Abstract
Transcription factors (TFs) have been difficult to identify by bioinformatics, due to the heterogenous nature of the domains they are composed of. Therefore, we have developed a simple and generally applicable screening system for the identification of transcriptional activators, based on the presence of a functional transactivation domain (TAD). The system utilizes a retroviral vector to express a cDNA library as fusion genes with the yeast gal4 DNA-binding domain. This retroviral library is transduced into a murine NIH3T3-based reporter cell line carrying a stable integrated gal4 promoter–green fluorescent protein reporter gene. cDNA inserts encoding a functional TAD reconstitute a chimeric TF that activates the reporter gene. After fluorescence activated cell sorting (FACS) and expansion of GFP-positive cells, the responsible cDNA inserts are retrieved. From a cDNA library of cytokine-stimulated human umbilical vein endothelial cells (HUVEC), a number of known as well as potentially novel TFs were isolated, demonstrating the suitability of the system. The identification of other factors that are currently not associated with transcriptional regulation suggest additional functions for these proteins. Moreover, our results have focused attention on signaling pathways that have not been recognized previously in the context of endothelial cell biology.
INTRODUCTION
Transcription factors (TFs) are key regulatory molecules that control the expression of genes and are involved in a wide variety of cellular functions; they also represent valuable targets for therapeutic interference. Their domain architecture includes at least a DNA-binding domain (DBD) that mediates the binding to sequence-specific DNA elements in the promoter region of genes, and a transactivation domain (TAD) that can interact with the basal transcription machinery. In many cases, additional domains mediate, e.g. homo- or heterodimerization of members of TF families, interaction with other TFs, or the binding of co-activators or low molecular weight ligands, e.g. steroid hormones.
Due to this domain structure, DNA binding and transactivating functions can be physically separated and domains exchanged between TFs. This has formed the basis for the development of a number of genetic tools, including the yeast one- and two-hybrid systems, and the path-detect trans-reporting system (1,2). By fusing a TF’s TAD to a heterologous DBD, usually the yeast gal4 DBD, transactivation is uncoupled from DNA binding and can be studied separately from other regulatory events that are necessary for activation, e.g. nuclear translocation (3).
The prediction of function based on primary sequence data obtained from the human and other genomes relies to a large extent on sequence homology and the definition of protein domains. For TFs, this has been difficult, due to the heterogeneity and lack of common structural elements of their domains. This is particularly true for TADs, whereas some of the DBDs, such as the helix–turn–helix or forkhead domains, are to a large extent indicative for TFs (4,5). Other DBDs, however, like leucine zippers or certain subfamilies of the zinc finger class, can also mediate protein–protein interactions. Therefore, we have developed a functional screen to identify TFs and transcriptional co-activators by virtue of their transactivating properties.
MATERIALS AND METHODS
Construction of the retroviral vector and cDNA library
A cDNA library was generated from poly(A+) RNA isolated from human umbilical vein endothelial cells (HUVEC) that had been stimulated with a mixture of TNFα, IL-1α and LPS for 0, 2 and 6 h. cDNA was synthesized using oligo random priming (Stratagene) followed by ligation of BstXI adapters (6). The size selected (>1.0 kb) cDNA was ligated into a derivative of the retroviral vector pBABE (7) that has been modified by insertion of the yeast gal4 DBD and by a polylinker containing two non-self complementary BstXI sites (pBG4B). The ligation was transformed into electrocompetent Escherichia coli XL-10, yielding a library of 2.2 × 107 independent clones, 90% of which had inserts with an average size of 1.3 kb. The library was packaged into retroviral particles using the Phoenix eco packaging cell line (8). As positive control, the nuclear factor kappa B (NF-κB) p65 TAD (3) was inserted into the same vector.
Generation of a gal4 promoter-dependent GFP reporter cell line
Murine NIH3T3 cells were transfected with a vector (pFR-EGFP) containing the gal4-dependent minimal promoter from pFR-Luc (Stratagene) where the luciferase gene was replaced by enhanced green fluorescent protein (EGFP) (9). Cells were selected with puromycin and several clones tested in transient transfections for GFP expression using the gal4–NF-κB TAD construct as positive control.
