Abstract
Huntington’s disease (HD) is a progressive neurodegenerative disorder with autosomal-dominant inheritance. The disease is caused by a CAG trinucleotide repeat expansion located in the first exon of the HD gene. The CAG repeat is highly polymorphic and varies from 6 to 37 repeats on chromosomes of unaffected individuals and from more than 30 to 180 repeats on chromosomes of HD patients. In this study, we show that the number of CAG repeats in the HD gene can be determined by restriction of the DNA with the endonuclease EcoP15I and subsequent analysis of the restriction fragment pattern by electrophoresis through non-denaturing polyacrylamide gels using the ALFexpress DNA Analysis System. CAG repeat numbers in the normal (30 and 35 repeats) as well as in the pathological range (81 repeats) could be accurately counted using this assay. Our results suggest that this high-resolution method can be used for the exact length determination of CAG repeats in HD genes as well as in genes affected in related CAG repeat disorders.
INTRODUCTION
Expansions of CAG trinucleotide repeats (CAG repeats) in coding regions of human genes cause neurodegenerative disorders by generating proteins with elongated polyglutamine (polyQ) stretches. This group of disorders includes Huntington’s disease (HD), dentatorubral pallidoluysian atrophy, spinal bulbar muscular atrophy and the spinocerebellar ataxia (SCA) types 1, 2, 3, 6 and 7 (1–3).
The HD gene (IT15 gene), which encodes huntingtin, a 350 kDa protein of unknown function, is located on the human chromosome 4 and consists of 67 exons. The disease-causing mutation is a CAG repeat expansion located within exon 1 of the HD gene (HD exon1). The CAG repeat is translated into a polyQ stretch. The disease manifests itself when the polyQ stretch exceeds the critical length of 37 glutamines (pathological threshold), whereas 8–35 glutamine residues in huntingtin are tolerated by neuronal cells. Experimental evidence has been presented that huntingtin fragments with polyQ tracts in the pathological range (more than 37 glutamines), but not in the normal range (20–32 glutamines), form high molecular weight protein aggregates with a fibrillar morphology in vitro and in cell culture model systems (4,5). In addition, inclusions with aggregated N-terminally truncated huntingtin protein were detected in HD transgenic mice carrying a CAG repeat expansion of 115–156 units and in HD patient brains (6,7), suggesting that the process of aggregate formation is important for the progression of HD. The mechanisms, however, by which the elongated polyQ sequences in huntingtin cause dysfunction and neurodegeneration are not yet understood (1,8,9).
Unaffected individuals have repeat numbers of up to 30, while individuals at a high risk of developing HD carry more than 37 CAG repeats. Individuals with 30–37 repeats have a high risk of passing on repeats in the affected size range to their offspring (10–12). The accurate determination of the number of CAG repeats is required for the DNA-based predictive testing of at-risk individuals. To date, CAG repeat length determination is based on polymerase chain reaction (PCR) amplification of genomic DNA using primers flanking the CAG repeat region in the IT15 gene, and subsequent electrophoretic separation of the products in denaturing polyacrylamide gels (13).
PCR amplifications of the CAG repeat region have primarily been performed by incorporating [α-32P]dNTPs, or using 32P or fluorescently end-labeled primers. Sizing of fluorescently end-labeled amplification products was performed in various Applied Biosystems DNA sequencers (14–22). The method of separation of amplification products involves capillary electrophoresis or denaturing polyacrylamide gel electrophoresis. Recently, Williams et al. (13) have undertaken a comparative analysis of sizing CAG repeat sequences of the IT15 gene using radioactive and fluorescent PCR amplification and subsequently slab gel and capillary electrophoresis for the separation of the PCR products. They found that the mobility of CAG repeat stretches containing amplification products of the IT15 gene is greater using capillary electrophoresis than using slab gel electrophoresis. The mobility difference increased with the length of the CAG repeat. By using an allele ladder for sizing CAG repeats as a calibration system, the number of CAG repeats in different HD alleles could be determined with high accuracy. However, the length determination of the amplicon could be hampered by deletions and insertions in the surrounding of the CAG repeat region.
