Abstract
Dietary gluten proteins from wheat, rye and barley are the primary triggers for the immuno-pathogenesis of Celiac Sprue, a widespread immune disease of the small intestine. Recent molecular and structural analyses of representative gluten proteins, most notably α- and γ-gliadin proteins from wheat, have improved our understanding of these pathogenic mechanisms. In particular, based on the properties of a 33-mer peptide, generated from α-gliadin under physiological conditions, a link between digestive resistance and inflammatory character of gluten has been proposed. Here we report three lines of investigation in support of this hypothesis. First, biochemical and immunological analysis of deletion mutants of α-2 gliadin confirmed that the DQ2 restricted T cell response to the α-2 gliadin are directed towards the epitopes clustered within the 33-mer. Second, proteolytic analysis of a representative γ-gliadin led to the identification of another multivalent 26-mer peptide that was also resistant to further gastric, pancreatic and intestinal brush border degradation, and was a good substrate of human transglutaminase 2 (TG2). Analogous to the 33-mer, the synthetic 26-mer peptide displayed markedly enhanced T cell antigenicity compared to monovalent control peptides. Finally, in silico analysis of the gluten proteome led to the identification of at least 60 putative peptides that share the common characteristics of the 33-mer and the 26-mer peptides. Together, these results highlight the pivotal role of physiologically generated, proteolytically stable, TG2-reactive, multivalent peptides in the immune response to dietary gluten in Celiac Sprue patients. Prolyl endopeptidase treatment was shown to abolish the antigenicity of both the 33-mer and the 26-mer peptides, and was also predicted to have comparable effects on other proline-rich putatively immunotoxic peptides identified from other polypeptides within the gluten proteome.
INTRODUCTION
Celiac Sprue, also known as Celiac Disease or gluten-sensitive enteropathy, is a widespread autoimmune disease of the small intestine caused by the intake of gluten proteins from common food sources such as wheat, rye and barley. In genetically susceptible patients the exposure of gluten to the small intestine induces a CD4+ T cell mediated inflammatory response, leading to destruction of the intestinal villous structure 1–3. Clinical symptoms of Celiac Sprue include fatigue, chronic diarrhea, abdominal distension, weight loss due to malabsorption, and extraintestinal symptoms including anemia, psychiatric syndromes and neurological symptoms 3. Untreated Celiac Sprue is also associated with an elevated risk of infertility, bone disorders and intestinal malignancies 4, 5. Despite of the wide prevalence (0.5–1%) and serious clinical implications of the disease, currently the only effective therapy is strict dietary abstinence from these common food grains.
The principal toxic components of wheat gluten belong to a family of closely related proline and glutamine rich proteins called gliadins. When genetically predisposed individuals who express the human leukocyte antigen (HLA) DQ2 (or DQ8) are exposed to certain gliadin epitopes 6–9, these epitopes are presented on the surfaces of antigen presenting cells, which in turn stimulate proliferation of gluten-responsive DQ2 (or DQ8) restricted CD4+ T cells in the small intestinal mucosa of Celiac Sprue patients. To date at least 17 distinct DQ2 restricted T cell epitopes have been identified from gluten proteins found in disease associated grains such as wheat, rye, barley and possibly oats 8–15. Interestingly, most of these epitopes have to undergo a transglutaminase 2 (TG2) mediated deamidation of certain glutamine to glutamate residues to elicit a T cell response 6, 7.
The identification of an exceptionally immunoreactive 33-mer peptide, LQLQPFPQPQLPYPQPQLPYPQPQLPYPQPQPF (underlined Q residues correspond to transglutaminase deamidation sites) from α-2 gliadin led us to propose that highly antigenic gluten epitopes share at least four properties 12. First, all antigenic gluten epitopes identified so far are located in proline-rich regions of gliadin 16. Proline-rich peptides are protected from proteolysis by gastric, pancreatic and intestinal brush border membrane enzymes, so they have an opportunity to build up to high concentrations in the small intestine. The structural basis for gastrointestinal stability of certain gluten peptides has been investigated 10, 12, 17. Second, the most inflammatory gluten epitopes are also preferred substrates for human transglutaminase 2 (TG2). The molecular logic for TG2 catalyzed deamidation of gluten epitopes has been elucidated 18, 19 Third, the DQ2 molecule appears to have unique features enabling the molecule to bind and present proline rich gluten peptides with glutamate residues to CD4+ T cells in the intestinal mucosa 8, 20, 21. Finally, it was also proposed that highly antigenic gluten epitopes occur within the context of long, multivalent peptides 12.
As a further test of this hypothesis, in this study we sought to verify that the 33-mer peptide from α-2 gliadin is indeed the principal contributor to this protein’s immunotoxicity. Using experimental and computational approaches, we also attempted to identify analogous peptides from other sub-families of gliadin proteins such as γ-gliadins, as well as other families of grains such as barley and rye. The results of these studies are reported here.
In addition to providing a new perspective on Celiac Sprue pathogenesis, the link between digestive resistance and gluten toxicity has also presented an opportunity for countering the toxic effects of gluten in Celiac Sprue patients through the use of an oral prolyl endopeptidase (PEP). For example, a PEP from Flavobacterium meningosepticum was shown to complement pancreatic proteases and the brush border enzymes to detoxify selected peptides from gluten 12, 17 as well as whole gluten 22. Identification of additional highly inflammatory peptides such as the 33-mer can therefore be expected to expand the panel of primary screening targets for isolation or engineering of new therapeutically useful PEPs.
MATERIALS AND METHODS
The peptides LQLQPFPQPQLPYPQPQLPYPQPQLPYPQPQPF, and FLQPQQPFPQQPQQPYPQQPQQPFPQ were synthesized by solid-phase peptide synthesis. Trypsin (T-1250), chymotrypsin (C-4754), carboxypeptidase (C-0261) (all from bovine pancreas), elastase from porcine pancreas were from Sigma (St. Louis, MO). Prolyl endopeptidase from Flavobacterium meningosepticum was obtained from US Biological (Marble Head, MA). Enzyme substrates were purchased from Sigma except for Z-Gly-Pro-p-nitroanilide (Bachem, Switzerland). All chemicals were from Sigma unless otherwise noted.
Preparation of recombinant gliadins
Gliadin genes including α-2 gliadin and γ-5 gliadin were cloned in pET28a plasmid (Novagen) and introduced via transformation into E. coli BL21(DE3) (Novagen). Transformed cells were grown in LB media containing 50 μg/ml of kanamycin at 37 ºC until OD600 reached 0.8. The expression of protein was induced with the addition of 0.4 mM isopropyl β-D-thiogalactoside (Sigma) and the cultures were further incubated at 37ºC for 20 hours. The cells were centrifuged at 3600 rpm for 30 minutes. The pellet was resuspended in 15 ml of disruption buffer (200 mM sodium phosphate; 200 mM NaCl; 2.5 mM DTT; 1.5 mM benzamidine; 2.5 mM EDTA; 2 mg/L pepstatin; 2 mg/L leupeptin; 30% v/v glycerol) and lysed by sonication (Branson Sonifier, 1 min; output control set to 6). After centrifugation at 45000g for 45 min, the supernatant was discarded and the pellet containing gliadin protein was resuspended in 50 ml of 7M urea in 50 mM Tris Cl (pH = 8.0). The suspension was again centrifuged at 45000g for 45 min and the supernatant was harvested for purification. The supernatant was incubated with 1 ml of nickel-nitrilotriacetic acid resin (Ni-NTA; Qiagen) overnight and then batch-loaded on a column with 2 ml of Ni-NTA. The column was washed with 7M urea in 50 mM Tris Cl (pH = 8.0) and protein was eluted with 200 mM imidazole, 7 M urea in 50 mM Tris Cl (pH = 4.5). The fractions containing gliadin were pooled together and concentrated 100-fold using a 10,000 MWCO concentrator (Amicon). A final concentration of 70% ethanol solution and two volumes of 1.5M NaCl were added to precipitate the protein overnight at 4 ºC. The precipitate was collected by centrifugation at 45000 g for 30 min, rinsed in water, and re-centrifuged. The final purification step was developed with reverse-phase HPLC. The Ni-NTA purified protein fractions were pooled in 7 M urea buffer and injected onto a Vydac (Hesperia, CA) polystyrene reverse-phase column (i.d. 4.6 mm × 25 cm) with the starting solvent (30% of solvent B: 1:1 HPLC-grade acetonitrile/isopropanol : 0.1% TFA). Solvent A was an aqueous solution with 0.1% TFA. The separation gradient extended from 30–100% of solvent B over 120 min at a flow rate of 0.8 ml/min. The protein solution was dried by lyophilization.
Mutagenesis of gliadin
Deletion mutations were engineered in the α-2 gliadin gene in pET28a in two steps. First, two polymerase chain reactions were performed to amplify the flanking gene fragments to the DNA piece to be deleted. Then the two PCR pieces were ligated to omit the deleted DNA. In order for the ligation in the second step to occur, the primers were designed to include an EcoRI restriction site on the ends of the two ligating DNAs with the minimal mutation of the wild-type sequence. For the deletion of fragment Gln35→Gln104 (Fig. 2), the following primers were designed: for the DNA flanking from the left, 5’-CATATGGTTAGAGTTCCAGTGCCAC-3’ (sense), 5’-CTGAATTCCTGGAAATTGTTGTTGTTGTACCAATGG-3’ (antisense); for the DNA flanking from the right, 5’-CGAATTCAACCACAACAACCAATTTCGCAGCAGCAG-3’ (sense), 5’-CAAGCTTCATCGATAGTTAGTACCGAAGATG-3’ (antisense). The following primers were synthesized for the deletion of fragment Leu57→Phe89 (Fig. 2): for the DNA flanking from the left, 5’-CATATGGTTAGAGTTCCAGTGCCACAATTG-3’ (sense), 5’-CTGAATTCGTGGTTGTTGTGATGGAAATGGTTGCGG-3’ (antisense); for the DNA flanking from right, 5’-CGAATTCAACAACCATATCCACAATCGCAACCACAG-3’ (sense), 5’-CAAGCTTCATCGATAGTTAGTACCGAAGATG-3’ (antisense). Residue Pro91 was converted to Ile to accommodate restriction sites. The PCR reactions were carried out using Pfu polymerase, the α-2 gliadin pET28a plasmid template, the sense and antisense primers. The reaction mixture (50μl) also contained 5μl of DMSO, 200μM of dNTP mix. The temperature profiles were 94°C for 5 min, annealing using a temperature step-down feature to decrease from 80°C to 65°C in 15 cycles, and then 72°C for 1 min in 20 cycles. The amplified PCR products were gel purified using a QIAquick gel extraction kit (Qiagen) and ligated using T4 ligase at 4°C overnight.
Figure 2.
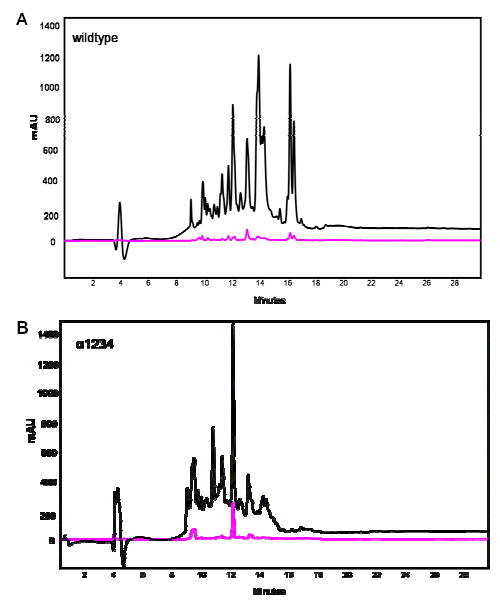
Gastric/pancreatic proteolysis of the wild-type α-2 gliadin (A) and its deletion mutant α1234 (B), illustrated by HPLC with UV detection at 215 nm (black traces) and 280 nm (magenta traces). The 33-mer resulted from the wildtype digestion eluted at a retention time of approximately 16 minutes, which is missing in the deletion mutant digestion.
Proteolysis of Gliadins
Purified recombinant α-2-gliadin, its mutants, and γ-5-gliadin in lyophilized forms were weighed and dissolved in 0.01M hydrocholoric acid. Proteins were incubated at 37°C with pepsin (protease to protein w/w ratio is 1:100, Sigma) at pH 2.0 for 20 min. The reaction mixture was then pH adjusted to 7.0 in 50mM phosphate buffer and supplemented with trypsin (1:100, Sigma), chymotrypsin (1:100, Sigma), elastase (1:500, Sigma) and carboxypeptidase (1:100, Cortex Biochem) at 37°C for various time periods.
Preparation of Rat Brush Border Enzymes (BBM) and BBM Digestion of Peptides
The rat jejunum was removed from the small intestine of adult female rats and further processed as previously described with slight variations 23. The jejunum was rinsed with 0.9% NaCl. Mucosal scrapings (~1 g) were homogenized in 10 ml of 5 mM EDTA in 5 mM histidine imidazole, pH 7.6 in a Dounce homogenizer and type B pestle (15 vertical strokes). The homogenate was centrifuged at 45000g for 30 min. The pellet was rehomogenized (15 vertical strokes) in 15 ml of 5 mM histidine imidazole, 0.5 mM EDTA, 250 mM sorbitol, 12.5 mM NaCl, pH 7.6. The homogenate was further diluted to 25 ml with the same buffer and centrifuged at 45000g for 30 min. The pellet was resuspended in 10 ml of 50 mM mannitol, 2 mM Tris-HCl, pH 7.1, and homogenized with two 30-s bursts of a Polytron homogenizer at Setting 4. Then CaCl2 was added to a final concentration of 10 mM, and the suspension was stirred at 4 °C for 30 min and centrifuged at 2000g for 10 min. The supernatant containing the partially purified BBM enzymes was harvested by centrifuging at 45000g for 30 min. The BBM pellet was stored in 1 ml of 5 mM histidine imidazole, 0.5 mM EDTA, 100 mM NaCl, pH 7.1. Marker enzyme assays were performed. Total protein concentration was determined by the Lowry method. Specific activities of alkaline phasphatase, dipeptidyl peptidase IV (DPP IV), aminopeptidase N (APN) were determined as previously described 10.
Stable peptic products identified from previous gastric/pancreatic digestions were synthesized (New England Peptides) and purified. BBM digestion mixtures of these peptides consisted of 100 μM of peptide, 10 mM Cbz-tryptophan (internal control for quantification), and a final concentration of BBM proteins of 12.5 ng/μl at 37 °C from 30 min up to 20 hours. The identification of the digestive products was obtained with LC-MS methods described above.
Deamidation by Tissue Transglutaminase
Human TG2 was expressed and purified according to previous work 11. The activity of the TG2-catalyzed deamidation reaction was analyzed using a spectrophotometric method.
Liquid chromatography-coupled mass spectrometry analysis
To identify the stable hydrolysis products from each enzymatic digestion, the reaction mixtures were analyzed by LC-MS (SpectraSystem, ThermoFinnigan) using a reversed-phase column (Vydac 259VHP54115). The mobile phase flow rate was 0.2 ml/min, beginning with 0% acetonitrile (in water, 0.1% formic acid and 0.025% trifluoroacetic acid) and achieving 95% acetonitrile over 40 min. The LC-separated peptides were further fragmented and detected by electrospray ionization mass spectrometry. The compounds defined by their m/z values were detected by their ionization intensities. The amino acid sequences of the proteolytic products were determined from the MS-MS fragmentation pattern.
Gluten specific T cells and T cell proliferation assays
Gluten reactive T cell lines (TCL) and T cell clones (TCC) were established from intestinal biopsy samples of DQ2 positive Celiac patients mainly as described 8. The following TCLs and TCCs were used in this study: TCL432.1.3 12, TCL437.1.1 12 , TCL432.1.4 12, TCL422.02.4.2 12, TCL462.1, TCL480.1, TCC430.1.134 15, TCC387.3 and TCC387.19 15. Antigen presenting cells (DR3-DQ2 homozygous, Epstein-Barr virus transformed B-lymphoblast cell lines) were irradiated 75 gray, seeded at 7.5 x 104 cells per well, incubated overnight with deamidated α-2 peptides or γ-35 peptides of interest, with or without prolyl endopeptidase (PEP) and BBM treatment in U-bottomed 96 well plates, before addition of T cells (2.5 to 5 x 104 cells per well). T cell proliferation was measured as [3H] thymidine incorporation after 48–72 h.
