Abstract
Uteroglobin-related protein 2 (UGRP2) is thought to play a role in inflammation and/or epithelial cell differentiation in the lung. Induction of Ugrp2 mRNA expression by epidermal growth factor (EGF) and transforming growth factor α was examined using mouse transformed lung Clara cell-derived mtCC cells. The EGF-induced increase of Ugrp2 occurred at the transcriptional level that required the EGF receptor and the activation of the ERK-MAPK and phosphoinositide-3 kinase pathways.
Keywords: Uteroglobin-related protein 2, Epidermal growth factor, Transforming growth factor α, Lung epithelial cells
Abbreviations: UGRP2, Uteroglobin-related protein 2; mtCC, mouse transformed lung Clara cell-derived cell line
1. Introduction
Uteroglobin-related protein (UGRP) 2, also called high in normal-1 or secretoglobin 3A1 (SCGB3A1), was originally identified as a putative growth inhibitory cytokine gene that is inactivated by methylation in the majority of human breast carcinomas [1]. Independently, UGRP2 was found as a homologous gene to UGRP1 that is a downstream target gene for a homeodomain transcription factor, T/EBP/NKX2.1 [2,3]. In humans, UGRP2 is highly expressed in the trachea, lung, salivary gland, prostate, esophagus, duodenum and mammary gland [1]. In the latter, the expression is specifically found in the luminal mammary epithelial cells of small ducts and lobules [1]. In the adult mouse, UGRP2 is primarily expressed in the trachea and lung, and weakly expressed in the heart, stomach and small intestine [4]. Consistent with the importance of UGRP2 in human breast carcinomas, UGRP2 is expressed in the mouse mammary gland, but its expression is weak compared to that in trachea and lung [4].
Based on amino acid sequences, UGRP1 and UGRP2 belong to a gene superfamily of the Uterogloglobin/Clara cell secretory proteins (UG/CCSP), offcially termed secretoglobins (SCGB), thus referring to UGRP1 and UGRP2 as SCGB3A2 and SCGB3A1, respectively [2–4]. UGRP1 is thought to play a role in lung inflammation [2,5,6]. Further, UG/CCSP, a prototypical protein of the SCGB gene superfamily, to which UGRP1 and UGRP2 are distantly related, exhibits several anti-inflammatory and anti-chemotactic activities [7]. The mouse Ugrp2 gene is localized at chromosome 11B1 [3], a homologous region to 5q31-q35 in human [8], in which many genes encoding inflammatory cytokines such as interleukin-3, -4, -5, -13 and colony-stimulating factor 2 are located. These facts together with the sites of UGRP2 expression suggest that UGRP2 may play a role in the regulation of inflammation and/or differentiation of epithelial cells [1,4]. However, little is known about the physiological function of UGRP2.
Both epidermal growth factor (EGF) and transforming growth factor α (TGFα) are potent mitogens for most epithelial tissues. Binding of EGF and TGFα to the receptor EGFR promotes dimerization of EGFR, which stimulates intrinsic tyrosine kinase activity, resulting in autophosphorylation of the receptor [9]. The tyrosine autophosphorylated EGFRs exhibit increased binding affnities to src homology 2 regions of substrate molecules. Substrate binding to the receptors is followed by phosphorylation of tyrosine residues on the substrate molecules, which in turn results in the activation of a variety of downstream signaling cascades [10,11]. Activation of the adaptor molecule, Grb2 occurs through its direct binding to the activated EGFR [12]. The Grb2/Sos-1 pathway is well characterized and is responsible for activation of the ERK-MAPK pathway, which ultimately leads to the regulation of gene transcription [13]. Phosphoinositide-3 kinase (PI3K) is also a substrate for activated EGFR [14,15]. Several PI3K isoforms are commonly found that play roles in a number of signal transduction pathways such as activation of PKB/Akt [16].