Retroviral transduction, fluorescence activated cell sorting (FACS) and analysis
Retroviral supernatants (SNs) were applied to the reporter cell line as described (10), and 3 days later EGFP-positive cells sorted by FACS directly into 96-well plates using a FACS Vantage (Becton Dickinson Immunocytometry Systems) FACS sorter. RNA was isolated from individual clones from confluent 6-well plates using the High Pure RNA isolation kit (Roche), inserts retrieved by RT–PCR using the OneStep RT–PCR kit (Qiagen) and sequenced. Primers used for RT–PCR and sequencing were pbmn3, 5′-aaggtcaaagacagttgact-3′, and pbmn4, 5′-tgaccttgatctgaacttct-3′, located in the gal4 DBD and 3′ region flanking the cloning site, respectively, of the pBG4B vector.
Luciferase assay
PCR fragments from positive clones were digested with BstXI and inserted into the pBG4B vector, sequenced to control the correct orientation, and transfected into cells together with pFR-Luc as well as RSV-βgal as internal control. Assays were performed as described and luciferase values normalized for β-gal expression (11).
RESULTS
Strategy of the cloning system
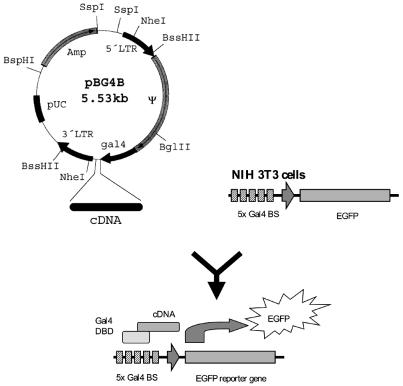
We have developed a retroviral expression cloning system capable of detecting TADs that utilizes two components (Fig. 1). First, a retroviral vector (pBG4B) was constructed by inserting the yeast gal4 DBD into pBABE (7). Inserts from a cDNA library in this vector are expressed as C-terminal fusion genes with the yeast gal4 DBD. The second component is a reporter cell line derived from murine NIH3T3 cells that were stably transfected with a gal4-dependent promoter driving an EGFP gene. A cDNA from cytokine-stimulated HUVEC was generated in pBG4B, packaged into retroviral particles and transduced into the reporter cell line. cDNAs that encoded a functional TAD lead to activation of the reporter gene, and EGFP-positive cells were isolated by FACS and expanded, allowing the isolation by RT–PCR and sequence analysis of the integrated inserts (Fig. 2).
Figure 1.
Strategy of the TF cloning system. cDNA from cytokine- stimulated EC was ligated into the BstXI sites of the retroviral expression vector pBG4B, a derivative of pBABE, leading to expression of the inserts as fusion genes with the yeast gal4 DBD. Plasmid DNA from this library was transfected into Phoenix ecotropic packaging cells, and retrovirus applied to a murine NIH3T3 reporter cell line that carries a gal4-dependent EGFP reporter gene. EGFP-positive cells were isolated by FACS, expanded, and the cDNA inserts retrieved by RT–PCR.
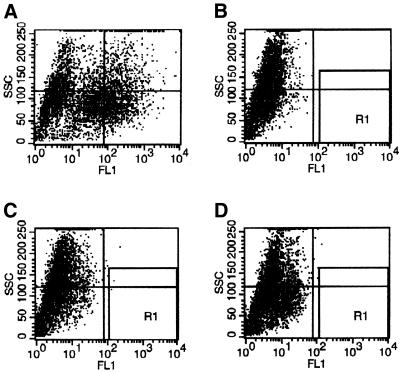
Figure 2.
