Abstract
To explore the basis of apparent conformational heterogeneity of cytochrome P450 3A4 (CYP3A4) the kinetics of dithionite-dependent reduction was studied in solution, in proteoliposomes and in Nanodiscs. In CYP3A4 oligomers in solution the kinetics obeys a three-exponential equation with similar amplitudes of each of the phases. Addition of substrate (bromocriptine) displaces the phase distribution towards the slow phase at the expense of the fast one, while the middle phase remains unaffected. The fraction reduced in the fast phase, either with or without substrate, is represented by the low-spin heme-protein only, while the slow-reducible fraction is enriched in the high-spin CYP3A4. Upon monomerization by 0.15% Emulgen-913, or by incorporation into Nanodiscs or into large proteoliposomes with a high lipid-to-protein (L/P) ratio (726:1 mol/mol), the kinetics observed in the absence of substrate becomes very rapid and virtually monoexponential. In Nanodiscs and in lipid-rich liposomes bromocriptine decreases the rate of reduction via appearance of the second (slow) phase, the amplitude of which reaches 100% at saturating bromocriptine. In contrast, in P450-rich liposomes (L/P= 112 mol/mol), where the surface molar density of the enzyme is comparable to that observed in liver microsomes, CYP3A4 behaves similar to that observed in solution. These results suggest that in CYP3A4 oligomers in solution and in the membrane the enzyme is distributed between two persistent . conformers with different accessibility of the heme for the reductant ( anion monomer). One of the apparent conformers exists in a substrate-dependent equilibrium between two states with different rate constants of reduction by dithionite, while the second conformer shows no response to substrate binding.
Keywords: cytochrome P450 3A4, bromocriptine, substrate binding, cooperativity, conformers, spin equilibrium, oligomers, dithionite, liposomes, Nanodiscs
Abbreviations and textual footnotes: CYP3A4, cytochrome P450 3A4; Hepes, N-[2-Hydroxyethylpiperazine-N′-[2-ethanesulfonic acid]; DTT, dithiothreitol; EDTA, ethylenediaminetetraacetic acid
In recent years there has been increasing interest in cooperativity of cytochromes P450. Studies of the mechanistic basis of cooperativity are especially important to understand the functional properties of cytochrome P450 3A4 (CYP3A4), the major human P450 enzyme, which is responsible for metabolism of a broad range of substrates of pharmacological and toxicological interest (1). Extensive studies of homo- and heterotropic cooperativity in CYP3A4 provide convincing evidence that a single molecule of this enzyme is capable of binding at least two substrate molecules (2–6). However, as the number and complexity of observations on mutual effects of various substrates on metabolism by CYP3A4 increases, it is becoming difficult to explain all known facts with any model involving two (7–10) or even three (5, 6, 11, 12) substrate binding sites within the same enzyme molecule. Rather, accumulating evidence of persistent conformational heterogeneity of CYP3A4 suggests that complete understanding of the mechanisms of cooperativity requires consideration of divergence of the enzyme pool between several functionally diverse, stable populations (4, 13–5). These data implicitly suggest slow transitions between CYP3A4 conformers, so that the distribution of the enzyme between populations remains unchanged within the time frame of the experiments. The cause of such apparent “freezing” of conformational states of the enzyme remains puzzling. From our perspective, the formation of P450 oligomers in the microsomal membrane and in reconstituted systems may be proposed as the most viable explanation for such persistent heterogeneity (4).
The ability of microsomal cytochromes P450 to form oligomers both in solution (16–8) and in membranes (19–24) is well known. Furthermore, there are several indications of the formation of mixed oligomers of different P450 species in the endoplasmic reticulum (22, 23). Although the co-existence of different P450 species in the membrane is primarily considered in terms of their competition for NADPH-cytochrome P450 reductase (25–28), some authors have recognized direct association between P450 enzymes as an important determinant of the functional properties of the microsomal monooxygenase (26, 29–31). In addition, functional heterogeneity of a single P450 species caused by oligomerization appears to be revealed in the kinetics of CO binding (32, 33), ability to be phosphorylated by cAMP-dependent protein kinase (34), and interactions with substrates (35, 36). However, despite increasing interest in homo- and heterooligomerization of cytochromes P450 (30, 31, 37–40), its functional importance appears underestimated.
Important insight into structural and functional consequences of oligomerization was derived from studies on pressure-induced transitions of CYP2B4 in solution and in membranes (41–43). We found that only about 65–70% of the ferrous carbonyl complex of this oligomeric protein in solution is susceptible to a pressure-induced P450 → P420 transition. The same non-uniform barotropic behavior was also observed for the oligomers of ferric CYP2B4, where only about 30–5% of the hemoprotein appears to be involved in substrate binding and subsequent spin transitions, being at the same time insensitive to pressure-induced inactivation (42). As these irregularities disappear upon P450 monomerization, we suggested that the oligomerization of CYP2B4 gives rise to important differences between subunits in their ability to interact with substrates, water accessibility of the heme pocket, and sensitivity to pressure-induced inactivation (42–44). Recently we demonstrated similar behavior for human CYP3A4 in solution and in recombinant yeast microsomes and suggested a role in the mechanisms of cooperativity in this enzyme (4).
To explore functional consequences of oligomerization and their possible involvement in the mechanisms of CYP3A4 cooperativity, we elected to examine the kinetics of reduction by dithionite. Sodium hydrosulfite, Na2S2O4, which is commonly known as sodium dithionite, is a versatile reducing agent widely used to prepare the reduced forms of electron transfer proteins. In particular, reduction of cytochrome P450 by dithionite is used in routine quantitative determinations of the heme protein. Studies of the kinetics of reduction by dithionite were employed to probe apparent conformational transitions and changes in the ligand sphere of the heme iron in such heme-containing enzymes as bovine heart cytochrome c oxidase (45, 46), cytochrome bo from E. coli (47, 48), and some other biological electron carriers (49–52). For most heme proteins studied it was shown that the mechanism of reduction involves the dithionite anion monomer as the reducing agent (53–58). Detailed analysis of the kinetics of reduction of P450cam by dithionite was published by Hintz and Peterson (59), who found the mechanism of this reaction to involve as the reducing species. The kinetics of reduction of substrate-free P450cam obeys simple pseudo-first order kinetics, and the binding of substrate (camphor) drastically decreases the rate constant of reduction. This observation was explained as evidence of restricted access of the charged reducing species, the dithionite anion monomer, to the heme iron of cytochrome P450, especially in the substrate-bound state. Kinetics of dithionite-dependent reduction of microsomal cytochrome P450 2B4, both in solution and in proteoliposomes was shown to obey an equation of the sum of two exponentials with 60–70% of the heme protein reduced in the fast phase (60, 61). The distribution of the phases showed no dependence on temperature (61) or addition of substrate (benzphetamine) (D. R. Davydov, unpublished observation). As this biphasicity disappears upon dissolution of the enzyme oligomers or proteoliposomes by detergent, it was suggested to reflect some peculiarities of the oligomer architecture, resulting in inequality of the subunits in conformation and/or orientation (61).
As these results appear to be highly relevant to the hypothesis on the involvement of apparent conformational heterogeneity in the cooperativity of CYP3A4 (4, 13–15), we decided to investigate the effect of the state of oligomerization of CYP3A4 on the kinetics of reduction by dithionite at different concentrations of substrate. Recent methodological advances in biochemical spectroscopy and nanotechnology enhance considerably the informativeness and reliability of this approach. In this study we employ a rapid scanning stop-flow technique to resolve the processes of reduction of the low- and high-spin states of the hemoprotein. We also take advantage of the recently developed methodology for incorporation of CYP3A4 into nanoscale, discoidal lipid bilayer particles (Nanodiscs), which provides purely monomeric cytochrome P450 in a membrane environment (12), avoiding undesirable effects of detergents on the heme protein. To probe the relevance of our results to cytochrome P450 oligomers in phospholipid bilayers we also compare kinetics of dithionite-dependent reduction of CYP3A4 in solution and in Nanodiscs with that observed in large unilamellar proteoliposomes with various surface molar densities of the enzyme.
Experimental Procedures
Materials
Bromocriptine mesylate, 3,4-dihydroxybenzoic (protocatechuic) acid, protocatechuate 3,4-dioxygenase from Pseudomonas sp., 3-sn-phosphatidic acid (PA) sodium salt from egg yolk lecithin, 3-sn-phosphatidylethanolamine (PE) from bovine brain, and L-a-phosphatidycholine (PC) were the products of Sigma Chemicals (St. Louis, MO). Emulgen-913 was obtained from Kao Atlas Co. (Japan). Octyl-β-D-glucopyranoside (octylglucoside, OG) was obtained from Fluka (Switzerland). 2-Decanoyl-1-(O-(11-(4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-propionyl)amino)undecyl)-sn-glycero-3-phosphocholine (BODIPY-PC) was a product of Molecular Probes Inc. (Eugene, OR). All other chemicals were of ACS grade and were used without further purification.
Expression and purification of CYP3A4
Wild-type CYP3A4 was expressed as a His-tagged protein in Escherichia coli TOPP3 cells and purified using Ni-NTA metal affinity resin from Qiagen Inc. (Valencia, CA) as described earlier (62, 63).
Preparation of proteoliposomes
Proteoliposomes were prepared using the octylglucoside/dialysis technique according to the procedure described by Schwartz and co-authors (24, 64) with some modifications. We used a 2:1:0.6 mixture of PC, PE and PA with addition of 10 μg of BODYPY-PC per 1 mg of lipid mixture. Lipids (10 mg) were mixed as chloroform solutions, and the solvent was removed by evaporation under a stream of nitrogen gas and subsequent drying under vacuum for 2 hours. The suspension of lipids in 2.5 mL of 100 mM HEPES, 150 mM KCl, 0.5 mM EDTA, 1 mM DTT containing 20% (v/v) glycerol, pH 7.4 (Buffer A) was prepared by short and gentle sonication in an ultrasonic bath at room temperature. After addition of a required amount of CYP3A4 (3.6 and 0.9 nmol/mg lipid for Sample I and Sample II respectively) and mixing on a Vortex mixer, the suspension was incubated at room temperature for 45 minutes and diluted to a final volume of 9 mL. Solid OG was added to a concentration of 0.43%, and the mixture was incubated at 4°C until complete solubilization of the lipid. The mixture was dialyzed at 4°C under constant gentle bubbling of argon gas against three changes of 250 mL portions of buffer A, each containing 1.5 mL of CALBIOSORB™ hydrophobic adsorbent (EMD Biosciences, Inc., La Jolla, CA). After 72 hours of dialysis (24 hours per each portion of the buffer) the mixture was further diluted by equal volume of the same buffer containing no glycerol and centrifuged at 45,000 rpm for 5 hours. The pellet was resuspended in 100 mM Na-Hepes buffer, pH 7.4, containing 0.5 mM EDTA, 1 mM DTT, and 10% glycerol and stored at −80ºC. The diameter of proteoliposomes was assessed by negative staining electron microscopy. The molar ratio of phospholipids to cytochrome P450 was determined based on the amplitude of the absorbance peak of BODIPY-PC using an extinction coefficient at 506 nm of 71 mM−1cm−1 (65). The content of phospholipids was also determined from inorganic phosphorus in chlorophorm/methanol extracts mineralized with perchloric acid (66).