The aim of the present study was to investigate the mechanisms governing regulation of Ugrp2 expression by EGF and TGFα. We demonstrate that both EGF and TGFα stimulate the expression of Ugrp2 through a mechanism dependent on the ERK-MAPK and PI3K pathways.
2. Material and methods
2.1. Materials
Recombinant murine EGF, TGFα and HGF were purchased from PeproTech Inc. (Rocky Hill, NJ). Murine basic FGF (bFGF), actinomycin D (ActD), SB203580, SP600125 and PD098059 were obtained from SIGMA (St. Louis, MO), and LY294002 and AG1478 were from Calbiochem (San Diego, CA). Antibodies against murine ERK1 and ERK2 (ERK1/2), and phospho-ERK (Y204) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA), and Akt and phospho-Akt (Ser473) from Cell Signaling Technology (Beverly, MA).
2.2. Cell culture
Mouse transformed Clara cells (mtCC) were maintained in Dulbecco’s modified Eagle’s medium (Biosource, Camarillo, CA) supplemented with 10% fetal bovine serum (GEMINI Bio-Products, Woodland, CA) and Antibiotic–Antimycotic containing 100 units/ml penicillin G sodium, 100 μg/ml streptomycin sulfate and 250 ng/ml amphotericin B (Invitrogen Life technology, Carlsbad, CA).
2.3. RT-PCR
To prevent genomic DNA contamination, total RNAs were treated with RNase-free DNase I (Ambion, Austin, TX). For cDNA synthesis, total RNAs were first incubated at 70 °C for 10 min, and chilled on ice. The cDNA synthesis reactions were carried out in a final volume of 20 μl containing RNA, 4 μl of 5× first strand synthesis buffer, 1 μl of mixture of four deoxynucleotide triphosphates (10 mM each), 2 μl of 0.1 M dithiothreitol, and 1 μl of 500 ng/μl N6 random hexamer. After incubation at 37 °C for 2 min, 200 U of Superscript II reverse transcriptase (Invitrogen Life technologies) was added, and the incubation was continued at 37 °C for 60 min, which was then subjected to PCR using Advantage 2 Taq DNA polymerase (BD Biosciences, San Jose, CA) under the following conditions: denaturation at 94 °C for 30 s, annealing at 60 °C for 30 s, and extension at 72 °C for 30 s, 20 cycles for 18S and 30 cycles for mouse EGFR. Oligonucleotide primers used for RT-PCR were as follows: mouse 18S, 5′-CGGCTACCACATCCAAGGAA-3′ and 5′-ATTGGAGCTGGAATTACCGC-3′; mouse EGFR, 5′-GGCGTTGGAAAAGAAAGT-3′ and 5′-GATGGGGTTGTTGCTGAATCG-3′.
2.4. Northern blot analysis
Five μg of total RNAs extracted from mtCC cells was electrophoresed on a 1% agarose gel containing 0.22 M formaldehyde and blotted onto GeneScreen Plus nylon membranes (Perkin–Elmer Life Sciences, Boston, MA). Filters were serially hybridized with mouse Ugrp2 and 18S cDNA probes. Hybridization was performed in Perfect Hybridization solution (SIGMA) overnight at 68 °C. The membrane was washed twice with 2 × SSC containing 0.1% SDS at 68 °C for 30 min, once with 2 × SSC at 68 °C for 30 min, followed by exposure to a storm phosphoimager screen (Molecular Dynamics, Sunnyvale. CA).