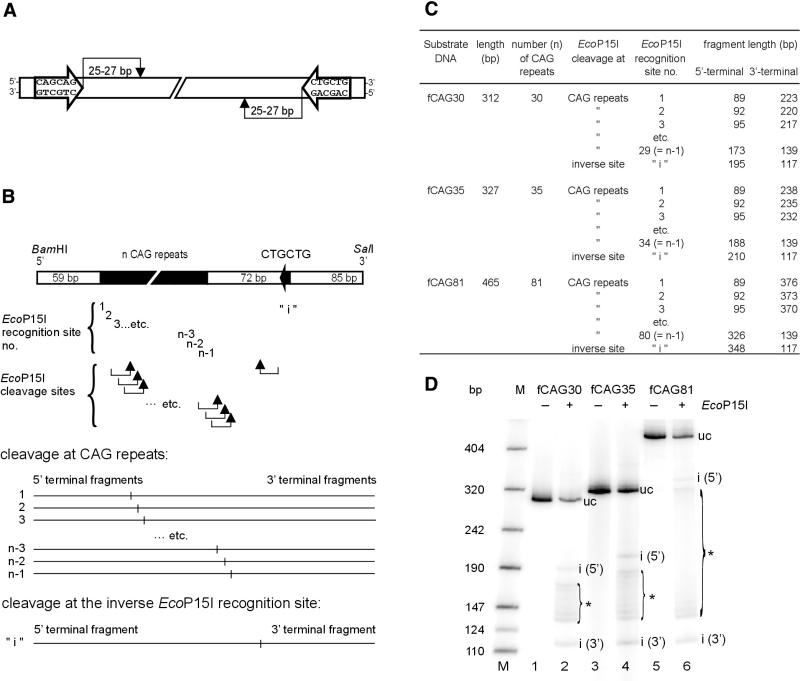
In this study we present an alternative approach to count the number of CAG repeats in the IT15 gene, which is based on digestion of the test DNA with the multifunctional heterooligomeric type III restriction-modification enzyme EcoP15I (for reviews see 23–27). The recognition sequence for EcoP15I consists of two copies of the asymmetric sequence 5′-CAGCAG present on opposite strands of the DNA double helix. Cleavage occurs 25–27 bp downstream of one of these sites (Fig. 1A). The distance between the two sites can be up to 3.5 kb (28).
Figure 1.
EcoP15I cleavage of DNA substrates that contain CAG repeats of different lengths. (A) Recognition and cleavage sites of restriction endonuclease EcoP15I in the DNA molecule. For DNA restriction, the enzyme needs two 5′-CAGCAG sequences being inversely oriented in the double-stranded DNA. Cleavage occurs 25–27 bp downstream of one of the two inverted sites (see Introduction). (B) General chart of the used DNA substrates that contain a number of n CAG repeats and an additional inverse EcoP15I recognition site 5′-CTGCTG at a distance of 72 bp from the 3′-end of the CAG repeats. Two CAG trinucleotides correspond to one EcoP15I recognition site 5′-CAGCAG. Thus, an expansion of n CAG trinucleotides results in a series of (n-1) EcoP15I recognition sites that overlap by 3 bp. Arrows indicate the EcoP15I cleavage sites 25–27 bp downstream of the various recognition sites. In the lower part of the chart the fragment patterns expected from cleavage either at one of the CAG repeats or at the inverse EcoP15I recognition site ‘i’ are schematically shown. (C) Calculated lengths of the DNA fragments expected from EcoP15I cleavage of the DNA substrates fCAG30, fCAG35 and fCAG81. Fragment lengths are assigned to the corresponding EcoP15I recognition sites nos 1 to (n-1) within the CAG repeats or to the inverse site ‘i’. (D) EcoP15I cleavage patterns of the DNA substrates fCAG30, fCAG35 and fCAG81. The DNA substrates were radioactively labeled at both ends. Each substrate (50 fmol) was incubated in the presence or absence of a 10-fold molar excess of EcoP15I enzyme over DNA substrate at 37°C for 30 min. Cleavage was analyzed on a non-denaturing 5% (w/v) polyacrylamide gel as described in the Materials and Methods. The ladders of EcoP15I cleavage fragments were marked by brackets. M, molecular weight marker; uc, uncleaved DNA substrates; i (5′), 5′-terminal DNA fragment after EcoP15I cleavage at the inverse EcoP15I recognition site; i (3′), 3′-terminal DNA fragment after EcoP15I cleavage at the inverse EcoP15I recognition site; *, DNA fragments generated by EcoP15I cleavage at EcoP15I recognition sites nos 1 to (n-1) within the CAG repeats.
We could show that HD exon1 DNA is a substrate of EcoP15I. Because of the overlapping recognition sites within the CAG repeat, EcoP15I digestion of DNA fragments bearing HD exon1 generates a ladder of restriction fragments, whose number corresponds to the number of CAG repeats. Making use of this fact we have developed an EcoP15I cleavage assay to determine the number of CAG repeats in HD exon1 DNA on the basis of the resulting DNA fragment pattern.
MATERIALS AND METHODS
Restriction enzymes
EcoP15I was expressed and purified as described (29). All other restriction and DNA-modifying enzymes were obtained from New England Biolabs.
Preparation and labeling of DNA substrates that contain CAG repeats of different lengths
To analyze EcoP15I cleavage of HD exon1 we used the plasmids pCAG30, pCAG35 and pCAG81 that have been constructed by the insertion of exon 1 of the HD gene containing 30 (pCAG30), 35 (pCAG35) and 81 (pCAG81) CAG repeats, respectively, into the BamHI and SalI sites of the vector pGEX-5X-1 (30). Large-scale purification of the plasmids following standard procedures was performed after transformation in Escherichia coli TG1 cells. As CAG repeats tend to be unstable during propagation in E.coli cells, their number was verified by DNA sequencing.