Preparation of Nanodiscs containing CYP 3A4
Soluble nanoscale membrane bilayer particles (Nanodisc) containing CYP3A4 were obtained by a detergent-removal technique from the mixture of CYP3A4, phospolipid and a membrane scaffold protein (MSP1D1) as previously described (12) (67).
Determination of the effect of detergent on the velocity of sedimentation of CYP3A4
Sedimentation velocity experiments were carried out using a Beckman-Coulter XL-A analytical ultracentrifuge with absorbance optics and an An60-Ti rotor at 20°C and 60,000 rpm and monitoring of absorbance at 417 nm. The analysis were carried out in 100 mM HEPES, 1 mM EDTA and 1mM DTT at pH 7.4, with or without 0.15% Emulgen 913. Scans were analyzed with the continuous sedimentation coefficient (c(s)) and molecular weight (c(M)) distribution methods (68) using the SedFit program (www.analyticalultracentrifugation.com). For this analysis we used the value of the partial specific volume of cytochrome P450 of 0.78 cm3/g, as determined by Wendel and co-authors by sedimentation velocity in the variable density media (69).
Experimental
Titration and scanning stop-flow kinetic experiments were performed using a rapid scanning two-channel MC2000-2 CCD-spectrometer (Ocean Optics, Inc., Dunedin, FL) equipped with a custom-modified CUV-ALL-UV thermostated cell holder (Ocean Optics) and an OSRAM 64614 UV-enhanced tungsten halogen lamp (OSRAM, Germany) as a light source. In the stop-flow experiments this instrument was combined with a SF-MiniMixer Stopped-Flow apparatus (KinTek Corporation Austin, TX) with a 5 mm path length quartz cell connected to the mixer with 3 cm long Teflon® tubing. Dead time of the mixer was less then 4 ms. Computer control of the mixer, which allowed us to attain a synchronization between the mixing and scanning, was achieved using an ACL-7120 digital I/O and timing board (ADLink Technology, Taiwan). Custom-designed software was used for data acquisition. The above configuration allowed us to collect a series of spectra in the 340–800 nm range with a time interval ≥4 ms starting at the exact moment of mixing. Typically, in each stop-flow experiment we collected 96 spectra in the 380–600 nm region using a double time-base scheme, so that the time interval between the spectra increased 10–20 times after the first 32 measurements. All experiments were carried out in 100 mM Na-HEPES buffer, pH 7.4, containing 1 mM dithiothreitol and 1 mM EDTA. The spectral baseline was recorded with the buffer in the sample cell. The temperature was 25 °C, and the concentration of dithionite in the stop-flow cell was 12.5 mM if not otherwise indicated. Both cytochrome P450 and sodium dithionite solutions were prepared using buffer saturated with CO by bubbling for 10 minutes. In the experiments on the dependence of the reduction kinetics on the concentration of dithionite the media was supplemented with an oxygen-scavenging system consisting of 0.1 unit/mL of protocatechuate dioxygenase and 1 mM protocatechuic acid.
Data Processing
In our analysis of the series of spectra obtained in kinetic experiments we implemented principal component analysis (PCA) (42, 70) to increase the signal-to-noise ratio and improve the accuracy of the assay. Only the signal represented by the first two principal components, which usually cover over 99.5% of the observed spectral changes, was taken into further analysis. Higher-order components containing no detectable bands of cytochrome P450 were discarded. To interpret the observed transitions in terms of the changes in concentration of P450 species we used a least-squares fitting of the spectra to the set of the spectral standards of pure high-spin and low-spin ferric P450 (4) and CO-bound ferrous P450 and P420 states of CYP3A4. The latter two spectral standards were resolved from experiments on pressure-induced P450-to-P420 transitions in CO-bound ferrous CYP3A4, which will be described elsewhere. This analysis gave us the curves representing time-dependent changes in the concentrations of the ferric low-spin, ferric high-spin and the ferrous carbonyl complexes of P450 and P420 states of the heme protein. These kinetic curves were fitted to a multiexponential equation:
where C0, Ct, and C∝ are the concentrations of the compound at the beginning of the reaction, at time t and at an infinite time respectively; Fi and ki are the fraction and the rate constant of i-th exponential phase. The number of exponents (n) varied from 1 to 3 depending on the case. In bromocriptine titration experiments the curves reflecting the substrate-induced spin shift were fitted to the equation for the equilibrium of binary association at comparable concentrations of interacting species ((71), p 73, eq. II-53). In all cases the fitting was made by non-linear regression using a combination of Nelder-Mead and Marquardt algorithms using SPECTRALAB program (42).
The estimates of the spin state of CYP3A4 given in this study were calculated from the amplitudes of the kinetic curves of reduction of the high-spin and low-spin P450 determined as described above.
Results
Kinetics of CYP3A4 reduction by dithionite in oligomers in solution
The process of dithionite reduction of CYP3A4 in solution is illustrated in Figure 1. Kinetic curves of the appearance of the reduced P450-CO complex were found to be multiexponential (Fig. 1). Three exponential terms were needed to avoid systematic deviation of the fitting curves from the experimental data. The square correlation coefficient for this fitting was always higher than 0.995 and in most cases as high as 0.9995 or above. As shown in Table 1, which summarizes the kinetic parameters of reduction in various systems, in solution with no substrate added each of the three phases of P450 reduction comprises about one-third of the total enzyme. The kinetics of disappearance of the low-spin ferric heme protein in the absence of substrate also obeys three-exponential kinetics exhibiting rate constants similar to those deduced from reduction of the total P450, and fractions of the fast, middle and slow phases equal to 0.42 ± 0.07, 0.31 ± 0.05, and 0.27 ± 0.05, respectively. In contrast, the kinetic curves of the reduction of the high-spin fraction of the enzyme (Fig. 1c,d) obey a bi-exponential equation with the fraction of the slow phase as high as 0.83 ± 0.1 and rate constants similar to those of the second and third phases of the total P450 reduction (Table 1). Thus, the heme protein reduced in the first phase was represented by the low-spin fraction only, and the spin state of the slow-reducible enzyme was shifted considerably towards the high-spin state. The high-spin content of the total heme protein was determined to be 14.2 ± 6.8% at 25 °C in the absence of substrate, while the high-spin content in the enzyme reduced in the slow phase was as high as 33 ± 10%. This difference in spin equilibrium between fast- and slow-reducible heme protein resulted in the time-dependent displacement of the position of the trough of ferric P450 in the difference spectra of reduction, as illustrated in Fig. 1b.
Fig. 1.
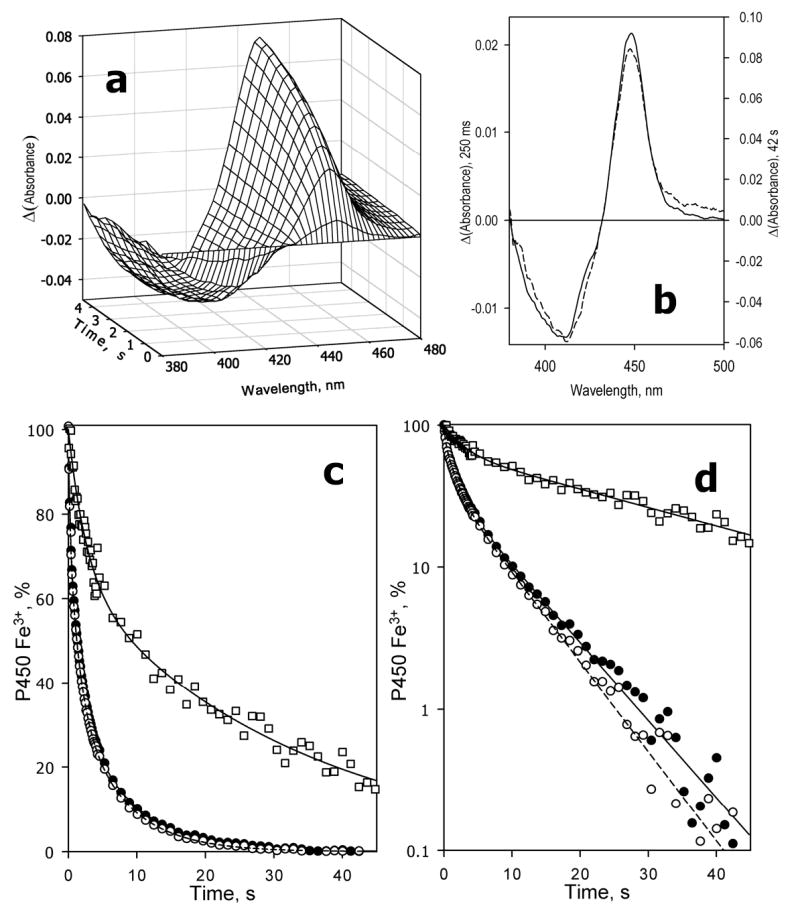
Kinetics of dithionite-dependent reduction of CYP3A4 in oligomers in solution. Conditions: 3 μM 3A4, 12.5 mM sodium dithionite, CO-saturated 0.1 M Na-Hepes buffer, pH 7.4, 1 mM DTT, 1 mM EDTA, 1 mM protochatechuic acid, 0.1 U/mL protocatechuate dioxygenase, 25 °C. Spectra recorded in a stop-flow cell with 5 mm optical path length. a: Changes in absorbance in the Soret region during the initial 5 seconds of reaction. The first spectrum shown corresponds to the time of 60 ms after mixing, and the following spectra were taken in 400 ms time intervals. The spectrum measured at time of origin is subtracted. b: The difference spectra of reduction measured at 250 ms (dashed line) and 42 s (solid line) after mixing. The spectra are scaled by linear least squares algorithm to have similar amplitudes in the graph. c,d: Kinetics of reduction of the total pool (filled circles, solid line), low-spin (empty circles, dashed line) and high-spin (squares, solid line) of CYP3A4 in linear (c) and semi-logarithmic (d) coordinates. The lines show the results of the fitting of the data to the equation of the sum of three (total heme protein and the low-spin fraction) or two (high-spin fraction) exponents. The data were scaled according to the fitting results to represent a percent decrease in the content of the respective states of the ferric enzyme.
Table 1.