2.5. Western blot analysis
mtCC cells were washed with phosphate-buffered saline and lysed in radio-immuno-protein-assay buffer (20 mM Tris–HCl, 150 mM NaCl, 0.1% SDS, 0.1% sodium deoxycholate, 1% Triton X-100, 2 mM EDTA, 10 mM NaF and 1 mM sodium orthovanadate) with Protease Inhibitor Cocktail Tablets (Complete Mini; Roche Applied Science, Indianapolis, IN). The protein lysates mixed with SDS sample loading buffer containing β-mercaptoethanol were electrophoresed on a 10% SDS–polyacrylamide gel containing 0.8% bis(N,N′-methylene-bisacrylamide), followed by electrotransfer to an Immobilon-P membrane (Millipore). Membranes were blocked with Tris-buffered saline (TBS) containing Tween 20 (TBS-Tween; 20 mM Tris–HCl, 150 mM NaCl, 0.1% Tween 20) with 5% skimmed milk, and were incubated with first antibody in TBS-Tween. Membranes were washed with TBS-Tween 20 and incubated with horseradish peroxidase-conjugated secondary antibody (Santa Cruz) for enhanced chemiluminescence detection using Western Lighting Chemiluminescence Reagent Plus (Perkin–Elmer Life Sciences). Image visualization was carried out by exposure to Scientific Imaging Film (Kodak, Rochester, NY).
3. Results
3.1. EGF and TGFα induce Ugrp2 mRNA expression
To study the effect of several mitogens on the levels of Ugrp1 and Ugrp2 mRNA expression, mtCC cells were used. These cells, derived from necrotic tumor tissues of lungs obtained from transgenic mice expressing the simian virus 40 large T antigen gene under the control of Ug/Ccsp promoter [17,18], constitutively expresses high and modest levels of Ugrp1 and Ugrp2 mRNAs, respectively. When treated with 50 ng/ml of EGF, HGF, TGFα, and bFGF for 12 h, both EGF and TGFα robustly induced Ugrp2 mRNA levels, while Ugrp1 expression stayed at similar levels regardless of addition of mitogens except for a slightly enhanced expression after EGF treatment (Fig. 1). These results suggest that the effect of EGF and TGFα is Ugrp2-specific, and thus only Ugrp2 regulation by these mitogens was studied further.
Fig. 1.
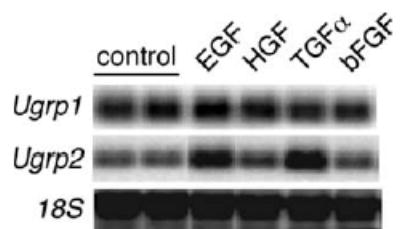
Induction of Ugrp1 and Ugrp2 mRNAs by EGF and TGFα. mtCC cells were treated with 50 ng/ml of selected growth factors (EGF, HGF, TGFα and bFGF) for 12 h. Total cellular RNAs were extracted and mRNA for Ugrp1, Ugrp2 and 18S were examined by Northern blot analysis.
3.2. Induction of Ugrp2 mRNA by EGF occurs in a time- and dose-dependent manner
When mtCC cells were treated with different concentrations of EGF and TGFα for 24 h, the slight increase of Ugrp2 mRNA levels was seen at 6.25 ng/ml for both mitogens (Fig. 2A). EGF induction appeared to plateau with 25 ng/ml while induction by TGFα continued with concentrations of up to 50 ng/ml. A time dependent effect of EGF and TGFα on the induction of Ugrp2 mRNA was further examined using a fixed concentration of 50 ng/ml EGF and TGFα In both cases, significant increase of Ugrp2 mRNA was detected at 6 h after the addition of the mitogen, and levels continued to increase up to 24 h (Fig. 2B). These results suggest that time-dependent increase of Ugrp2 mRNA expression occurs with similar kinetics for both EGF and TGFα. To examine whether the increase of Ugrp2 mRNA expression by treatment with EGF is due to transcriptional control, mtCC cells were pre-treated with the RNA synthesis inhibitor, ActD, followed by treatment with EGF. ActD abrogated the EGF-induced increase of Ugrp2 mRNA expression (Fig. 2C), suggesting that the increase of Ugrp2 mRNA expression by EGF is mediated at the transcriptional level.
Fig. 2.
