Abstract
For the production of DNA microarrays from PCR products, purification of the the DNA fragments prior to spotting is a major expense in cost and time. Also, a considerable amount of material is lost during this process and contamination might occur. Here, a protocol is presented that permits the manufacture of microarrays from unpurified PCR products on aminated surfaces such as glass slides coated with the widely used poly(l-lysine) or aminosilane. The presence of primer molecules in the PCR sample does not increase the non-specific signal upon hybridisation. Overall, signal intensity on arrays made of unpurified PCR products is 94% of the intensity obtained with the respective purified molecules. This slight loss in signal, however, is offset by a reduced variation in the amount of DNA present at the individual spot positions across an array, apart from the considerable savings in time and cost. In addition, a larger number of arrays can be made from one batch of amplification products.
INTRODUCTION
DNA microarrays have become a widely used commodity in biological and biomedical research. For many applications, PCR products are attached to the support as probe molecules. For global analyses on highly complex arrays with many thousands of probe molecules especially (see for example 1–3), preparation of the PCR products is a major part of the microarray production in terms of both time and cost involved. In the production process, the DNA fragments that result from the PCR are generally checked on agarose gels, concomitantly obtaining an estimate of the DNA concentration. After purification, the DNA is taken up in a volume of spotting buffer that is smaller than the original volume during PCR, thereby increasing the DNA concentration. Usually, no accurate determination of the DNA concentration is performed, since on microarrays mostly relative measurements with two differently labelled targets are performed. In addition, a levelling in the DNA amounts present at the various spot positions can be obtained if the individual PCR products are at a concentration above the binding capacity of the array surface (4).
Purification of the PCR products requires several steps as well as expensive consumables if, for example, purification is achieved by filter- or column-based techniques. Alternatively, mere precipitation avoids this specific cost factor but is work-intensive and inefficient. Here, we describe a procedure that avoids purification altogether; only the volume of the PCR products is reduced by evaporation to increase the DNA concentration. Additional pipetting steps are avoided and no DNA is lost. Because of the latter, more microarrays can be produced from a single PCR amplification and higher concentrations can be used during spotting, thereby reducing variation in the DNA amounts across a microarray. Additionally, the procedure simplifies the quality check of the PCR fragments on agarose gels and provides the means for an uncomplicated and thus routinely applicable analysis of the spotting quality on all microarrays.
MATERIALS AND METHODS
DNA sources
About 5200 human cDNA clones of the IMAGE library (5) were obtained from the RZPD Resource Centre (Berlin, Germany). Some 21 000 random shotgun clones representing the genome of Trypanosoma brucei were provided by Najib El-Sayed of the Institute for Genomic Research (TIGR, Rockville, USA). Nearly 4550 shotgun clones covering the entire genome of Pseudomonas putida as a minimal tiling path were obtained from Helmut Hilbert of Qiagen (Hilden, Germany). PCR products for some 21 000 predicted open reading frames (ORFs) of Drosophila melanogaster were produced directly from genomic DNA. The template for some 7300 ORF-specific PCR products of Candida albicans was strain SC5314 (Can14).
PCR amplification
PCR amplifications were performed in 384- or 96-well microtitre plates. For PCR on the cDNA and shotgun clones, 0.2 µM of the respective, vector-specific primer pairs d(TCA CACAGGAAACAGCTATGAC) and d(GTAAAACGACGGCCAGTG) (human clones), d(TTGTAAAACGACGGCCAGTG) and d(GCGGATAACAATTTCACACAGGA) (T.brucei) or d(TCGGATCCACTAGTAACG) and d(GGCCGCCAGTGTGATG) (P.putida) (all from Interactiva, Ulm, Germany) were used. The reactions were started by inoculating 25 or 100 µl of PCR mix, usually in 10 mM Tris–HCl, pH 8.3, 2.25 mM MgCl2, 50 mM KCl, 0.2 mM each dATP, dTTP, dGTP and dCTP, 1.5 M betaine, 0.1 mM cresol red and 2 U Taq polymerase, with a few Escherichia coli cells transferred from a growth culture using a plastic 384- or 96-pin gadget (Genetix, New Milton, UK). The plates were incubated for 3 min at 94°C, before 35 cycles of denaturation at 94°C for 30 s, annealing at 51°C for 30 s and elongation at 72°C for 90 s were performed, followed by a final elongation phase at 72°C for 10 min. In some cases, the PCR was performed without betaine. The Drosophila ORFs were initially amplified on 100 ng genomic DNA with some 43 000 gene-specific primers, all of which contained one of several common tag sequences of 15 nt length at their 5′-ends. Subsequent re-amplification was carried out using the fitting primer pair. PCR products of C.albicans ORFs were produced on 20 ng genomic DNA with 7300 specific primer pairs.