FACS analysis. The gal4–EGFP NIH3T3 reporter cell line was transduced with retrovirus containing either the positive control, the empty vector or the cDNA library, and 3 days later analyzed for EGFP expression. (A) Positive control (p65-NF-κB TAD fused to the gal4 DBD); (B) negative control, non-transduced NIH3T3-Gal4–EGFP cells; (C) HUVEC cDNA library; (D) empty vector. Gates were set as indicated (R1) to definitely exclude GFP-negative cells. After transduction with the cDNA library (C), ∼1 in 10 000 cells (0.01%) scored positive, in the vector control (D) 0.0012%.
We first tested this concept using a positive control, the strong NF-κB p65 TAD (3) inserted into the retroviral vector. After transduction, EGFP-positive cells could be readily distinguished from control cells by FACS analysis (Fig. 2A and B). In order to determine the background of GFP-positive cells after retroviral transduction, we applied in a separate experiment retroviral SN from the packaged empty pBG4B vector. The empty vector yielded 18 positives per 1.5 × 106 sorted cells, in contrast, the same amount of retroviral SN from the cDNA library yielded 151 GFP-positive cells per 1.5 × 106 (Fig. 2C and D), indicating that ∼12% of the GFP-positive cells in an experiment are non-specific, possibly resulting from integration of the retroviral vector in close proximity to the reporter gene (see Discussion). When applying retroviral SN from the HUVEC library, we noticed in our initial experiments multiple integrations of cDNAs in the positive clones, due to a too high concentration of the retroviral SN. In subsequent experiments, the retroviral preparation was therefore diluted, and at a score of 1 positive per 10 000–20 000 cells in the FACS analysis we found single integration of inserts (Fig. 2C). Sorted cells were expanded into individual clones for further analysis. It appeared that isolation of RNA followed by RT–PCR was superior to PCR from genomic DNA to re-isolate integrated cDNAs. Subsequent sequence analysis of inserts from positive clones and comparison with databases revealed the identity of the isolated cDNAs, as shown in Table 1. They can be grouped into cognate and potential novel TFs, as well as false positives. In a series of experiments, a total of ∼10 million cells were transduced with the retroviral cDNA library and sorted by FACS, and between 1/10 000 and 1/20 000 cells scored GFP-positive. After expansion into individual clones, 69 of them were further analyzed. A summary of these experiments is given in Table 2.
Table 1. Genes identified in the TF screen: (A) cognate TFs, (B) potentially novel TFs and (C) false positives.
| Name | Accession no. | Remarks |
|---|---|---|
| A | ||
| Notch-1 (3×) | AF308602 | TF, vascular development |
| MAML-1 | AF221759 | Co-activator of NOTCH |
| HSF-1 | HUMHSF1 | Heat shock TF |
| RelA | NM_021975 | NF-κB p65 subunit, TF involved in inflammation and apoptosis |
| Fus | HSFUSA | RelA co-activator |
| Sox-7 (3×) | BC004299 | Sry related HMG-box related TF, involved in wnt signaling |
| MondoA | AF312918 | bHLH TF, Max interacting |
| STAT-6 (2×) | XM_043112 | TF, interferon signaling |
| STAT-3 | AF029311 | TF, interferon signaling |
| Foxo1a | NM_002015 | TF, forkhead domain |
| B | ||
| Hypothetical | Hs11_9491_22_27_7 | Proline-rich, forkhead domain |
| Hypothetical | Hs19_11453_22_26_1 | Proline-rich, ankyrin repeat |
| Eric-1 | HSA243997 | TACC (transforming acidic coiled-coil) domain present in centrosomal proteins |
| E7/Hakai | AF167441 | Proline-rich, C2H2 and C2HC4 zinc fingers |
| Junction Plakoglobin | BC011865 | Involvement in β-catenin/wnt signaling |
| TFG (3×) | NM_006070 | TRK fused gene, transforming, glutamine-rich |
| VE-cadherin | X79981 | Acidic region, membrane associated |
| Atrophin | XM_006637 | Polyglutamine stretch |
| BCAR-1 | AJ242987 | Proline-rich, breast cancer anti-estrogen resistence protein |
| Zyxin | XM_056979 | Contains LIM domain |
| Peflin | AB026628 | Penta-EF-hand Ca2+ binding domains |
| Reticuloalbin | BC010120 | Penta-EF-hand Ca2+ binding domains |
| HNRPF | NM_004966 | RNA binding protein; RRM motif can also bind single-strand DNA |
| C | ||
| von Willebrand factor | XM_006947 | Stop codon |
| Metallothionein-1 | XM_165657 | Antisense |
| PAI-1 | AH002922 | Antisense |
| RP11-4H20 | AL445543 | Putative protein, out of frame |
| MORF | NM_012330 | Out of frame |
| NUMB | NM_003744 | Out of frame |
| MMP-1 | NM_002421 | Antisense/stop codon |
| Empty or re-arranged vector |
Table 2. Breakdown of cell numbers.