Gluten Protein Database Analysis
Gluten protein sequences (α-gliadins, α gliadin precursors, γ-gliadins, glutenins, hordeins, secalins and avenins) were obtained from NCBI protein and nucleotide databases. Computational prediction of proteolytic fragments by chymotrypsin, trypsin and pancreatic elastase was performed with GCG SeqWeb software (Accelrys, CA). Chymotrypsin cleavage was set to occur at the C-terminal of F, W, Y, M, L; trypsin cleavage is after K and R; pancreatic elastase cleavages after A, G, S, V; carboxypeptidase A (non-specific but prefers aromatic and long side-chain amino acids) combined with carboxypeptidase B (preferentially cleaves K and R) were predicted to cleave from C-terminal end of a polypeptide until the PX sequence. Homology alignment of proteins and their expected stable peptides was performed with BLASTP provided by NCBI. TG2 glutamine deamidation sites were predicted according to the recognition motifs that identifies non-terminal Gln in QXP, or QXP(F,Y,W,M,L,I,V) or QXX(F,Y,W,M,L,I,V) but not QP or QXXP 18. DQ2 binding motifs were predicted as follows: P1 – F,Y,W, I, L,M,S,P or Q; P2 – not P; P3 – any; P4 – not P; P5 – any; P6 – P, E, Q or D; P7 – not P; P8 – any; P9 – not P 24.
RESULTS
Expression, Purification and Digestion of Recombinant α-2 Gliadin
The yield (2–3 mg/L) and purity of the Ni-NTA purified gliadin protein were adequate for further studies. Purified α-2 gliadin was subjected to proteolysis under conditions simulating gastric and pancreatic conditions, as described in the Experimental Methods section. The resulting peptide fragments were analyzed by LC/MS/MS. The resistant QLQPFPQPQLPYPQPQLPYPQPQLPYPQPQPF (and its N-terminal pyroglutamate counterpart) was present in the reaction products and calculated to be in stoichiometric amount with the protein prior to digestion.
α-2 Gliadin in T Cell Proliferation
The mixture of proteolyzed fragments from recombinant α-2 gliadin was used to test six individual DQ2-restricted T cell lines (TCL432.1.3, TCL437.1, TCL432.1.4, TCL422.02.4.2, TCL462.1 and TCL480.1) in a proliferation assay. The digested recombinant α-2 gliadin peptides induce a potent T cell proliferation response comparable to what we have previously observed with the 33-mer (an example can be seen in Figure 4). The EC50 for the proteolyzedmixture of α-2 gliadin peptides was found to be comparable to the 33-mer (Table 1). This suggested that the 33-mer sequence in the α-2 gliadin protein contributed most, if not all, of this gliadin’s antigenic properties.
Figure 4.
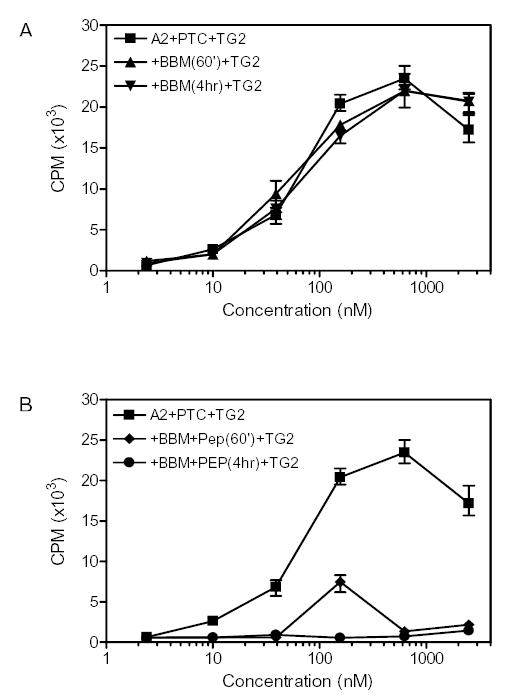
Peptidase catalyzed breakdown of α-2 gliadin. The results of T cell line TCL 422.02.4.2 are shown and similar results were obtained with the T cell lines TCL432.1.4, TCL462.1 and TCL480.1. (A) Brush border membrane (BBM) treatment of α-2 gliadin did not result in efficient antigenic detoxification; (B) BBM and PEP treatment of α-2 gliadin efficiently abrogated T cell responses.
Table 1.
EC50 values of the α-2 gliadin peptide mixture, the 33mer and a control peptide for the epitope FPQPQLPYPQP.
| α -2 gliadin | 33-mer | Control peptide | |
|---|---|---|---|
| EC50 (nM) | 180 ± 50 | 175 ± 40 | 1000 |
α-2 Gliadin Mutants
Earlier studies involving a panel of overlapping synthetic peptides suggested that a cluster of three overlapping epitopes, designated αI, αII and αIII gliadin epitopes, are the primary immunogenic sequences in the α-2 gliadin protein 12, 25. As an alternative strategy for assessing the immunological importance of these epitopes, two deletion mutants for α-2 gliadin were constructed (Figure 1). Deletion mutant α-1234 was designed to exclude the 33-mer sequence only, whereas the deletion mutant α-1278 excludes the 33-mer sequence and additional N-terminal residues QQPFPPQQPYPQPQPFPSQQPY and C-terminal residues RPQQPYPQSQPQYSQ, sequences analogous to known T cell epitopes DQ2-γ-III and DQ2-γ-VI 26 and possibly including a disease-specific T cell epitope 27.
Figure 1.

Protein sequences of the α-2 gliadin deletion mutants. (A) Deletion mutant A1234. (B) Deletion mutant A1278. The peptide sequence in red is the proteolytically resistant 33-mer.
Gastric/pancreatic digestions were performed on wild-type α-2 gliadin as well as both deletion mutants. The resulting proteolytic mixtures were compared via reversed phase HPLC, as shown in Fig. 2A for the wild-type protein and in Fig. 2B for deletion mutant α-1234. The 33-mer fragment at retention time of 16 min was present in the wild-type digestion mixture, but not in the mutant digestion mixture. The remaining peptides could be found in both mixtures. Peptide fragments at retention times of 14–15 min from the deletion mutant were found in lower quantities compared to those from the wild-type gliadin, presumably due to a more thorough digestion of these shorter peptides. Similar results were also obtained in the case of deletion mutant α-1278 (data not shown).
In order to determine whether the remaining peptides from the two digested mutants retain T cell stimulatory capacity, these peptide mixtures were assayed against four patient-derived polyclonal T cell lines. As seen in Figure 3, the α-2 gliadin deletion mutants elicit virtually no response in the polyclonal T cell lines tested of these DQ2 positive Celiac Sprue patients, suggesting that the 33-mer peptide is in fact the sole source of α-2 gliadin toxicity. Aside from the 3 epitopes present in the 33-mer (α-I, α-II and α-III), there is another α-gliadin epitope, FRPQQPYPQ (Glia-α20, see supplementary Table S1). Since the Phe residue of the 9-mer core region of this epitope is part of the 33-mer sequence, deletion mutants void of the 33-mer would be unlikely to stimulate Glia-α20-specific T cells.
Figure 3.
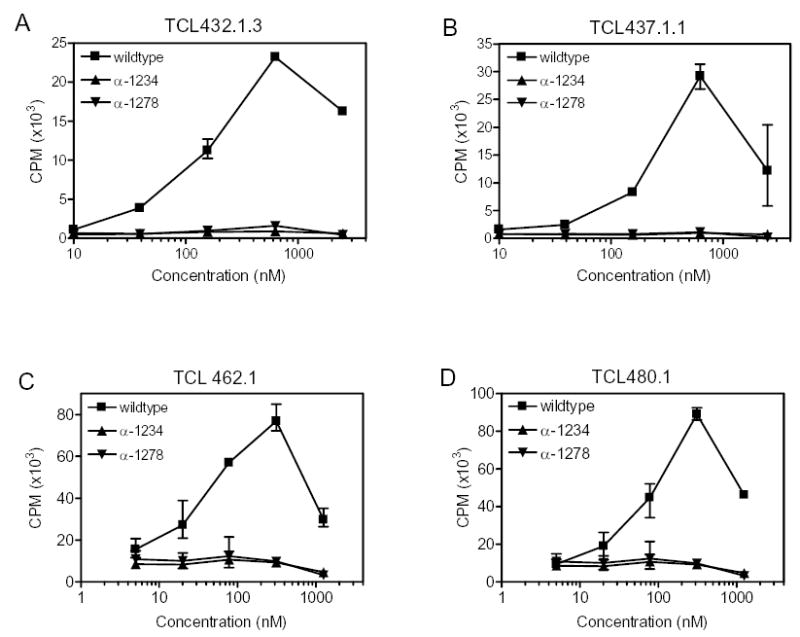
Stimulation of four HLA-DQ2 restricted T cell lines derived from biopsies of Celiac Sprue patients by the PTCEC (pepsin, trypsin, chymotrypsin, elastase, carboxypeptidase B) and brush-border membranedigestive peptide mixtures from α-2 gliadin deletion mutants α-1234 and α-1278. No T cell stimulation was elicited. Error bars indicate the observed range within the triplicates.
Table S1 (A).
Known prolamin epitopes recognized by intestinal T cells of Celiac Sprue patients.
| T cell epitope | Source | Native sequence of 9-mer core region of epitope4 | Ref. | |
|---|---|---|---|---|
| α–I | α-gliadin, recombinant1 | PFPQPQLPY | Crystal5 | Arentz-Hansen, 2000 |
| α–III | α -gliadin, recombinant1 | PYPQPQLPY | TCC6 | Arentz-Hansen, 2002 |
| α–II | α -gliadin, recombinant1 | PQPQLPYPQ | DQ-TCC7 | Arentz-Hansen, 2000 |
| Glia-α 20 | α–gliadin, peptide2 | FRPQQPYPQ | TCC6 | Vader 2002,Vader, 2003 |
| γ–I | γ–gliadin, natural3 | PQQSFPQQQ | DQ-TCC7 | Sjöström, 1998 |
| γ–II (Glia-γ 30) | γ–gliadin, natural3 | IQPQQPAQL | DQ-TCC 7 | Vader 2002, Vader, 2003; Qiao, 2005 |
| γ–III | γ–gliadin, recombinant1 | QQPQQPYPQ | DQ-TCC7 | Arentz-Hansen, 2002; Qiao, 2005 |
| γ–IV | γ–gliadin, recombinant1 | SQPQQQFPQ | DQ-TCC7 | Arentz-Hansen, 2002; Qiao, 2005 |
| γ–VIa | γ–gliadin, peptide2 | QQPFPQQPQ | TCC6 | Qiao, 2005 |
| γ–VIb | γ–gliadin, peptide2 | QQPYPQQPQ | TCC6 | Qiao, 2005 |
| γ–VIIa | γ–gliadin, peptide2 | PQPQQQFPQ | TCC6 | Qiao, 2005 |
| γ–VIIb | γ–gliadin, peptide2 | QQPQQPFPQ | TCC6 | Qiao, 2005 |
| Glia-γ 2 | γ–gliadin, unknown | PYPQQPQQP | Vader, 2003 | |
| Glu-5 (Var1) | Not defined, natural3 | QIPQQPQQF | TCC6 | Vader 2002,Vader, 2003 |
| Glu-5 (Var2) | Not defined, natural3 | QLPQQPQQF | TCC6 | Vader 2002,Vader, 2003 |
| Glt-17 (Var1) | LMW-glutenin, peptide2 | PFSQQQQPV | TCC6 | Vader 2002,Vader, 2003 |
| Glt-17 (Var2) | LMW-glutenin, peptide2 | PFSQQQQPI | TCC6 | Vader 2002,Vader, 2003 |
| Hor-α 2/Sec-α 2 | Hordein/Secalin2 | PQPQQPFPQ | TCL8 | Vader, 2003 |
| Hor-α 9/Sec-α 9 | Hordein/Secalin2 | PFPQPQQPF | TCL8 | Vader, 2003 |
| Avenin-I (Av-α 9A) | Avenin2,3 | PYPEQQEPF | TCC6 | Vader, 2003, Arentz-Hansen, 2004 |
| Avenin-II (Av-α 9B) | Avenin2 | PYPEQQQPF | TCL8 | Vader, 2003, Arentz- Hansen, 2004 |
Identified from fragments of recombinant gliadin protein digests.
Identified from panels of synthetic peptides.
Identified from fragments of natural gliadin or gluten protein digests.
Core 9-mer region interacting with DQ2 is shown. T cells usually require additional flanking residues for recognition. Glutamine residues deamidated by tissue transglutaminase are underlined.
Defined by the crystal structure of the peptide DQ2 complex.
Register defined from minimal fragment recognized by specific T cell clones and binding analysis of lysine substituted analogs.
Register defined from minimal fragment recognized by specific T cell clones and compliance with the DQ2 binding motif.
Register defined from recognition by T cell lines and homology with other well characterized epitopes.
Prolyl endopeptidase activity
Earlier studies have shown that a prolyl endopeptidase (PEP) enzyme is able to accelerate breakdown of the synthetic 33-mer peptide. In light of the above results, we wished to confirm if comparable PEP mediated detoxification of the entire α-2 gliadin protein could also be achieved. Recombinant α-2 gliadin was pretreated with gastric and pancreatic proteases. The digestive products were then treated with rat brush border enzymes for further breakdown. Ordinarily, the small intestinal brush-border membrane enzymes rapidly break down the products of gastric and pancreatic digestion into single amino acids, dipeptides or tripeptides 28. However, their action on the relatively long, proline-rich products of peptic/pancreatic digestion of gluten is at best sluggish. Whereas the brush border enzymes failed to detoxify pretreated recombinant α-2 gliadin (Figure 4A), Flavobacterium PEP addition was able to detoxify the α-2 gliadin protein in a dose-dependent fashion (Figure 4B).
γ-5 Gliadin
Recombinant γ-5 gliadin (GenBank accession number AJ416339 16) was expressed and purified. The protein was proteolyzed with gastric and pancreatic enzymes under physiological conditions. In comparison to the α-2 gliadin, the peptide fragments are not as stable as the 33-mer (data not shown). The main observable peptide products were separated and determined by LC/MS/MS. The most stable fragment was the 26-mer FLQPQQPFPQQPQQPYPQQPQQPFPQ (γ-5 (26–51); Figure 5), another Pro- and Gln- rich peptide containing multiple T cell epitopes.
Figure 5.
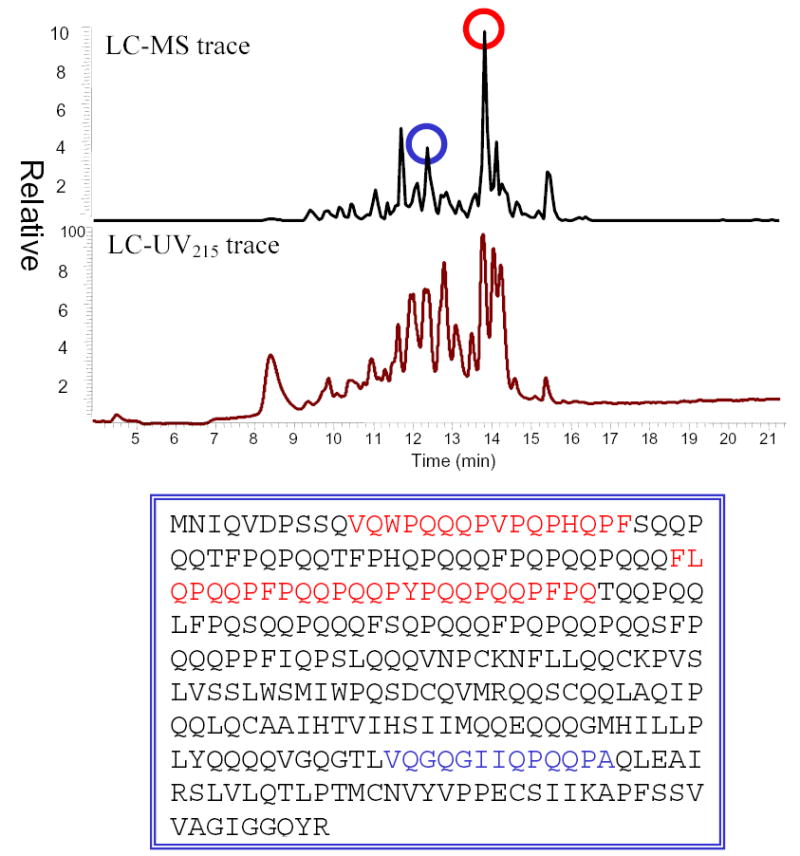
LC-MS analysis of γ-5 gliadin proteolysis. The gliadin protein was treated with pepsin, trypsin, chymotrypsin, elastase, and carboxypeptidase. The blue- and red- colored peptides in the protein sequence are found present in the product mixture.