Quality check
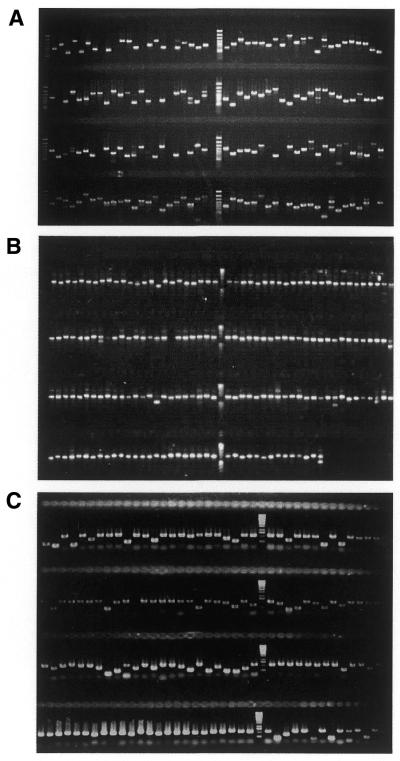
The DNA concentration of PCR products was determined by incubation with the fluorophore PicoGreen (Molecular Probes, Eugene, USA) according to the manufacturer’s recommendations. The DNA fragments were routinely checked on agarose gels stained with ethidium bromide (see Fig. 1).
Figure 1.
Agarose gel separation of PCR fragments. Typical results from amplifications of (A) human IMAGE cDNA clones and (B) shotgun clones of the P.putida genomic library and (C) the secondary amplification of Drosophila ORFs are shown.
Purification of PCR products
Purification was performed by isopropanol precipitation (6) or in MultiScreen filter plates (Millipore, Bedford, USA) as recommended by the manufacturer. The DNA was taken up in 450 mM NaCl, 45 mM sodium citrate, pH 7.5 (3× SSC), supplemented with 1.5 M betaine. During this process, the average DNA concentration of the samples was adjusted to a value of 250 ng/µl.
Microarray spotting
For unpurified DNA, the volume of each PCR sample was usually reduced to a quarter by evaporation before being used directly for microarray spotting. Alternatively, evaporation was completed, if there had been no betaine in the PCR mix, and the pellet was taken up in a quarter to a fifth of the original volume of 3× SSC plus 1.5 M betaine. Spotting was done with an SDDC-2 Micro-Arrayer from Engineering Services Inc. (Toronto, Canada) or a MicroGrid-II (BioRobotics, Cambridge, UK) and SMP3 pins (TeleChem International Inc., Sunnyvale, USA) onto amino-derivatised glass slides as described in detail earlier (4). Storage of unused PCR product was at –20 or –80°C.
RNA isolation
Total RNA was extracted from the respective organism by homogenising cells in the presence of liquid nitrogen. The still frozen powder was instantly taken up in RNA-Clean solution (ThermoHybaid, Ulm, Germany) or the equivalent TRIzol reagent (Invitrogen, Karlsruhe, Germany). RNA was extracted following the manufacturer’s instructions and in some cases purified further by precipitation with an equal volume of 5 M LiCl, 0.1 M Tris–HCl, pH 7.4. The quality of the RNA was confirmed by denaturing gel electrophoresis or using an Agilent 2100 Bioanalyser (Agilent Technologies, Waldbronn, Germany). The concentration of the total RNA was measured with the RiboGreen fluorescence assay (Molecular Probes, Eugene, USA).
Target labelling and hybridisation
Labelling of total RNA was by reverse transcription in the presence of 0.5 mM each dATP, dGTP and dTTP, 0.2 mM dCTP and 0.1 mM Cy3- or Cy5-labelled dCTP (Amersham Biosciences, Freiburg, Germany). Alternatively, labelling was done by incorporation of aminoallyl-modified dUTP (Sigma, Munich, Germany) and subsequent incorporation of monofunctional Cy3- or Cy5-N-succinimide esters (Amersham Biosciences) according to the manufacturer’s recommendations. For hybridisations on the Pseudomonas microarray, genomic DNA was labelled by random priming (7) in the presence of 25 µM Cy3- or Cy5-labelled dCTP.
