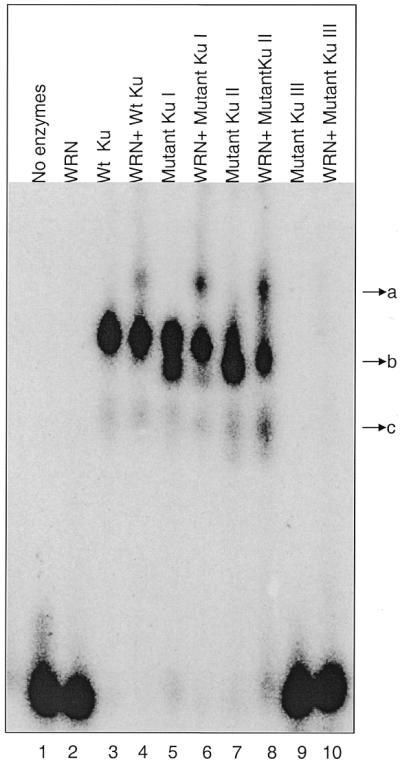
DNA binding of mutant Ku–WRN complex determined by gel mobility shift assay. The same radiolabeled linear partial duplex DNA probe used for exonuclease assay (Fig. 8A) was incubated with either different Ku mutants or wild-type Ku in the presence or absence of full-length WRN (5 ng each). The binding mixtures were resolved by electrophoresis on a 4% polyacrylamide gel, followed by visualization with a phosphorimager. Lane 1, substrate; lane 2, WRN; lane 3, wild-type Ku heterodimer; lane 4, wild-type Ku + WRN; lane 5, Ku80ΔC + KU70; lane 6, Ku80ΔC + Ku70 with WRN; lane 7, Ku80 + Ku70ΔN; lane 8, Ku80 + Ku70ΔN with WRN; lane 9, Ku80 + Ku70ΔCΔN; lane 10, Ku80 + Ku70ΔCΔN with WRN. Binding of Ku with DNA is represented by band c. Band b represents binding of more than one Ku molecule to DNA. Addition of WRN to the Ku–DNA complex produces another shift (band a).
DISCUSSION
In this study we have mapped the physical and functional interactions between the Ku heterodimer and WRN and shown that WRN physically associates with each of the two distinct subunits of the Ku heterodimer. An N-terminal fragment of WRN interacts with Ku70, while a C-terminal fragment of WRN interacts with Ku80. This is shown using two different approaches: far western analysis with purified proteins and immunoprecipitation experiments using purified WRN fragments and Ku subunits expressed in insect cells. Our results are in sharp contrast to a previous study, using in vitro translated proteins, arguing that WRN associates with Ku only through an interaction between the N-terminus of WRN and a portion of Ku80 (12). These different results might be due to misfolding of Ku subunits in the in vitro expression systems that presumably does not occur when Ku is expressed in living cells.
The exact cellular roles of WRN helicase and exonuclease are not yet clear, but recent studies from several laboratories indicate that WRN might play an important role in general DNA metabolism. Regulation of WRN catalytic activities seems to be important in maintaining genomic stability. Several important cellular proteins interact with WRN through its C- or N-terminus and some of them modulate its catalytic activities. For example, p53 interacts directly with the C-terminus of WRN and this interaction inhibits WRN exonuclease activity (8). The C-terminus of WRN also interacts with human FEN1 and stimulates its cleavage activity on a flap substrate (9). The N-terminus of WRN physically interacts with PCNA (22), which forms a ring around duplex DNA. This stabilizes the association of DNA polymerase δ with DNA during replication and increases DNA polymerase δ processivity (23).
Our observations here as well as another recent study (11) demonstrate that the N-terminus of WRN interacts with Ku. Further, we have presented evidence in this report that both the C- and N-termini of WRN interact distinctly with the Ku70 and Ku80 subunits, respectively. Recent characterization of the structure of Ku indicates that this factor similarly forms a ring around DNA helices, much as does PCNA, and suggests that Ku and PCNA might act as interchangeable processivity factors for WRN activity (24).