To generate 5′-end-labeled fragments, pCAG30, pCAG35 and pCAG81 were linearized with BamHI and labeled with Cy5-dCTP (100 µM; Amersham Pharmacia Biotech), dATP, dGTP, dTTP (100 µM each; New England Biolabs) using the Klenow fragment (3′–5′ exo–). After heat inactivation of the Klenow enzyme and removal of non-incorporated nucleotides by gel filtration using Probe Quant G-50 micro columns (Amersham Pharmacia Biotech) the linearized plasmids were digested with SalI generating DNA fragments of 312 bp (fCAG30), 327 bp (fCAG35) and 465 bp (fCAG81) in length. The labeled fragments were purified from agarose gels using the Qiagen Gel Extraction Kit (Qiagen). 3′-End-labeled fragments were prepared in the same way, with the difference that in this case the plasmids were linearized with SalI prior to labeling and only afterwards digested with BamHI. DNA fragments labeled radioactively at both ends were prepared as described above using [α-32P]dCTP (Hartmann Analytic) after simultaneous digestion of the plasmids with BamHI and SalI. The DNA marker VIII (Roche Molecular Biochemicals) was either Cy5- or radioactively labeled and purified by gel filtration as described above.
EcoP15I cleavage assays and analysis of cleavage pattern
The radioactively or fluorescence-labeled DNA substrates fCAG30, fCAG35 or fCAG81 (50 fmol) were cleaved with various amounts of EcoP15I in 10 mM Tris–HCl pH 8.0, 10 mM KCl, 10 mM MgCl2, 0.1 mM EDTA, 0.2 mM dithiothreitol, 50 µg/ml bovine serum albumin and 2 mM ATP for 30 min at 37°C in a total volume of 10 µl. In case of the radioactively labeled fragments EcoP15I cleavage was stopped by adding EDTA to a final concentration of 20 mM. Reaction products were separated in a non-denaturing 5% (w/v) polyacrylamide gel and analyzed using a phosphorimager (PhosphoImager type SI, ImageQuant 5.0 software; Amersham Pharmacia Biotech). EcoP15I cleavage of the fluorescence-labeled DNA substrates was stopped by adding 2 µl STOP-solution (95% formamide, 20 mM EDTA, pH 8.3, 50 mg/ml dextranblue 2 000 000). DNA fragments were separated and analyzed on a non-denaturing 7% (w/v) polyacrylamide gel using the ALFexpress DNA Analysis System. Cleavage was quantified by use of the AlleliX 1.0 software (Amersham Pharmacia Biotech).
RESULTS
EcoP15I cleavage of DNA substrates with CAG repeats results in ‘ladders’ of DNA fragments
To investigate whether EcoP15I can recognize and cleave the CAG repeats present in the HD gene the HD exon1 fragments fCAG30, fCAG35 and fCAG81, which were radioactively labeled at both ends, were incubated with a 10-fold excess of EcoP15I. The cleavage reactions were analyzed by polyacrylamide gel electrophoresis.
As mentioned above, EcoP15I cleavage requires the presence of two 5′-CAGCAG sequences that are located on opposite strands of the DNA. In the case of HD exon1 the CAG repeat of ‘n’ triplets length displays (n-1) EcoP15I recognition sites on one DNA strand. A single inverse EcoP15I recognition site on the opposite strand, termed ‘i’, is located 72 bp downstream of the CAG repeat (Fig. 1B). EcoP15I cleaves 25–27 bp downstream of one of the two recognition sites. Thus, in the case of HD exon1 cleavage can occur either in the vicinity of the inverse site ‘i’ or 25–27 bp downstream of any of the (n-1) recognition sequences formed by the n CAG repeats. If a linear HD exon1 DNA substrate is used, such cleavage should generate n cleavage products of different length (Fig. 1B). The calculated lengths of the various putative EcoP15I cleavage products expected for substrates fCAG30, fCAG35 and fCAG81 are shown in Figure 1C. We considered only those putative DNA cleavage fragments that possess an end label; ‘internal’ DNA fragments, which result from cleavage at multiple sites, cannot disturb the determinations as they do not contain labeling.
As expected, cleavage of the different DNA substrates with a 10-fold excess of EcoP15I generated characteristic ‘ladders’ of DNA fragments (Fig. 1D). In addition to DNA fragments resulting from cleavage at the ‘i’ site we found multiple DNA fragments generated by cleavage within the CAG repeats. The upper end of those ‘ladders’ corresponds to the 5′-terminal cleavage fragment generated by cleavage at the EcoP15I recognition site ‘n-1’, which represents the recognition site proximal to ‘i’. The corresponding 3′-terminal cleavage product represents the lower end of the ‘ladder’. The cleavage fragments in the center of the ‘ladder’ are generated by cleavage at the EcoP15I recognition sites nos (n-2), (n-3) etc. Bands representing these fragments are less intense than those representing the ‘ladder’ ends. Fragments that result from cleavage at the EcoP15I recognition site nos 1, 2, etc., were not detectable. This suggests that under our experimental conditions EcoP15I cleaved preferentially at recognition sites located close to the ‘i’ recognition site, while more distant sites are less efficiently recognized and cleaved.