Radioactively labelled cDNA mixtures were synthesised from 0.2–2 µg total RNA. Reverse transcription was performed by adding ∼50 µCi [α-33P]dCTP (Amersham Biosciences) to 20 µl of 10 mM each dATP, dGTP and dTTP and 66 µM unlabelled dCTP. The reaction was incubated at 37°C for 2 h.
Samples were purified with the QIAquick PCR purification kit (Qiagen, Hilden, Germany), dried in vacuum and dissolved in 4 µl of hybridisation buffer (50% formamide, 3× SSC, 1% SDS, 5× Denhardt’s reagent and 5% dextran sulphate) (8) per square centimetre of coverslip. During hybridisation, the slides were kept in a humidified chamber (TeleChem International Inc.) at 42°C for 16 h. Subsequently, the slides were washed in 2× SSC, 0.1% SDS for 2 min, followed by 2 min in 1× SSC, rinsed briefly in 0.2× SSC and dried by centrifugation at 500 r.p.m. for 5 min.
Data analysis
Fluorescence signals were detected on a ScanArray 5000 unit (Packard, Billerica, USA) and analysed with the GenePix software package (Axon Instruments, Union City, USA). Slides hybridised with radioactively labelled samples were covered with scintillator foil (Zinsser Analytics, Frankfurt, Germany) and the disintegrations were measured for 6–24 h with a MicroImager (Biospace Mesures SA, Paris, France). Subsequent analysis of the microarray data was performed with software algorithms written in MATLAB (Mathworks, Natick, USA), which is described in detail elsewhere (9,10; www.dkfz-heidelberg.de/tbi/services/mchips).
RESULTS
Amplification and spotting process
In the analysis, PCR products from several sources were used as probes on the microarrays. While the human cDNAs varied in size, the shotgun clones of T.brucei and P.putida had undergone a size selection to a length of ∼2 kb (Fig. 1). In all three cases, amplification was performed with vector-specific primers of 16–22 nt. The reactions were started by inoculation of the ready mixed PCR ingredients with a small amount of the relevant E.coli clone. The Drosophila probes were produced by direct amplification from genomic DNA of selected coding regions of ∼500 bp. The initial PCR products were then used as template in secondary reactions. For these, common 15 nt long primers were used, whose complements were present as tags at the ends of all genomic primers. Generation of probes for the C.albicans ORFs was performed on genomic DNA by PCR with gene-specific primer pairs only. Slightly adapted versions of the amplification protocol were used for the various biological systems.
Because of the presence of cresol red and betaine in the PCR buffer, the DNA samples could be loaded onto agarose gels directly after PCR amplification, without addition of a loading buffer. The cresol red made the samples visible during the loading process, while the betaine substituted for the glycerol that is usually added to make the sample sink into the gel slot.
Prior to spotting, the DNA was concentrated by reducing by evaporation the volume of the reaction to about a quarter. The presence of betaine was again useful in this respect, since the rate of evaporation decreases with increasing betaine concentration, eventually coming to a halt at 6.8 M betaine (4). Due to this effect, there was no need for a tight control of the evaporation process if the PCR buffer had been supplemented with 1.5 M betaine. However, PCR samples generated without betaine could also be spotted without purification. In this case, evaporation was completed and the pellet taken up in an appropriate volume of 3× SSC supplemented with 1.5 M betaine.
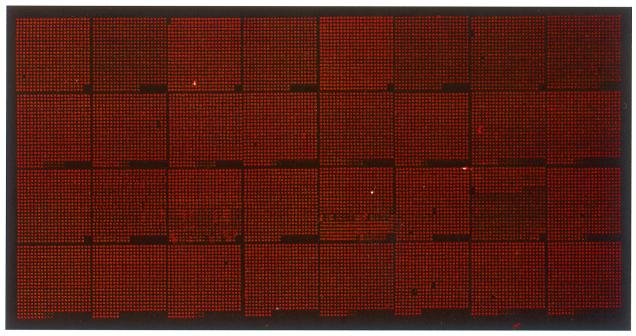
Taking advantage of the cresol red a second time, the quality of the actual spotting process could also be visualised by imaging the slides in the Cy3 channel of the laser scanner prior to the post-processing step (Fig. 2). The positional accuracy of the spotting and the spot shape and homogeneity were routinely checked by this procedure. Upon post-processing of the slide, necessary to block the remaining amino functions, the cresol red was washed off entirely, therefore not causing any background signal in the hybridisation.
Figure 2.