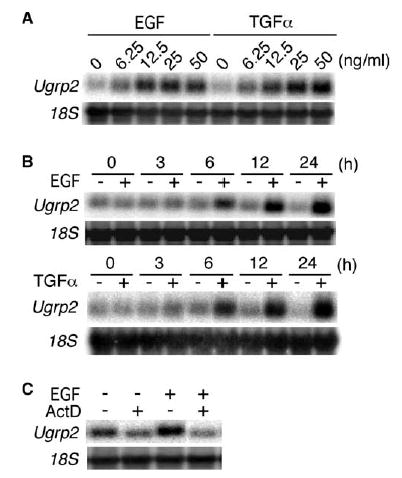
Dose- and time-dependent increase of Ugrp2 mRNA by EGF occurs at the transcriptional level. (A) Dose effects of EGF and TGFα. mtCC cells were treated with 6.25, 12.5, 25 and 50 ng/ml EGF or TGFα for 24 h. (B) Time course analysis of EGF and TGFα effects. mtCC cells were treated with 50 ng/ml EGF or TGFα for 3, 6, 12 or 24 h. (C) EGF induces Ugrp2 mRNA at the transcriptional level. mtCC cells were pre-treated with 5 μg/ml ActD for 1 h, followed by treatment with 25 ng/ml EGF in the presence of ActD for 6 h. Expression of Ugrp2 and 18S was detected by Northern blot analysis.
3.3. Induction of Ugrp2 mRNA by EGF is through EGFR
Both EGF and TGFα start a signaling cascade through their binding to a shared EGFR, a cell surface transmembrane protein with tyrosine kinase activity [19,20]. In order to study the mechanism of EGF-induced increase of Ugrp2 expression, the presence of EGFR was first examined using RT-PCR analysis (Fig. 3A). The results clearly demonstrate that EGFR is present in mtCC cells. When cells were treated with AG1478, a selective inhibitor of EGFR tyrosine kinase activity [21] before the addition of EGF, the increase of Ugrp2 mRNA by EGF was completely inhibited (Fig. 3B). These results suggest that the induction of Ugrp2 mRNA by EGF occurs through EGFR activation.
Fig. 3.
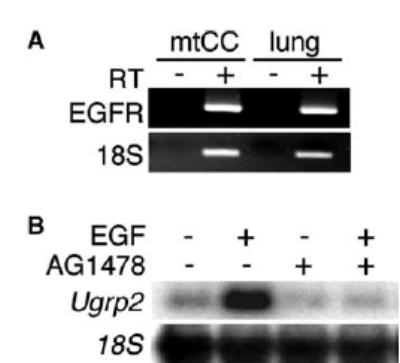
Induction of Ugrp2 mRNA by EGF requires activation of EGFR. (A) RT-PCR analysis for the presence of EGFR in mtCC cells. The size of PCR product is 397 bp for EGFR and 193 bp for 18S. RT(−) is shown as a negative control. RNAs extracted from fetal lung were used as a positive control. (B) AG1478, which is a selective inhibitor of EGFR phosphorylation inhibits the induction of Ugrp2 by EGF. mtCC cells were pre-treated with 1 μM AG1478 for 1 h followed by treatment with 50 ng/ml EGF in the presence of AG1478 for 6 h. Expression of Ugrp2 and 18S was detected by Northern blot analysis.