However, these results show clearly that EcoP15I recognizes and cleaves the multiple overlapping sites within the CAG repeat of HD exon1 and thereby generates ‘ladders’ of DNA fragments of different lengths.
Optimization of EcoP15I cleavage reactions
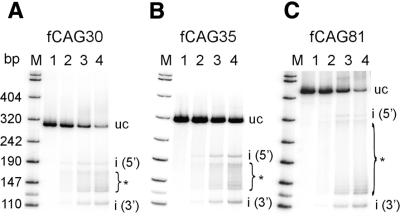
To optimize the cleavage reaction we tested whether EcoP15I cleavage efficiency can be influenced by the variation of the molar ratio of EcoP15I enzyme and DNA substrate. For this purpose, different amounts of EcoP15I were incubated with a given molarity of DNA substrate. We found that cleavage of fCAG30, fCAG35 and fCAG81 depended highly on the EcoP15I enzyme to DNA substrate ratio (Fig. 2). An increased enzyme to substrate ratio enhanced the efficiency of cleavage. However, even in presence of a 20-fold excess of enzyme versus substrate, not all theoretically possible cleavage products could be identified. Instead those CAG repeats proximal to the inverse site [recognition site nos (n-1), (n-2), etc.] were preferentially cleaved.
Figure 2.
Influence of EcoP15I concentration on cleavage efficiency and cleavage pattern. The 32P-labeled DNA fragments fCAG30 (A), fCAG35 (B) or fCAG81 (C) were cleaved with increasing amounts of EcoP15I (for cleavage conditions and analysis see Fig. 1D). Lane 1, without EcoP15I; lanes 2–4, molar ratios of EcoP15I enzyme to DNA substrate of 1, 10 or 20. For abbreviations see legend to Figure 1.
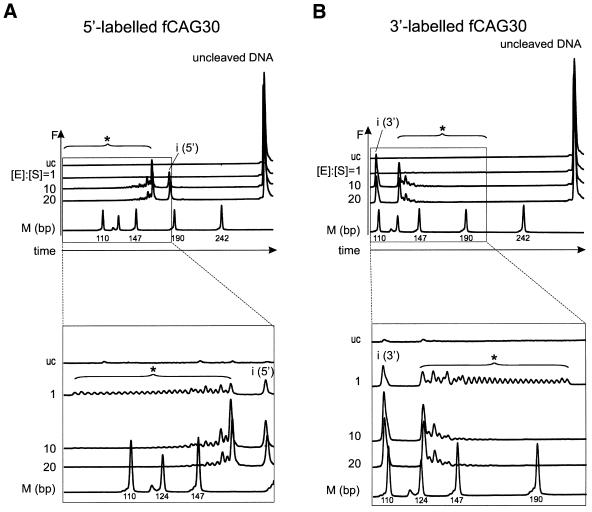
At an equimolar ratio of EcoP15I enzyme to DNA substrate, cleavage efficiency was too small to analyze the generated fragment pattern in detail using our initial experimental set up. We therefore searched for a sensitive high-resolution method to separate and detect EcoP15I cleavage fragments. We chose the ALFexpress DNA Analysis System, a half-automated DNA sequencer, for our analysis. This system allows high-resolution separation of DNA cleavage fragments and exhibits similar sensitivity as a phosphorimager (28,31). To apply this procedure, we labeled the DNA substrates with the fluorescent dye Cy5 instead of the 32P radioisotope. Either the 5′- or the 3′-end of the DNA substrate fCAG30 was Cy5-labeled to distinguish 5′- and 3′-cleavage fragments of similar lengths (Fig. 1C). According to the results mentioned above (Fig. 2), at a 10- or 20-fold molar excess of EcoP15I enzyme over DNA substrate, EcoP15I preferably cleaved at the inverse EcoP15I recognition site ‘i’ and at the recognition site nos (n-1), (n-2), etc. (Fig. 3A and B; [E]:[S] = 10, [E]:[S] = 20). However, at an equimolar ratio of enzyme to substrate we could count exactly 29 DNA fragments in the ‘ladder’ (Fig. 3A and B; [E]:[S] = 1); this number corresponded to the 29 overlapping EcoP15I recognition sites within the 30 CAG repeats of fCAG30. This result demonstrates that every theoretically possible EcoP15I recognition site within a sequence of 30 CAG repeats is indeed recognized and cleaved by EcoP15I. Thus, we could define the conditions for EcoP15I cleavage allowing an exact determination of the CAG repeat number. EcoP15I cleavage of DNA substrates labeled at their 5′- or 3′-end exhibited comparable patterns (Fig. 3). Therefore, we decided to use only 3′-end-labeled DNA fragments for our next investigations.
Figure 3.