Kinetic parameters of dithionite-dependent reduction of the total pool and the high-spin fraction of CYP3A4 in various systems at no substrate added and in the presence of bromocriptine.*
| Total CYP3A4 | High-spin fraction | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| System | Bromocriptine, μM | k1 s−1 | k2 s−1 | k3 s−1 | F1 | F2 | F3 | kfast, s−1 | kslow, s−1 | Fslow |
| Oligomers in solution | 0 | 3.3 ± 0.6 | 0.41 ± 0.07 | 0.081 ± 0.01 | 0.33 ± 0.07 | 0.35 ± 0.05 | 0.32 ± 0.05 | 0.48 ±0.17 | 0.074 ±0.02 | 0.83 ±0.10 |
| 32 | 3.5 ± 0.4 | 0.23 ± 0.10 | 0.043 ± 0.02 | 0.06 ± 0.02 | 0.37 ± 0.06 | 0.58 ± 0.07 | 0.29 ±0.12 | 0.043 ±0.02 | 0.71 ±0.13 | |
| Monomers (0.15% Emulgen-913) | 0 | 0.44 ± 0.01 | 1.0 | 0.56 ±0.17 | 0 | |||||
| 24 | 0.37 ± 0.01 | 1.0 | 0.47 ±0.03 | 0 | ||||||
| Nanodisc- incorporated CYP3A4 | 0 | 6.2 ± 0.6 | 0.59 ± 0.14 | 0.84 ± 0.03 | 0.16 ± 0.03 | 6.2 ± 0.6 | 0.72 ±0.30 | 0.3 ±0.10 | ||
| 24 | 4.4 ± 1.8 | 0.14 ± 0.01 | 0.03 ± 0.07 | 0.97 ± 0.07 | 0.11 ±0.01 | 1.0 | ||||
| P450-rich proteoliposomes (Sample I) | 0 | 8.8 ± 1.6 | 2.2 ± 0.2 | 0.20 ± 0.05 | 0.22 ± 0.09 | 0.52 ± 0.09 | 0.25 ± 0.07 | 1.9 ±0.5 | 0.14 ±0.06 | 0.7 ±0.16 |
| 6 | 3.9 ± 0.1 | 0.92 ± 0.42 | 0.12 ± 0.01 | 0.11 ± 0.01 | 0.21 ± 0.05 | 0.66 ± 0.13 | 0.10 ±0.01 | 1.0 | ||
| Lipid-rich proteoliposomes (Sample II) | 0 | 2.0 ± 0.1 | 1.0 | 1.9 ± 0.1 | 0 | |||||
| 32 | 0.050 ± 0.02 | 1.0 | 0.045 ±0.01 | 1.0 | ||||||
The values given in the Table were obtained by averaging the result of 3–12 individual measurements and the “±” values show the confidence interval calculated for p = 0.05.
In corroboration of the validity of the three-exponential approximation of the reduction kinetics, the distribution of the phases was independent of the concentration of dithionite in the range probed (0.8–50 mM). All three constants are linearly related to the square root of the dithionite concentration in the 0.8–12.5 mM range (Fig. 2). Thus, similar to P450cam (59), CYP3A4 is reduced by dithionite anion-monomer ( ), not by the dithionite anion itself. At higher concentrations of the reductant the value of k1 becomes independent of the dithionite concentration, thus revealing a change in the rate-limiting step of the fastest phase of reduction. Linear approximation of the dependences shown in Fig. 2 gives the values of the rate constants of 29.5 ± 1.7 s−1M−0.5, 3.18 ± 0.18 s−1M−0.5, and 0.46 ± 0.08 s−1M−0.5 for each of the three phases respectively. Interestingly, linear approximation of the dependence of k3 on the square root of the dithionite concentration yields an important offset at zero dithionite concentration, which may indicate an extremely slow fourth phase of reduction with a rate constant independent of the dithionite concentration. This putative fourth phase, which cannot be resolved by our multiexponential fitting, appears to be limited by some transition in the protein represented by a first-order process with the rate constant of 0.035 ± 0.01 s−1. Thus, our results on the kinetics of dithionite-dependent reduction of CYP3A4 in solution clearly demonstrate a persistent heterogeneity of the enzyme. The enzyme pool appears to be distributed between several different conformers with different rate constants of reduction by dithionite anion-radical.
Fig. 2.
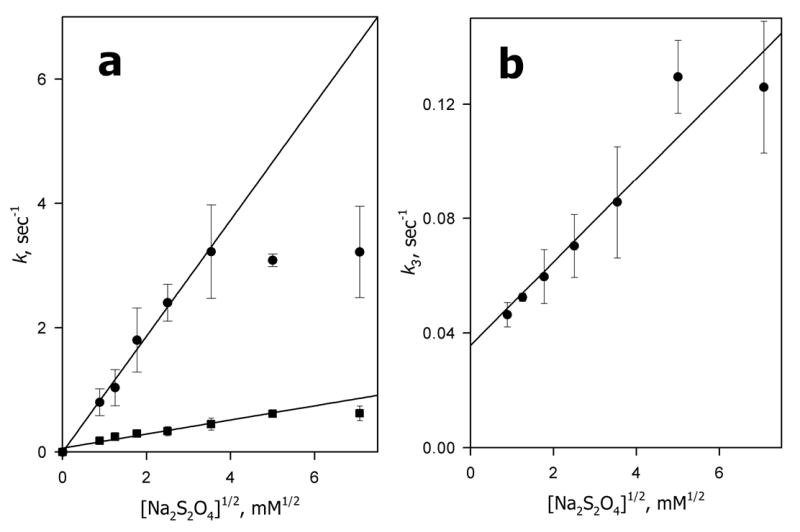
Dependence of the pseudo-first order rate constants of CYP3A4 reduction on the square root of dithionite concentration. Panel a shows the dependencies for k1 (circles) and k2 (squares). Data for k3 are shown in panel b. The points shown correspond to averages of 3–6 measurements. Error bars represent the confidence interval (p = 0.05). Concentrations of dithionite varied from 0.78 to 50 mM. Other conditions as indicated in Fig. 1.
Effect of CYP3A4 interactions with bromocriptine on the kinetics of reduction
In agreement with previous findings (4, 70), addition of bromocriptine to CYP3A4 caused an important increase in the content of the high-spin heme protein, which reaches 63 ± 7% at saturating bromocriptine (32 μM). This substrate-induced spin shift was accompanied by a drastic decrease in the overall rate of reduction (Fig. 3). This effect is achieved by an increase in the amplitude of the third phase of the reduction at the expense of the first phase, while the changes in the fraction of the middle phase were marginal (Fig. 4a).
Fig. 3.
Effect of bromocriptine on the kinetics of reduction of the total pool (a) and the high-spin fraction (b) of CYP3A4 in the oligomers in solution. The kinetic curves were recorded at no substrate added (circles) and in the presence of 1 μM (squares) and 6 μM (diamonds) bromocriptine. The data are plotted in semilogarithmic coordinates. Lines show the results of fitting of the data to the equation of sum of three (a) or two (b) exponents. Conditions: 3 μM 3A4, 12.5 mM sodium dithionite, CO-saturated 0.1 M Na-Hepes buffer, pH 7.4, 1 mM DTT, 1 mM EDTA, 25 °C.
Fig. 4.
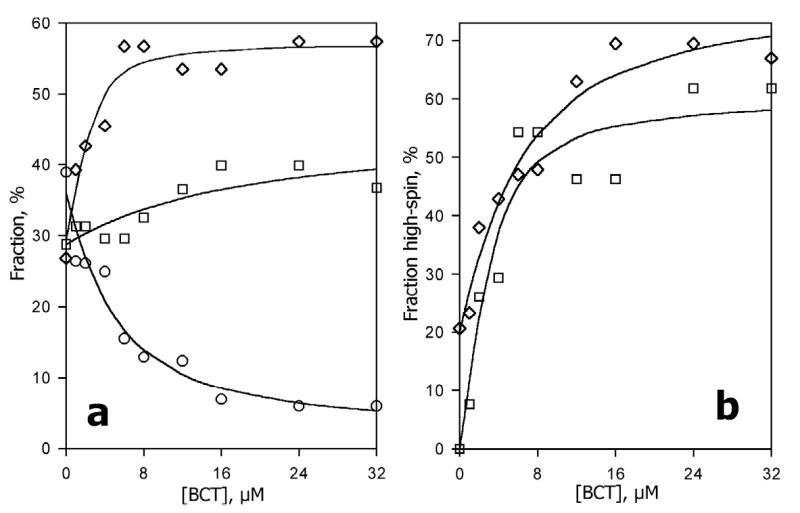
Effect of bromocriptine on the kinetics of reduction of CYP3A4 in the oligomers in solution. a: The fractions of the fast phase (F1, circles), middle phase (F2, squares) and the slow phase (F3, diamonds) plotted versus bromocriptine concentration. b: The content of the high-spin P450 in the fractions reduced in the middle (squares) and slow (diamonds) phase. Lines shown in panels a and b were obtained by fitting the respective data sets to the equation for the equilibrium of binary association. The values were obtained from the fitting of the kinetic curves of dithionite-dependent reduction. Conditions as indicated in Fig. 3.
In contrast to important changes in the kinetics of the reduction of the total P450 pool, the effect of bromocriptine on the reduction of the high-spin P450 was rather modest. Addition of substrate caused some decrease in the fraction of the slow phase (Table 1). Superposition of the phase distribution in the kinetics of the reduction of the low-spin, high-spin and the total heme protein allows us to calculate the content of the high-spin P450 reduced in each of three phases in the total CYP3A4. The fraction of CYP3A4 reduced in the first (fastest) phase is represented by the low-spin heme protein only in the whole range of bromocriptine concentrations studied. In contrast to low-spin fast-reducible P450, the heme-protein reduced in the second and third phases revealed a substrate-dependent equilibrium between the high-spin and low-spin states, as illustrated in in Fig. 4a,b. The dependencies of the high-spin content in P450 reduced in the second and third phases may be fitted to an equation for the equilibrium of binary association at comparable concentrations of enzyme and substrate. This fitting gives the values of the apparent dissociation constant of P450-bromocriptine complex of 1.0 ± 0.6 μM and 4.3 ± 1.3 μM for the hemeprotein reduced in the second and third phases, respectively.
Effect of monomerization of the protein by Emulgen-913 on the kinetics of reduction
By analogy to suggestions in earlier studies of CYP2B4 (61), we may assume that the heterogeneity of the CYP3A4 pool revealed in the kinetics of dithionite-dependent reduction mey be caused by a non-equivalency of the subunits in protein oligomers. Sedimentation velocity experiments at 1–3 μM CYP3A4 in the absence of any detergent showed large aggregates with sedimentation coefficients of 8.2–12.6 S (data not shown). Continous molecular weight distribution (c(M)) analysis (68) reveals at least three major sedimenting species with molecular masses of 215, 290 and 450 kDa. Addition of 0.15% Emulgen-913 resulted in complete dissociation of these oligomers, as evidenced by a single sedimentation band centered at 3.4 S. According to c(M) analysis this band represents a species with molecular mass of 56 kDa, which corresponds to a monomer of CYP3A4. Therefore, we assume that addition of 0.15% Emulgen-913 results in complete monomerization of the heme protein.