3.4. ERK-MAPK and PI3K pathways are responsible for the induction of Ugrp2 mRNA by EGF
As typical of growth factors and their receptors, ligand binding to EGFR initiates activation of ERK-MAPK and PI3K pathways. To understand whether ERK-MAPK and PI3K pathways are activated in mtCC cells upon EGF stimulation, phosphorylation of ERK1/2 and Akt was examined for each pathway, respectively. Treatment with the MEK inhibitor, PD098059 and the PI3K inhibitor, LY294002 totally abolished EGF-induced ERK1/2 and Akt phosphorylation (Fig. 4A and B). Interestingly, small amount of activated ERK1/2 was constitutively observed without any treatment, which was also abolished by MEK inhibitor, PD098059 (Fig. 4A). Further, both EGF-induced and constitutive expression of Ugrp2 were partially and almost completely inhibited by PD098059 and LY294002, respectively (Fig. 4C). The level of Ugrp2 expression did not seem to differ by the addition of both inhibitors together as compared to LY294002 alone, suggesting that the latter pathway may be dominant. In order to determine whether other classical MAPKs such as p38 kinase and JNK affect on the induction of Ugrp2 mRNA, cells were treated with the p38 kinase inhibitor, SB203580 and the JNK inhibitor, SP600125. Neither inhibitors affected EGF-induced induction of Ugrp2 expression (Fig. 4D). These results suggest that the induction of Ugrp2 mRNA by EGF occurs through the ERK-MAPK and PI3K pathways, the latter being dominant.
Fig. 4.
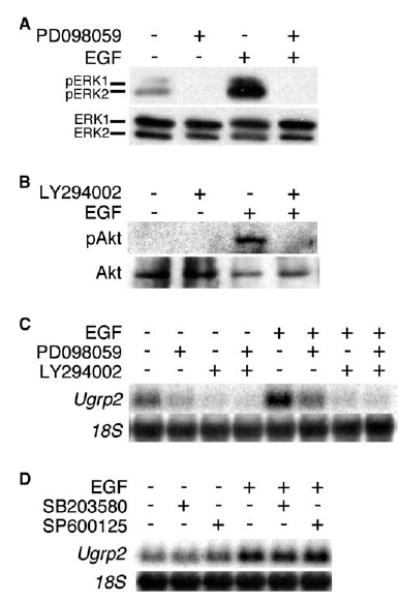
Induction of Ugrp2 mRNA by EGF requires activation of both ERK-MAPK and PI3K pathways. (A) ERK1/2 activation in mtCC cells by EGF stimulation. mtCC cells were treated with 25 ng/ml EGF for 5 min. Activation of ERK1/2 was measured by the presence of phosphorylated ERK1/2 (Y204) bands. The total ERK was detected by using anti-ERK antibody. (B) Akt activation in mtCC cells by EGF stimulation. mtCC cells were treated with 25 ng/ml EGF for 30 min. Activation of Akt was measured by the presence of phosphorylated Akt (Ser473) band. The total Akt was detected by using anti-Akt antibody. (C) Both ERK-MAPK and PI3K pathway inhibitors inhibit Ugrp2 mRNA induction by EGF. mtCC cells were pre-treated with 10 μM PD098059 (MEK inhibitor) and/or 10 μM LY294002 (PI3K inhibitor) for 1 h, followed by treatment of 25 ng/ml EGF in the presence of either or both inhibitors for 6 h. Expression of Ugrp2 and 18S was detected by Northern blot analysis. (D) Neither p38 kinase nor JNK pathway inhibitors inhibit Ugrp2 mRNA induction by EGF. mtCC cells were pre-treated with 10 μM SB203580 or 10 μM SP600125 for 1 h, followed by treatment of 25 ng/ml EGF in the presence of either inhibitors for 6 h. Expression of Ugrp2 and 18S was detected by Northern blot analysis.
4. Discussion
The present study demonstrates that Ugrp2 mRNA can be regulated by EGF and TGFα through EGFR activation. EGF activates both ERK-MAPK and PI3K pathways, leading to the upregulation of Ugrp2 gene expression in mtCC cells. Both the ERK-MAPK inhibitor, PD098059 and the PI3K inhibitor, LY294002 alone or in combination, partially or completely blocked EGF-induced expression of Ugrp2, suggesting that Ugrp2 transcriptional activation may require the cooperation of two or more transcription factors activated by distinct signal transduction pathways.