Like the 33-mer, this peptide was a good substrate of human TG2, as judged by a standard kinetic assay 18 (data not shown). It also contains multiple copies of known T cell epitopes (Figure 6). In T cell proliferation assays, the TG2-treated 26-mer peptide was compared with its smaller peptide fragments, including PEQPFPEQPEQ (DQ2-γ-VI epitope, core epitope is underlined), FPEQPEQPYPEQ (epitope DQ2-γ-III), TG2-treated LQPQQPFPQQPQQPYPQQPQ (containing both DQ2-γ-III and DQ2-γ-VI), and TG2-treated QFPQTQQPQQPFPQPQQTFP (epitope DQ2-γ-VII).15 In all cases, the 26-mer peptide exhibited greater antigenicity compared to its shorter derivatives (Figure 7). The EC50 value of the 26-mer is ca. 5-fold less than the single DQ2-γ-VI epitope PEQPFPEQPEQ, 30-fold less than the DQ2-γ-III epitope FPEQPEQPYPEQ, and 2–3 fold less than the longer peptides LQPQQPFPQQPQQPYPQQPQ or QFPQTQQPQQPFPQPQQTFP. To evaluate whether the antigenicity of the 26-mer could be reduced or eliminated with exogenous PEP, the peptide was co-treated with rat-derived brush-border-membrane enzymes and PEP. After a 2 h incubation, the antigenicity (as measured by the increase of EC50) was reduced by 10 to 100 fold in all of the T cell clones tested (Figure 7D).
Figure 6.
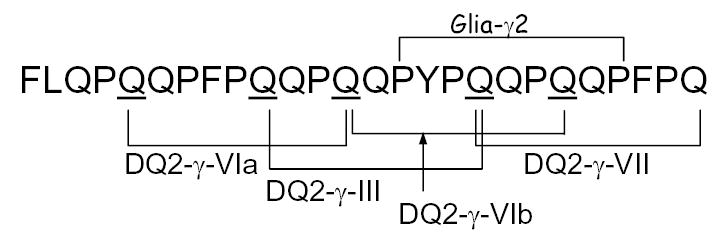
Known γ-gliadin epitopes are clustered in the 26-mer.
Figure 7.
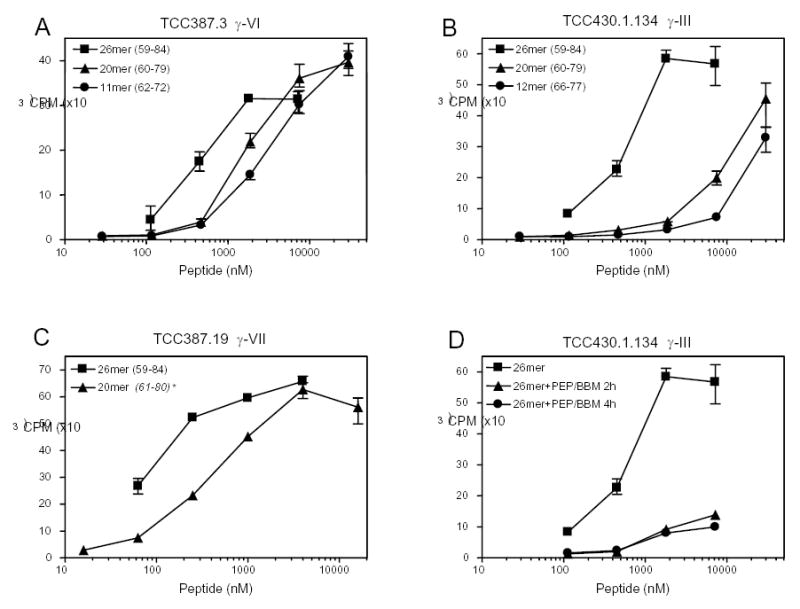
T cell stimulation assay of the 26-mer fragment from γ-5 gliadin. T cell clones were tested with the 26-mer and control shorter peptides. (A) TCC387.3 (reactive to the DQ2-γ-VI epitope) was tested with TG2-treated 26-mer, TG2-treated LQPQQPFPQQPQQPYPQQPQ (“20mer”);, and the reference mono-epitope peptide PEQPFPEQPEQ (“11mer”), (B) TCC430.1.134 (reactive to the DQ2-γ-III epitope) was tested with TG2-treated 26-mer, TG2-treated LQPQQPFPQQPQQPYPQQPQ (“20mer”), and the reference mono-epitope peptide FPEQPEQPYPEQ (“12mer”); (C) TCC387.19 (reactive to DQ2-γ-VII epitope) was tested with the TG2-treated 26-mer and TG2-treated QFPQTQQPQQPFPQPQQTFP (“20mer”); (D) TCC430.1.134 (reactive to DQ2-γ-III) was stimulated with TG2-treated 26-mer, with and without rat intestinal brush border membrane (BBM) and prolyl endopeptidase (PEP) incubation. Numbers in the brackets refer to the positioning of the peptides within the γ-5 gliadin. * This peptide is derived from the 1507333A γ-gliadin (M36999) and identical to sequence 61–80.
Computational Analysis of Gluten Proteins in the Protein Sequence Database
Although our experimental analysis thus far has focused on one representative α-gliadin and one representative γ-gliadin protein, wheat gluten is a complex mixture of hundreds of homologous gliadin and glutenin proteins. (Similarly, barley and rye contain hordein and secalin proteins, respectively, which are homologous to gliadins and are toxic to Celiac Sprue patients.) Therefore, guided by our experimental data, we performed a thorough search of the NCBI protein and nucleotide database to identify other potentially metastable and inflammatory peptides that might be expected to be released as a result of duodenal breakdown of dietary gluten from wheat, rye or barley by the major pancreatic proteases (trypsin, chymotrypsin, elastase, and carboxypeptidases A and B). For the purposes of this analysis, the effect of pepsin on gluten proteins was not considered, due to variability in gastric residence time and food texture; however, since pepsin cleavage specificity is similar to chymotrypsin in gluten (data not shown) 29, there is reasonable confidence in the comprehensiveness of the analysis.
Overall, the sequences of 157 gluten proteins were analyzed, including α-gliadins, α gliadin precursors, γ-gliadins, wheat glutenins, secalins and hordeins. Since peptide size is a major factor that influences the subsequent proteolytic kinetics of the products of pancreatic protease digestion, we mostly focused our analysis on peptides with 18 or more residues. A few exceptions (accession numbers CAA26449, AAQ63849, AAQ63848, AAQ63847, AAQ63842) correspond to hordeins and secalins, whose longest peptide fragments contain only 17 amino acids (Table S1).
Of the 157 proteins analyzed, 128 proteins revealed the presence of one or more putative proteolytic product with 18 or more residues. 58 proteins have two such peptides encoded within their sequence, whereas 35 proteins have three or more such peptides. Out of a total of 271 such peptides, 147 peptides (from 100 gluten proteins) are longer than 25 residues, and 34 peptides (from 34 proteins) are longer than 35 residues (i.e. ca. 10% of the length of a typical gliadin polypeptide). In addition to their length, the high proline content of these peptides is presumably critical to their proteolytic resistance; the average peptide in Table S1 is comprised of ~30% Pro residues.
The potential T cell antigenicity of these peptides can be appreciated by the fact that 51% of the identified peptides contain one or more epitopes known to be recognized by inflammatory T cells from Celiac Sprue patients (Table S1(A)). Furthermore, analogous to the 33-mer from α-2 gliadin and the 26-mer from γ-5 gliadin, 30% of the identified peptides are potentially multivalent (based on known epitopes). An extreme example is the 1507333A γ-gliadin (also designated as sequence M36999), which is predicted to yield a 68-residue peptide spanning at least 4 putative epitopes, PFPQPQQTF (homologous to the known DQ2-α-I epitope PFPQPQLPY), QQPQQPFPQ (DQ2-γ-VII epitope), FPQQPQLPFPQQ (homologous to the known DQ2-γ-III epitope QQPQQPYPQ), and PFPQPQQPQ (also homologous to DQ2-α-I). Indeed, 20-mer synthetic peptides tiling across the entire amino acid sequence of M36999 have been synthesized and assayed against multiple polyclonal T cell lines from DQ2 positive Celiac patients 16. All peptides containing the above four epitopes were found to be antigenic. Another very long peptide was found in a γ-gliadin (AAK84772) as a 92-mer. It contains at least four T- cell epitopes (4 copies of the known DQ2-γ-VII epitope QQPQQPFPQ), and has yet to be tested for bioavailability and T cell activity. Altogether, of the 21 Celiac-specific gluten epitopes reported to date (Table S1(A)), 14 can be observed in one or more of the putative proteolytically stable peptides identified through our computational analysis. These findings provide a vivid testimony to our earlier proposal that the proteolytic stability and pro-inflammatory character of gluten are intimately correlated.
It is also interesting to note that not all gluten proteins appear to contain long (i.e. proteolytically stable), epitope bearing peptides. For example, several α-gliadins and α-gliadin precursors are predicted to release either a 19-residue peptide fragment QQQPFPPQQPYPQPQPFPS that lacks any intact T cell epitope, or considerably shorter putative monovalent peptides such as RPQQPYPQPQPQY (α-10, not shown in the Supplementary Tables) and QPFPRPQLPYPQPQPF (residues 60–75 in α-7 gliadin). Through the use of pepsin or chymotrypsin digested recombinant α-gliadins, some of these proteins have been shown to exhibit greatly diminished T cell antigenicity 25. Presumably this reflects the higher susceptibility, and consequently reduced half-life, of the corresponding physiologically generated peptides in the duodenum to intestinal brush border membrane proteolysis. Similarly, proteolytically stable but non-antigenic peptides may also be pathogenically interesting, as discussed below. For example, although α-1 and α-6 gliadin are predicted to release peptide QQPYPQPQPFPPQLPYPQTQPFPPQQPYPQPQPQYPQPQQPI, there is no known T cell epitope in this sequence 25.
Careful analysis of Table S1 also reveals some relatively minor but noteworthy discrepancies between our experimental and computational data. For example, experimental studies with recombinant α-2 and γ-5 gliadin polypeptides has led to the identification of the 33-mer and 26-mer peptides, whereas in silico analysis as outlined above reveals the closely related but distinct putative sequences, QPFPQPQLPYPQPQLPYPQPQLPYPQPQPF and QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ, respectively. In both cases the observed discrepancies arise due to the relatively unpredictable specificity of chymotrypsin. Apparently, chymotrypsin is unable to cleave N-terminal residues (LQL from the 33-mer and FL from the 26-mer), either due to the strong secondary structure content of the peptides 12 or due to end effects. Conversely, chymotrypsin is able to cleave the C-terminal TQQPQ from the computationally predicted analog of the 26-mer, presumably due to its lower but well-known specificity for Gln in the S1 site. Notwithstanding these differences, the strong overall agreement between the experimental and computational results validates our in silico method for identifying long, proteolytically resistant, and multivalent peptides.
In addition to screening the putative peptides in Table S1 for experimentally established T cell epitope sequences, we also screened for putative TG2 deamidation sites and other likely but as yet unproven DQ2-binding epitopes (Table S2). For this, we used TG2 18 and DQ2 15, 24 recognition algorithms (see Materials and Methods for details) that have recently been proposed in the literature. Over 100 as yet unreported 9-residue sequences, which represent the core region of T cell epitopes of typically 10-14 residues with TG2 deamidation sites at the P4, P6 or P7 positions, were identified from the sequences of the proteolytically stable fragments in Table S1. The core regions of new putative DQ2 epitopes are tabulated in Table 2, and the complete data set is summarized in Table S2. Notably, this in silico method was able to identify many new epitopes in hordeins and secalins (from barley and rye, respectively), which are known to be toxic grains for Celiac Sprue patients but have not been studied extensively so far. At the same time, a few discrepancies between experimental and bioinformatic data are apparent. For example, although two new epitopes were predicted in the peptide QQPYPQPQPFPPQLPYPQTQPFPPQQPYPQPQPQYPQPQQPI from α-1 and α-6 gliadin (9-mer core sequences: PQPQPQYPQ and QYPQPQQPI), Proteolytic digests of the α-1 and α-6 gliadin failed to stimulate a limited number of polyclonal T cell lines. The precise reason for this discrepancy remains to be established, and could either reflect a cryptic proteolytic site in the peptide sequence, or it may highlight the need for further testing of these gliadins against a wider panel of T cell lines from Celiac Sprue patients.
Table S2.
Computational predictions of physiological DQ2 antigenic peptides from gluten proteins in wheat, barley, and rye. Peptide sequences are from pancreatic proteolysis of individual gluten polypeptides catalyzed by chymotrypsin, trypsin, elastase and carboxypeptidases. The predicted TG2 deamidated sites are underlined. The DQ2 binding motifs and core region of 9 residues of known T cell epitopes (Table S1(A)) are in bold. For details, see text. Each peptide’s residue positions from the original protein sequence obtained from NCBI Protein database are indicated. The accession number of each protein is indicated in their names.