As shown in Fig. 5, kinetics of reduction of CYP3A4 in the presence of 0.15% Emulgen-913 becomes monophasic, and addition of 24 μM bromocriptine had virtually no effect on the kinetics (Table 1). Therefore, monomerization of the enzyme by detergent eliminates the heterogeneity and prevents the substrate-induced transition observed in the enzyme oligomers. However, besides monomerization, non-ionic detergents may affect substrate binding, as Triton N101, Emulgen-913 and similar detergents are metabolized by CYP3A4 and exhibit rather high affinity (Km = 39 μM for Triton N101) (72).
Fig. 5.
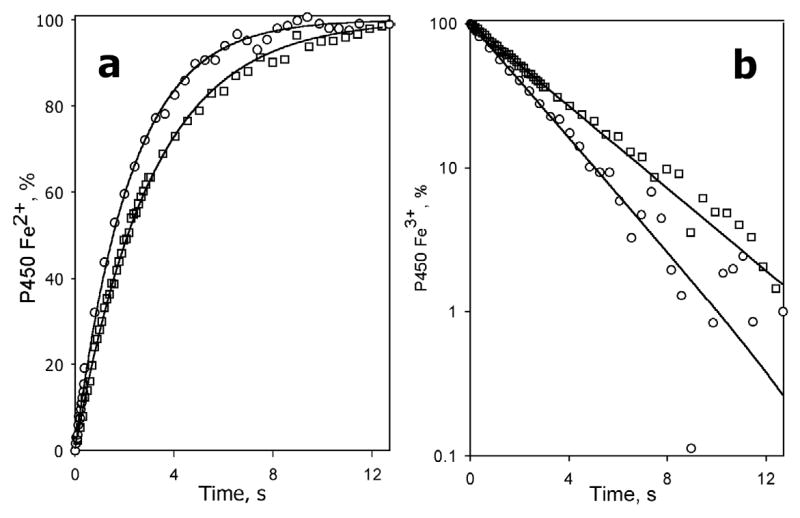
Kinetics of dithionite-dependent reduction of monomeric CYP3A4 in solution containing Emulgen-913 at no substrate added (circles) and in the presence of 12 μM bromocriptine (squares) in linear (a) and semilogarithmic (b) coordinates. Lines show the results of fitting of the data to a mono-exponential equation. The buffer contained 0.15% Emulgen-913. Other conditions as indicated in Fig. 3.
Kinetics of dithionite-dependent reduction of CYP3A4 incorporated into Nanodiscs
To discriminate the effect of monomerization from possible specific effects of Emulgen-913 it was important to probe a detergent-free monomeric system. For this purpose, we studied the kinetics of dithionite-dependent reduction of CYP3A4 incorporated in a soluble nanoscale membrane bilayer, or Nanodisc (12). The Nanodisc system, obtained by self-assembly from the mixture of P450, phospholipid, and a membrane scaffold protein, represents a homogeneous monomeric population of CYP3A4 embedded in small discoidal lipoprotein particles of uniform size (12).
In the absence of substrate the kinetics of reduction of CYP3A4 in Nanodiscs was extremely fast and obeys a bi-exponential equation with 84 ± 3% protein reduced in the fast phase (Table 1, Fig. 6a,b). In much the same way as in P450 oligomers, the fractions reduced in the fast and slow phase differed in the position of spin equilibrium. Thus, the heme protein reduced in the fast phase contained only 15.6 ± 6 % high-spin state, whereas the high-spin content in the slow phase was 40 ± 2%.
Fig. 6.
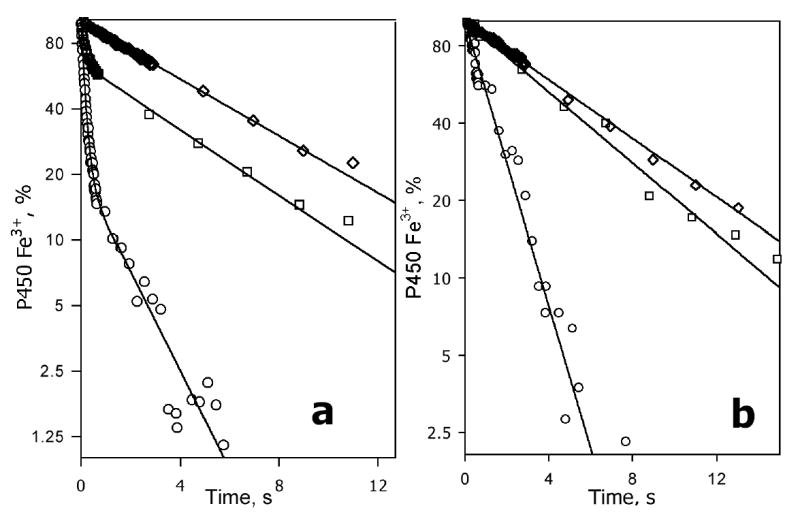
Effect of bromocriptine on the kinetics of reduction of the total pool (a) and the high-spin fraction (b) of CYP3A4 incorporated into Nanodiscs. The kinetic curves were recorded at no substrate added (circles) and in the presence of 6 μM (squares) and 24 μM (diamonds) bromocriptine. The data are plotted in semilogarithmic coordinates. Lines show the results of fitting of the data to the equation of sum of two exponents (a) or mono-exponential (b) equation. The reaction mixture contained 2.5 μM Nanodisc-incorporated CYP3A4. Other conditions as indicated in Fig. 3.
Similar to oligomers in solution, addition of bromocriptine drastically decreased the rate of reduction by increasing the fraction reduced in the slow phase (Fig. 6a,b; Fig. 7). At 24 μM bromocriptine, where the content of the high-spin state of P450 reaches 60 ± 10%, the fraction of the slow phase of the reduction (Fslow) became as high as 0.97 ± 0.07, making the kinetics essentially mono-exponential. The dependence of Fslow on the concentration of bromocriptine fits the equation for the equilibrium of bimolecular association with KD = 0.5 μM (Fig. 7), which is consistent with the value determined by spectrophotometric titration of CYP3A4 in Nanodiscs by bromocriptine (KD = 0.39 ± 0.15 μM). In the presence of any concentration of bromocriptine tested (1–24 μM) the kinetics of reduction of the high-spin protein obeys a mono-exponential equation with the rate constant equal to that of the slow phase of the total reduction process (Fig 6a,b). Thus, consistent with the effect of monomerization of 3A4 by detergent, incorporation of the enzyme into Nanodiscs eliminates its heterogeneity. However, in contrast to the behavior in the presence of Emulgen, the effect of substrate on the kinetics of the dithionite-dependent reduction is retained in Nanodiscs. CYP3A4 in Nanodiscs behaves as a single population, where the interaction with bromocriptine results in a conversion from a fast-reducible, predominately low-spin substrate-free state to slowly-reducible, predominately high-spin substrate-bound form.
Fig. 7.
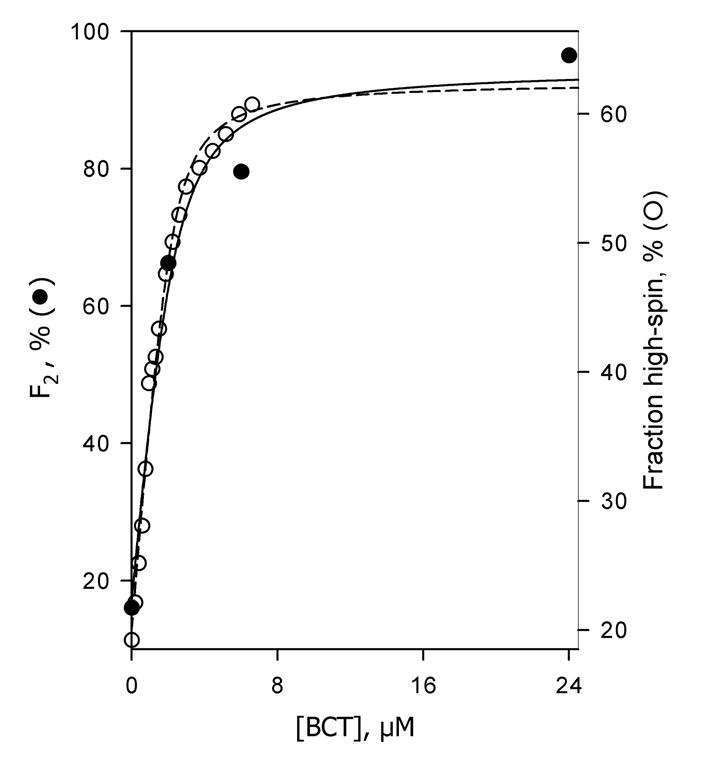
Bromocriptine-induced changes in the fraction of the slow phase of dithionite-dependent reduction (Fslow, filled circles) and in the high-spin content (empty circles) in CYP3A4 incorporated into Nanodiscs. The data on the high-spin content were obtained by titration of Nanodisc-incorporated CYP3A4 by bromocriptine. Lines show the results of fitting the respective data sets to the equation for the equilibrium of binary association. Reaction mixture contained 2.5 μM Nanodisc-incorporated CYP3A4. Other conditions as indicated in Fig. 3.
Kinetics of dithionite-dependent reduction of CYP3A4 in proteoliposomes
The above data clearly indicate that the oligomerization of P450 in solution results in a divergence of the heme protein pool into two fractions differing in the rates of dithionite-dependent reduction, position of spin equilibrium, and response to substrate binding. To probe the relevance of this conclusion in P450 oligomers in model membranes we studied the kinetics of CYP3A4 reduction in proteoliposomes using two samples having a lipid-to-protein ratio (L/P) of 112:1 and 726:1 mol/mol. These preparations are designated as Samples I and II, respectively. As demonstrated by electron microscopy, in both cases the average diameter of the liposome vesicles was around 200 nm (data not shown). Assuming the partial surface of the lipid bilayer per one molecule of phospholipids to be of 75 Å2 (73), the number of P450 molecules per vesicle is around 1500 in Sample I and around 230 in Sample II. Assuming that CYP3A4 exists in equilibrium between monomeric and oligomeric states in the proteoliposomal membrane, we may expect considerably lower degree of oligomerization in Sample II, compared with Sample I.