| A | |
| Total number of cells sorted | 10 million |
| Number of cells scored positive in FACS | 500 |
| Number of clones analyzed | 69 |
| Known TFs | 15 |
| Potentially novel TFs | 40 |
| False positives | 14 |
| B | |
| cDNA library | 151 |
| Empty vector | 18 |
(A) The total number of cells in an experiment is given, the transduction efficiency is estimated between 20 and 50% depending on the experiment. False positives include inserts in antisense orientation, short (<5 amino acids) open reading frame, or empty vector (see Table 1). (B) In a separate experiment, the number of GFP-positives obtained after transduction of 1.5 × 106 cells with the library and with the empty vector was determined.
Analysis of isolated cDNAs
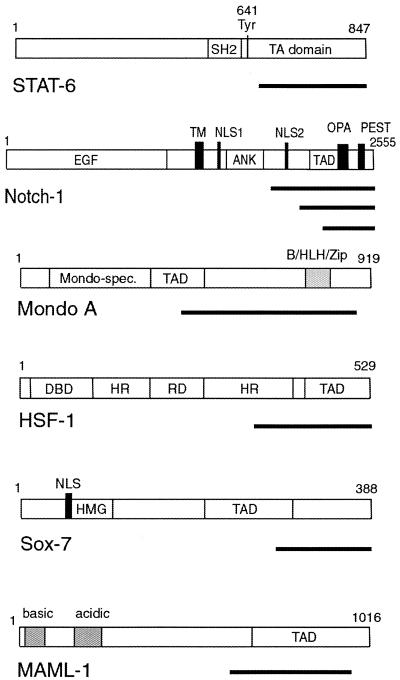
Cognate TFs that were isolated include, for example, STAT-6, Notch-1, MondoA, HSF-1, the human homolog of Sox7 as well as the transcriptional co-activator MAML-1. In those cases where the TF has been characterized with respect to its domain structure, the isolated fragment contained always at least part of the published TAD, further demonstrating the specificity of the screening system (Fig. 3). Secondly, a number of cDNAs were isolated that encoded either novel proteins or proteins not described as TFs. Upon sequence analysis of the full-length clones, some of them contained a potential DBD, often in the N-terminal part. These include, for example, the hypothetical protein encoded by Hs11_9491_22_27_7 and E7/Hakai, the latter being recently described as a E-cadherin-interacting protein (12). The others did not show any apparent DBD, which may be either due to the fact that classes of DBD exist that have not yet been recognized as such, or that these proteins form a functional TF only after binding to another protein that provides a DBD (as in the case of, for example, wnt signaling), or represent transcriptional co-activators which do not have their own DBD. Some clones contained inserts in the antisense orientation, e.g. metallothionein. When translated, it gave rise to a short proline-rich open reading frame, which is most likely responsible for its transactivating properties (see Discussion). Furthermore, a number of clones (e.g. von Willebrand factor) did not contain inserts with an open reading frame of significant length; it is unclear how they were able to activate the reporter. We speculated that in these cases the retrovirus may have inserted in close proximity to the integrated EGFP reporter and caused direct activation of the EGFP gene by the retroviral long terminal repeats.