Quality assessment of the spotting process. DNA representing the ORFs of C.albicans was spotted. The signal produced by cresol red was visualised in the Cy3 channel of the laser scanner prior to the post-processing step, during which the signal disappeared entirely.
Effect on signal intensities
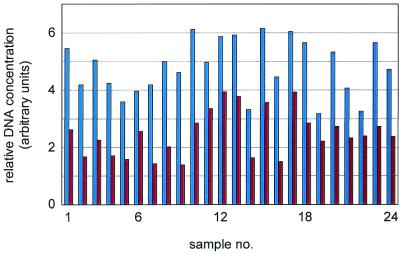
For direct comparison, PCR products were split immediately after amplification. Half of each reaction was purified. The resulting DNA pellets were dissolved in 3× SSC spotting buffer supplemented with betaine. During this process, the DNA concentration was adjusted to an average value of ∼250 ng/µl, at which saturation of the binding capacity of the support is reached under the spotting conditions used (4). Some 20–50% of the initial DNA amount was lost during the purification processes (Fig. 3). The other half of each PCR sample was not purified but only evaporated. By reducing the volume to a quarter, the DNA concentration in the samples increased accordingly to an average value that was again above the binding capacity of the slide surface.
Figure 3.
DNA loss during purification. The amounts of DNA present before (left bars) and after purification (right bars) of 24 randomly chosen PCR products of P.putida are shown. On average, 49% of the material was lost during this particular purification process.
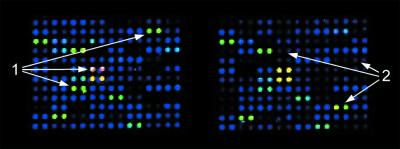
Purified and unpurified DNA fragments were immobilised next to each other on the same arrays, for side-by-side evaluation of their performance in hybridisation (Fig. 4). Comparisons were done on microarrays made from the different kinds of PCR products and organisms mentioned above. The presence of the primer molecules in the unpurified PCR samples did not increase the non-specific signal in the spots upon hybridisation, indicating that binding to the support is a relatively rare, randomly occurring event along a DNA strand, thus selecting against the attachment of the short primer molecules. Comparing hybridisations with complex samples made from various sources and labelled both directly by incorporation of fluorophore-labelled nucleotides or indirectly by addition of aminoallyl-modified dUTP and subsequent attachment of the dye, the average signal intensity on microarrays made from unpurified PCR products was found to be 94% of that obtained on purified material. However, fluctuation in the amount of DNA present in individual spots was more profound on microarrays made of purified DNA. A few PCR products were missing after the purification process that were present when unpurified material was spotted. While the overall signal was slightly lower with unpurified DNA, no such prediction could be made for individual PCR products. Although all processes were performed in a multi-well format, thus minimising differences in the processing of individual PCR products, a particular DNA fragment could be found in higher yields with either process (Fig. 4).
Figure 4.
False-colour image of signal intensities on a DNA microarray used for comparison. PCR products made from human cDNA clones had been split in half and processed with (left) and without purification (right). Spotted onto one microarray, they were subjected to hybridisation with a complex, Cy5-labelled sample made from total RNA. The arrows indicate individual spots which produced stronger signals in the respective field. Differences between the two patterns could clearly be attributed to the purification process, since the two patterns remained basically the same on microarrays of one production batch. Similar data were obtained with other probe molecules.