Analysis of the EcoP15I cleavage pattern of fCAG30 using the ALFexpress DNA Analysis System. DNA substrate fCAG30 was Cy5-labeled either (A) at the 5′-end or (B) at the 3′-end and incubated with EcoP15I at molar ratios of enzyme [E] to DNA substrate [S] of 1, 10 or 20. Cleavage products were separated on non-denaturing 7% (w/v) polyacrylamide gels and analyzed in the ALFexpress DNA Analysis System. Output data show DNA fragments as fluorescence intensity F in arbitrary units. The upper panels show all cleavage products and uncleaved DNA substrate. The lower panels are a close up of the cleavage products. For abbreviations see legend to Figure 1. The molecular weight marker fragments contained two HpaII ends and were therefore Cy5-labeled at both ends. In contrast, fCAG30 was labeled at only one end. As Cy5 is known to change the electrophoretic mobility of DNA fragments (42), the marker could not be used as an absolute but as a relative length standard.
Counting CAG repeat numbers in the pathological range
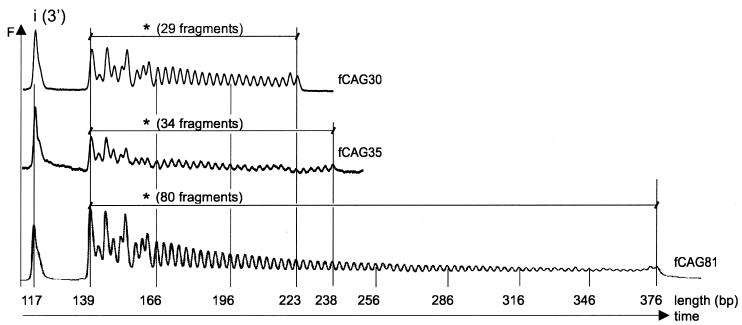
After the correct determination of a CAG repeat length in the normal range as present in fragment fCAG30, we wanted to test our refined method for determining repeat lengths in the borderline (30–39) and pathological range (more than 39). For this purpose fragments fCAG35 and fCAG81 were cleaved with EcoP15I using an equimolar ratio of enzyme to DNA. The cleavage products, Cy5-labeled at their 3′-ends, were analyzed with the ALFexpress DNA Analysis System. Cleavage patterns for the three substrates fCAG30, fCAG35 and fCAG81 are shown in Figure 4. We obtained ‘ladders’ of 29, 34 and 80 DNA fragments, respectively, and an additional fragment of equal size [i(3′)] for each substrate. Thus, EcoP15I cleaved all three DNA substrates at the inverse EcoP15I recognition site and at each of the overlapping EcoP15I recognition sites (5′-CAGCAG) within the CAG repeats. According to our results presented above, EcoP15I preferred recognition sites located proximal to the inverse recognition site (nos n-1, n-2, etc.) over more distant ones (nos 1, 2, etc.). This preference is most likely caused by the faster formation of the active enzyme–DNA complex when two EcoP15I recognition sites are located close to each other.
Figure 4.
Counting of CAG repeats in fCAG30, fCAG35 and fCAG81 at an equimolar enzyme to substrate ratio. The DNA substrates fCAG30, fCAG35 and fCAG80 were Cy5-labeled at their 3′-ends. EcoP15I cleavage fragments were separated on non-denaturing 7% (w/v) polyacrylamide gels and analyzed using the ALFexpress DNA Analysis System. Output data show DNA fragments as fluorescence intensity F in arbitrary units. Cleavage by EcoP15I resulted in a ladder of DNA fragments in 3 bp steps from 139 to 223 bp for fCAG30, 139 to 238 bp for fCAG35 and 139 to 376 bp for fCAG81. In addition, a 117 bp DNA fragment was observed due to the cleavage at the inverse EcoP15I recognition site ‘i(3′)’.
These results validate the high-resolution analysis of the EcoP15I cleavage products of exon 1 of the HD gene as a suitable method for the exact quantification of the CAG repeat numbers in the normal as well as in the pathological range.
DISCUSSION
In this study we have demonstrated that the analysis of the EcoP15I cleavage patterns of fluorescence-labeled HD exon 1 DNA fragments using the ALFexpress DNA Analysis system allows the exact determination of the number of CAG triplet repeats present in the HD gene. Using this approach we could correctly detect extensions of 30, 35 and 81 CAG repeats. Therefore, our approach is useful for the exact determination of the CAG repeat numbers in the normal (up to 30), borderline (30–39) and pathological (more than 39) ranges. The application of EcoP15I cleavage of fluorescence-labeled DNA substrates combined with half-automated DNA sequencers offers numerous advantages over existing procedures for the quantification of CAG repeats. Our non-radioactive method exhibits high-resolution quality, is time saving, not hazardous and prevents isotopic waste (13,19,22,28,31).
To apply our assay to the analysis of clinical samples, PCR amplification of the HD gene exon 1 area as already described (e.g. 15,16,19,20) could be used to obtain DNA substrates on the basis of genomic DNA. Using our novel approach for CAG repeat quantification, 50 fmol specific DNA substrate, an amount usually insufficient for DNA sequencing, allowed analysis. Compared with methods based on the sizing of PCR fragments our approach offers different advantages: we do not need internally sequenced DNA standards, which are time consuming to produce and, even more important, our procedure is not affected by possible deletions or insertions in the surrounding of the CAG repeat region as long as the inverse EcoP15I site (‘i’) is present in the DNA fragment. As we directly count the EcoP15I-generated fragments of the ‘ladder’ to determine the exact number of CAG repeats, exact sizing of the CAG-repeat containing DNA fragments is not necessary.