Figure 3.
Domain structures of selected TFs. Black bars represent the region contained in the cDNA inserts that were isolated. For some of the TFs, several independent clones were identified.
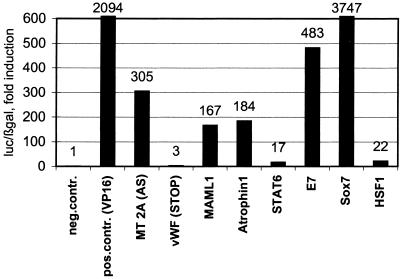
Therefore, to confirm the transactivating properties in a transient transfection that excludes effects of the integration site, inserts from several clones were isolated by PCR, (re-) inserted into the pBG4B vector and transiently transfected into cells together with pFR-Luc (a gal4-dependent luciferase reporter vector corresponding to the EGFP reporter used in the screen). Except in the case of von Willebrand factor, the transactivating properties could be confirmed (Fig. 4).
Figure 4.
Luciferase assay. cDNA inserts from EGFP-positive clones were recovered by RT–PCR, subcloned into the pBG4B vector and transiently transfected into NIH3T3 cells together with the gal4–Luc reporter as well as RSV-βgal as internal control. Positive control, gal4–VP-16; negative control, empty gal4 vector; MT, metallothionein; vWF, von Willebrand factor. Luciferase values were normalized for β-galactosidase expression.
DISCUSSION
The method for the isolation of TFs that is presented here is based on the detection of functional TADs in a cDNA library. The presence of a TAD is usually indicative for TFs or transcriptional co-activators, however, their absence does not exclude the possibility that the respective protein is a TF, since the TAD may be provided by another protein as is the case in TCF/β-catenin (13). TADs are still poorly characterized, and have therefore been difficult to identify by sequence comparison. Two features, proline-rich regions and polyglutamine stretches, have been associated with TADs (14–16). Both types of sequences have been found in our screen, proline-rich regions, for example, in RelA/p65 NF-κB, Notch-1, MondoA, STAT-6, and in the human homolog of Sox-7. The proline-rich domain is probably responsible for the identification of E7/Hakai and Hs11_9491_22_27_7. However, a high proline content alone appears not to predict transactivating properties, since a vast number of proteins exist that share this feature, but are unrelated to transcription/transactivation, e.g. the small proline-rich proteins that are part of the UV stress response; none of these were isolated in our screen. Other proline-rich motifs have been characterized and found to have other functions, namely binding to specific protein domains: PPXP to WW domains, PXXP to SH3 domains and FPPPP to EVH1 domains (17–19). In contrast, the identified TADs contain rather long regions with a proline content in the range of 8–20%. Polyglutamine stretches have been associated with transcriptional regulation, they are contained, for example, in NFAT-5, in the TATA-binding and CREB-binding proteins (15,16), and also in Notch-1. Extended polyglutamine stretches in huntingtin can interact with CREB (20). They are present in atrophin (21) and may therefore be responsible for the identification of this protein in our screen. Still, the structural basis for a TAD remains unclear. In a conceptually similar approach, the group of Ptashne (22,23) has identified E.coli genomic fragments with transactivating properties using yeast as a screening system. They found ∼1% of the fragments analyzed to display transactivating properties, however, all of them contained an acidic region. This may be due to the fact that acidic regions are favored in yeast with regard to transactivation, since proline- and glutamine-rich TADs do not function (or only under certain conditions) in this organism (24).
While several clones that we have isolated are cognate TFs, and demonstrate the feasibility of our system, others have well established functions in the cell that are unrelated to transcription. It cannot be ruled out that some of them represent artefacts in the sense that regions in proteins may have transactivating properties, although the protein has no transactivating function due to, for example, different compartmentalization. However, it should be noted that proteins may have more than one function in the cell; even their apparent subcellular localization may not be indicative. Examples include Notch-1 that is a transmembrane receptor from which the active TF is proteolytically cleaved (25). Furthermore, zyxin, a well characterized α-actinin- and VASP-interacting protein that localizes to sites of cell adhesion, and the zyxin-related protein LPP, have been reported to shuttle between cytoplasm and nucleus or even have transactivating properties, respectively, suggesting additional (nuclear) functions for these proteins (26,27).