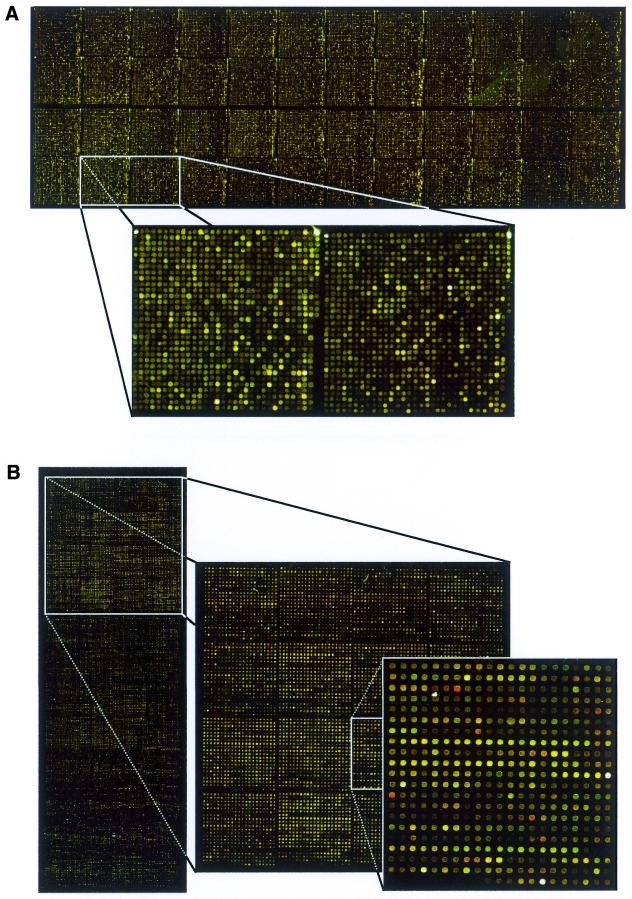
In competitive hybridisations with two samples, one labelled with Cy3, the other with Cy5, no difference in the resulting ratios could be observed. We also subjected microarrays to quantitative analyses with 33P-labelled target molecules. As with the fluorescent dyes, there were no differences in the behaviour of arrays made of purified or unpurified PCR products. Most of our microarray production is now performed without purification of the PCR products (Fig. 5). Also, in routine production, reproducibility on these microarrays is identical to results obtained with purified material and no adverse effects to the spotting equipment were noticed.
Figure 5.
Hybridisations to complex DNA microarrays made from unpurified PCR products. Typical results are presented for two different systems. (A) A microarray containing the potential ORFs of D.melanogaster plus appropriate controls, each spotted twice at different locations of the slide. In total, about 46 000 PCR fragments were applied to each microarray slide. Hybridisation was with two samples of genomic DNA labelled with Cy3 and Cy5, respectively. (B) A microarray made from the 5200 human IMAGE clones. Supplemented with PCR products of another library, this microarray contains some 12 000 different fragments of genes that had been found to be differentially transcribed in a variety of cancer tissues. Two-dye hybridisation was performed with target material representing total RNA of two human cell culture preparations.
DISCUSSION
Alternatives to microarrays made of PCR products exist in the form of oligonucleotide arrays. However, for many applications in the field of biological research, PCR products are still the more convenient probe molecule. While oligomer probes might take over eventually for transcript analyses, PCR fragments are advantageous or even essential for many other studies, such as assays on non-coding DNA regions or the interaction of proteins and double-stranded DNA, for example, which cannot be performed easily on other array formats.
Performance of the PCR amplification and spotting of the resulting DNA fragments form the work-intensive core of microarray production. Although systems like filter-bottom microtitre plates provide an effective means for the purification of PCR products, the time and cost involved are considerable. In addition, the purification process itself needs to be well established and quality controlled in order to produce reliable results (11). Even then, precious material gets lost during this process. To circumvent all this, we established a procedure of spotting PCR products without purification. Only the DNA concentration in the probe samples is increased by a reduction in the volume, since the PCR usually yields less than the ∼250 ng/µl needed to reach saturation of the binding capacity of the microarray surface. The presence of betaine is helpful at the stage of PCR, for its positive effect on amplification efficiency (12,13), in addition to its significance for spot quality (4).
Using the concentrated PCR buffer directly as spotting solution, no increase in the clogging of spotting pins was observed when the PCR products were kept at room temperature for some time prior to spotting, thereby avoiding the potential formation of salt crystals in the capillaries during the process. The presence of cresol red permitted an immediate quality control of the spotting. No significant variations could be detected even during the spotting of batches of 70–100 microarrays of several tens of thousands of features each.
Although present in ∼5-fold molar excess, the primer oligonucleotides do not seem to compete well with the PCR products for binding sites on the aminated support. This is not surprising, however. The size difference is ∼100-fold in favour of the PCR products and the attachment is a cooperative process. However, even if a small number of primer molecules are still bound to the support during hybridisation, they will not contribute significant signal, due to their substantially lower duplex stability. Also, they are kinetically much disadvantaged compared to the PCR products.
In our hands, the procedure proved very robust, irrespective of the type or source of the DNA dealt with, and analyses that would otherwise have been beyond our financial ability were possible, without compromising data quality. As a matter of fact, since chip production is performed more readily, more microarrays were used in experiments, thus providing a better statistical basis for interpretation of the results.
Acknowledgments
ACKNOWLEDGEMENTS
We thank other members of our laboratory and partners collaborating on the mentioned projects for fruitful discussions. This work was funded by the German Ministry for Education and Research (BMBF).