A short central region of Ku70 and Ku80 mediates heterodimerization (25). The Ku70 heterodimerization domain (Ku70ΔCΔN) paired with full-length Ku80 fails to stimulate WRN exonuclease activity, yet it can interact with WRN though a Ku80–C-WRN interaction. In contrast, a truncation of the C-terminal 178 amino acids of Ku80 paired with full-length Ku70 disrupts the interaction with the C-terminal fragment of WRN, but does not affect interaction with the N-WRN fragment or the ability of Ku to stimulate exonuclease activity. The interaction between Ku80 and WRN is thus not sufficient for stimulation of exonuclease activity. Rather, the inability of the Ku70ΔNΔC/Ku80 heterodimer to stimulate exonuclease activity indicates that either the ability of Ku to bind DNA (missing in Ku70ΔNΔC/Ku80; Fig. 5A) or the Ku70–N-WRN interaction is functionally critical.
The C-terminal domain of Ku80 is required for interaction with DNA-PKcs (26) as well as with the C-terminal domain of WRN. Recent work from this laboratory also indicates that DNA-PKcs inactivates WRN exonuclease activity by phosphorylation (27). We suggest that the interaction between Ku80 and C-WRN may help WRN evade this DNA-PKcs-dependent inactivation, allowing WRN to compete with the ability of DNA-PKcs to interact with Ku.
The physical interaction between N-WRN and Ku70 enhances the nucleolytic processing of DNA ends by WRN. This interaction brings the N-terminus of WRN into close proximity to the DNA. For NHEJ, the excess sequences beyond the region of microhomology should then be trimmed off. Thus, it is tempting to speculate that WRN exonuclease activity may be necessary in processing DNA ends before ligation, and we propose that interaction between WRN and Ku is important to facilitate the nucleolytic processing of ends often observed prior to NHEJ-mediated DSB repair. A recent study showed greater fragmentation of chromosomes in WS cells after ionizing radiation (28), further strengthening this possibility. Other recent studies support the notion that WRN could be involved in recombination (29). WS cells are hypersensitive to some kinds of DNA damage that cause DNA interstrand cross-links (30) and we have recently found that WS cells are deficient in the processing of psoralen-induced DNA interstrand cross-links (31). Thus, WRN may participate in recombinational repair via both homologous and non-homologous rejoining. The ability of Ku to facilitate WRN exonuclease bypass of certain types of oxidative DNA base lesions could suggest additional flexibility in the processing of ends that might be important for repair of radiation-induced DNA breaks.
While Ku and WRN are likely involved in DNA repair, they may also participate in other processes. For example, a study has shown that Ku participates in transcription (32,33) and we have previously reported that WRN protein participates in transcription, both in vitro and in vivo (34). Ku also locates at the telomeric ends (35), where its interaction with WRN may be biologically important. The WRN homolog sgs1 from Saccharomyces cerevisiae also locates to telomeric ends (36).
Interactions between WRN and several cellular proteins strongly suggest that WRN plays an important role in cellular metabolism, including transcription, replication, repair and/or recombination. A better understanding of WRN function is necessary to address how a loss of WRN causes WS cellular and clinical phenotypes, including increased risk of cancer and age-associated diseases.
Acknowledgments
ACKNOWLEDGEMENTS
We thank W. Reeves (University of Florida) for the gift of baculovirus to express Ku70 trucation mutants. We thank Drs Opresko and Harrigan for helpful suggestions and Jason Piotrowski for technical assistance. A portion of this work was supported by NIH grant CA 84442-01 to D.A.R.