| NAME | POSITION | COMPUTATIONAL RESULTS | # EPITOPES |
|---|---|---|---|
| > CAB76964_Alpha-2 Gliadin | 60 – 89 | QPFPQPQLPYPQPQLPYPQPQLPYPQPQPF | >3 |
| > CAC94871_Gamma-5 Gliadin | 28 – 56 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 |
| 61 – 89 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | 3 | |
| > CAB76961_Alpha-8 Gliadin | 34 – 52 | QQQPFPPQQPYPQPQPFPS | |
| > CAB76958_Alpha-4 Gliadin | 34 – 52 | QQQPFPPQQPYPQPQPFPS | |
| > CAB76962_Alpha Gliadin-10 | none | ||
| > CAB76956_Alpha-7 Gliadin | 34 – 52 | QQQPFPPQQPYPQPQPFPS | |
| > CAB76955_Alpha-9 Gliadin | 34 – 52 | QQQPFPPQQPYPQPQPFPS | |
| > CAB76957_Alpha-1 Gliadin | 35 – 53 | QQEQFPPQQPYPHQQPFPS | |
| 54 – 95 | QQPYPQPQPFPPQLPYPQTQPFPPQQPYPQPQPQYPQPQQPI | 3 | |
| > CAB76954_Alpha-6 Gliadin | 54 – 95 | QQPYPQPQPFPPQLPYPQTQPFPPQQPYPQPQPQYPQPQQPI | 3 |
| > CAB76960_Alpha-3 Gliadin | 34 – 52 | QQQPFPPQQPYPQPQPFPS | |
| > CAB76959_Alpha-5 Gliadin | 34 – 52 | QQQPFPPQQPYPQPQPFPS | |
| > AAA17741_Alpha Gliadin | 53 – 71 | QQQPFPPQQPYPQPQPFPS | |
| > CAA10257_Alpha Gliadin | 53 – 71 | QQQPFPPQQPYPQPQPFPS | |
| > BAA12318_Alpha Gliadin | 33 – 51 | QQQPFPPQQPYPQPQPFPS | |
| > T06500_Alpha Gliadin A-IV Precursor | 79 – 101 | QPFPQPQLPYPQPQLPYPQPQPF | >3 |
| 53 – 71 | QQQPFPPQQPYPQPQPFPS | ||
| > AAA96276_Alpha Gliadin | 53 – 70 | QQQPFPPQQPYPQPQFPS | |
| > D22364_alpha-gliadin precursor ( clone A735) | 53 – 71 | QQQPFPPQQPYPQPQPFPS | |
| 79 – 106 | QPFPQPQPFPPQLPYPQPQLPYPQPQPF | 2 | |
| > P04727_Alpha-gliadin clone PW8142 precursor | 50 – 68 | QQQQFPPQQPYPQPQPFPS | |
| > P04725_Alpha-gliadin A-V precursor | 53 – 71 | QQQQFPPQQPYPQPQPFPS | |
| 79 – 98 | QPFPQPQPFPPQLPYPQPQS | ||
| > AAA96522_Alpha Gliadin | 50 – 68 | QQQQFPPQQPYPQPQPFPS | |
| > AAA96524_Alpha Gliadin | none | ||
| > AAA96523_Alpha Gliadin | 53 – 71 | QQQPFPPQQPYPQPQPFPS | |
| > AAQ63862_Gamma Gliadin | 28 – 51 | QQPQQIFPQPQQTFPHQPQQPFPQ | 2 |
| 53 – 73 | QQPQQPFPQPQQQFPQPQQPQ | >3 | |
| > AAQ63865_Gamma Gliadin | 28 – 56 | QQPQQIFPQPQQTFPHQPQQQFPQPQQPQ | >3 |
| 64 – 94 | QPFPQQPQQPYPQQPQQPFPQTQQPQQPFPQ | >3 | |
| > AAQ63864_Gamma Gliadin | 28 – 61 | QQPQQIFPQPQQPYPQQPQQPFPQTQQPQQPFPQ | >3 |
| > AAF42989_Gamma Gliadin | 26 – 44 | QQPQQTFPQPQQTFPHQPQ | 1 |
| 59 – 120 | QPQQPFPQQPQQPFPQTQQPQQPFPQQPQQPFPQTQQPQQPFPQQPQQPFPQTQQPQQPFPL | >3 | |
| 123 – 143 | QQPQQPFPQPQQQLPQPQQPQ | >3 | |
| > AAK84779_Gamma Gliadin | 45 – 63 | QQPQQTFPQPQQTFPHQPQ | 1 |
| 79 – 140 | QPQQPFPQQPQQPFPQTQQPQQPFPQQPQQPFPQTQQPQQPFPQQPQQPFPQTQQPQQPFPQ | >3 | |
| 142 – 162 | QQPQQPFPQPQQQLPQPQQPQ | >3 | |
| > CAB75404_Gamma-1 Gliadin | 28 – 46 | QQPQQTFPQPQQTFPHQPQ | 1 |
| 60 – 89 | IQPQQPQQTYPQRPQQPFPQTQQPQQPFPQ | >3 | |
| 91 – 111 | QQPQQPFPQPQQQFPQPQQPQ | >3 | |
| > CAC94868_Gamma-2 Gliadin | 28 – 56 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | 3 |
| 61 – 89 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| > AAK84773_Gamma Gliadin | 30 – 78 | QWPQQQQPFPQPQQPQQPFPQPQQPQLPFPQQPQQPFPQPQQPQQPFPQ | >3 |
| 80 – 103 | QQPQQPLPQPQQPQQPFPQQQQPL | >3 | |
| > AAK84780_Gamma Gliadin | 1 – 38 | QPFPQPQQTFPQQPQLPFPQQPQQPFPQPQQPQQPFPQ | >3 |
| 40 – 60 | QQPQQPFPQPQQQFPQPQQPQ | >3 | |
| > M36999_Gamma Gliadin | 60 – 127 | HHQPQQTFPQPQQTYPHQPQQQFPQTQQPQQPFPQPQQTFPQQPQLPFPQQPQQPFPQPQQPQQPFPQ | >3 |
| 129 – 149 | QQPQQPFPQPQQQFPQPQQPQ | >3 | |
| > AAQ63868_Hordein ( leaf) | none | ||
| > S07975_hordein B3 ( clone pB7) | 23 – 47 | QPQQPQPFPQQPIPQQPQPYPQQPQ | >3 |
| > CAA60681_hordein B1 | 20 – 51 | QQQPFPQQPIPQQPQPYPQQPQPYPQQPFPPQ | >3 |
| >1103203A_hordein | 23 – 47 | QPQQPQPFPQQPIPQQPQPYPQQPQ | >3 |
| > P80198_Gamma-hordein 3 | 36 – 82 | QQEPEQPYPQQQPLPQQQPFPQQPQLPHQHQFPQQLPQQQFPQQMPL | >3 |
| > BAB78747_Glutenin | 41 – 61 | HHHQQQPIQQQPHQFPQQQPC | 2 |
| > BAB78749_Glutenin | none | ||
| > AAS66085_glutenin pGM107 | none | ||
| > AAV92073_Glutenin ( in press) | none | ||
| > AAV92049_Glutenin ( in press) | none | ||
| > AAV92036_Glutenin ( in press) | none | ||
| > AAV91997_Glutenin | 150 – 168 | QQPPFPQQHQQFPQQQIPV | 3 |
| > AAS10189_Glutenin | none | ||
| > S18236_secalin clone pSec2B | 76 – 100 | PIQPQQPFPQQPQQPFPQPQQQLPL | >3 |
| 101 – 118 | QPQQPFPQPQQPIPQQPQ | 3 | |
| 121 – 150 | FPQQPQRPEQQFPQQPQQIIPQQTQQPFPL | >3 | |
| 228 – 255 | QQPQRPQQPFPQQPQQIIPQQPQQPFPL | >3 | |
| > S18235_secalin clone pSec1B | 63 – 100 | QPQQPFPQPQQPTPIQPQQPFPQRPQQPFPQPQQQLPL | >3 |
| 121 – 150 | FPQQPQRPEQQFPQQPQQIIPQQTQQPFPL | >3 | |
| 228 – 255 | QQPQRPQQPFPQQPQQIIPQQPQQPFPL | >3 | |
| > CAA26449_secalin unnamed | 5 – 21 | FPLPQQPFPQPQQPFPQ | >3 |
| > AAB58403_secalin omega | 63 – 91 | QPQQPFPQPQQPTPIQPQQPFPQQPQQPF | >3 |
| 121 – 150 | FPQQPQRPEQQFPQQPQQIIPQQTQQPFPL | >3 | |
| 187 – 208 | QPQQPFPQQPEQIIPQQPQQPS | 2 | |
| 228 – 255 | QQPQRPQQPFPQQPQQIIPQQPQQPFPL | >3 | |
| > AAG35598_secalin omega | 68 – 119 | PQPQQPYPQQPFPQQPQQPYPQQPQQPFPQQPQQPYPQQPQQQFPQQPQQPV | >3 |
| 141 – 159 | QQFPQQPQQPFPQQPQQPV | 2 | |
| 165 – 185 | QQPFPQTQQPQQPFPQPQQPQ | >3 | |
| 138 – 160 | QQPQQPFPQPQQPQQPFPQPQPQ | 3 | |
| > AV86084_secalinGlutenin | 152 – 170 | QQPPFPQQHQQFPQQQIPV | 3 |
| > AV86085_secalinGlutenin | 77 – 95 | QQPPFPQQHQQFPQQQIPV | 3 |
| > AAT74547_omegaGliadin | 39 – 56 | HQQQPFPQQPYPQQPYPS | >4 |
| 91 – 116 | QPQQPFPQQPQQPQQPFPQPQQPFPW | >3 | |
| 132 – 154 | QPQQPFPQQPQQPFPQPQLPFPQ | >3 | |
| 271 – 297 | FPQQPQQPFPPTTTKPFPQQPQQPFPL | >3 | |
| 234 – 252 | QPQQPFPQQPQQPFPQQPQ | >3 | |
| > AAT01617_omegaGliadin | none | ||
| > AAG17702_omegaGliadin | 82 – 116 | TQPQQPTPIQPQQPFPQQPQQPQQPFPQPQQPFPW | >3 |
| 132 – 154 | QPQQPFPQQPQQPFPQPQLPFPQ | >3 | |
| 171 – 193 | QPQQPFPQQPQQPFPQPQQPIPV | >3 | |
| 235 – 253 | QPQQPFPQQPQQPFPQQPQ | >3 | |
| > S52390_D_hordein precursor | none | ||
| > S18350_hordein | 84 – 106 | QPQQPFPQQPQQPLPRPQQPFPW | >3 |
| 107 – 140 | QPQQPFPQPQEPIPQQPQQPFPQQPQQPFPQQPQ | >3 | |
| 208 – 227 | QPQPFPQQPQQPLPQPQQPF | >3 | |
| > T06211_C-hordein precursor | 84 – 106 | QPQQPFPQQPQQPLPRPQQPFPW | >3 |
| 137 – 160 | QQPQQPFPQQPQQPFPQPQQPFPW | >3 | |
| 217 – 235 | QPQQPFPQQPQQPFPQQPQ | >3 | |
| > S20519_B-hordein | 20 – 48 | QQQPFPQQPFPQQPQPYPQQPQPYPQQPF | >3 |
| 49 – 83 | QPQQPFPQQTIPQQPQPYPQQPFPPQQEFPQQPPF | >3 | |
| > S07365_B1-hordein | 20 – 59 | QQQPFPQQPIPQQPQPYPQQPQPYPQQPFPPQQPFPQQPV | >3 |
| 60 – 83 | PQQPQPYPQQPFPPQQPFPQQPPF | 1 | |
| > P17990_hordeinGamma 1 | 49 – 69 | QQQFPQPQQPFPQQPQQPFPQ | >3 |
| 76 – 94 | QQPQHQFPQPTQQFPQRPL | 3 | |
| >P06471_B3-hordein | 23 – 47 | QPQQPQPFPQQPIPQQPQPYPQQPQ | >3 |
| >P06470_B1-hordein | 20 – 59 | QQQPFPQQPIPQQPQPYPQQPQPYPQQPFPPQQPFPQQPV | >3 |
| 60 – 82 | PQQPQPYPQQPFPPQQPFPQQPPF | 1 | |
| >AAU06229_hordeinB | 32 – 78 | PWPQQPFPQQPYPQQPQPYPQQPQPYPQQPYPQQPQPYPQQPQPYPS | >3 |
| >AAU06228_hordeinB | 32 – 52 | PWPQQPFPQQPYPQQPQPYPQ | >3 |
| >AAU06227_hordeinB | 32 – 62 | PWPQQPFPQQPYPQQPQPYPQQPQPYPQQPY | >3 |
| 63 – 85 | LPQPQPYPQQPQPYPQQPQPYPS | >3 | |
| >AAQ63849_hordeinGamma3 | 50 – 66 | QQQQLPQQHPFPQQMPL | 1 |
| >AAQ63848_hordeinGamma3 | 67 – 83 | QQQQLPQQHPFPQQMPL | 1 |
| >AAQ63847_hordeinGamma3 | 67 – 83 | QQQQLPQQHPFPQQMPL | 1 |
| >AAQ63842_hordeinGamma3 | 67 – 83 | QQQQLPQQHPFPQQMPL | 1 |
| >AAP31051_D-hordein | none | ||
| >T04474_B1-hordein | 20 – 51 | QQQPFPQQPIPQQPQPYPQQPQPYPQQPFPPQ | >3 |
| >BAA11642_D-hordein | none | ||
| >AAB28161_C-hordein | 84 – 106 | QPQQPFPQQPQQPLPRPQQPFPW | >3 |
| 136 – 159 | QQPQQPFPQQPQQPFPQPQQPFPW | >3 | |
| 216 – 234 | QPQQPFPQQPQQPFPQQPQ | >3 | |
| >1604464A_gammaHordein | 48 – 68 | QQQFPQPQQPFPQQPQQPFPQ | >3 |
| 76 – 94 | QQPQHQFPQPTQQFPQRPL | 3 | |
| >AAA92333_C-hordein | 92 – 117 | QPQPPQQPFPQPQQPNPQQPQQPFPR | >3 |
| 124 – 146 | PQQPQQPFPQQPQQPFPQPQQPF | >3 | |
| 194 – 210 | QQPQQTIPQQPQQPFPL | 2 | |
| 211 – 229 | QPQQPFPQQPQQPLPQQPQ | 3 | |
| 242 – 259 | QPQQPFPQPQPFPQEQPQ | ||
| >CAA59104_D-hordein | none | ||
| >CAA26889_hordein unnamed | 20 – 59 | QQQPFPQQPIPQQPQPYPQQPQPYPQQPFPPQQPFPQQPV | >3 |
| 60 – 83 | PQQPQPYPQQPFPPQQPFPQQPPF | 1 | |
| >CAA42642_hordein unnamed | 84 – 106 | QPQQPFPQQPQQPLPRPQQPFPW | >3 |
| 107 – 140 | QPQQPFPQPQEPIPQQPQQPFPQQPQQPFPQQPQ | >3 | |
| 208 – 227 | QPQPFPQQPQQPLPQPQQPF | >3 | |
| >CAA48209_hordeinD | none | ||
| >CAA51204_gamma3-hordein | 33 – 79 | QQEPEQPYPQQQPLPQQQPFPQQPQLPHQHQFPQQLPQQQFPQQMPL | >3 |
| >CAI35909_alpha-precursor | 68 – 86 | QQQPFPPQQPYPQPQPFPS | |
| >T06282_alpha-precursor | none | ||
| >AAA96525_alpha | none | ||
| >EEWTG_gamma-precursor | 46 – 74 | QQPQQIFPQPQQTFPHQPQQQFPQPQQPQ | >3 |
| >P21292_gamma-precursor | 60 – 126 | HQPQQTFPQPQQTYPHQPQQQFPQTQQPQQPFPQPQQTFPQQPQLPFPQQPQQPFPQPQQPQQPFPQ | >3 |
| 129 – 149 | QQPQQPFPQPQQQFPQPQQPQ | >3 | |
| >P06659_gamma-precursor | 46 – 74 | QQPQQIFPQPQQTFPHQPQQQFPQPQQPQ | >3 |
| 82 – 112 | QPFPQQPQQPYPQQPQQPFPQTQQPQQPFPQ | >3 | |
| >P04730_gamma | none | ||
| >P08453_gamma-precursor | 46 – 67 | QQPQQTFPQPQQTFPHQPQQQV | 1 |
| 79 – 140 | QPQQPFPQQPQQPFPQTQQPQQPFPQQPQQPFPQTQQPQQPFPQQPQQPFPQTQQPQQPFPQ | >3 | |
| 142 – 162 | QQPQQPFPQPQQQLPQPQQPQ | >3 | |
| >P04729_gamma-precursor | none | ||
| >P08079_gamma | 46 – 74 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | 3 |
| 79 – 107 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| >AAQ63863_gamma | 64 – 94 | QPFPQQPQQPYPQQPQQPFPQTQQPQQPFPQ | >3 |
| >AAQ63861_gamma | 28 – 57 | QQPQQIFPQPQQTFPHQPQQQFPQPQQPQ | >3 |
| 64 – 94 | QPFPQQPQQPYPQQPQQPFPQTQQPQQPFPQ | >3 | |
| >AAQ63860_gamma | 28 – 46 | QQPQQTFPQPQQTFPHQPQ | 1 |
| 60 – 93 | QPQQPFPQQPQQTYPQRPQQPFPQTQQPQQPFPQ | >3 | |
| 96 – 116 | QQPQQPFPQPQQQFPQPQQPQ | >3 | |
| >AAQ63859_gamma | 36 – 64 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 |
| >AAQ63858_gamma | 28 – 46 | QQPQQTFPQPQQTFPHQPQ | 1 |
| 61 – 122 | QPQQPFPQQPQQPFPQTQQPQQPFPQQPQQPFPQTQQPQQPFPQQPQQPFPQTQQPQQPFPQ | >3 | |
| 124 – 144 | QQPQQPFPQPQQQLPQPQQPQ | >3 | |
| >AAQ63857_gamma | 28 – 46 | QQPQQIFPQPQQTFPHQPQ | 2 |
| 60 – 85 | QPQQPFPQQPQQPFPQTQQPQQPFPQ | >3 | |
| 88 – 108 | QQPQQPFPQPQQQFPQPQQPQ | >3 | |
| >AAQ63856_gamma | 28 – 56 | QQPQQIFPQPQQTFPHQPQQQFPQPQQPQ | >3 |
| 64 – 94 | QPFPQQPQQPYPQQPQQPFPQTQQPQQPFPQ | >3 | |
| >JS0402_gamma | 46 – 64 | QQPQQTFPQPQQTFPHQPQ | 1 |
| 79 – 140 | QPQQPFPQQPQQPFPQTQQPQQPFPQQPQQPFPQTQQPQQPFPQQPQQPFPQTQQPQQPFPQ | >3 | |
| 142 – 162 | QQPQQPFPQPQQQLPQPQQPQ | >3 | |
| >JA0153_gamma | 60 – 127 | HHQPQQTFPQPQQTYPHQPQQQFPQTQQPQQPFPQPQQTFPQQPQLPFPQQPQQPFPQPQQPQQPFPQ | >3 |
| 129 – 149 | QQPQQPFPQPQQQFPQPQQPQ | >3 | |
| >CAA54925_gamma | 40 – 107 | HHQPQQTFPQPEQTYPHQPQQQFPQTQQPQQPFPQPQQTFPQQPQLPFPQQPQQPFPQPQQPQQPFPQ | >3 |
| 109 – 129 | QQPQQPFPQPQQQFPQPQQPQ | >3 | |
| >AAN32705_gamma | none | ||
| >CAC94870_gamma-4 | 28 – 56 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | 3 |
| 72 – 89 | QPYPQQPQQPFPQTQQPQ | 3 | |
| >CAC94869_gamma-3 | 28 – 56 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | 3 |
| 60 – 79 | LQPQQPFPQQPQQPYPQQPQ | >3 | |
| >AAK84778_gamma | 46 – 64 | QQPQQTFPQPQQTFPHQPQ | 1 |
| 78 – 112 | IQPQQPFPQQPQQTYPQRPQQPFPQTQQPQQPFPQ | >3 | |
| 114 – 134 | QQPQQPFPQPQQQFPQPQQPQ | >3 | |
| >AAK84777_gamma | 60 – 127 | HHQPQQTFPQPQQTYPHQPQQQFPQTQQPQQPFPQPQQTFPQQPQLPFPQQPQQPFPQPQQPQQPFPQ | >3 |
| 129 – 149 | QQPQQPFPQPQQQFPQPQQPQ | >3 | |