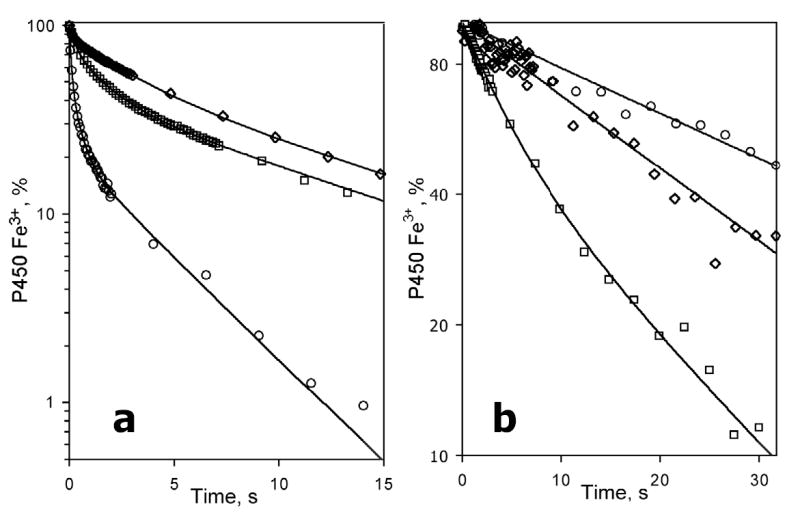
Kinetics of dithionite-dependent reduction of CYP3A4 in Sample I was close to that observed in oligomers in solution. In the absence of substrate kinetic curves obey an equation of the sum of three exponents with the fractions of 0.22 ± 0.09, 0.52 ± 0.09, and 0.25 ± 0.07 (Table 1). Addition of bromocriptine does not eliminate the multiphasicity, but causes a redistribution of the fractions towards the third phase (Table 1, Fig. 8). At 6 μM bromocriptine the fraction of the third phase became as high as 0.65 ± 0.12 at the expense of decreased fractions of the first and the second phases (Table 1).
Fig. 8.
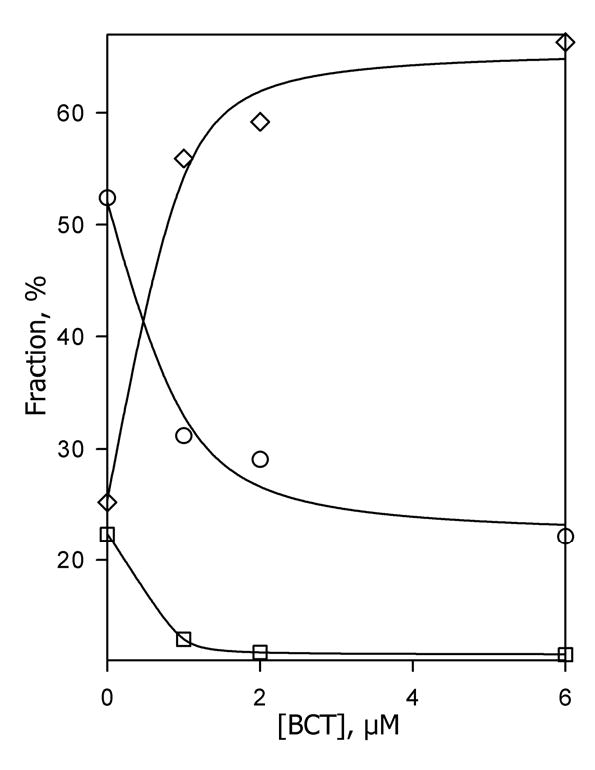
Effect of bromocriptine on the kinetics of reduction of CYP3A4 in P450-rich proteoliposomes (Sample I). The fractions of the fast phase (F1, circles), middle phase (F2, squares) and the slow phase (F3, diamonds) plotted versus bromocriptine concentration. Lines represent the results of fitting of the respective data sets to the equation for the equilibrium of binary association. The values were obtained from the fitting of the kinetic curves of dithionite-dependent reduction of 3 μM suspension of the P450-rich liposomes (Sample I). Other conditions as indicated in Fig. 3.
The behavior of Sample II was considerably different. At no substrate added the kinetics of reduction was mono-exponential with a rate constant of 2.0 ± 0.1 s−1 (Fig. 9). Similar to Nanodiscs, the addition of bromocriptine caused conversion to the second (slow) phase, the amplitude of which was proportional to the degree of saturation of the enzyme with substrate (Fig. 10). At 32 μM bromocriptine the fraction of the slow phase reached 1.0, so that the kinetics exhibited a single exponential (slow) phase (Table 1). Also, as in Nanodiscs, in the presence of bromocriptine the fractions reduced in the fast and slow phases were strictly different in spin state. At any concentration of the substrate probed (1–32 μM) the kinetics of reduction of the high-spin P450 followed a mono-exponential equation with a rate constant equal to that of the slow phase of the total reduction process.
Fig. 9.
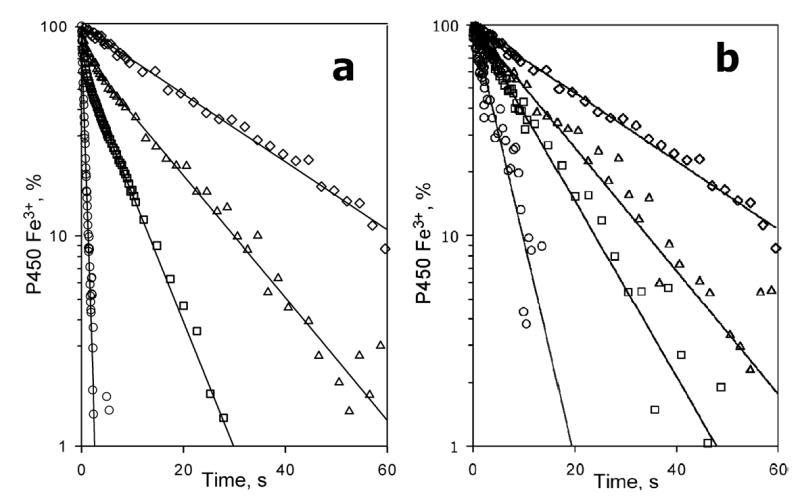
Effect of bromocriptine on the kinetics of reduction of the total pool (a) and the high-spin fraction (b) of CYP3A4 incorporated into the lipid-rich proteoliposomes (Sample II). Kinetic curves were recorded at no substrate added (circles) and in the presence of 8 μM (squares), 16 μM (triangles) and 32 μM (diamonds) bromocriptine. The data are plotted in semilogarithmic coordinates. Lines show the results of fitting of the data to the equation of sum of two exponents (a) or mono-exponential equation (b). Reaction mixture contained 3 μM of CYP3A4 in lipid-rich liposomes (Sample II). Other conditions as indicated in Fig. 3.
Fig. 10.
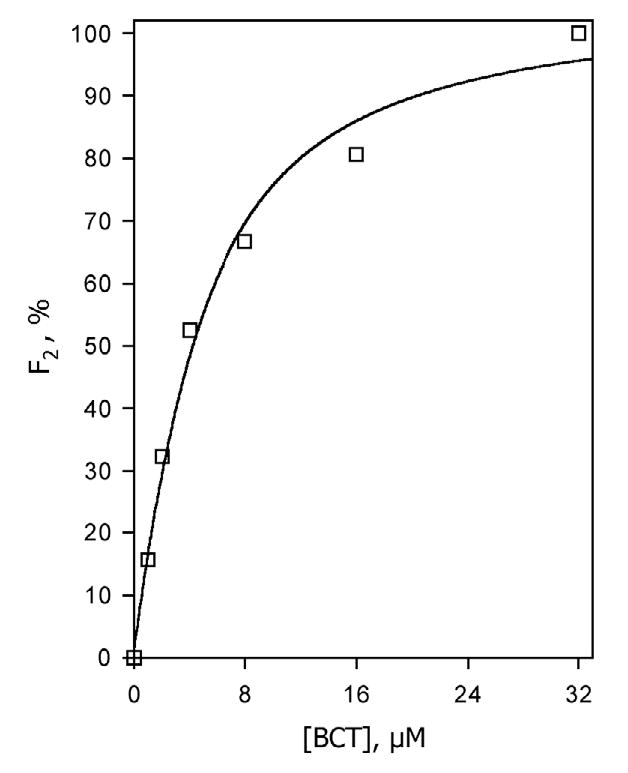
Bromocriptine-induced changes in the fraction of the slow phase of dithionite-dependent reduction (Fslow,) of CYP3A4 in lipid-rich liposomes (Sample II). Lines show the results of the data fitting to the equation for the equilibrium of binary association. Other conditions as indicated in Fig. 3.
Discussion
The study of dithionite-dependent reduction of CYP3A4 has provided an important link between the oligomeric state of the enzyme and its functional properties. Reduction of CYP3A4 in oligomers in solution exhibited complex three-exponential kinetics with similar amplitudes of the three phases. Addition of bromocriptine resulted in the conversion of the protein reduced in the fast phase into a slowly reducible form, such that the increase in the fraction of the slow phase appears to be proportional to the degree of enzyme saturation with substrate. The fraction of the middle phase remained almost unchanged. Importantly, the fraction reduced in the fast phase was always represented by the low-spin P450 only, and the slowly reducible fraction was highly enriched in the high-spin heme protein. These results suggest that, similar to P450cam (59), binding of the substrate to CYP3A4 results in a transition . that decreases the accessibility of the heme for the reductant, dithionite anion monomer . However, this conclusion appears to be correct only for about 2/3 of the heme protein pool, which is reduced in the first (fast) and the third (slow) phase. The fraction reduced in the middle phase remains unchanged upon addition of substrate. The substrate-induced spin shift observed in this fraction suggests that it is capable of substrate binding but does not undergo any substrate-induced transition in accessibility of the heme moiety. Our results strongly confirm the point of view that the term “spin equilibrium” is inapplicable to the pool of CYP3A4 taken as a whole, but that there are several stable sub-populations of the enzyme with different position of spin equilibrium.
Abolishment of the heterogeneity of the CYP3A4 pool upon solubilization with Emulgen-913 confirms that this heterogeneity reflects a non-equivalency of the subunits in the CYP3A4 oligomers. To discriminate the effect of monomerization from possible specific effects of Emulgen-913 we studied the kinetics of dithionite-dependent reduction of CYP3A4 incorporated in a soluble nanoscale membrane bilayer, or Nanodisc (12). Consistent with results in the detergent-containing system, incorporation of CYP3A4 into Nanodiscs eliminated the heterogeneity of the heme protein revealed by dithionite reduction. Kinetics of reduction in Nanodiscs in the absence of substrate was close to mono-exponential, and addition of bromocriptine caused appearance of the slow phase, the amplitude of which was proportional to the concentration of the enzyme-substrate complex. (Fig. 7). This behavior reveals the same regularities as described by Hintz and Peterson for P450cam, where the kinetics of reduction was monophasic both in substrate-free and camphor-bound enzyme, but the binding of camphor resulted in a 4-fold decrease in the rate constant of reduction (59).
In order to approach the question whether the heterogeneity of CYP3A4 observed in solution takes place in the microsomal membrane as well, we studied the kinetics of dithionite-dependent reduction of the enzyme incorporated into large unilamellar liposomes with different lipid-to-protein ratio. A decrease in the fraction of rotationally immobile (aggregated) P450 with increasing lipid-to-protein ratio in proteoliposomes and rabbit liver microsomes was shown by rotational diffusion measurements in the laboratory of Kawato (20, 74). These authors demonstrated that in proteoliposomes with L/P weight ratio of 1 (~60:1 mol/mol) about 35% of the P450 had decreased rotational mobility, while increase in the L/P ratio to 10 (~600 mol/mol) decreased the immobile fraction to 7% (20). Thus, assuming CYP3A4 to exist in equilibrium between monomeric and oligomeric states in the proteoliposomal membrane, we may expect the degree of oligomerization to be considerably lower in our liposomal Sample II, compared with Sample I. In fact, we observed a striking similarity between the diluted liposomal preparation (Sample II) and CYP3A4 incorporated into Nanodiscs. We may conclude therefore that the effect of incorporation into Nanodiscs or diluted liposomal membranes on the kinetics of dithionite-dependent reduction of CYP3A4 is caused by elimination of oligomers.