Some of the identified TFs have focused attention on signaling pathways that have been only poorly studied in EC. They include the function of Sox7, a member of a gene family characterized by a 79 amino acid DBD designated HMG box. Sox family TFs have been implicated in developmental processes. However, Sox7 has until now not been described to be expressed in cells of the vasculature. Interestingly, it forms a subgroup with Sox17 and Sox18, due to sequence similarity and overlap of expression; Sox18 mutations are the underlying cause of cardiovascular and hair follicle defects in ragged (ra) mice (28). Sox7 has been described to interfere with wnt signaling (29).
MAML-1, the human homolog of the Drosophila mastermind, is a co-activator of Notch (25) and has also not been described in endothelial cells (EC). Notch signaling plays a major role during development of the vascular system, as targeted disruption of Notch-1, and in particular Notch-1/Notch-4, displayed severe defects in angiogenic vascular remodeling that affected the embryo, the yolk sac and the placenta (30). In rats, Notch-1 has been shown to be upregulated in vascular cells after balloon angioplasty (31).
Foxo1a/FKHR is a member of the forkhead TF family involved in apoptosis (32). It is a target of Akt, a phosphatidylinositol 3-kinase-stimulated serine/threonine kinase that activates BAD, GSK3 and caspase-9. Phosphorylated Foxo1a is retained in the cytoplasm via binding to 14-3-3 proteins. Interruption of Akt signaling leads to dephoshorylation of Foxo1a, its translocation to the nucleus and expression of Foxo1a-dependent genes, including FasL. In vascular cells, apoptosis plays an important role during angiogenesis, remodeling or in degenerative diseases such as diabetic microangiopathies, but the role of Foxo1a has not been investigated in the vascular context.
In summary, we have developed a generally applicable expression cloning strategy in mammalian cells for the isolation of transcriptional activators by virtue of their transactivating properties. In addition to cognate TFs that play well documented roles in EC, others were identified that will focus attention on signaling pathways, the functions of which have not been established in terminally differentiated or cytokine-stimulated EC. The ability to identify TFs from a given cell type or tissue will clearly help in assigning functions to previously uncharacterized genes.
Acknowledgments
ACKNOWLEDGEMENTS
We wish to thank G. Nolan for providing the pBABE vector and Phoenix packaging cell line, and S. Renner for help with FACS. This work was supported by a grant from the Austrian Science Foundation (SFB-5-12) to R.d.M.
REFERENCES
- 1.Fields S. and Song,O. (1989) A novel genetic system to detect protein-protein interactions. Nature, 340, 245–246. [DOI] [PubMed] [Google Scholar]
- 2.Inouye C., Remondelli,P., Karin,M. and Elledge,S. (1994) Isolation of a cDNA encoding a metal response element binding protein using a novel expression cloning procedure: the one hybrid system. DNA Cell Biol., 13, 731–742. [DOI] [PubMed] [Google Scholar]
- 3.Schmitz M.L., dos Santos Silva,M.A., Altmann,H., Czisch,M., Holak,T.A. and Baeuerle,P.A. (1994) Structural and functional analysis of the NF-kappa B p65 C terminus. An acidic and modular transactivation domain with the potential to adopt an alpha-helical conformation. J. Biol. Chem., 269, 25613–25620. [PubMed] [Google Scholar]