| >AAK84776_gamma-13 | 46 – 91 | QQPQHTFPQPQQTCPHQPQQQFPQPQQPQQPFPQPQQPQQPFPQPQ | >3 |
| 94 – 117 | QLPFPQQPQQPLPQPQQPQQPFPQ | >3 | |
| >AAK84775_gamma | 1 – 28 | PQQPQLPFPQQPQQPFPQPQQPQQPFPQ | >3 |
| 30 – 50 | QQPQQPFPQPQQQFPQPQQPQ | >3 | |
| >AAK84774_gamma | 47 – 65 | QQPQQIFPQPQQTFPHQPQ | 2 |
| FPQPQQTFPHQPQQQFPQPQQPQQPFPQQPQQQFPQPQQPQQPFPQQPQQQFPQPQQPQQPFPQPQQPQLPFPQQPQQPFPQPQQPQQP | >3 | ||
| 68 – 159 | FPQ | ||
| 161 – 184 | QQPQQPLPQPQQPQQPFPQQQQPL | >3 | |
| >AAK84772_gamma | 47 – 65 | QQPQQIFPQPQQTFPHQPQ | 2 |
| FPQPQQTFPHQPQQQFPQPQQPQQPFPQQPQQQFPQPQQPQQPFPQQPQQQFPQPQQPQQPFPQPQQPQLPFPQQPQQPFPQPQQPQQP | >3 | ||
| 68 – 159 | FPQ | ||
| 161 – 184 | QQPQQPLPQPQQPQQPFPQQQQPL | >3 | |
| >AAD30440_gamma | 46 – 74 | QQPQQIFPQPQQTFPHQPQQQFPQPQQPQ | >3 |
| 79 – 107 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| >CAC10619_gamma | 20 – 37 | QTFPHQPQQQFPQPQQPQ | 2 |
| 48 – 70 | HQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| 77 – 103 | QQPQQPYPQQPQQPFPQTQQPQQQFPQ | >3 | |
| >CAC11097_gamma | 29 – 76 | FPQPQQPQQQFPQPQQPQQPFPQPQQPQLPFPQQPFPQPQQPQQPFPQ | >3 |
| 78 – 98 | QQPQQPFPQPQQQFPQPQQPQ | >3 | |
| >CAC11096_gamma | 29 – 76 | FPQPQQPQQQFPQPQQPQQPFPQPQQPQLPFPQQPFPQPQQPQQPFPQ | >3 |
| 78 – 98 | QQPQQPFPQPQQQFPQPQQPQ | >3 | |
| >CAC11099_gamma | 17 – 45 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | 3 |
| 50 – 78 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| >CAC11098_gamma | 14 – 42 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | 3 |
| 47 – 75 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| >CAC11089_gamma | 30 – 58 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | 3 |
| 63 – 91 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| >CAC11088_gamma | 15 – 43 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | 3 |
| 48 – 76 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| >CAC11087_gamma | 25 – 53 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | 3 |
| 58 – 86 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| >CAC11080_gamma | 30 – 58 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | 3 |
| 63 – 91 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| >CAC11079_gamma | 30 – 58 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | 3 |
| 63 – 91 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| > CAC11078_gamma | 30 – 58 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | 3 |
| 63 – 91 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| > CAC11065_gamma | 46 – 74 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | 3 |
| 79 – 107 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| > CAC11064_gamma | 27 – 55 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | 3 |
| 60 – 88 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| > CAC11057_gamma | 23 – 51 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | 3 |
| 56 – 84 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| > CAC11056_gamma | 17 – 45 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | 3 |
| 50 – 78 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| > CAC11055_gamma | 19 – 47 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | 3 |
| 52 – 80 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| > CAC10624_gamma | 13 – 35 | PQPQQTFPHQQQQQQFPQPQQPQ | >3 |
| 40 – 68 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| 75 – 104 | QQPQQPYPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| 111 – 137 | QQPQQPYPQQPQQPFPQTQQPQQQFPQ | >3 | |
| > CAC10623_gamma | 13 – 34 | PQPQQTFPHQPQQQFPQPQQPQ | 2 |
| 39 – 67 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| 74 – 103 | QQPQQPYPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| 110 – 136 | QQPQQPYPQQPQQPFPQTQQPQQQFPQ | >3 | |
| > CAC10622_gamma | 8 – 36 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | 3 |
| 41 – 69 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| 76 – 102 | QQPQQPYPQQPQQPFPQTQQPQQQFPQ | >3 | |
| > CAC10620_gamma | 20 – 37 | QTFPHQPQQQFPQPQQPQ | 2 |
| 48 – 70 | HQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| 77 – 103 | QQPQQPYPQQPQQPFPQTQQPQQQFPQ | >3 | |
| > CAC10633_gamma | 9 – 37 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | 3 |
| 42 – 70 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| > CAC10632_gamma | 42 – 70 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 |
| > CAC10631_gamma | 7 – 35 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | 3 |
| 40 – 68 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| > CAC10630_gamma | 9 – 37 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | 3 |
| 42 – 65 | QPQQPQQPYPQEPQQPFPQTQQPQ | >3 | |
| 77 – 97 | QQPQQQFPQPQQQFPQPQQPQ | >3 | |
| > CAC10629_gamma | 42 – 70 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 |
| > CAC10628_gamma | 9 – 37 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | 3 |
| 42 – 65 | QPQQPQQPYPQEPQQPFPQTQQPQ | >3 | |
| 77 – 97 | QQPQQQFPQPQQQFPQPQQPQ | >3 | |
| > CAC10627_gamma | 9 – 37 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | 3 |
| 42 – 65 | QPQQPQQPYPQEPQQPFPQTQQPQ | >3 | |
| 72 – 92 | QQPQQQFPQPQQQFPQPQQPQ | >3 | |
| > CAC10626_gamma | 46 – 74 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | 3 |
| 79 – 107 | QPQQPFPQQQQQPYPQQPQQPFPQTQQPQ | >3 | |
| > CAC10625_gamma | 46 – 74 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | 3 |
| 79 – 107 | QPQQPFPQQQQQPYPQQPQQPFPQTQQPQ | >3 | |
| > CAC10617_gamma | 30 – 60 | QKPQQTFPQPQQTFPFPHQPQQQFPQPQQPQ | >3 |
| 65 – 88 | QPQQPQQPYPQQPQQTFPQTQQPQ | >3 | |
| > CAC10616_gamma | 30 – 58 | QQPQQTFPQPQQTFPHQPQQQIPQPQQPQ | 3 |
| 63 – 91 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| 98 – 124 | QQPQQPYPQQPQQPFPQTQQPQQQFPQ | >3 | |
| > CAC10594_gamma | 30 – 58 | QQPQQTFPQPQQTFPHQPQQQIPQPQQPQ | 3 |
| 63 – 91 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| 98 – 124 | QQPQQPYPQQPQQPFPQTQQPQQQFPQ | >3 | |
| > CAC10593_gamma | 30 – 58 | QQPQQTFPQPQQTFPHQPQQQIPQPQQPQ | 3 |
| 63 – 91 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| 98 – 124 | QQPQQPYPQQPQQPFPQTQQPQQQFPQ | >3 | |
| > AAD30556_gamma | 46 – 74 | QQPQQIFPQPQQTFPHQPQQQFPQPQQPQ | >3 |
| 79 – 107 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| > AAF42989_gamma | 27 – 45 | QQPQQTFPQPQQTFPHQPQ | 1 |
| 60 – 121 | QPQQPFPQQPQQPFPQTQQPQQPFPQQPQQPFPQTQQPQQPFPQQPQQPFPQTQQPQQPFPQ | >3 | |
| 123 – 143 | QQPQQPFPQPQQQLPQPQQPQ | >3 | |
| > BAA11251_gamma-precursor | 27 – 45 | QQPQQTFPQPQQTFPHQPQ | 1 |
| 59 – 91 | PQQPFPQQPQQTYPQRPQQPFPQTQQPQQPFPQ | >3 | |
| 95 – 115 | QQPQQPFPQPQQQFPQPQQPQ | >3 | |
| > 1507333A_gamma | 59 – 126 | HHQPQQTFPQPQQTYPHQPQQQFPQTQQPQQPFPQPQQTFPQQPQLPFPQQPQQPFPQPQQPQQPFPQ | >3 |
| 128 – 148 | QQPQQPFPQPQQQFPQPQQPQ | >3 | |
| > 1209306A_gamma | none | ||
| > CAA37491_gamma | 60 – 126 | HQPQQTFPQPEQTYPHQPQQQFPQTQQPQQPFPQPQQTFPQQPQLPFPQQPQQPFPQPQQPQQPFPQ | >3 |
| 129 – 149 | QQPQQPFPQPQQQFPQPQQPQ | >3 | |
| > AAA34289_gamma | 46 – 64 | QQPQQTFPQPQQTFPHQPQ | 1 |
| 79 – 140 | QPQQPFPQQPQQPFPQTQQPQQPFPQQPQQPFPQTQQPQQPFPQQPQQPFPQTQQPQQPFPQ | >3 | |
| > AAA34288_gamma | 46 – 74 | QQPQQIFPQPQQTFPHQPQQQFPQPQQPQ | >3 |
| 82 – 112 | QPFPQQPQQPYPQQPQQPFPQTQQPQQPFPQ | >3 | |
| > AAA34287_gamma | none | ||
| > AAA34286_gamma | none | ||
| > AAA34285_gamma-precursor | none | ||
| > AAA34274_gamma-precursor | 46 – 74 | QQPQQIFPQPQQTFPHQPQQQFPQPQQPQ | >3 |
| 82 – 112 | QPFPQQPQQPYPQQPQQPFPQTQQPQQPFPQ | >3 | |
| > AAA34272_gamma-precursor | 60 – 127 | HHQPQQTFPQPQQTYPHQPQQQFPQTQQPQQPFPQPQQTFPQQPQLPFPQQPQQPFPQPQQPQQPFPQ | >3 |
| 129 – 149 | QQPQQPFPQPQQQFPQPQQPQ | >3 |
Table 2.
Predicted novel HLA-DQ2 restricted T cell epitopes. The native 9-mer sequences of the core regions are displayed. Typically, the entire T cell epitopes span 10–14 residues and contain glutamate residues converted from glutamine by tissue transglutaminase mediated deamidation.
| Epitopes | Mainly found in | Epitopes | Mainly found in |
|---|---|---|---|
| QYPQPQQPI | alpha gliadin | PQHQFPQPT | hordein |
| IFPQPQQTF | gamma gliadin | PQPFPQQPI | hordein |
| PFPQPQQPQ | gamma gliadin | PQPFPQQPQ | hordein |
| PFPQPQQQF | gamma gliadin | PQPQEPIPQ | hordein |
| PFPQPQQQL | gamma gliadin | PQPQQPNPQ | hordein |
| PFPQPQQTF | gamma gliadin | PQPTQQFPQ | hordein |
| PFPQQPQQQ | gamma gliadin | PQPYPQQPF | hordein |
| PFPQQPQQT | gamma gliadin | PQPYPQQPQ | hordein |
| PFPQQQQPL | gamma gliadin | PQPYPQQPY | hordein |
| PFPQTQQPQ | gamma gliadin | PQQLPQQQF | hordein |
| PHQQQQQQF | gamma gliadin | PQQQFPQQM | hordein |
| PLPQPQQPQ | gamma gliadin | PRPQQPFPW | hordein |
| PQPQQPQQQ | gamma gliadin | PWPQQPFPQ | hordein |
| PQPQQQLPQ | gamma gliadin | PYPQQPQPY | hordein |
| PQQQFPQPQ | gamma gliadin | QEPIPQQPQ | hordein |
| PQQQFPQTQ | gamma gliadin | QFPQPQQPF | hordein |
| PQQQIPQPQ | gamma gliadin | QLPQQQFPQ | hordein |
| PQQQLPQPQ | gamma gliadin | QQPFPPQQE | hordein |
| PQQQQPFPQ | gamma gliadin | QQPFPQQPF | hordein |
| PQTQQPQQQ | gamma gliadin | QQPFPQQPI | hordein |
| QFPQPQQPQ | gamma gliadin | QQPFPQQPV | hordein |
| QFPQTQQPQ | gamma gliadin | QQPFPQQPY | hordein |
| QIPQPQQPQ | gamma gliadin | QQPFPQQTI | hordein |
| QLPFPQQPF | gamma gliadin | QQPLPQQPQ | hordein |
| QLPFPQQPQ | gamma gliadin | QQPNPQQPQ | hordein |
| QQPQLPFPQ | gamma gliadin | QQPQHQFPQ | hordein |
| QQPQQPFPL | gamma gliadin | QQPQLPHQH | hordein |
| QQPQQPLPQ | gamma gliadin | QQPQQPFPR | hordein |
| QQPQQQFPQ | gamma gliadin | QQPQQPLPR | hordein |
| QQQQFPQPQ | gamma gliadin | QQQQLPQQH | hordein |
| QQQQQQFPQ | gamma gliadin | QQTIPQQPQ | hordein |
| QQTFPQQPQ | gamma gliadin | PFPQQPYPQ | hordein, omega gliadin |
| QQTYPQRPQ | gamma gliadin | PQPQQPFPW | hordein, omega gliadin |
| QRPQQPFPQ | gamma gliadin | QQPIPQQPQ | hordein, secalin |
| QTQQPQQPF | gamma gliadin | IQPQQPFPQ | secalin |
| QWPQQQQPF | gamma gliadin | PFPQPQQPT | secalin |
| PIQQQPHQF | glutenin | PFPQQPQQI | secalin |
| PQQHQQFPQ | glutenin | PQPQQPIPQ | secalin |
| QFPQQQIPV | glutenin | PQPQQPTPI | secalin |
| QQPIQQQPH | glutenin | PQPQQQLPL | secalin |
| PFPQPQLPF | omega gliadin | PYPQQPFPQ | secalin |
| PFPQPQQPI | omega gliadin | PYPQQPQQQ | secalin |
| PQPQLPFPQ | omega gliadin | QFPQQPQQI | secalin |
| PQPQQPIPV | omega gliadin | QHQQFPQQQ | secalin |
| PYPQQPYPS | omega gliadin | QQIIPQQPQ | secalin |
| QQPYPQQPY | omega gliadin | QQPFPQRPQ | secalin |
| PFPQPQEPI | hordein | QQPQRPEQQ | secalin |
| PFPQPQQPN | hordein | QQPYPQQPF | secalin |
| PFPQQPFPQ | hordein | QQQFPQQPQ | secalin |
| PFPQQPIPQ | hordein | QQTQQPFPL | secalin |
| PFPQQPQPY | hordein | QRPEQQFPQ | secalin |
| PIPQQPQPY | hordein | PQPQQPYPQ | secalin, gamma gliadin |
| PLPRPQQPF | hordein | ||
| PLPQPQQPF | hordein |
Finally, as a control, we performed equivalent in silico analysis on several other sequenced families of dietary proteins that are non-toxic to Celiac Sprue patients, including avenins (from oats) bovine myoglobin (from meat), chicken ovalbumin (from eggs), bovine casein and lactoglobulin (from milk). Remarkably, none of these dietary proteins were predicted to release peptide products longer than 10 amino acids as a result of pancreatic proteolysis (Table S3).
Table S3.