Similar behavior of CYP3A4 oligomers in solution and in P450-rich liposomes (Sample I) suggests a common effect of P450 oligomerization in the membrane and in solution on the kinetics of dithionite-dependent reduction. In much the same way as in oligomers in solution, in the liposomal Sample I the kinetics of reduction in the absence of substrate revealed three exponential phases, although the fraction of the middle phase was as high as 0.52 ± 0.9 (compared to 0.35 ± 0.05 in CYP3A4 oligomers). The response of this liposomal system to addition of bromocriptine was more complex than that observed in oligomers in solution in that the interactions with substrate resulted in increase in the amplitude of the slow phase at the expense of both the fast and the middle phase. Nevertheless, interactions with substrate did not abolish the multiphasicity of the reduction (Fig. 8). Differences in the response of the P450-rich liposomes compared with oligomers in solution may reflect the effect of substrate on equilibrium of the oligomerization of the heme protein in the liposomes. Thus, in contrast to tight and essentially irreversible aggregation (oligomerization) in solution, the heme protein in the membranes exists in equilibrium between the monomeric and oligomeric states, which may be displaced upon substrate binding. The effect of substrate on the oligomerization equilibrium is supported by the observation of Greinert and co-workers that binding of benzphetamine to CYP2B4 in the proteoliposomal membrane decreases rotational mobility of the heme protein, which is indicative of substrate-induced oligomerization (19, 75).
The data presented in this report are consistent with a hypothetical scheme of CYP3A4 transitions shown in Fig. 11. We hypothesize that in the oligomers of CYP3A4 in solution and in P450-rich proteoliposomes the hemoprotein is distributed between two populations. Two forms of the heme protein — one reduced in the first phase (designated by (1) in Fig. 11) and one reduced in the third phase ((3) in Fig. 11), constitute the first population, which is shown on Fig. 11 by unfilled polygons. The fast-reducible state (1) is represented by the low-spin enzyme only. The content of the high spin heme protein in the slow-reducible form (3) changes from 20% in substrate-free enzyme to about 75% in the complex with bromocriptine (Fig. 4b). Binding of bromocriptine favors the formation of the slow form, so that at the saturation with the substrate the fast form almost vanishes. The behavior of this population of the enzyme is similar to that of P450cam, CYP3A4 incorporated into Nanodiscs, or CYP3A4 in diluted proteoliposomal membranes. The second population is designated by gray-filled rectangles in Fig. 11. It constitutes about 1/3 of the total enzyme in the oligomers in solution and is represented by the form reduced in the middle phase ((2), Fig. 11). It is also capable of binding substrate and exhibits a substrate-induced spin transition but fails to undergo any change affecting the rate of reduction. In Nanodiscs and in diluted proteoliposomal membrane the oligomers of CYP3A4 are absent, and the whole pool of enzyme behaves similarly to the population of the enzyme reduced in the first and third phases in the oligomers in solution. The postulated un-equivalency of the subunits of the oligomer may be caused by some peculiarities of their organization, causing a difference in orientation and/or conformation of the molecules of the enzyme. A crucial effect of substrate binding on the kinetics of reduction of the first population of CYP3A4 in oligomers, which also affects the whole pool of the monomeric P450 in Nanodiscs and diluted membranes, indicates that the interactions of the enzyme with substrate result in an important transition resulting in a restricted accessibility of the heme to anion monomer. We hypothesize that due to some steric constrains in the oligomer, the population of the enzyme reduced in the middle phase is incapable of this transition.
Fig. 11.
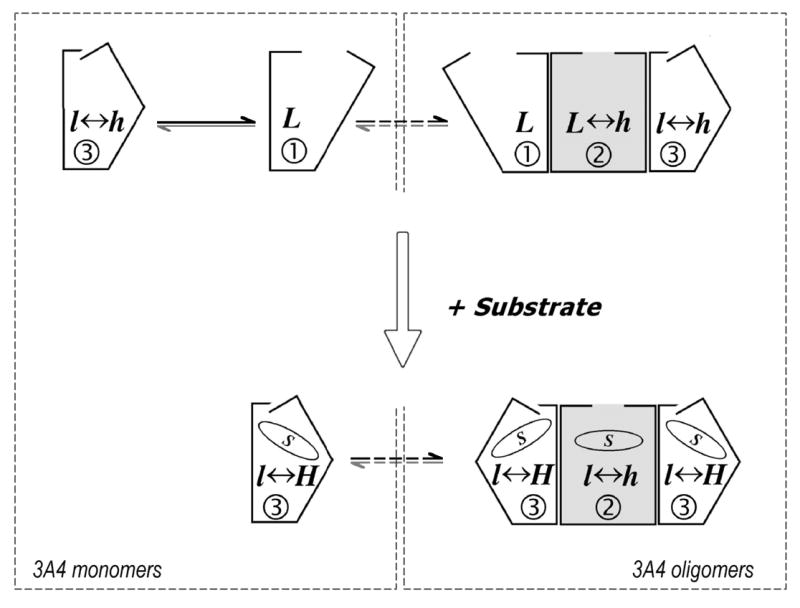
A suggested scheme of conformational transitions and oligomerization in CYP3A4, which explains the heterogeneity of the enzyme observed in the kinetics of dithionite dependent reduction. The whole scheme represents the situation expected in protein-rich liposomes and in microsomes. The left dashed rectangle outlines the transitions taking place in a monomeric system, such as CYP3A4 incorporated into Nanodiscs and the enzyme in lipid-rich liposomes. The right rectangle outlines the states observed in the oligomer in solution. Encircled digits symbolize the phase of the total hemeprotein reduction corresponding to each conformational state of the enzyme. Letters “L” and “H” symbolize high- and low-spin states of P450 respectively and the displacement of spin equilibrium in each of P450 conformations is symbolized by capitalization of these letters. Dashed arrows show slow transitions between monomeric and oligomeric states.
Representation of P450 oligomers as trimers (Fig. 11) provides the simplest way to explain the 1:1:1 distribution of the phases of reduction kinetics in CYP3A4 in solution. Trimers are also consistent with a 2:1 distribution of the exponential phases of dithionite-dependent reduction in CYP2B4 and 2:1 distribution of apparent conformers reported in the pressure-perturbation studies of CYP2B4 and CYP3A4 (4, 41, 42). The trimeric state is also supported by electron microscopy of the hexamers of CYP2B4 and CYP1A2, which were shown to be organized as dimers of trimers (18, 76). Nevertheless, the hypothesis that the elementary size of the P450 oligomer is a trimer should be considered as provisional. Other possibilities, such as representation of P450 oligomers as a mixture of dimers of different architecture, appear to be plausible as well.
The results presented here corroborate the observation of the heterogeneity of CYP3A4 in our high-pressure spectroscopy experiments (4), where we found that only about 70% of the heme protein in solution and about 50% in the membrane is susceptible to a pressure-induced P450→P420 transition. Our conclusions are also in good agreement with the results of Koley and co-workers who demonstrated that CYP3A4 in the microsomal membrane is represented by several populations with different accessibility of the heme for CO and different substrate specificity (13–15). Results of these authors suggest that the modulation of CYP3A4 distribution between these populations by allosteric ligands, such as α-naphthoflavone (ANF), is involved in the mechanisms of heterotropic cooperativity of CYP3A4. We suggest that the divergence of CYP3A4 into different populations mentioned by Koley and co-authors is caused by the oligomerization of the enzyme in the membranes. Thus, taking into account the observation that the interactions of CYP3A4 with ANF enhance activity of the enzyme by increasing the pool of active P450 molecules (13, 14), we infer that the phenomena of heterotropic cooperativity observed with ANF may reflect an effect of this allosteric ligand on the organization of CYP3A4 oligomers and/or the equilibrium of CYP3A4 oligomerization in the membrane. These effects will be considered in a forthcoming study.
Acknowledgments
The authors are grateful to Professor Vsevolod I. Popov and Mrs. Julie W. Ven (Department of Pathology, UTMB) for help in electron microscopy of proteoliposomes. We are also indebted to Dr. Christopher Chin (Sealy Center for Structural Biology, UTMB) for his assistance in sedimentation experiments.
Footnotes
This research was supported by NIH grants GM54995 (JRH), GM33775 (SGS), GM31756 (SGS) and Center grant ES06676 (JRH).