Our approach to count CAG repeats should be applicable to the analysis of further hereditary neurodegenerative diseases caused by expanded CAG repeats. As mentioned before, the precondition for the application of this method is the presence of an inverse EcoP15I recognition site (5′-CTGCTG) located up to 3.5 kb downstream of the CAG repeats. Such inverse EcoP15I recognition sites in distances of ∼0.2–1.7 kb to the CAG repeats are present in the genes involved in spinal bulbar muscular atrophy and SCA types 1, 2 and 7 (accession nos. M27423, X79204, U70323, AJ000517). As EcoP15I can functionally interact with other type III restriction endonucleases such as EcoP1I (32), an inverse EcoP1I site (5′-GGTCT) in the 3′ region of the CAG repeats could be used for DNA cleavage in the absence of an effective inverse EcoP15I site. Moreover, in order to apply our assay to count the number of CAG repeats in related CAG repeat diseases, an inverse EcoP15I recognition site could be introduced via PCR in the DNA substrate to be analyzed.
The SCA2 gene, which, in its mutated form, causes SCA type 2, is characterized by CAG repeats that are polymorphic in length and interrupted by CAA triplets (15). CAA, as CAG, encodes glutamine but cannot be directly detected by our EcoP15I cleavage approach. In the case of the SCA2 gene, however, CAA interruptions in the CAG repeats could be indirectly detected because of gaps in the EcoP15I cleavage ‘ladder’.
Recently it has been shown that SCA type 10 is linked to the expansion of ATTCT pentanucleotide repeats (33). This pentameric repeat sequence resembles the recognition sequences of type III restriction endonucleases HinfIII (ATTCG), EcoP1I (GGTCT) or StyLTI (CTCTG) (34). As new restriction enzymes with additional sequence specificities are still being discovered, enzymes with ATTCT specificity could be among them. Therefore, assays similar to the one presented in our study could be designed for the diagnosis of diseases caused by other sequence repeats.
The expansion of certain repeat regions has also been observed in lower organisms such as yeast (35) and prokaryotes (36). In the yeast Candida albicans many repeats consist of CAG and/or CAA stretches, which result in polyQ accumulation on the protein level resembling the situation in human neurodegenerative diseases (37). In prokaryotes, repeat sequences (repetitions of the same nucleotide or of di-, tri-, tetra- or pentanucleotides) have been found to be involved in switching on and off the expression of certain genes (38,39). Repeat sequences are potentially involved in gene expression modulation as, e.g. the CACAG pentanucleotide repeat in Pasteurella haemolytica (40) or the AGTC tetranucleotide repeat in Haemophilus influenzae (41). Curiously enough, these repeat sequences occur in those open reading frames, which exhibit clear homologies to genes of type III restriction-modification enzymes.
Acknowledgments
ACKNOWLEDGEMENTS
We wish to thank Ursula Scherneck, Heike Goehler and Katja Schweiger for their technical help, sequencing of the DNA substrates and preparation of exon 1 constructs, respectively. The support of A. Meisel and K. Ruscher in using the ALFexpress DNA Analysis System is gratefully acknowledged. We are obliged to Anja Dröge for critical reading of the manuscript. This work was supported by Deutsche Forschungsgemeinschaft (grant KR1293/1-3), Fonds der chemischen Industrie, Sonnenfeld-Stiftung, Konsul Karl und Dr Gabriele Sandmann Stiftung, and Humboldt University Medical School.
REFERENCES
- 1.Wanker E.E. (2000) Protein aggregation and pathogenesis of Huntington’s disease: mechanisms and correlations. Biol. Chem., 381, 937–942. [DOI] [PubMed] [Google Scholar]
- 2.Gusella J.F. and MacDonald,M.E. (2000) Molecular genetics: unmasking polyglutamine triggers in neurodegenerative disease. Nature Rev. Neurosci., 1, 109–115. [DOI] [PubMed] [Google Scholar]
- 3.Usdin K. and Grabczyk,E. (2000) DNA repeat expansions and human disease. Cell. Mol. Life Sci., 57, 914–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scherzinger E., Sittler,A., Schweiger,K., Heiser,V., Lurz,R., Hasenbank,R., Bates,G.P., Lehrach,H. and Wanker,E.E. (1999) Self-assembly of polyglutamine-containing huntingtin fragments into amyloid-like fibrils: implications for Huntington’s disease pathology. Proc. Natl Acad. Sci. USA, 96, 4604–4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waelter S., Boeddrich,A., Lurz,R., Scherzinger,E., Lueder,G., Lehrach,H. and Wanker,E.E. (2001) Accumulation of mutant huntingtin fragments in aggresome-like inclusion bodies as a result of insufficient protein degradation. Mol. Biol. Cell, 12, 1393–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davies S.W., Turmaine,M., Cozens,B.A., DiFiglia,M., Sharp,A.H., Ross,C.A., Scherzinger,E., Wanker,E.E., Mangiarini,L. and Bates,G.P. (1997) Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell, 90, 537–548. [DOI] [PubMed] [Google Scholar]