REFERENCES
- 1.Oshima J. (2000) The werner syndrome protein: an update. Bioessays, 22, 894–901. [DOI] [PubMed] [Google Scholar]
- 2.Martin G.M. (1978) Genetic syndromes in man with potential relevance to the pathobiology of aging. Birth Defects, 14, 5–39. [PubMed] [Google Scholar]
- 3.Yu C.E., Oshima,J., Fu,Y.H., Wijsman,E.M., Hisama,F., Alisch,R., Matthews,S., Nakura,J., Miki,T., Ouais,S. et al. (1996) Positional cloning of the Werner’s syndrome gene. Science, 272, 258–262. [DOI] [PubMed] [Google Scholar]
- 4.Bohr V.A., Cooper,M., Orren,D., Machwe,A., Piotrowski,J., Sommers,J., Karmakar,P. and Brosh,R. (2000) Werner syndrome protein: biochemical properties and functional interactions. Exp. Gerontol., 35, 695–702. [DOI] [PubMed] [Google Scholar]
- 5.Shen J.C., Gray,M.D., Oshima,J. and Loeb,L.A. (1998) Characterization of Werner syndrome protein DNA helicase activity: directionality, substrate dependence and stimulation by replication protein A. Nucleic Acids Res., 26, 2879–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Machwe A., Ganunis,R., Bohr,V.A. and Orren,D.K. (2000) Selective blockage of the 3′→5′ exonuclease activity of WRN protein by certain oxidative modifications and bulky lesions in DNA. Nucleic Acids Res., 28, 2762–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brosh R.M. Jr, Orren,D.K., Nehlin,J.O., Ravn,P.H., Kenny,M.K., Machwe,A. and Bohr,V.A. (1999) Functional and physical interaction between WRN helicase and human replication protein A. J. Biol. Chem., 274, 18341–18350. [DOI] [PubMed] [Google Scholar]
- 8.Brosh R.M. Jr, Karmakar,P., Sommers,J.A., Yang,Q., Wang,X.W., Spillare,E.A., Harris,C.C. and Bohr,V.A. (2001) p53 modulates the exonuclease activity of Werner syndrome protein. J. Biol. Chem., 276, 35093–35102. [DOI] [PubMed] [Google Scholar]
- 9.Brosh R.M. Jr, von Kobbe,C., Sommers,J.A., Karmakar,P., Opresko,P.L., Piotrowski,J., Dianova,I., Dianov,G.L. and Bohr,V.A. (2001) Werner syndrome protein interacts with human flap endonuclease 1 and stimulates its cleavage activity. EMBO J., 20, 5791–5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooper M.P., Machwe,A., Orren,D.K., Brosh,R.M., Ramsden,D. and Bohr,V.A. (2000) Ku complex interacts with and stimulates the Werner protein. Genes Dev., 14, 907–912. [PMC free article] [PubMed] [Google Scholar]
- 11.Li B. and Comai,L. (2000) Functional interaction between Ku and the werner syndrome protein in DNA end processing. J. Biol. Chem., 275, 39800. [DOI] [PubMed] [Google Scholar]
- 12.Li B. and Comai,L. (2001) Requirements for the nucleolytic processing of DNA ends by the Werner syndrome protein-Ku70/80 complex. J. Biol. Chem., 276, 9896–9902. [DOI] [PubMed] [Google Scholar]
- 13.Orren D.K., Machwe,A., Karmakar,P., Piotrowski,J., Cooper,M.P. and Bohr,V.A. (2001) A functional interaction of Ku with Werner exonuclease facilitates digestion of damaged DNA. Nucleic Acids Res., 29, 1926–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Featherstone C. and Jackson,S.P. (1999) Ku, a DNA repair protein with multiple cellular functions? Mutat. Res., 434, 3–15. [DOI] [PubMed] [Google Scholar]
- 15.Yannone S.M., Roy,S., Chan,D.W., Murphy,M.B., Huang,S., Campisi,J. and Chen,D.J. (2001) Werner syndrome protein is regulated and phosphorylated by DNA-dependent protein kinase. J. Biol. Chem., 276, 38242–38248. [DOI] [PubMed] [Google Scholar]
- 16.Nick McElhinny S.A., Snowden,C.M., McCarville,J. and Ramsden,D.A. (2000) Ku recruits the XRCC4-ligase IV complex to DNA ends. Mol. Cell. Biol., 20, 2996–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu Y., Seidl,K.J., Rathbun,G.A., Zhu,C., Manis,J.P., van der Stoep,N., Davidson,L., Cheng,H.L., Sekiguchi,J.M., Frank,K. et al. (1997) Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity, 7, 653–665. [DOI] [PubMed] [Google Scholar]
- 18.Orren D.K., Brosh,R.M.,Jr, Nehlin,J.O., Machwe,A., Gray,M.D. and Bohr,V.A. (1999) Enzymatic and DNA binding properties of purified WRN protein: high affinity binding to single-stranded DNA but not to DNA damage induced by 4NQO. Nucleic Acids Res., 27, 3557–3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J., Dong,X., Myung,K., Hendrickson,E.A. and Reeves,W.H. (1998) Identification of two domains of the p70 Ku protein mediating dimerization with p80 and DNA binding. J. Biol. Chem., 273, 842–848. [DOI] [PubMed] [Google Scholar]