Computational proteolysis analysis on avenins (from oats) and other dietary proteins including myoglobin, ovalalbumin, casein and lactoglobulin.
| Name | Position | Chymotrypsin, Trypsin, Elastase, Carboxypeptidases Prediction | Length |
|---|---|---|---|
| >1411172A_avenin | 13–22 | QPYPEQQEPF | 10 |
| >S06455_avenin-precursor | 61–68 | DFPITWPW | 8 |
| >AAA32716_avenin | 32–41 | QPYPEQQEPF | 10 |
| >AAA32715_avenin | 28–38 | QPYPEQQQPI | 10 |
| >AAA32714_avenin | 28–36 | QPYPEQQPF | 9 |
| >AAA32713_avenin | 61–68 | DFPITWPW | 8 |
| >1502200A_avenin | 61–68 | DFPITWPW | 8 |
| >AAB32025_avenin | 9–19 | QPYPEQQQPI | 10 |
| >MYBO_bovine-myoglobin | none | 0 | |
| >NP_990483_ovalbumin-chicken | 89–97 | 0 | |
| >NP_851351_betaCasein-bovine | 51–60 | 0 | |
| 160–170 | HQPHQPLPPT | 10 | |
| >NP_776719_kappaCasein-bovine | 22–31 | QEQNQEQPI | 9 |
| 138–147 | 0 | ||
| >NP_776354_betaLactoglobulin-bovine | none | 0 |
DISCUSSION
The recent discovery of an unusually long and potent T cell proliferation agent, the 33-mer peptide from α-2 gliadin, highlighted the importance of understanding the molecular fate of gluten under physiological conditions. In particular, the role of proteolytic resistance in gluten antigenicity became clear. The 33-mer peptide has become a useful probe for DQ2 binding 30, 31, for peptidase development 32 and for studying the transport of peptide and Celiac pathogenesis at the intestinal level 33. The goals of the current work were therefore two-fold. On one hand we sought to verify the centrality of the 33-mer in the immunotoxicity of α-2 gliadin via deletion mutagenesis. On the other hand, we wished to identify similar peptides from other gluten proteins, especially the closely related γ-gliadins.
Our studies verified that the 33-mer peptide is the single crucial antigenic component in α-2 gliadin. The digestion mixture of α-2 gliadin was comparably potent to an equivalent amount of the synthetic 33-mer peptide, and 10–20 times more potent than any individual T cell epitope present in the 33-mer sequence. Two deletion mutants of α-2 gliadin lacking the 33-mer were engineered, one excluding only the 33-mer sequence and the other deleting the 33-mer as well as additional C-terminal residues. The resulting proteins were no longer toxic. Taken together with earlier data, the 33-mer was thus confirmed as the major Celiac-toxic segment of α-2 gliadin.
The existence of other long, proteolytically resistant, antigenic peptides in gluten proteins was anticipated from early in silico analysis using the 33-mer sequence as a probe 14. To obtain direct experimental evidence for this proposal, here we have identified a 26-mer peptide, FLQPQQPFPQQPQQPYPQQPQQPFPQ, from γ-5 gliadin. This peptide is not as resistant to pancreatic proteolysis as the 33-mer peptide, presumably due to its PQ↓Q repeats, which are weakly susceptible to chymotrypsin cleavage. Like the 33-mer however, the 26-mer peptide is highly resistant to intestinal brush border membrane proteolysis. It is also multivalent; thus far three epitopes, EQPFPEQPE (DQ2-γ-VI), EQPEQPYPE (DQ2-γ-III), EQPEQPFPQ (DQ2-γ-VII) have been identified in the 26-mer 16. Similar to the 33-mer, the intact 26-mer is more antigenic compared to its smaller monovalent counterparts. Finally, analogous to the 33-mer, the 26-mer is susceptible to PEP breakdown, as illustrated in Fig. 7D.
In an attempt to integrate available biochemical and immunological information to derive a global picture of the physiological immunotoxicity of dietary gluten for Celiac Sprue patients, we conducted an in-depth computational analysis of the gluten proteome. Our analysis revealed that long, proteolytically resistant fragments were widespread within the α-gliadin, γ-gliadin, glutenin, hordein and secalin protein families of wheat, rye and barley. In contrast, analogous investigations on sequences of avenins (from oats) and a range of proteins present in bovine myoglobin, chicken ovalbumin, bovine casein and lactoglobulin (from meat, eggs or milk) showed no peptide products longer than 10 amino acids (Table S2). Their susceptibility to proteolysis corresponds to their low proline content (an average of 6% in avenins, 2% in myoglobin, 3% in ovalbumin, 12% in casein and 4% in lactoglobulin). Only avenin and casein proteins produced digestion fragments of 8–10 residues. Notably, these avenin fragments contain T cell epitopes recognized by intestinal T cells of oats-intolerant Celiac Sprue patients 14. Gluten proteins known to be highly antigenic, such as α-2 gliadin, γ-5 gliadin and the M36999 γ-gliadin, appear to release long, proteolytically stable, multivalent peptides as a result of duodenal proteolysis. In contrast, gluten proteins with limited T cell antigenicity seem to be incapable of generating such long, epitope-bearing peptides. Due to the rapid intestinal brush-border membrane proteolysis of peptides shorter than 10 amino acids 12, the antigenic peptides released from these gluten proteins are likely to be rapidly destroyed by the peptidases of the intestinal brush border membrane.
The putative existence in gluten of long, proteolytically stable peptides that lack any known T cell antigenicity bears some reflection. Perhaps this suggests that the epitope repertoire of gluten responsive T cells in Celiac Sprue patients has not been thoroughly mapped as yet. Alternatively, there may exist non-T cell mediated mechanisms that contribute to the overall gluten enteropathy in Celiac Sprue patients. A few such mechanisms have been proposed based upon the identification of gluten peptides that lack TH1 reactivity, but are capable of triggering inflammatory reactions in intestinal biopsies derived from Celiac Sprue patients 34–37. For example, the 13-residue sequence, LGQQQPFPPQQPY, is found in putative physiological peptides (LG)QQQPFPPQQPYPQPQPFPS among α-gliadins, and is known to elicit an innate immune response. However, in no case has a primary molecular target been identified that interacts with the gluten peptide. It is possible that identification of such targets would require use of the physiological forms of these peptides. Finally, gluten sensitivity is occasionally diagnosed in patients who do not have Celiac Sprue 38. It is conceivable that one or more of these proteolytically resistant peptides plays a crucial role in eliciting a pathological response in those patients. Regardless of the precise mechanism of pathogenicity, peptide length and a high proline content are likely to be common denominators of gluten intolerance. If so, the use of an oral prolyl endopeptidase may be appropriate therapy for such patients, as proposed recently 14, 17, 18, 20, 21.
The limitations of the above computational analysis of the physiological fate of gluten must be acknowledged. For example, such analysis does not consider the effect of gastric pepsin on food. Similarly, it does not incorporate complexities resulting from the extensive disulfide crosslinked structure of glutenin, or other secondary structural features of gluten peptides that influence their proteolytic susceptibility. Ultimately, a thorough analysis of gluten proteolysis using experimental proteomic methods will be required in order to obtain a quantitatively accurate picture of the physiological process of gluten digestion. Notwithstanding these limitations, our results have clearly reinforced the notion that protease stability and immunotoxicity of gluten are intimately correlated. This has implications for protein engineering of wheat strains with reduced antigenicity for Celiac Sprue patients 26, 39, and for the development of high-throughput screening tools for identifying and engineering therapeutically useful enzymes 10, 12, 22, 32, 40.
Synopsis
The link between digestive resistance and inflammatory character of gluten in Celiac Sprue was further validated. Deletion mutations confirmed the central role of the 33-mer in T cell response. Proteolysis of γ-gliadin resulted in a 26-mer with enhanced T cell antigenicity. In silico analysis of the gluten proteome found over 60 putative physiological, stable, TG2-reactive, and multivalent peptides. Prolyl endopeptidase abolished the antigenicity of the 33-mer and the 26-mer peptides.
Graphic.
Table S1 (B).
Computational predictions of physiological DQ2- antigenic peptides from gluten proteins in wheat, barley, and rye. These peptide sequences are from pancreatic proteolysis of chymotrypsin, trypsin, elastase and carboxypeptidases. The known DQ-2 restricted T cell epitopes are bolded in the sequences. The α-2 and γ-5 gliadin peptides are colored red. Each peptide’s residue positions from the original protein sequence obtained from NCBI Protein database are indicated. The accession number of each protein is indicated in their names.
| Name | Position | Peptide Fragments with Chymotrypsin, Trypsin, Elastase, Carboxypeptidases Cleavage Predictions and Known T-Cell Epitopes | # Epitopes |
|---|---|---|---|
| > CAB76964_Alpha-2 Gliadin | 60 – 89 | QPFPQPQLPYPQPQLPYPQPQLPYPQPQPF | >3 |
| > CAC94871_Gamma-5 Gliadin | 28 – 56 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 |
| 61 – 89 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | ||
| > CAB76961_Alpha-8 Gliadin | 34 – 52 | QQQPFPPQQPYPQPQPFPS | |
| > CAB76958_Alpha-4 Gliadin | 34 – 52 | QQQPFPPQQPYPQPQPFPS | |
| > CAB76962_Alpha Gliadin-10 | none | ||
| > CAB76956_Alpha-7 Gliadin | 34 – 52 | QQQPFPPQQPYPQPQPFPS | |
| > CAB76955_Alpha-9 Gliadin | 34 – 52 | QQQPFPPQQPYPQPQPFPS | |
| > CAB76957_Alpha-1 Gliadin | 35 – 53 | QQEQFPPQQPYPHQQPFPS | |
| 54 – 95 | QQPYPQPQPFPPQLPYPQTQPFPPQQPYPQPQPQYPQPQQPI | ||
| > CAB76954_Alpha-6 Gliadin | 54 – 95 | QQPYPQPQPFPPQLPYPQTQPFPPQQPYPQPQPQYPQPQQPI | |
| > CAB76960_Alpha-3 Gliadin | 34 – 52 | QQQPFPPQQPYPQPQPFPS | |
| > CAB76959_Alpha-5 Gliadin | 34 – 52 | QQQPFPPQQPYPQPQPFPS | |
| > AAA17741_Alpha Gliadin | 53 – 71 | QQQPFPPQQPYPQPQPFPS | |
| > CAA10257_Alpha Gliadin | 53 – 71 | QQQPFPPQQPYPQPQPFPS | |
| > BAA12318_Alpha Gliadin | 33 – 51 | QQQPFPPQQPYPQPQPFPS | |
| > T06500_Alpha Gliadin A-IV Precursor | 79 – 101 | QPFPQPQLPYPQPQLPYPQPQPF | >3 |
| 53 – 71 | QQQPFPPQQPYPQPQPFPS | ||
| > AAA96276_Alpha Gliadin | 53 – 70 | QQQPFPPQQPYPQPQFPS | |
| > D22364_alpha-gliadin precursor (clone A735) | 53 – 71 | QQQPFPPQQPYPQPQPFPS | |
| 79 – 106 | QPFPQPQPFPPQLPYPQPQLPYPQPQPF | 2 | |
| > P04727_Alpha-gliadin clone PW8142 precursor | 50 – 68 | QQQQFPPQQPYPQPQPFPS | |
| > P04725_Alpha-gliadin A-V precursor | 53 – 71 | QQQQFPPQQPYPQPQPFPS | |
| 79 – 98 | QPFPQPQPFPPQLPYPQPQS | ||
| > AAA96522_Alpha Gliadin | 50 – 68 | QQQQFPPQQPYPQPQPFPS | |
| > AAA96524_Alpha Gliadin | none | ||
| > AAA96523_Alpha Gliadin | 53 – 71 | QQQPFPPQQPYPQPQPFPS | |
| > AAQ63862_Gamma Gliadin | 28 – 51 | QQPQQIFPQPQQTFPHQPQQPFPQ | |
| 53 – 73 | QQPQQPFPQPQQQFPQPQQPQ | 2 | |
| > AAQ63865_Gamma Gliadin | 28 – 56 | QQPQQIFPQPQQTFPHQPQQQFPQPQQPQ | |
| 64 – 94 | QPFPQQPQQPYPQQPQQPFPQTQQPQQPFPQ | >3 | |
| > AAQ63864_Gamma Gliadin | 28 – 61 | QQPQQIFPQPQQPYPQQPQQPFPQTQQPQQPFPQ | >3 |
| > AAF42989_Gamma Gliadin | 26 – 44 | QQPQQTFPQPQQTFPHQPQ | |
| 59 – 120 | QPQQPFPQQPQQPFPQTQQPQQPFPQQPQQPFPQTQQPQQPFPQQPQQPFPQTQQPQQPFPL | >3 | |
| 123 – 143 | QQPQQPFPQPQQQLPQPQQPQ | 1 | |
| > AAK84779_Gamma Gliadin | 45 – 63 | QQPQQTFPQPQQTFPHQPQ | |
| 79 – 140 | QPQQPFPQQPQQPFPQTQQPQQPFPQQPQQPFPQTQQPQQPFPQQPQQPFPQTQQPQQPFPQ | >3 | |
| 142 – 162 | QQPQQPFPQPQQQLPQPQQPQ | 1 | |
| > CAB75404_Gamma-1 Gliadin | 28 – 46 | QQPQQTFPQPQQTFPHQPQ | |
| 60 – 89 | IQPQQPQQTYPQRPQQPFPQTQQPQQPFPQ | 1 | |
| 91 – 111 | QQPQQPFPQPQQQFPQPQQPQ | 2 | |
| > CAC94868_Gamma-2 Gliadin | 28 – 56 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | |
| 61 – 89 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| > AAK84773_Gamma Gliadin | 30 – 78 | QWPQQQQPFPQPQQPQQPFPQPQQPQLPFPQQPQQPFPQPQQPQQPFPQ | 3 |
| 80 – 103 | QQPQQPLPQPQQPQQPFPQQQQPL | 1 | |
| > AAK84780_Gamma Gliadin | 1 – 38 | QPFPQPQQTFPQQPQLPFPQQPQQPFPQPQQPQQPFPQ | 2 |
| 40 – 60 | QQPQQPFPQPQQQFPQPQQPQ | 2 | |
| > M36999_Gamma Gliadin | 60 – 127 | HHQPQQTFPQPQQTYPHQPQQQFPQTQQPQQPFPQPQQTFPQQPQLPFPQQPQQPFPQPQQPQQPFPQ | 3 |