- 4.Miller C.P., McGehee,R.E.,Jr and Habener,J.F. (1994) IDX-1: a new homeodomain transcription factor expressed in rat pancreatic islets and duodenum that transactivates the somatostatin gene. EMBO J., 13, 1145–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weigel D., Seifert,E., Reuter,D. and Jackle,H. (1990) Regulatory elements controlling expression of the Drosophila homeotic gene fork head. EMBO J., 9, 1199–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seed B. and Aruffo,A. (1987) Molecular cloning of the CD2 antigen, the T-cell erythrocyte receptor, by a rapid immunoselection procedure. Proc. Natl Acad. Sci. USA, 84, 3365–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morgenstern J.P. and Land,H. (1990) Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res., 18, 3587–3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hitoshi Y., Lorens,J., Kitada,S.I., Fisher,J., LaBarge,M., Ring,H.Z., Francke,U., Reed,J.C., Kinoshita,S. and Nolan,G.P. (1998) Toso, a cell surface, specific regulator of Fas-induced apoptosis in T cells. Immunity, 8, 461–471. [DOI] [PubMed] [Google Scholar]
- 9.Tsien R.Y. (1998) The green fluorescent protein. Annu. Rev. Biochem., 67, 509–544. [DOI] [PubMed] [Google Scholar]
- 10.Steinberger P., Szekeres,A., Wille,S., Stockl,J., Selenko,N., Prager,E., Staffler,G., Madic,O., Stockinger,H. and Knapp,W. (2002) Identification of human CD93 as the phagocytic C1q receptor (C1qRp) by expression cloning. J. Leukoc. Biol., 71, 133–140. [PubMed] [Google Scholar]
- 11.de Wet J.R., Wood,K.V., DeLuca,M., Helinski,D.R. and Subramani,S. (1987) Firefly luciferase gene: structure and expression in mammalian cells. Mol. Cell. Biol., 7, 725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujita Y., Krause,G., Scheffner,M., Zechner,D., Leddy,H.E., Behrens,J., Sommer,T. and Birchmeier,W. (2002) Hakai, a c-Cbl-like protein, ubiquitinates and induces endocytosis of the E-cadherin complex. Nature Cell Biol., 4, 222–231. [DOI] [PubMed] [Google Scholar]
- 13.Hecht A., Litterst,C.M., Huber,O. and Kemler,R. (1999) Functional characterization of multiple transactivating elements in β-catenin, some of which interact with the TATA-binding protein in vitro. J. Biol. Chem., 274, 18017–18025. [DOI] [PubMed] [Google Scholar]
- 14.Sprenger-Haussels M. and Weisshaar,B. (2000) Transactivation properties of parsley proline-rich bZIP transcription factors. Plant J., 22, 1–8. [DOI] [PubMed] [Google Scholar]
- 15.Hebinck A., Dalski,A., Engel,H., Mattei,M., Hawken,R., Schwinger,E. and Zuhlke,C. (2000) Assignment of transcription factor NFAT5 to human chromosome 16q22.1, murine chromosome 8D and porcine chromosome 6p1.4 and comparison of the polyglutamine domains. Cytogenet. Cell Genet., 90, 68–70. [DOI] [PubMed] [Google Scholar]
- 16.Kazantsev A., Preisinger,E., Dranovsky,A., Goldgaber,D. and Housman,D. (1999) Insoluble detergent-resistant aggregates form between pathological and nonpathological lengths of polyglutamine in mammalian cells. Proc. Natl Acad. Sci. USA, 96, 11404–11409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sudol M., Chen,H.I., Bougeret,C., Einbond,A. and Bork,P. (1995) Characterization of a novel protein-binding module—the WW domain. FEBS Lett., 369, 67–71. [DOI] [PubMed] [Google Scholar]
- 18.Ren R., Mayer,B.J., Cicchetti,P. and Baltimore,D. (1993) Identification of a ten-amino acid proline-rich SH3 binding site. Science, 259, 1157–1161. [DOI] [PubMed] [Google Scholar]