- 7.DiFiglia M., Sapp,E., Chase,K.O., Davies,S.W., Bates,G.P., Vonsattel,J.P. and Aronin,N. (1997) Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science, 277, 1990–1993. [DOI] [PubMed] [Google Scholar]
- 8.Tobin A.J. and Signer,E.R. (2000) Huntington’s disease: the challenge for cell biologists. Trends Cell Biol., 10, 531–536. [DOI] [PubMed] [Google Scholar]
- 9.Perutz M.F. (1999) Glutamine repeats and neurodegenerative diseases: molecular aspects. Trends Biochem. Sci., 24, 58–63. [DOI] [PubMed] [Google Scholar]
- 10.Andrew S.E., Goldberg,Y.P. and Hayden,M.R. (1997) Rethinking genotype and phenotype correlations in polyglutamine expansion disorders. Hum. Mol. Genet., 6, 2005–2010. [DOI] [PubMed] [Google Scholar]
- 11.Duyao M., Ambrose,C., Myers,R., Novelletto,A., Persichetti,F., Frontali,M., Folstein,S., Ross,C., Franz,M., Abbott,M. et al. (1993) Trinucleotide repeat length instability and age of onset in Huntington’s disease. Nature Genet., 4, 387–392. [DOI] [PubMed] [Google Scholar]
- 12.Snell R.G., MacMillan,J.C., Cheadle,J.P., Fenton,I., Lazarou,L.P., Davies,P., MacDonald,M.E., Gusella,J.F., Harper,P.S. and Shaw,D.J. (1993) Relationship between trinucleotide repeat expansion and phenotypic variation in Huntington’s disease. Nature Genet., 4, 393–397. [DOI] [PubMed] [Google Scholar]
- 13.Williams L.C., Hegde,M.R., Herrera,G., Stapleton,P.M. and Love,D.R. (1999) Comparative semi-automated analysis of (CAG) repeats in the Huntington disease gene: use of internal standards. Mol. Cell Probes, 13, 283–289. [DOI] [PubMed] [Google Scholar]
- 14.Andrew S.E., Goldberg,Y.P., Kremer,B., Telenius,H., Theilmann,J., Adam,S., Starr,E., Squitieri,F., Lin,B., Kalchman,M.A. et al. (1993) The relationship between trinucleotide (CAG) repeat length and clinical features of Huntington’s disease. Nature Genet., 4, 398–403. [DOI] [PubMed] [Google Scholar]
- 15.Choudhry S., Mukerji,M., Srivastava,A.K., Jain,S. and Brahmachari,S.K. (2001) CAG repeat instability at SCA2 locus: anchoring CAA interruptions and linked single nucleotide polymorphisms. Hum. Mol. Genet., 10, 2437–2446. [DOI] [PubMed] [Google Scholar]
- 16.Ishii T., Sato,S., Kosaki,K., Sasaki,G., Muroya,K., Ogata,T. and Matsuo,N. (2001) Micropenis and the AR gene: mutation and CAG repeat-length analysis. J. Clin. Endocrinol. Metab., 86, 5372–5378. [DOI] [PubMed] [Google Scholar]
- 17.Le H., Fung,D. and Trent,R.J. (1997) Applications of capillary electrophoresis in DNA mutation analysis of genetic disorders. Mol. Pathol., 50, 261–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mangiarini L., Sathasivam,K., Mahal,A., Mott,R., Seller,M. and Bates,G.P. (1997) Instability of highly expanded CAG repeats in mice transgenic for the Huntington’s disease mutation. Nature Genet., 15, 197–200. [DOI] [PubMed] [Google Scholar]
- 19.Pelotti S., Maiolini,E., Bini,C., Rimondi,S., Luiselli,D. and Pappalardo,G. (2001) Automated fluorescence analysis of CAG repeats at the human androgen receptor gene (HUMARA): evaluation of polymorphism in an Italian sample and report of a new allele. Am. J. Forensic Med. Pathol., 22, 55–57. [DOI] [PubMed] [Google Scholar]
- 20.Wallerand H., Remy-Martin,A., Chabannes,E., Bermont,L., Adessi,G.L. and Bittard,H. (2001) Relationship between expansion of the CAG repeat in exon 1 of the androgen receptor gene and idiopathic male infertility. Fertil. Steril., 76, 769–774. [DOI] [PubMed] [Google Scholar]
- 21.Warner J.P., Barron,L.H. and Brock,D.J. (1993) A new polymerase chain reaction (PCR) assay for the trinucleotide repeat that is unstable and expanded on Huntington’s disease chromosomes. Mol. Cell Probes, 7, 235–239. [DOI] [PubMed] [Google Scholar]
- 22.Warner J.P., Barron,L.H., Goudie,D., Kelly,K., Dow,D., Fitzpatrick,D.R. and Brock,D.J. (1996) A general method for the detection of large CAG repeat expansions by fluorescent PCR. J. Med. Genet., 33, 1022–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bickle T.A. and Krüger,D.H. (1993) Biology of DNA restriction. Microbiol. Rev., 57, 434–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dryden D.T., Murray,N.E. and Rao,D.N. (2001) Nucleoside triphosphate-dependent restriction enzymes. Nucleic Acids Res., 29, 3728–3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krüger D.H., Kupper,D., Meisel,A., Reuter,M. and Schroeder,C. (1995) The significance of distance and orientation of restriction endonuclease recognition sites in viral DNA genomes. FEMS Microbiol. Rev., 17, 177–184. [DOI] [PubMed] [Google Scholar]
- 26.Krüger D.H. and Reuter,M. (1999) Host-controlled modification and restriction. In Granoff,A. and Webster,R.G. (eds), Encyclopedia of Virology. Academic Press, San Diego, pp. 758–763.