The MAPK family of serine/threonine kinases is well known to be composed of ERKs, JNKs and p38 kinases [22]. Activation of EGFR also induces JNK activation in a Rac-dependent manner [23], and p38 activation in a Src- and/or Fyn-dependent manner [24]. The induction of Ugrp2 mRNA by EGFR activation does not seem to be through the p38 kinase and JNK pathways because no effect was observed on the Ugrp2 mRNA levels by treatment of cells with their specific inhibitors. Further, these inhibitors did not affect constitutive levels of Ugrp2 expression while MEK and PI3K inhibitors suppressed constitutive Ugrp2 expression. In particular, the PI3K inhibitor, LY294002 almost completely suppressed constitutive expression of Ugrp2. This complete suppression was also observed for EGF-induced increase of Ugrp2 expression. This suggests that PI3K pathway is dominant over ERK-MAPK pathway for constitutive as well as EGF-induced expression of Ugrp2. Constitutive Ugrp2 expression may be partly accounted for by low levels of endogenous ERK1/2 activation observed in control cells, which was abolished by MEK inhibitor. On the other hand, Akt phosphorlylation was not observed in control cells without EGF stimulation. There remains the possibility that a minute amount of phosphorylated Akt is also present in cells that cannot be detected by western blotting due to antibody sensitivity.
Although it remains to be determined whether other pathways are involved in the regulation of Ugrp2 mRNA expression, the ERK-MAPK and PI3K pathways are currently responsible for Ugrp2 gene regulation (Fig. 5). There is a molecular basis for cross talk between ERK-MAPK and PI3K pathways at the level of Raf and Akt, and ERK-MAPK pathway is activated by Akt through c-Raf activation [25]. Whether this cross talk plays a role in the activation of Ugrp2 gene and whether this is related to the apparent dominancy of the PI3K pathway over ERK-MAPK pathway need to be determined.
Fig. 5.
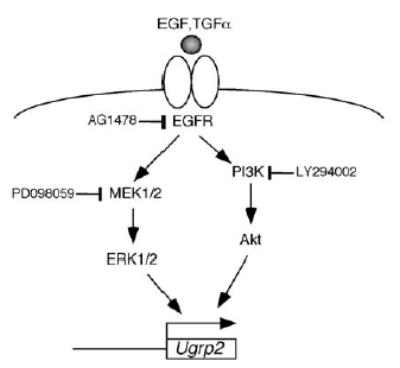
Signaling pathways leading to the Ugrp2 upregulation. EGF/TGFα activates ERK-MAPK and PI3K pathways leading to Ugrp2 activation. Inhibitors at particular steps are shown.
Based on the pattern of Ugrp2 mRNA expression in terminally differentiated airway epithelial cells of adult and developing mouse embryos, an association between UGRP2 and the terminally differentiated epithelial phenotype was suggested [4]. Thus, the induction of Ugrp2 mRNA was correlated with upregulation of Muc2, a mucinous phenotype marker gene and downregulation of the squamouse cell marker gene, transglutaminase I during retinoic acid-induced mucinous differentiation of primary normal human bronchial epithelial cells [4]. Despite these observations, the precise physiological and functional role of UGRP2 in the airways is not fully understood. Whether the regulation of UGRP2 expression by EGF/TGFα is involved in the development of terminally differentiated airway epithelial phenotypes awaits further studies.
In summary, the expression of Ugrp2 is enhanced by both EGF and TGFα through the mechanism dependent on the ERK-MAPK and PI3K pathways.
Acknowledgments
We thank Dr. Francisco DeMayo for providing mtCC cells and Dr. Frank Gonzalez for his critical review of the manuscript. A. Yamada was partially supported by a postdoctoral fellowship from the Japanese Society for the Promotion of Science.