- 19.Niebuhr K., Ebel,F., Frank,R., Reinhard,M., Domann,E., Carl,U.D., Walter,U., Gertler,F.B., Wehland,J. and Chakraborty,T. (1997) A novel proline-rich motif present in ActA of Listeria monocytogenes and cytoskeletal proteins is the ligand for the EVH1 domain, a protein module present in the Ena/VASP family. EMBO J., 16, 5433–5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nucifora F.C. Jr, Sasaki,M., Peters,M.F., Huang,H., Cooper,J.K., Yamada,M., Takahashi,H., Tsuji,S., Troncoso,J., Dawson,V.L., Dawson,T.M. and Ross,C.A. (2001) Interference by huntingtin and atrophin-1 with cbp-mediated transcription leading to cellular toxicity. Science, 291, 2423–2428. [DOI] [PubMed] [Google Scholar]
- 21.Wood J.D., Nucifora,F.C.,Jr, Duan,K., Zhang,C., Wang,J., Kim,Y., Schilling,G., Sacchi,N., Liu,J.M. and Ross,C.A. (2000) Atrophin-1, the dentato-rubral and pallido-luysian atrophy gene product, interacts with ETO/MTG8 in the nuclear matrix and represses transcription. J. Cell Biol., 150, 939–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma J. and Ptashne,M. (1987) A new class of yeast transcriptional activators. Cell, 51, 113–119. [DOI] [PubMed] [Google Scholar]
- 23.Ruden D.M., Ma,J., Li,Y., Wood,K. and Ptashne,M. (1991) Generating yeast transcriptional activators containing no yeast protein sequences. Nature, 350, 250–252. [DOI] [PubMed] [Google Scholar]
- 24.Kunzler M., Braus,G.H., Georgiev,O., Seipel,K. and Schaffner,W. (1994) Functional differences between mammalian transcription activation domains at the yeast GAL1 promoter. EMBO J., 13, 641–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu L., Aster,J.C., Blacklow,S.C., Lake,R., Artavanis-Tsakonas,S. and Griffin,J.D. (2000) MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nature Genet., 26, 484–489. [DOI] [PubMed] [Google Scholar]
- 26.Petit M.M., Fradelizi,J., Golsteyn,R.M., Ayoubi,T.A., Menichi,B., Louvard,D., Van de Ven,W.J. and Friederich,E. (2000) LPP, an actin cytoskeleton protein related to zyxin, harbors a nuclear export signal and transcriptional activation capacity. Mol. Biol. Cell, 11, 117–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nix D.A. and Beckerle,M.C. (1997) Nuclear-cytoplasmic shuttling of the focal contact protein, zyxin: a potential mechanism for communication between sites of cell adhesion and the nucleus. J. Cell Biol., 138, 1139–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pennisi D., Bowles,J., Nagy,A., Muscat,G. and Koopman,P. (2000) Mice null for sox18 are viable and display a mild coat defect. Mol. Cell. Biol., 20, 9331–9336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takash W., Canizares,J., Bonneaud,N., Poulat,F., Mattei,M.G., Jay,P. and Berta,P. (2001) SOX7 transcription factor: sequence, chromosomal localisation, expression, transactivation and interference with Wnt signalling. Nucleic Acids Res., 29, 4274–4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krebs L.T., Xue,Y., Norton,C.R., Shutter,J.R., Maguire,M., Sundberg,J.P., Gallahan,D., Closson,V., Kitajewski,J., Callahan,R., Smith,G.H., Stark,K.L. and Gridley,T. (2000) Notch signaling is essential for vascular morphogenesis in mice. Genes Dev., 14, 1343–1352. [PMC free article] [PubMed] [Google Scholar]
- 31.Lindner V., Booth,C., Prudovsky,I., Small,D., Maciag,T. and Liaw,L. (2001) Members of the Jagged/Notch gene families are expressed in injured arteries and regulate cell phenotype via alterations in cell matrix and cell–cell interaction. Am. J. Pathol., 159, 875–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brunet A., Bonni,A., Zigmond,M.J., Lin,M.Z., Juo,P., Hu,L.S., Anderson,M.J., Arden,K.C., Blenis,J. and Greenberg,M.E. (1999) Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell, 96, 857–868. [DOI] [PubMed] [Google Scholar]