Department of Pharmacology and Toxicology, The University of Texas Medical Branch, Galveston, 301 University Blvd., Texas 77555; Department of Biochemistry and Chemistry, Beckman Institute for Advanced Science and Technology, University of Illinois Urbana-Champaign, Urbana, Illinois 61801
References
- 1.Guengerich FP. Cytochrome P-450 3A4: regulation and role in drug metabolism. Annu Rev Pharmacol Toxicol. 1999;39:1–17. doi: 10.1146/annurev.pharmtox.39.1.1. [DOI] [PubMed] [Google Scholar]
- 2.Ekins S, Stresser DM, Williams JA. In vitro and pharmacophore insights into CYP3A enzymes. Trends Pharm Sci. 2003;24:161–166. doi: 10.1016/s0165-6147(03)00049-x. [DOI] [PubMed] [Google Scholar]
- 3.Yoon MY, Campbell AP, Atkins WM. “Allosterism” in the elementary steps of the cytochrome P450 reaction cycle”. Drug Metab Rev. 2004;36:219–230. doi: 10.1081/dmr-120033998. [DOI] [PubMed] [Google Scholar]
- 4.Davydov DR, Halpert JR, Renaud JP, Hui Bon Hoa G. Conformational heterogeneity of cytochrome P450 3A4 revealed by high pressure spectroscopy. Biochem Biophys Res Commun. 2003;312:121–130. doi: 10.1016/j.bbrc.2003.09.247. [DOI] [PubMed] [Google Scholar]
- 5.He YA, Roussel F, Halpert JR. Analysis of homotropic and heterotropic cooperativity of diazepam oxidation. Arch Biochem Biophys. 2003;409:92–101. doi: 10.1016/s0003-9861(02)00484-8. [DOI] [PubMed] [Google Scholar]
- 6.Hosea NA, Miller GP, Guengerich FP. Elucidation of distinct ligand binding sites for cytochrome P450 3A4. Biochemistry. 2000;39:5929–5939. doi: 10.1021/bi992765t. [DOI] [PubMed] [Google Scholar]
- 7.Korzekwa KR, Krishnamachary N, Shou M, Ogai A, Parise RA, Rettie AE, Gonzalez FJ, Tracy TS. Evaluation of atypical cytochrome P450 kinetics with two-substrate models: evidence that multiple substrates can simultaneously bind to cytochrome P450 active sites. Biochemistry. 1998;37:4137–4147. doi: 10.1021/bi9715627. [DOI] [PubMed] [Google Scholar]
- 8.Shou M, Grogan J, Mancewicz JA, Krausz KW, Gonzalez FJ, Gelboin HV, Korzekwa KR. Activation of CYP3A4 - evidence for the simultaneous binding of 2 substrates. Biochemistry. 1994;33:6450–6455. doi: 10.1021/bi00187a009. [DOI] [PubMed] [Google Scholar]
- 9.Ueng YF, Kuwabara T, Chun YJ, Guengerich FP. Cooperativity in oxidations catalyzed by cytochrome P450 3A4. Biochemistry. 1997;36:370–381. doi: 10.1021/bi962359z. [DOI] [PubMed] [Google Scholar]
- 10.Harlow GR, Halpert JR. Analysis of human cytochrome P450 3A4 cooperativity: construction and characterization of a site-directed mutant that displays hyperbolic steroid hydroxylation kinetics. Proc Natl Acad Sci USA. 1998;95:6636–6641. doi: 10.1073/pnas.95.12.6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galetin A, Clarke SE, Houston JB. Multisite kinetic analysis of interactions between prototypical CYP3A4 subgroup substrates: Midazolam, testosterone, and nifedipine. Drug Metab Disp. 2003;31:1108–1116. doi: 10.1124/dmd.31.9.1108. [DOI] [PubMed] [Google Scholar]
- 12.Baas BJ, Denisov IG, Sligar SG. Homotropic cooperativity of monomeric cytochrome P450 3A4 in a nanoscale native bilayer environment. Arch Biochem Biophys. 2004;430:218–228. doi: 10.1016/j.abb.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 13.Koley AP, Buters JTM, Robinson RC, Markowitz A, Friedman FK. Cytochrome P450 conformation and substrate interactions as probed by CO binding kinetics. Biochimie. 1996;78:706–713. doi: 10.1016/s0300-9084(97)82528-x. [DOI] [PubMed] [Google Scholar]
- 14.Koley AP, Buters JTM, Robinson RC, Markowitz A, Friedman FK. Differential mechanisms of cytochrome P450 inhibition and activation by alpha-naphthoflavone. J Biol Chem. 1997;272:3149–3152. doi: 10.1074/jbc.272.6.3149. [DOI] [PubMed] [Google Scholar]
- 15.Koley AP, Robinson RC, Markowitz A, Friedman FK. Drug-drug interactions: effect of quinidine on nifedipine binding to human cytochrome P450 3A4. Biochem Pharm. 1997;53:455–460. doi: 10.1016/s0006-2952(96)00836-2. [DOI] [PubMed] [Google Scholar]
- 16.Dean WL, Gray RD. Hydrodynamic properties of monomeric cytochrome P-450lm2 and cytochrome P-450lm4 in normal-octylglucoside solution. Biochem Biophys Res Commun. 1982;107:265–271. doi: 10.1016/0006-291x(82)91699-0. [DOI] [PubMed] [Google Scholar]
- 17.Wendel I, Behlke J, Janig GR. Hydrodynamic studies on the association of cytochrome P-450. Biomedica Biochimica Acta. 1983;42:633–640. [PubMed] [Google Scholar]
- 18.Tsuprun VL, Myasoedova KN, Berndt P, Sograf ON, Orlova EV, VYa C, Archakov AI, Skulachev VP. Quaternary structure of the liver microsomal cytochrome P-450. FEBS Lett. 1986;205:35–40. doi: 10.1016/0014-5793(86)80861-4. [DOI] [PubMed] [Google Scholar]
- 19.Greinert R, Finch SA, Stier A. Cytochrome P-450 rotamers control mixed-function oxygenation in reconstituted membranes. Rotational diffusion studied by delayed fluorescence depolarization. Xenobiotica. 1982;12:717–726. doi: 10.3109/00498258209038946. [DOI] [PubMed] [Google Scholar]
- 20.Kawato S, Gut J, Cherry RJ, Winterhalter KH, Richter C. Rotation of cytochrome P-450. I. Investigations of protein-protein interactions of cytochrome P-450 in phospholipid vesicles and liver microsomes. J Biol Chem. 1982;257:7023–7029. [PubMed] [Google Scholar]
- 21.Schwarz D, Pirrwitz J, Meyer HW, Coon MJ, Ruckpaul K. Membrane topology of microsomal cytochrome P-450: Saturation transfer EPR and freeze-fracture electron microscopy studies. Biochem Biophys Res Commun. 1990;171:175–181. doi: 10.1016/0006-291x(90)91373-z. [DOI] [PubMed] [Google Scholar]
- 22.Alston K, Robinson RC, Park SS, Gelboin HV, Friedman FK. Interactions among cytochromes P-450 in the endoplasmic reticulum. Detection. J Biol Chem. 1991;266:735–739. [PubMed] [Google Scholar]
- 23.Szczesna-Skorupa E, Mallah B, Kemper B. Fluorescence resonance energy transfer analysis of cytochromes P450 2C2 and 2E1 molecular interactions in living cells. J Biol Chem. 2003;278:31269–31276. doi: 10.1074/jbc.M301489200. [DOI] [PubMed] [Google Scholar]
- 24.Schwarz D, Chernogolov L, Kisselev P. Complex formation in vesicle-reconstituted mitochondrial cytochrome P450 systems CUP11A1 and CYP11B1 as evidenced by rotational diffusion experiments using EPR and ST-EPR. Biochemistry. 1999;38:9456–9464. doi: 10.1021/bi990584l. [DOI] [PubMed] [Google Scholar]
- 25.Cawley GF, Batie CJ, Backes WL. Substrate-dependent competition of different P450 isozymes for limiting NADPH-cytochrome P450 reductase. Biochemistry. 1995;34:1244–1247. doi: 10.1021/bi00004a018. [DOI] [PubMed] [Google Scholar]
- 26.Yamazaki H, Gillam EM, Dong MS, Johnson WW, Guengerich FP, Shimada T. Reconstitution of recombinant cytochrome P450 2C10 2C9 and comparison with cytochrome P450 3A4 and other forms: effects of cytochrome P450- P450 and cytochrome P450-b5 interactions. Arch Biochem Biophys. 1997;342:329–337. doi: 10.1006/abbi.1997.0125. [DOI] [PubMed] [Google Scholar]
- 27.Tan Y, Patten CJ, Smith T, Yang CS. Competitive interactions between cytochromes P450 2A6 and 2E1 for NADPH-cytochrome P450 oxidoreductase in the microsomal membranes produced by a baculovirus expression system. Arch Biochem Biophys. 1997;342:82–91. doi: 10.1006/abbi.1997.9995. [DOI] [PubMed] [Google Scholar]
- 28.Li DN, Pritchard MP, Hanlon SP, Burchell B, Wolf CR, Friedberg T. Competition between cytochrome P-450 isozymes for NADPH-cytochrome P-450 oxidoreductase affects drug metabolism. J Pharm Exp Ther. 1999;289:661–667. [PubMed] [Google Scholar]
- 29.Kaminsky LS, Guengerich FP. Cytochrome P-450 isozyme/isozyme functional interactions and NADPH-cytochrome P-450 reductase concentrations as factors in microsomal metabolism of warfarin. Arch Biochem Biophys. 1985;149:479–489. doi: 10.1111/j.1432-1033.1985.tb08950.x. [DOI] [PubMed] [Google Scholar]
- 30.Backes W, Kelley R. Organization of multiple cytochrome P450s with NADPH-cytochrome P450 reductase in membranes. Pharm Ther. 2003;98:221–233. doi: 10.1016/s0163-7258(03)00031-7. [DOI] [PubMed] [Google Scholar]
- 31.Davydov DR, Petushkova NA, Bobrovnikova EV, Knyushko TV, Dansette P. Association of cytochromes P450 1A2 and 2B4: Are the interactions between different P450 species involved in the control of the monooxygenase activity and coupling? Adv Exp Med Biol. 2001;500:335–338. doi: 10.1007/978-1-4615-0667-6_53. [DOI] [PubMed] [Google Scholar]
- 32.Davydov RM, Khanina OY, Iagofarov S, Uvarov VY, Archakov AI. Effect of lipids and substrates on the kinetics of binding of ferrocytochrome P-450 to CO. Biokhimiia. 1986;51:125–129. [PubMed] [Google Scholar]
- 33.Oertle M, Richter C, Winterhalter KH, Di Iorio EE. Kinetics of carbon monoxide binding to phenobarbital-induced cytochrome P-450 from rat liver microsomes: a simple bimolecular process. Proc Natl Acad Sci USA. 1985;82:4900–4904. doi: 10.1073/pnas.82.15.4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jansson I, Curti M, Epstein PM, Peterson JA, Schenkman JB. Relationship between phosphorylation and cytochrome P450 destruction. Arch Biochem Biophys. 1990;283:285–292. doi: 10.1016/0003-9861(90)90644-e. [DOI] [PubMed] [Google Scholar]
- 35.Hildebrandt P, Garda H, Stier A, Bachmanova GI, Kanaeva IP, Archakov AI. Protein-protein interactions in microsomal cytochrome P-450 isozyme LM2 and their effect on substrate binding. Eur J Biochem. 1989;186:383–388. doi: 10.1111/j.1432-1033.1989.tb15220.x. [DOI] [PubMed] [Google Scholar]
- 36.Kanaeva IP, Nikityuk OV, Davydov DR, Dedinskii IR, Koen YM, Kuznetsova GP, Skotselyas ED, Bachmanova GI, Archakov AI. Comparative study of monomeric reconstituted and membrane microsomal monooxygenase systems of the rabbit liver. II. Kinetic parameters of reductase and monooxygenase reactions. Arch Biochem Biophys. 1992;298:403–412. doi: 10.1016/0003-9861(92)90428-y. [DOI] [PubMed] [Google Scholar]
- 37.Davydov DR, Petushkova NA, Archakov AI, Hui Bon Hoa G. Stabilization of P450 2B4 by Its Association with P450 1A2 Revealed by High-Pressure Spectroscopy. Biochem Biophys Res Commun. 2000;276:1005–1012. doi: 10.1006/bbrc.2000.3596. [DOI] [PubMed] [Google Scholar]
- 38.Cawley GF, Zhang SX, Kelley RW, Backes WL. Evidence supporting the interaction of CYP2B4 and CYP1A2 in microsomal preparations. Drug Metab Disp. 2001;29:1529–1534. [PubMed] [Google Scholar]
- 39.Zangar RC, Davydov DR, Verma S. Mechanisms that regulate production of reactive oxygen species by cytochrome P450. Toxicol Appl Pharm. 2004;199:316–331. doi: 10.1016/j.taap.2004.01.018. [DOI] [PubMed] [Google Scholar]
- 40.Hazai E, Kupfer D. Interactions between CYP2C9 and CYP2C19 in reconstituted binary systems influence their catalytic activity: possible rationale for the inability of CYP2C19 to catalyze methoxychlor demethylation in human liver microsomes. Drug Metab Disp. 2005;33:157–164. doi: 10.1124/dmd.104.001578. [DOI] [PubMed] [Google Scholar]
- 41.Davydov DR, Knyushko TV, Hui Bon Hoa G. High pressure induced inactivation of ferrous cytochrome P-450 LM2 2B4 CO complex: evidence for the presence of two conformers in the oligomer. Biochem BiophysResCommun. 1992;188:216–221. doi: 10.1016/0006-291x(92)92372-5. [DOI] [PubMed] [Google Scholar]
- 42.Davydov DR, Deprez E, Hui Bon Hoa G, Knyushko TV, Kuznetsova GP, Koen YM, Archakov AI. High-Pressure-Induced Transitions in Microsomal Cytochrome P450 2B4 in Solution - Evidence for Conformational Inhomogeneity in the Oligomers. Arch Biochem Biophys. 1995;320:330–344. doi: 10.1016/0003-9861(95)90017-9. [DOI] [PubMed] [Google Scholar]
- 43.Davydov, D.R., Hui Bon Hoa, G. (1997) Pressure-induced transitions in cytochrome P450 2B4 as an evidence of it functional inhomogeneity in the proteoliposomal membranes. inHigh pressure applications in biochemistry, biophysics and biotechnology Proceedings of the XXXII EHPRG meeting (Heremans, K, Ed) pp 111–114, Leuven University Press.