| 129 – 149 | QQPQQPFPQPQQQFPQPQQPQ | 2 | |
| > AAQ63868_Hordein (leaf) | none | ||
| > S07975_hordein B3 (clone pB7) | 23 – 47 | QPQQPQPFPQQPIPQQPQPYPQQPQ | |
| > CAA60681_hordein B1 | 20 – 51 | QQQPFPQQPIPQQPQPYPQQPQPYPQQPFPPQ | |
| > 1103203A_hordein | 23 – 47 | QPQQPQPFPQQPIPQQPQPYPQQPQ | |
| > P80198_Gamma-hordein 3 | 36 – 82 | QQEPEQPYPQQQPLPQQQPFPQQPQLPHQHQFPQQLPQQQFPQQMPL | 1 |
| > BAB78747_Glutenin | 41 – 61 | HHHQQQPIQQQPHQFPQQQPC | |
| > BAB78749_Glutenin | none | ||
| > AAS66085_glutenin pGM107 | none | ||
| > AAV92073_Glutenin (in press) | none | ||
| > AAV92049_Glutenin (in press) | none | ||
| > AAV92036_Glutenin (in press) | none | ||
| > AAV91997_Glutenin | 150 – 168 | QQPPFPQQHQQFPQQQIPV | |
| > AAS10189_Glutenin | none | ||
| > S18236_secalin clone pSec2B | 76 – 100 | PIQPQQPFPQQPQQPFPQPQQQLPL | 2 |
| 101 – 118 | QPQQPFPQPQQPIPQQPQ | ||
| 121 – 150 | FPQQPQRPEQQFPQQPQQIIPQQTQQPFPL | ||
| 228 – 255 | QQPQRPQQPFPQQPQQIIPQQPQQPFPL | 1 | |
| > S18235_secalin clone pSec1B | 63 – 100 | QPQQPFPQPQQPTPIQPQQPFPQRPQQPFPQPQQQLPL | 2 |
| 121 – 150 | FPQQPQRPEQQFPQQPQQIIPQQTQQPFPL | 1 | |
| 228 – 255 | QQPQRPQQPFPQQPQQIIPQQPQQPFPL | ||
| > CAA26449_secalin unnamed | 5 – 21 | FPLPQQPFPQPQQPFPQ | |
| > AAB58403_secalin omega | 63 – 91 | QPQQPFPQPQQPTPIQPQQPFPQQPQQPF | |
| 121 – 150 | FPQQPQRPEQQFPQQPQQIIPQQTQQPFPL | ||
| 187 – 208 | QPQQPFPQQPEQIIPQQPQQPS | ||
| 228 – 255 | QQPQRPQQPFPQQPQQIIPQQPQQPFPL | 1 | |
| > AAG35598_secalin omega | 68 – 119 | PQPQQPYPQQPFPQQPQQPYPQQPQQPFPQQPQQPYPQQPQQQFPQQPQQPV | >3 |
| 141 – 159 | QQFPQQPQQPFPQQPQQPV | 2 | |
| 165 – 185 | QQPFPQTQQPQQPFPQPQQPQ | 1 | |
| 138 – 160 | QQPQQPFPQPQQPQQPFPQPQPQ | 2 | |
| > AAV86084_secalinGlutenin | 152 – 170 | QQPPFPQQHQQFPQQQIPV | |
| > AAV86085_secalinGlutenin | 77 – 95 | QQPPFPQQHQQFPQQQIPV | |
| > AAT74547_omegaGliadin | 39 – 56 | HQQQPFPQQPYPQQPYPS | |
| 91 – 116 | QPQQPFPQQPQQPQQPFPQPQQPFPW | 3 | |
| 132 – 154 | QPQQPFPQQPQQPFPQPQLPFPQ | 2 | |
| 271 – 297 | FPQQPQQPFPPTTTKPFPQQPQQPFPL | ||
| 234 – 252 | QPQQPFPQQPQQPFPQQPQ | 2 | |
| > AAT01617_omegaGliadin | none | ||
| > AAG17702_omegaGliadin | 82 – 116 | TQPQQPTPIQPQQPFPQQPQQPQQPFPQPQQPFPW | 3 |
| 132 – 154 | QPQQPFPQQPQQPFPQPQLPFPQ | 2 | |
| 171 – 193 | QPQQPFPQQPQQPFPQPQQPIPV | 2 | |
| 235 – 253 | QPQQPFPQQPQQPFPQQPQ | 2 | |
| > S52390_D_hordein precursor | none | ||
| > S18350_hordein | 84 – 106 | QPQQPFPQQPQQPLPRPQQPFPW | 1 |
| 107 – 140 | QPQQPFPQPQEPIPQQPQQPFPQQPQQPFPQQPQ | 2 | |
| 208 – 227 | QPQPFPQQPQQPLPQPQQPF | ||
| > T06211_C-hordein precursor | 84 – 106 | QPQQPFPQQPQQPLPRPQQPFPW | 1 |
| 137 – 160 | QQPQQPFPQQPQQPFPQPQQPFPW | 3 | |
| 217 – 235 | QPQQPFPQQPQQPFPQQPQ | 2 | |
| > S20519_B-hordein | 20 – 48 | QQQPFPQQPFPQQPQPYPQQPQPYPQQPF | 1 |
| 49 – 83 | QPQQPFPQQTIPQQPQPYPQQPFPPQQEFPQQPPF | ||
| > S07365_B1-hordein | 20 – 59 | QQQPFPQQPIPQQPQPYPQQPQPYPQQPFPPQQPFPQQPV | |
| 60 – 83 | PQQPQPYPQQPFPPQQPFPQQPPF | ||
| > P17990_hordeinGamma 1 | 49 – 69 | QQQFPQPQQPFPQQPQQPFPQ | 3 |
| 76 – 94 | QQPQHQFPQPTQQFPQRPL | ||
| > P06471_B3-hordein | 23 – 47 | QPQQPQPFPQQPIPQQPQPYPQQPQ | |
| > P06470_B1-hordein | 20 – 59 | QQQPFPQQPIPQQPQPYPQQPQPYPQQPFPPQQPFPQQPV | |
| 60 – 82 | QQPQPYPQQPFPPQQPFPQQPPF | ||
| > AAU06229_hordeinB | 32 – 78 | PWPQQPFPQQPYPQQPQPYPQQPQPYPQQPYPQQPQPYPQQPQPYPS 40 | 2 |
| > AAU06228_hordeinB | 32 – 52 | PWPQQPFPQQPYPQQPQPYPQ | 1 |
| > AAU06227_hordeinB | 32 – 62 | PWPQQPFPQQPYPQQPQPYPQQPQPYPQQPY | 1 |
| 63 – 85 | LPQPQPYPQQPQPYPQQPQPYPS | ||
| > AAQ63849_hordeinGamma3 | 50 – 66 | QQQQLPQQHPFPQQMPL | |
| > AAQ63848_hordeinGamma3 | 67 – 83 | QQQQLPQQHPFPQQMPL | |
| > AAQ63847_hordeinGamma3 | 67 – 83 | QQQQLPQQHPFPQQMPL | |
| > AAQ63842_hordeinGamma3 | 67 – 83 | QQQQLPQQHPFPQQMPL | |
| > AAP31051_D-hordein | none | ||
| > T04474_B1-hordein | 20 – 51 | QQQPFPQQPIPQQPQPYPQQPQPYPQQPFPPQ | |
| > BAA11642_D-hordein | none | ||
| > AAB28161_C-hordein | 84 – 106 | QPQQPFPQQPQQPLPRPQQPFPW | 1 |
| 136 – 159 | QQPQQPFPQQPQQPFPQPQQPFPW | 3 | |
| 216 – 234 | QPQQPFPQQPQQPFPQQPQ | 2 | |
| > 1604464A_gammaHordein | 48 – 68 | QQQFPQPQQPFPQQPQQPFPQ | 3 |
| 76 – 94 | QQPQHQFPQPTQQFPQRPL | ||
| > AAA92333_C-hordein | 92 – 117 | QPQPPQQPFPQPQQPNPQQPQQPFPR | |
| 124 – 146 | PQQPQQPFPQQPQQPFPQPQQPF | 3 | |
| 194 – 210 | QQPQQTIPQQPQQPFPL | ||
| 211 – 229 | QPQQPFPQQPQQPLPQQPQ | 1 | |
| 242 – 259 | QPQQPFPQPQPFPQEQPQ | ||
| > CAA59104_D-hordein | none | ||
| > CAA26889_hordein unnamed | 20 – 59 | QQQPFPQQPIPQQPQPYPQQPQPYPQQPFPPQQPFPQQPV | |
| 60 – 83 | PQQPQPYPQQPFPPQQPFPQQPPF | ||
| > CAA42642_hordein unnamed | 84 – 106 | QPQQPFPQQPQQPLPRPQQPFPW | 1 |
| 107 – 140 | QPQQPFPQPQEPIPQQPQQPFPQQPQQPFPQQPQ | 2 | |
| 208 – 227 | QPQPFPQQPQQPLPQPQQPF | ||
| > CAA48209_hordeinD | none | ||
| > CAA51204_gamma3-hordein | 33 – 79 | QQEPEQPYPQQQPLPQQQPFPQQPQLPHQHQFPQQLPQQQFPQQMPL | 1 |
| > CAI35909_alpha-precursor | 68 – 86 | QQQPFPPQQPYPQPQPFPS | |
| > T06282_alpha-precursor | none | ||
| > AAA96525_alpha | none | ||
| > EEWTG_gamma-precursor | 46 – 74 | QQPQQIFPQPQQTFPHQPQQQFPQPQQPQ | |
| > P21292_gamma-precursor | 60 – 126 | HQPQQTFPQPQQTYPHQPQQQFPQTQQPQQPFPQPQQTFPQQPQLPFPQQPQQPFPQPQQPQQPFPQ | 3 |
| 129 – 149 | QQPQQPFPQPQQQFPQPQQPQ | 2 | |
| > P06659_gamma-precursor | 46 – 74 | QQPQQIFPQPQQTFPHQPQQQFPQPQQPQ | |
| 82 – 112 | QPFPQQPQQPYPQQPQQPFPQTQQPQQPFPQ | >3 | |
| > P04730_gamma | none | ||
| > P08453_gamma-precursor | 46 – 67 | QQPQQTFPQPQQTFPHQPQQQV | |
| 79 – 140 | QPQQPFPQQPQQPFPQTQQPQQPFPQQPQQPFPQTQQPQQPFPQQPQQPFPQTQQPQQPFPQ | >3 | |
| 142 – 162 | QQPQQPFPQPQQQLPQPQQPQ | 1 | |
| > P04729_gamma-precursor | none | ||
| > P08079_gamma | 46 – 74 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | |
| 79 – 107 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| > AAQ63863_gamma | 64 – 94 | QPFPQQPQQPYPQQPQQPFPQTQQPQQPFPQ | >3 |
| > AAQ63861_gamma | 28 – 57 | QQPQQIFPQPQQTFPHQPQQQFPQPQQPQR | |
| 64 – 94 | QPFPQQPQQPYPQQPQQPFPQTQQPQQPFPQ | >3 | |
| > AAQ63860_gamma | 28 – 46 | QQPQQTFPQPQQTFPHQPQ | |
| 60 – 93 | QPQQPFPQQPQQTYPQRPQQPFPQTQQPQQPFPQ | 2 | |
| 96 – 116 | QQPQQPFPQPQQQFPQPQQPQ | 2 | |
| > AAQ63859_gamma | 36 – 64 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 |
| > AAQ63858_gamma | 28 – 46 | QQPQQTFPQPQQTFPHQPQ | |
| 61 – 122 | QPQQPFPQQPQQPFPQTQQPQQPFPQQPQQPFPQTQQPQQPFPQQPQQPFPQTQQPQQPFPQ | >3 | |
| 124 – 144 | QQPQQPFPQPQQQLPQPQQPQ | 1 | |
| > AAQ63857_gamma | 28 – 46 | QQPQQIFPQPQQTFPHQPQ | |
| 60 – 85 | QPQQPFPQQPQQPFPQTQQPQQPFPQ | 3 | |
| 88 – 108 | QQPQQPFPQPQQQFPQPQQPQ | 2 | |
| > AAQ63856_gamma | 28 – 56 | QQPQQIFPQPQQTFPHQPQQQFPQPQQPQ | |
| 64 – 94 | QPFPQQPQQPYPQQPQQPFPQTQQPQQPFPQ | >3 | |
| > JS0402_gamma | 46 – 64 | QQPQQTFPQPQQTFPHQPQ | |
| 79 – 140 | QPQQPFPQQPQQPFPQTQQPQQPFPQQPQQPFPQTQQPQQPFPQQPQQPFPQTQQPQQPFPQ | >3 | |
| 142 – 162 | QQPQQPFPQPQQQLPQPQQPQ | 1 | |
| > JA0153_gamma | 60 – 127 | HHQPQQTFPQPQQTYPHQPQQQFPQTQQPQQPFPQPQQTFPQQPQLPFPQQPQQPFPQPQQPQQPFPQ | 3 |
| 129 – 149 | QQPQQPFPQPQQQFPQPQQPQ | 2 | |
| > CAA54925_gamma | 40 – 107 | HHQPQQTFPQPEQTYPHQPQQQFPQTQQPQQPFPQPQQTFPQQPQLPFPQQPQQPFPQPQQPQQPFPQ | 3 |
| 109 – 129 | QQPQQPFPQPQQQFPQPQQPQ | 2 | |
| > AAN32705_gamma | none | ||
| > CAC94870_gamma-4 | 28 – 56 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | |
| 72 – 89 | QPYPQQPQQPFPQTQQPQ | 2 | |
| > CAC94869_gamma-3 | 28 – 56 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | |
| 60 – 79 | LQPQQPFPQQPQQPYPQQPQ | 3 | |
| > AAK84778_gamma | 46 – 64 | QQPQQTFPQPQQTFPHQPQ | |
| 78 – 112 | IQPQQPFPQQPQQTYPQRPQQPFPQTQQPQQPFPQ | 2 | |
| 114 – 134 | QQPQQPFPQPQQQFPQPQQPQ | 2 | |
| > AAK84777_gamma | 60 – 127 | HHQPQQTFPQPQQTYPHQPQQQFPQTQQPQQPFPQPQQTFPQQPQLPFPQQPQQPFPQPQQPQQPFPQ | 3 |
| 129 – 149 | QQPQQPFPQPQQQFPQPQQPQ | 2 | |
| > AAK84776_gamma-13 | 46 – 91 | QQPQHTFPQPQQTCPHQPQQQFPQPQQPQQPFPQPQQPQQPFPQPQ | 2 |
| 94 – 117 | QLPFPQQPQQPLPQPQQPQQPFPQ | 1 | |
| > AAK84775_gamma | 1 – 28 | PQQPQLPFPQQPQQPFPQPQQPQQPFPQ | 2 |
| 30 – 50 | QQPQQPFPQPQQQFPQPQQPQ | 2 | |
| > AAK84774_gamma | 47 – 65 | QQPQQIFPQPQQTFPHQPQ | |
| 68 – 159 | FPQPQQTFPHQPQQQFPQPQQPQQPFPQQPQQQFPQPQQPQQPFPQQPQQQFPQPQQPQQPFPQPQQPQLPFPQQPQQPFPQPQQPQQPFPQ | >3 | |
| 161 – 184 | QQPQQPLPQPQQPQQPFPQQQQPL | 1 | |
| > AAK84772_gamma | 47 – 65 | QQPQQIFPQPQQTFPHQPQ | |
| 68 – 159 | FPQPQQTFPHQPQQQFPQPQQPQQPFPQQPQQQFPQPQQPQQPFPQQPQQQFPQPQQPQQPFPQPQQPQLPFPQQPQQPFPQPQQPQQPFPQ | >3 | |
| 161 – 184 | QQPQQPLPQPQQPQQPFPQQQQPL | 1 | |
| > AAD30440_gamma | 46 – 74 | QQPQQIFPQPQQTFPHQPQQQFPQPQQPQ | |
| 79 – 107 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| > CAC10619_gamma | 20 – 37 | QTFPHQPQQQFPQPQQPQ | |
| 48 – 70 | HQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| 77 – 103 | QQPQQPYPQQPQQPFPQTQQPQQQFPQ | >3 | |
| > CAC11097_gamma | 29 – 76 | FPQPQQPQQQFPQPQQPQQPFPQPQQPQLPFPQQPFPQPQQPQQPFPQ | 2 |
| 78 – 98 | QQPQQPFPQPQQQFPQPQQPQ | 2 | |
| > CAC11096_gamma | 29 – 76 | FPQPQQPQQQFPQPQQPQQPFPQPQQPQLPFPQQPFPQPQQPQQPFPQ | 2 |
| 78 – 98 | QQPQQPFPQPQQQFPQPQQPQ | 2 | |
| > CAC11099_gamma | 17 – 45 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | |
| 50 – 78 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| > CAC11098_gamma | 14 – 42 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | |
| 47 – 75 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| > CAC11089_gamma | 30 – 58 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | |
| 63 – 91 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| > CAC11088_gamma | 15 – 43 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | |
| 48 – 76 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| > CAC11087_gamma | 25 – 53 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | |
| 58 – 86 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| > CAC11080_gamma | 30 – 58 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | |
| 63 – 91 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| > CAC11079_gamma | 30 – 58 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | |
| 63 – 91 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| > CAC11078_gamma | 30 – 58 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | |
| 63 – 91 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| > CAC11065_gamma | 46 – 74 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | |
| 79 – 107 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| > CAC11064_gamma | 27 – 55 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | |
| 60 – 88 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| > CAC11057_gamma | 23 – 51 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | |
| 56 – 84 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| > CAC11056_gamma | 17 – 45 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | |
| 50 – 78 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| > CAC11055_gamma | 19 – 47 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | |
| 52 – 80 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| > CAC10624_gamma | 13 – 35 | PQPQQTFPHQQQQQQFPQPQQPQ | |
| 40 – 68 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| 75 – 104 | QQPQQPYPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| 111 – 137 | QQPQQPYPQQPQQPFPQTQQPQQQFPQ | >3 | |
| > CAC10623_gamma | 13 – 34 | PQPQQTFPHQPQQQFPQPQQPQ | |
| 39 – 67 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| 74 – 103 | QQPQQPYPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| 110 – 136 | QQPQQPYPQQPQQPFPQTQQPQQQFPQ | >3 | |
| > CAC10622_gamma | 8 – 36 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | |
| 41 – 69 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| 76 – 102 | QQPQQPYPQQPQQPFPQTQQPQQQFPQ | >3 | |
| > CAC10620_gamma | 20 – 37 | QTFPHQPQQQFPQPQQPQ | |
| 48 – 70 | HQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| 77 – 103 | QQPQQPYPQQPQQPFPQTQQPQQQFPQ | >3 | |
| > CAC10633_gamma | 9 – 37 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | |
| 42 – 70 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| > CAC10632_gamma | 42 – 70 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 |
| > CAC10631_gamma | 7 – 35 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | |
| 40 – 68 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| > CAC10630_gamma | 9 – 37 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | |
| 42 – 65 | QPQQPQQPYPQEPQQPFPQTQQPQ | 1 | |