REFERENCES
- 1.Nielsen T.O., West,R.B., Linn,S.C., Alter,O., Knowling,M.A., O’Connell,J.X., Zhu,S., Fero,M., Sherlock,G., Pollack,J.R., Brown,P.O., Botstein,D. and van de Rijn,M. (2002) Molecular characterisation of soft tissue tumours: a gene expression study. Lancet, 359, 1301–1307. [DOI] [PubMed] [Google Scholar]
- 2.Hegde P., Qi,R., Gaspard,R., Abernathy,K., Dharap,S., Earle-Hughes,J., Gay,C., Nwokekeh,N.U., Chen,T., Saeed,A.I., Sharov,V., Lee,N.H., Yeatman,T.J. and Quackenbush,J. (2001) Identification of tumor markers in models of human colorectal cancer using a 19,200-element complementary DNA microarray. Cancer Res., 61, 7792–7797. [PubMed] [Google Scholar]
- 3.Pomeroy S.L., Tamayo,P., Gaasenbeek,M., Sturla,L.M., Angelo,M., McLaughlin,M.E., Kim,J.Y.H., Goumnerova,L.C., Black,P.M., Lau,C., Allen,J.C., Zagzag,D., Olson,J.M., Curran,T., Wetmore,C., Biegel,J.A., Poggio,T., Mukherjee,S., Rifkin,R., Califano,A., Stolovitzky,G., Louis,D.N., Mesirov,J.P., Lander,E.S. and Golub,T.R. (2002) Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature, 415, 436–442. [DOI] [PubMed] [Google Scholar]
- 4.Diehl F., Grahlmann,S., Beier,M. and Hoheisel,J.D. (2001) Manufacturing DNA-microarrays of high spot homogeneity and reduced background signal. Nucleic Acids Res., 29, e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hillier L., Lennon,G., Becker,M., Bonaldo,M.F., Chiapelli,B., Chissoe,S., Dietrich,N., DuBuque,T., Favello,A., Gish,W., Hawkins,M., Hultman,M., Kucuba,T., Lacy,M., Le,M., Le,N., Mardis,E., Moore,B., Morris,M., Parsons,J., Prange,C., Rifkin,L., Rohlfing,T., Schellenberg,K., Soares,M.B., Tan,F., Thierry-Meg,J., Trevaskis,E., Underwood,K., Wohldman,P., Waterston,R., Wilson,R. and Marra,M. (1996) Generation and analysis of 280.000 human expressed sequence tags. Genome Res., 6, 807–828. [DOI] [PubMed] [Google Scholar]
- 6.Sambrook J. and Russel,D.W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 7.Feinberg A.P. and Vogelstein,B. (1983) A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem., 132, 6–13. [DOI] [PubMed] [Google Scholar]
- 8.Welford S.M., Gregg,J., Chen,E., Garrison,D., Sorensen,P.H., Denny,C.T. and Nelson,S.F. (1998) Detection of differentially expressed genes in primary tumor tissues using representational differences analysis coupled to microarray hybridisation. Nucleic Acids Res., 26, 3059–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beißbarth T., Fellenberg,K., Brors,B., Arribas-Prat,R., Boer,J., Hauser,N.C., Scheideler,M., Hoheisel,J.D., Schütz,G., Poustka,A. and Vingron,M. (2000) Comparison and quality measures of DNA array hybridisation data. Bioinformatics, 16, 1014–1022. [DOI] [PubMed] [Google Scholar]
- 10.Fellenberg K., Hauser,N.C., Brors,B., Hoheisel,J.D. and Vingron,M. (2002) Microarray data warehouse allowing for the statistical analysis of experiment annotations. Bioinformatics, 18, 423–433. [DOI] [PubMed] [Google Scholar]
- 11.Hegde P., Qi,R., Abernathy,K., Gay,C., Dharap,S., Gaspard,R., Hughes,J.E., Snesrud,E., Lee,N. and Quackenbush,J. (2000) A concise guide to cDNA microarray analysis. Biotechniques, 29, 548–562. [DOI] [PubMed] [Google Scholar]
- 12.Mytelka D.S. and Chamberlin,M.J. (1996) Analysis and suppression of DNA polymerase pauses associated with a trinucleotide consensus. Nucleic Acids Res., 24, 2774–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henke W., Herdel,K., Jung,K., Schnorr,D. and Loening,S.A. (1997) Betaine improves the PCR amplification of GC-rich DNA sequences. Nucleic Acids Res., 25, 3957–3958. [DOI] [PMC free article] [PubMed] [Google Scholar]