- 44.Davydov DR, Knyushko TV, Kanaeva IP, Koen YM, Samenkova NF, Archakov AI, Hui Bon Hoa G. Interactions of cytochrome P450 2B4 with NADPH-cytochrome P450 reductase studied by fluorescent probe. Biochimie. 1996;78:734–743. doi: 10.1016/s0300-9084(97)82531-x. [DOI] [PubMed] [Google Scholar]
- 45.Cooper CE, Junemann S, Ioannidis N, Wrigglesworth JM. Slow ‘resting’forms of mitochondrial cytochrome c oxidase consist of two kinetically distinct conformations of the binuclear CuB/a3 centre--relevance to the mechanism of proton translocation. Biochim Biophys Acta. 1993;1144:149–160. doi: 10.1016/0005-2728(93)90167-e. [DOI] [PubMed] [Google Scholar]
- 46.Moody AJ, Cooper CE, Rich PR. Characterisation of ‘fast’ and ‘slow’ forms of bovine heart cytochrome-c oxidase. Biochim Biophys Acta. 1991;1059:189–207. doi: 10.1016/s0005-2728(05)80204-x. [DOI] [PubMed] [Google Scholar]
- 47.Cheesman MR, Watmough NJ, Pires CA, Turner R, Brittain T, Gennis RB, Greenwood C, Thomson AJ, Gennis RB, Greenwood C, Thomson AJ. Cytochrome bo from Escherichia coli: identification of haem ligands and reaction of the reduced enzyme with carbon monoxide. Biochem J. 1993;289:709–718. doi: 10.1042/bj2890709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moody AJ, Cooper CE, Gennis RB, Rumbley JN, Rich PR. Interconversion of fast and slow forms of cytochrome bo from Escherichia coli. Biochemistry. 1995;34:6838–6846. doi: 10.1021/bi00020a030. [DOI] [PubMed] [Google Scholar]
- 49.Coletta M, Costa H, Desanctis G, Neri F, Smulevich G, Turner DL, Santos H. pH dependence of structural and functional properties of oxidized cytochrome c’’ from Methylophilus methylotrophus. J Biol Chem. 1997;272:24800–24804. doi: 10.1074/jbc.272.40.24800. [DOI] [PubMed] [Google Scholar]
- 50.Catarino T, Coletta M, Legall J, Xavier AV. Kinetic study of the reduction mechanism for Desulfovibrio gigas cytochrome c3. Eur J Biochem. 1991;202:1107–1113. doi: 10.1111/j.1432-1033.1991.tb16477.x. [DOI] [PubMed] [Google Scholar]
- 51.Blackmore RS, Brittain T, Greenwood C. An analysis of the reaction kinetics of the hexahaem nitrite reductase of the anaerobic rumen bacterium Wolinella succinogenes. Biochem J. 1990;271:457–461. doi: 10.1042/bj2710457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DiSpirito AA, Balny C, Hooper AB. Conformational change accompanies redox reactions of the tetraheme cytochrome c-554 of Nitrosomonas europaea. Eur J Biochem. 1987;162:299–304. doi: 10.1111/j.1432-1033.1987.tb10600.x. [DOI] [PubMed] [Google Scholar]
- 53.Davies DM, Lawther JM. Kinetics and mechanism of electron transfer from dithionite to microsomal cytochrome b5 and to forms of the protein associated with charged and neutral vesicles. Biochem J. 1989;258:375–380. doi: 10.1042/bj2580375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aviram I, Sharabani M. Kinetic studies of the reduction of neutrophil cytochrome b-558 by dithionite. Biochem J. 1986;237:567–572. doi: 10.1042/bj2370567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arif Kazmi S, Mills MA, Pitluk ZW, Scott RA. Kinetics of dithionite reduction of the heme nonapeptide of cytochrome c. J Inorg Biochem. 1985;24:9–12. doi: 10.1016/0162-0134(85)85009-1. [DOI] [PubMed] [Google Scholar]
- 56.Favaudon V, Ferradini C, Pucheault J, Gilles L, Le Gall J. Kinetics of reduction of the cytochrome C3 from Desulvovibrio vulgaris. Biochem Biophys Res Commun. 1978;84:435–440. doi: 10.1016/0006-291x(78)90188-2. [DOI] [PubMed] [Google Scholar]
- 57.Creutz C, Sutin N. Kinetics of ligand-binding and oxidation-reduction reactions of cytochrome c from horse heart and Candida krusei. J Biol Chem. 1974;249:6788–6795. [PubMed] [Google Scholar]
- 58.Lambeth DO, Palmer G. The kinetics and mechanism of reduction of electron transfer proteins and other compounds of biological interest by dithionite. J Biol Chem. 1973;248:6095–6103. [PubMed] [Google Scholar]
- 59.Hintz MJ, Peterson JA. The kinetics of reduction of cytochrome P-450cam by reduced putidaredoxin. J Biol Chem. 1981;256:6721–6728. [PubMed] [Google Scholar]
- 60.Davydov DR, Kurganov BI. Comparative Study of the Reaction Kinetics of Cytochrome P-450 Reduction by NADPH-Cytochrome P-450 Reductase and Dithionite. Biokhimiia. 1982;47:1476–1482. [PubMed] [Google Scholar]
- 61.Davydov DR, Karyakin AV, Binas B, Kurganov BI, Archakov AI. Kinetic studies on reduction of cytochromes P-450 and b5 by dithionite. Eur J Biochem. 1985;150:155–159. doi: 10.1111/j.1432-1033.1985.tb09001.x. [DOI] [PubMed] [Google Scholar]
- 62.Domanski TL, Liu J, Harlow GR, Halpert JR. Analysis of four residues within substrate recognition site 4 of human cytochrome P450 3A4: role in steroid hydroxylase activity and alpha- naphthoflavone stimulation. Arch Biochem Biophys. 1998;350:223–232. doi: 10.1006/abbi.1997.0525. [DOI] [PubMed] [Google Scholar]
- 63.Domanski TL, He YA, Khan KK, Roussel F, Wang Q, Halpert JR. Phenylalanine and tryptophan scanning mutagenesis of CYP3A4 substrate recognition site residues and effect on substrate oxidation and cooperativity. Biochemistry. 2001;40:10150–10160. doi: 10.1021/bi010758a. [DOI] [PubMed] [Google Scholar]
- 64.Schwarz D, Gast K, Meyer HW, Lachmann U, Coon MJ, Ruckpaul K. Incorporation of the cytochrome P-450 monooxygenase system into large unilamellar liposomes using octylglucoside, especially for measurements of protein diffusion in membranes. Biochem Biophys Res Commun. 1984;121:118–125. doi: 10.1016/0006-291x(84)90695-8. [DOI] [PubMed] [Google Scholar]
- 65.Molecular Probes Inc. (2003) Handbook of Fluorescent Probes and Research Products, Web edition: http://www.probes.com.handbook/ Molecular Probes, Inc., ed., Eugene, Oregon.
- 66.Rouser G, Fleische S, Yamamoto A. 2-Dimensional thin layer chromatographic separation of polar lipids and determination of phospholipids by phosphorus analysis of spots. Lipids. 1970;5:494–496. doi: 10.1007/BF02531316. [DOI] [PubMed] [Google Scholar]
- 67.Denisov IG, Grinkova YV, Lazarides AA, Sligar SG. Directed self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size. J Am Chem Soc. 2004;126:3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 68.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wendel I, Behlke J, Janig GR. Determination of the partial specific volume of cytochrome P-450 as a model. Biomed Biochim Acta. 1983;42:623–631. [PubMed] [Google Scholar]
- 70.Renaud JP, Davydov DR, Heirwegh KPM, Mansuy D, Hui Bon Hoa G. Thermodynamic studies of substrate binding and spin transitions in human cytochrome P-450 3A4 expressed in yeast microsomes. Biochem J. 1996;319:675–681. doi: 10.1042/bj3190675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Segel, I. H. (1975) Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme Systems, Wiley-Interscience,. New York.
- 72.Hosea NA, Guengerich FP. Oxidation of nonionic detergents by cytochrome P450 enzymes. Arch Biochem Biophys. 1998;353:365–373. doi: 10.1006/abbi.1998.0659. [DOI] [PubMed] [Google Scholar]
- 73.Schulze HU, Staudinger H. Zur Struktur des endoplasmatischen Retikulums der Rattenleberzelle: Korrelation von morphometrischen und biochemischen Messwerten. Hoppe-Seylers Z Phys Chem. 1971;352:1675–1680. [PubMed] [Google Scholar]
- 74.Yamada M, Ohta Y, Bachmanova GI, Archakov AI, Hatta I, Kawato S. Effect of microsome-liposome fusion on the rotational mobility of cytochrome P450IIB4 in rabbit liver microsomes. J Inorg Biochem. 2001;83:261–268. doi: 10.1016/s0162-0134(00)00169-0. [DOI] [PubMed] [Google Scholar]
- 75.Greinert R, Finch SA, Stier A. Conformation and rotational diffusion of cytochrome P-450 changed by substrate binding. Bioscience Reports. 1982;2:991–994. doi: 10.1007/BF01122166. [DOI] [PubMed] [Google Scholar]
- 76.Myasoedova KN, Tsuprun VL. Cytochrome P-450: hexameric structure of the purified LM4 form. FEBS Letters. 1993;325:251–254. doi: 10.1016/0014-5793(93)81083-c. [DOI] [PubMed] [Google Scholar]