References
- 1.Krop IE, Sgroi D, Porter DA, Lunetta KL, LeVangie R, Seth P, Kaelin CM, Rhei E, Bosenberg M, Schnitt S, Marks JR, Pagon Z, Belina D, Razumovic J, Polyak K. HIN-1, a putative cytokine highly expressed in normal but not cancerous mammary epithelial cells. Proc Natl Acad Sci USA. 2001;98:9796–9801. doi: 10.1073/pnas.171138398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Niimi T, Keck-Waggoner CL, Popescu NC, Zhou Y, Levitt RC, Kimura S. UGRP1, a uteroglobin/Clara cell secretory protein-related protein, is a novel lung-enriched downstream target gene for the T/EBP/NKX2.1 homeodomain transcription factor. Mol Endocrinol. 2001;15:2021–2036. doi: 10.1210/mend.15.11.0728. [DOI] [PubMed] [Google Scholar]
- 3.Niimi T, Copeland NG, Gilbert DJ, Jenkins NA, Srisodsai A, Zimonjic DB, Keck-Waggoner CL, Popescu NC, Kimura S. Cloning, expression, and chromosomal localization of the mouse gene (Scgb3a1, alias Ugrp2) that encodes a member of the novel uteroglobin-related protein gene family. Cytogenet Genome Res. 2002;97:120–127. doi: 10.1159/000064067. [DOI] [PubMed] [Google Scholar]
- 4.Porter D, Lahti-Domenici J, Torres-Arzayus M, Chin L, Polyak K. Expression of high in normal-1 (HIN-1) and uteroglobin related protein-1 (UGRP-1) in adult and developing tissues. Mech Dev. 2002;114:201–204. doi: 10.1016/s0925-4773(02)00056-4. [DOI] [PubMed] [Google Scholar]
- 5.Chiba Y, Kusakabe T, Kimura S. Decreased expression of uteroglobin-related protein 1 in inflamed mouse airways is mediated by IL-9. Am J Physiol Lung Cell Mol Physiol. 2004;287:L1193–L1198. doi: 10.1152/ajplung.00263.2004. [DOI] [PubMed] [Google Scholar]
- 6.Chiba Y, Srisodsai A, Supavilai P, Kimura S. Interleukin-5 reduces the expression of uteroglobin-related protein (UGRP) 1 gene in allergic airway inflammation. Immunol Lett. 2005;97:123–129. doi: 10.1016/j.imlet.2004.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mukherjee AB, Kundu GC, Mantile-Selvaggi G, Yuan CJ, Mandal AK, Chattopadhyay S, Zheng F, Pattabiraman N, Zhang Z. Uteroglobin: a novel cytokine. Cell Mol Life Sci. 1999;55:771–787. doi: 10.1007/s000180050331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Searle AG, Peters J, Lyon MF, Hall JG, Evans EP, Edwards JH, Buckle VJ. Chromosome maps of man and mouse IV. Ann Hum Genet. 1989;53 (Pt 2):89–140. doi: 10.1111/j.1469-1809.1989.tb01777.x. [DOI] [PubMed] [Google Scholar]
- 9.Cadena DL, Gill GN. Receptor tyrosine kinases. FASEB J. 1992;6:2332–2337. doi: 10.1096/fasebj.6.6.1312047. [DOI] [PubMed] [Google Scholar]
- 10.Carpenter G. Receptor tyrosine kinase substrates: src homology domains and signal transduction. FASEB J. 1992;6:3283–3289. doi: 10.1096/fasebj.6.14.1385243. [DOI] [PubMed] [Google Scholar]
- 11.Ullrich A, Schlessinger J. Signal transduction by receptors with tyrosine kinase activity. Cell. 1990;61:203–212. doi: 10.1016/0092-8674(90)90801-k. [DOI] [PubMed] [Google Scholar]
- 12.Buday L, Downward J. Epidermal growth factor regulates p21ras through the formation of a complex of receptor, Grb2 adapter protein, and Sos nucleotide exchange factor. Cell. 1993;73:611–620. doi: 10.1016/0092-8674(93)90146-h. [DOI] [PubMed] [Google Scholar]