- 20.Brosh R.M. Jr, Li,J.L., Kenny,M.K., Karow,J.K., Cooper,M.P., Kureekattil,R.P., Hickson,I.D. and Bohr,V.A. (2000) Replication protein A physically interacts with the Bloom’s syndrome protein and stimulates its helicase activity. J. Biol. Chem., 275, 23500–23508. [DOI] [PubMed] [Google Scholar]
- 21.Singleton B.K., Torres-Arzayus,M.I., Rottinghaus,S.T., Taccioli,G.E. and Jeggo,P.A. (1999) The C terminus of Ku80 activates the DNA-dependent protein kinase catalytic subunit. Mol. Cell. Biol., 19, 3267–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lebel M., Spillare,E.A., Harris,C.C. and Leder,P. (1999) The Werner syndrome gene product co-purifies with the DNA replication complex and interacts with PCNA and topoisomerase I. J. Biol. Chem., 274, 37795–37799. [DOI] [PubMed] [Google Scholar]
- 23.Prelich G., Tan,C.K., Kostura,M., Mathews,M.B., So,A.G., Downey,K.M. and Stillman,B. (1987) Functional identity of proliferating cell nuclear antigen and a DNA polymerase-delta auxiliary protein. Nature, 326, 517–520. [DOI] [PubMed] [Google Scholar]
- 24.Walker J.R., Corpina,R.A. and Goldberg,J. (2001) Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature, 412, 607–614. [DOI] [PubMed] [Google Scholar]
- 25.Osipovich O., Durum,S.K. and Muegge,K. (1997) Defining the minimal domain of Ku80 for interaction with Ku70. J. Biol. Chem., 272, 27259–27265. [DOI] [PubMed] [Google Scholar]
- 26.Gell D. and Jackson,S.P. (1999) Mapping of protein–protein interactions within the DNA-dependent protein kinase complex. Nucleic Acids Res., 27, 3494–3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karmakar P., Piotrowski,J., Brosh,R.M.,Jr, Sommers,J.A., Miller,S.P., Cheng,W.H., Snowden,C.M., Ramsden,D.A. and Bohr,V.A. (2002) Werner protein is a target of DNA-dependent protein kinase in vivo and in vitro, and its catalytic activities are regulated by phosphorylation. J. Biol. Chem., 277, 18291–18302. [DOI] [PubMed] [Google Scholar]
- 28.Grigorova M., Balajee,A.S. and Natarajan,A.T. (2000) Spontaneous and X-ray-induced chromosomal aberrations in Werner syndrome cells detected by FISH using chromosome-specific painting probes. Mutagenesis, 15, 303–310. [DOI] [PubMed] [Google Scholar]
- 29.Oshima J., Huang,S., Pae,C., Campisi,J. and Schiestl,R.H. (2002) Lack of WRN results in extensive deletion at nonhomologous joining ends. Cancer Res., 62, 547–551. [PubMed] [Google Scholar]
- 30.Poot M., Yom,J.S., Whang,S.H., Kato,J.T., Gollahon,K.A. and Rabinovitch,P.S. (2001) Werner syndrome cells are sensitive to DNA cross-linking drugs. FASEB J., 15, 1224–1226. [DOI] [PubMed] [Google Scholar]
- 31.Bohr V.A., Souza,P.N., Nyaga,S.G., Dianov,G., Kraemer,K., Seidman,M.M. and Brosh,R.M.,Jr (2001) DNA repair and mutagenesis in Werner syndrome. Environ. Mol. Mutagen., 38, 227–234. [DOI] [PubMed] [Google Scholar]
- 32.Yoo S. and Dynan,W.S. (1998) Characterization of the RNA binding properties of Ku protein. Biochemistry, 37, 1336–1343. [DOI] [PubMed] [Google Scholar]
- 33.Woodard R.L., Lee,K.J., Huang,J. and Dynan,W.S. (2001) Distinct roles for Ku protein in transcriptional reinitiation and DNA repair. J. Biol. Chem., 276, 15423–15433. [DOI] [PubMed] [Google Scholar]
- 34.Balajee A.S., Machwe,A., May,A., Gray,M.D., Oshima,J., Martin,G.M., Nehlin,J.O., Brosh,R., Orren,D.K. and Bohr,V.A. (1999) The Werner syndrome protein is involved in RNA polymerase II transcription. Mol. Biol. Cell, 10, 2655–2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsu H.L., Gilley,D., Blackburn,E.H. and Chen,D.J. (1999) Ku is associated with the telomere in mammals. Proc. Natl Acad. Sci. USA, 96, 12454–12458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson F.B., Marciniak,R.A., McVey,M., Stewart,S.A., Hahn,W.C. and Guarente,L. (2001) The Saccharomyces cerevisiae WRN homolog Sgs1p participates in telomere maintenance in cells lacking telomerase. EMBO J., 20, 905–913. [DOI] [PMC free article] [PubMed] [Google Scholar]