- 27.Redaschi N. and Bickle,T.A. (1996) DNA restriction and modification systems. In Neidhardt,F.C. (ed.), Escherichia coli and Salmonella: Cellular and Molecular Biology. ASM Press, Washington, DC, pp. 773–781.
- 28.Mücke M., Reich,S., Möncke-Buchner,E., Reuter,M. and Krüger,D.H. (2001) DNA cleavage by type III restriction-modification enzyme EcoP15I is independent of spacer distance between two head to head oriented recognition sites. J. Mol. Biol., 312, 687–698. [DOI] [PubMed] [Google Scholar]
- 29.Meisel A., Mackeldanz,P., Bickle,T.A., Krüger,D.H. and Schroeder,C. (1995) Type III restriction endonucleases translocate DNA in a reaction driven by recognition site-specific ATP hydrolysis. EMBO J., 14, 2958–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scherzinger E., Lurz,R., Turmaine,M., Mangiarini,L., Hollenbach,B., Hasenbank,R., Bates,G.P., Davies,S.W., Lehrach,H. and Wanker,E.E. (1997) Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell, 90, 549–558. [DOI] [PubMed] [Google Scholar]
- 31.Ruscher K., Reuter,M., Kupper,D., Trendelenburg,G., Dirnagl,U. and Meisel,A. (2000) A fluorescence based non-radioactive electrophoretic mobility shift assay. J. Biotechnol., 78, 163–170. [DOI] [PubMed] [Google Scholar]
- 32.Kunz A., Mackeldanz,P., Mücke,M., Meisel,A., Reuter,M., Schroeder,C. and Krüger,D.H. (1998) Mutual activation of two restriction endonucleases: interaction of EcoP1 and EcoP15. Biol. Chem., 379, 617–620. [PubMed] [Google Scholar]
- 33.Matsuura T., Yamagata,T., Burgess,D.L., Rasmussen,A., Grewal,R.P., Watase,K., Khajavi,M., McCall,A.E., Davis,C.F., Zu,L. et al. (2000) Large expansion of the ATTCT pentanucleotide repeat in spinocerebellar ataxia type 10. Nature Genet., 26, 191–194. [DOI] [PubMed] [Google Scholar]
- 34.Roberts R.J. and Macelis,D. (2001) REBASE—restriction enzymes and methylases. Nucleic Acids Res., 29, 268–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Field D. and Wills,C. (1996) Long, polymorphic microsatellites in simple organisms. Proc. R. Soc. Lond. B Biol. Sci., 263, 209–215. [DOI] [PubMed] [Google Scholar]
- 36.van Belkum A., Scherer,S., van Alphen,L. and Verbrugh,H. (1998) Short-sequence DNA repeats in prokaryotic genomes. Microbiol. Mol. Biol. Rev., 62, 275–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Field D. and Wills,C. (1996) Long, polymorphic microsatellites in simple organisms. Proc. R. Soc. Lond. B Biol. Sci., 263, 209–215. [DOI] [PubMed] [Google Scholar]
- 38.van Belkum A., Scherer,S., van Alphen,L. and Verbrugh,H. (1998) Short-sequence DNA repeats in prokaryotic genomes. Microbiol. Mol. Biol. Rev., 62, 275–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Belkum A. (1999) Short sequence repeats in microbial pathogenesis and evolution. Cell Mol. Life Sci., 56, 729–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryan K.A. and Lo,R.Y. (1999) Characterization of a CACAG pentanucleotide repeat in Pasteurella haemolytica and its possible role in modulation of a novel type III restriction-modification system. Nucleic Acids Res., 27, 1505–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Bolle X., Bayliss,C.D., Field,D., van de Ven,T., Saunders,N.J., Hood,D.W. and Moxon,E.R. (2000) The length of a tetranucleotide repeat tract in Haemophilus influenzae determines the phase variation rate of a gene with homology to type III DNA methyltransferases. Mol. Microbiol., 35, 211–222. [DOI] [PubMed] [Google Scholar]
- 42.Tu O., Knott,T., Marsh,M., Bechtol,K., Harris,D., Barker,D. and Bashkin,J. (1998) The influence of fluorescent dye structure on the electrophoretic mobility of end-labeled DNA. Nucleic Acids Res., 26, 2797–2802. [DOI] [PMC free article] [PubMed] [Google Scholar]