- 13.Gutkind JS. The pathways connecting G protein-coupled receptors to the nucleus through divergent mitogen-activated protein kinase cascades. J Biol Chem. 1998;273:1839–1842. doi: 10.1074/jbc.273.4.1839. [DOI] [PubMed] [Google Scholar]
- 14.Otsu M, Hiles I, Gout I, Fry MJ, Ruiz-Larrea F, Panayotou G, Thompson A, Dhand R, Hsuan J, Totty N, Smith AD, Morgan SJ, Courtneidge SA, Parker PJ, Waterfield MD. Characterization of two 85 kd proteins that associate with receptor tyrosine kinases, middle-T/pp60c-src complexes, and PI3-kinase. Cell. 1991;65:91–104. doi: 10.1016/0092-8674(91)90411-q. [DOI] [PubMed] [Google Scholar]
- 15.Skolnik EY, Margolis B, Mohammadi M, Lowenstein E, Fischer R, Drepps A, Ullrich A, Schlessinger J. Cloning of PI3 kinase-associated p85 utilizing a novel method for expression/cloning of target proteins for receptor tyrosine kinases. Cell. 1991;65:83–90. doi: 10.1016/0092-8674(91)90410-z. [DOI] [PubMed] [Google Scholar]
- 16.Burgering BM, Coffer PJ. Protein kinase B (c-Akt) in phosphatidylinositol-3-OH kinase signal transduction. Nature. 1995;376:599–602. doi: 10.1038/376599a0. [DOI] [PubMed] [Google Scholar]
- 17.Magdaleno SM, Wang G, Jackson KJ, Ray MK, Welty S, Costa RH, DeMayo FJ. Interferon-gamma regulation of Clara cell gene expression: in vivo and in vitro. Am J Physiol. 1997;272:L1142–L1151. doi: 10.1152/ajplung.1997.272.6.L1142. [DOI] [PubMed] [Google Scholar]
- 18.Magdaleno SM, Wang G, Mireles VL, Ray MK, Finegold MJ, DeMayo FJ. Cyclin-dependent kinase inhibitor expression in pulmonary Clara cells transformed with SV40 large T antigen in transgenic mice. Cell Growth Differ. 1997;8:145–155. [PubMed] [Google Scholar]
- 19.Derynck R, Roberts AB, Winkler ME, Chen EY, Goeddel DV. Human transforming growth factor-alpha: precursor structure and expression in E. coli. Cell. 1984;38:287–297. doi: 10.1016/0092-8674(84)90550-6. [DOI] [PubMed] [Google Scholar]
- 20.Derynck R. Transforming growth factor alpha. Cell. 1988;54:593–595. doi: 10.1016/s0092-8674(88)80001-1. [DOI] [PubMed] [Google Scholar]
- 21.Levitzki A, Gazit A. Tyrosine kinase inhibition: an approach to drug development. Science. 1995;267:1782–1788. doi: 10.1126/science.7892601. [DOI] [PubMed] [Google Scholar]
- 22.Kyriakis JM, Avruch J. Protein kinase cascades activated by stress and inflammatory cytokines. Bioessays. 1996;18:567–577. doi: 10.1002/bies.950180708. [DOI] [PubMed] [Google Scholar]
- 23.Fanger GR, Johnson NL, Johnson GL. MEK kinases are regulated by EGF and selectively interact with Rac/Cdc42. EMBO J. 1997;16:4961–4972. doi: 10.1093/emboj/16.16.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frey MR, Golovin A, Polk DB. Epidermal growth factor-stimulated intestinal epithelial cell migration requires Src family kinase-dependent p38 MAPK signaling. J Biol Chem. 2004;279:44513–44521. doi: 10.1074/jbc.M406253200. [DOI] [PubMed] [Google Scholar]
- 25.Zimmermann S, Moelling K. Phosphorylation and regulation of Raf by Akt (protein kinase B) Science. 1999;286:1741–1744. doi: 10.1126/science.286.5445.1741. [DOI] [PubMed] [Google Scholar]