| 77 – 97 | QQPQQQFPQPQQQFPQPQQPQ | 1 | |
| > CAC10629_gamma | 42 – 70 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 |
| > CAC10628_gamma | 9 – 37 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | |
| 42 – 65 | QPQQPQQPYPQEPQQPFPQTQQPQ | 1 | |
| 77 – 97 | QQPQQQFPQPQQQFPQPQQPQ | 1 | |
| > CAC10627_gamma | 9 – 37 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | |
| 42 – 65 | QPQQPQQPYPQEPQQPFPQTQQPQ | 1 | |
| 72 – 92 | QQPQQQFPQPQQQFPQPQQPQ | 1 | |
| > CAC10626_gamma | 46 – 74 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | |
| 79 – 107 | QPQQPFPQQQQQPYPQQPQQPFPQTQQPQ | 3 | |
| > CAC10625_gamma | 46 – 74 | QQPQQTFPQPQQTFPHQPQQQFPQPQQPQ | |
| 79 – 107 | QPQQPFPQQQQQPYPQQPQQPFPQTQQPQ | 3 | |
| > CAC10617_gamma | 30 – 60 | QKPQQTFPQPQQTFPFPHQPQQQFPQPQQPQ | |
| 65 – 88 | QPQQPQQPYPQQPQQTFPQTQQPQ | 2 | |
| > CAC10616_gamma | 30 – 58 | QQPQQTFPQPQQTFPHQPQQQIPQPQQPQ | |
| 63 – 91 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| 98 – 124 | QQPQQPYPQQPQQPFPQTQQPQQQFPQ | >3 | |
| > CAC10594_gamma | 30 – 58 | QQPQQTFPQPQQTFPHQPQQQIPQPQQPQ | |
| 63 – 91 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| 98 – 124 | QQPQQPYPQQPQQPFPQTQQPQQQFPQ | >3 | |
| > CAC10593_gamma | 30 – 58 | QQPQQTFPQPQQTFPHQPQQQIPQPQQPQ | |
| 63 – 91 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| 98 – 124 | QQPQQPYPQQPQQPFPQTQQPQQQFPQ | >3 | |
| > AAD30556_gamma | 46 – 74 | QQPQQIFPQPQQTFPHQPQQQFPQPQQPQ | |
| 79 – 107 | QPQQPFPQQPQQPYPQQPQQPFPQTQQPQ | >3 | |
| > AAF42989_gamma | 27 – 45 | QQPQQTFPQPQQTFPHQPQ | |
| 60 – 121 | QPQQPFPQQPQQPFPQTQQPQQPFPQQPQQPFPQTQQPQQPFPQQPQQPFPQTQQPQQPFPQ | >3 | |
| 123 – 143 | QQPQQPFPQPQQQLPQPQQPQ | 1 | |
| > BAA11251_gamma-precursor | 27 – 45 | QQPQQTFPQPQQTFPHQPQ | |
| 59 – 91 | PQQPFPQQPQQTYPQRPQQPFPQTQQPQQPFPQ | 2 | |
| 95 – 115 | QQPQQPFPQPQQQFPQPQQPQ | 2 | |
| > 1507333A_gamma | 59 – 126 | HHQPQQTFPQPQQTYPHQPQQQFPQTQQPQQPFPQPQQTFPQQPQLPFPQQPQQPFPQPQQPQQPFPQ | 3 |
| 128 – 148 | QQPQQPFPQPQQQFPQPQQPQ | 2 | |
| > 1209306A_gamma | none | ||
| > CAA37491_gamma | 60 – 126 | HQPQQTFPQPEQTYPHQPQQQFPQTQQPQQPFPQPQQTFPQQPQLPFPQQPQQPFPQPQQPQQPFPQ | 3 |
| 129 – 149 | QQPQQPFPQPQQQFPQPQQPQ | 2 | |
| > AAA34289_gamma | 46 – 64 | QQPQQTFPQPQQTFPHQPQ | |
| 79 – 140 | QPQQPFPQQPQQPFPQTQQPQQPFPQQPQQPFPQTQQPQQPFPQQPQQPFPQTQQPQQPFPQ | >3 | |
| > AAA34288_gamma | 46 – 74 | QQPQQIFPQPQQTFPHQPQQQFPQPQQPQ | |
| 82 – 112 | QPFPQQPQQPYPQQPQQPFPQTQQPQQPFPQ | >3 | |
| > AAA34287_gamma | none | ||
| > AAA34286_gamma | none | ||
| > AAA34285_gamma-precursor | none | ||
| > AAA34274_gamma-precursor | 46 – 74 | QQPQQIFPQPQQTFPHQPQQQFPQPQQPQ | |
| 82 – 112 | QPFPQQPQQPYPQQPQQPFPQTQQPQQPFPQ | >3 | |
| > AAA34272_gamma-precursor | 60 – 127 | HHQPQQTFPQPQQTYPHQPQQQFPQTQQPQQPFPQPQQTFPQQPQLPFPQQPQQPFPQPQQPQQPFPQ | 3 |
| 129 – 149 | QQPQQPFPQPQQQFPQPQQPQ | 2 |
Acknowledgments
This research is supported by a grant (DK063158) from the NIH and by the Research Council of Norway. L.S. is a recipient of a Stanford Graduate Fellowship. We thank Marie K. Johannesen for excellent technical assistance.
References
- 1.Schuppan D. Current concepts of celiac disease pathogenesis. Gastroenterology. 2000;119(1):234–42. doi: 10.1053/gast.2000.8521. [DOI] [PubMed] [Google Scholar]
- 2.Sollid LM. Coeliac disease: dissecting a complex inflammatory disorder. Nat Rev Immunol. 2002;2(9):647–55. doi: 10.1038/nri885. [DOI] [PubMed] [Google Scholar]
- 3.Sollid LM. Molecular basis of celiac disease. Annu Rev Immunol. 2000;18:53–81. doi: 10.1146/annurev.immunol.18.1.53. [DOI] [PubMed] [Google Scholar]
- 4.Maki M, Collin P. Coeliac disease. Lancet. 1997;349(9067):1755–9. doi: 10.1016/S0140-6736(96)70237-4. [DOI] [PubMed] [Google Scholar]
- 5.Corrao G, Corazza GR, Bagnardi V, Brusco G, Ciacci C, Cottone M, Sategna Guidetti C, Usai P, Cesari P, Pelli MA, Loperfido S, Volta U, Calabro A, Certo M. Mortality in patients with coeliac disease and their relatives: a cohort study. Lancet. 2001;358(9279):356–61. doi: 10.1016/s0140-6736(01)05554-4. [DOI] [PubMed] [Google Scholar]
- 6.Molberg O, McAdam SN, Korner R, Quarsten H, Kristiansen C, Madsen L, Fugger L, Scott H, Noren O, Roepstorff P, Lundin KE, Sjostrom H, Sollid LM. Tissue transglutaminase selectively modifies gliadin peptides that are recognized by gut-derived T cells in celiac disease. Nat Med. 1998;4(6):713–7. doi: 10.1038/nm0698-713. [DOI] [PubMed] [Google Scholar]
- 7.van de Wal Y, Kooy Y, van Veelen P, Pena S, Mearin L, Papadopoulos G, Koning F. Selective deamidation by tissue transglutaminase strongly enhances gliadin-specific T cell reactivity. J Immunol. 1998;161(4):1585–8. [PubMed] [Google Scholar]
- 8.Arentz-Hansen H, Korner R, Molberg O, Quarsten H, Vader W, Kooy YM, Lundin KE, Koning F, Roepstorff P, Sollid LM, McAdam SN. The intestinal T cell response to alpha-gliadin in adult celiac disease is focused on a single deamidated glutamine targeted by tissue transglutaminase. J Exp Med. 2000;191(4):603–12. doi: 10.1084/jem.191.4.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson RP, Degano P, Godkin AJ, Jewell DP, Hill AV. In vivo antigen challenge in celiac disease identifies a single transglutaminase-modified peptide as the dominant A-gliadin T-cell epitope. Nat Med. 2000;6(3):337–42. doi: 10.1038/73200. [DOI] [PubMed] [Google Scholar]
- 10.Hausch F, Shan L, Santiago NA, Gray GM, Khosla C. Intestinal digestive resistance of immunodominant gliadin peptides. Am J Physiol Gastrointest Liver Physiol. 2002;283(4):G996–G1003. doi: 10.1152/ajpgi.00136.2002. [DOI] [PubMed] [Google Scholar]
- 11.Piper JL, Gray GM, Khosla C. High selectivity of human tissue transglutaminase for immunoactive gliadin peptides: implications for celiac sprue. Biochemistry. 2002;41(1):386–93. doi: 10.1021/bi011715x. [DOI] [PubMed] [Google Scholar]
- 12.Shan L, Molberg O, Parrot I, Hausch F, Filiz F, Gray GM, Sollid LM, Khosla C. Structural basis for gluten intolerance in celiac sprue. Science. 2002;297(5590):2275–9. doi: 10.1126/science.1074129. [DOI] [PubMed] [Google Scholar]
- 13.Sjostrom H, Lundin KE, Molberg O, Korner R, McAdam SN, Anthonsen D, Quarsten H, Noren O, Roepstorff P, Thorsby E, Sollid LM. Identification of a gliadin T-cell epitope in coeliac disease: general importance of gliadin deamidation for intestinal T-cell recognition. Scand J Immunol. 1998;48(2):111–5. doi: 10.1046/j.1365-3083.1998.00397.x. [DOI] [PubMed] [Google Scholar]
- 14.Arentz-Hansen H, Fleckenstein B, Molberg O, Scott H, Koning F, Jung G, Roepstorff P, Lundin KE, Sollid LM. The molecular basis for oat intolerance in patients with celiac disease. Plos Med. 2004;1(1):e1. doi: 10.1371/journal.pmed.0010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qiao, S. W.; Bergseng, E.; Molberg, Ø.; Jung, G.; Fleckenstein, B.; Sollid, L. M., Refining the rules of gliadin T cell epitope binding to the disease associated DQ2 molecule in celiac disease; importance of proline spacing and glutamine deamidation. Journal of Immunology, in press 2005. [DOI] [PubMed]
- 16.Arentz-Hansen H, McAdam SN, Molberg O, Fleckenstein B, Lundin KE, Jorgensen TJ, Jung G, Roepstorff P, Sollid LM. Celiac lesion T cells recognize epitopes that cluster in regions of gliadins rich in proline residues. Gastroenterology. 2002;123(3):803–9. doi: 10.1053/gast.2002.35381. [DOI] [PubMed] [Google Scholar]
- 17.Piper JL, Gray GM, Khosla C. Effect of prolyl endopeptidase on digestive-resistant gliadin peptides in vivo. J Pharmacol Exp Ther. 2004 doi: 10.1124/jpet.104.068429. [DOI] [PubMed] [Google Scholar]
- 18.Vader LW, de Ru A, van der Wal Y, Kooy YM, Benckhuijsen W, Mearin ML, Drijfhout JW, van Veelen P, Koning F. Specificity of tissue transglutaminase explains cereal toxicity in celiac disease. J Exp Med. 2002;195(5):643–9. doi: 10.1084/jem.20012028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fleckenstein B, Molberg O, Qiao SW, Schmid DG, von der Mulbe F, Elgstoen K, Jung G, Sollid LM. Gliadin T cell epitope selection by tissue transglutaminase in celiac disease. Role of enzyme specificity and pH influence on the transamidation versus deamidation process. J Biol Chem. 2002;277(37):34109–16. doi: 10.1074/jbc.M204521200. [DOI] [PubMed] [Google Scholar]
- 20.Vader W, Stepniak D, Kooy Y, Mearin L, Thompson A, van Rood JJ, Spaenij L, Koning F. The HLA-DQ2 gene dose effect in celiac disease is directly related to the magnitude and breadth of gluten-specific T cell responses. Proc Natl Acad Sci U S A. 2003;100(21):12390–5. doi: 10.1073/pnas.2135229100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quarsten H, Molberg O, Fugger L, McAdam SN, Sollid LM. HLA binding and T cell recognition of a tissue transglutaminase-modified gliadin epitope. Eur J Immunol. 1999;29(8):2506–14. doi: 10.1002/(SICI)1521-4141(199908)29:08<2506::AID-IMMU2506>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 22.Marti T, Molberg O, Li Q, Gray GM, Khosla C, Sollid LM. Prolyl endopeptidase-mediated destruction of T cell epitopes in whole gluten: chemical and immunological characterization. J Pharmacol Exp Ther. 2005;312(1):19–26. doi: 10.1124/jpet.104.073312. [DOI] [PubMed] [Google Scholar]
- 23.Ahnen DJ, Santiago NA, Cezard JP, Gray GM. Intestinal aminooligopeptidase. In vivo synthesis on intracellular membranes of rat jejunum. J Biol Chem. 1982;257(20):12129–35. [PubMed] [Google Scholar]
- 24.Kim CY, Quarsten H, Bergseng E, Khosla C, Sollid LM. Structural basis for HLA-DQ2-mediated presentation of gluten epitopes in celiac disease. Proc Natl Acad Sci U S A. 2004;101(12):4175–9. doi: 10.1073/pnas.0306885101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arentz-Hansen EH, McAdam SN, Molberg O, Kristiansen C, Sollid LM. Production of a panel of recombinant gliadins for the characterisation of T cell reactivity in coeliac disease. Gut. 2000;46(1):46–51. doi: 10.1136/gut.46.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Molberg O, Uhlen AK, Jensen T, Flaete NS, Fleckenstein B, Arentz-Hansen H, Raki M, Lundin KE, Sollid LM. Mapping of gluten T-cell epitopes in the bread wheat ancestors: implications for celiac disease. Gastroenterology. 2005;128(2):393–401. doi: 10.1053/j.gastro.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 27.Vader W, Kooy Y, Van Veelen P, De Ru A, Harris D, Benckhuijsen W, Pena S, Mearin L, Drijfhout JW, Koning F. The gluten response in children with celiac disease is directed toward multiple gliadin and glutenin peptides. Gastroenterology. 2002;122(7):1729–37. doi: 10.1053/gast.2002.33606. [DOI] [PubMed] [Google Scholar]
- 28.Erickson RH, Kim YS. Digestion and absorption of dietary protein. Annu Rev Med. 1990;41:133–9. doi: 10.1146/annurev.me.41.020190.001025. [DOI] [PubMed] [Google Scholar]
- 29.Konigsberg W, Goldstein J, Hill RJ. The structure of human hemoglobin. VII. The digestion of the beta chain of human hemoglobin with pepsin. J Biol Chem. 1963;238:2028–33. [PubMed] [Google Scholar]
- 30.Qiao SW, Bergseng E, Molberg O, Xia J, Fleckenstein B, Khosla C, Sollid LM. Antigen presentation to celiac lesion-derived T cells of a 33-mer gliadin peptide naturally formed by gastrointestinal digestion. J Immunol. 2004;173(3):1757–62. doi: 10.4049/jimmunol.173.3.1757. [DOI] [PubMed] [Google Scholar]
- 31.Xia J, Sollid LM, Khosla C. Equilibrium and kinetic analysis of the unusual binding behavior of a highly immunogenic gluten peptide to HLA-DQ2. Biochemistry. 2005;44(11):4442–9. doi: 10.1021/bi047747c. [DOI] [PubMed] [Google Scholar]
- 32.Shan L, Marti T, Sollid LM, Gray GM, Khosla C. Comparative biochemical analysis of three bacterial prolyl endopeptidases: implications for coeliac sprue. Biochem J. 2004;383(Pt 2):311–8. doi: 10.1042/BJ20040907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matysiak-Budnik T, Candalh C, Dugave C, Namane A, Cellier C, Cerf-Bensussan N, Heyman M. Alterations of intestinal transport and processing of gliadin peptides in celiac disease. Gastroenterology. 2003;125(3):696–707. doi: 10.1016/s0016-5085(03)01049-7. [DOI] [PubMed] [Google Scholar]
- 34.Maiuri L, Ciacci C, Ricciardelli I, Vacca L, Raia V, Auricchio S, Picard J, Osman M, Quaratino S, Londei M. Association between innate response to gliadin and activation of pathogenic T cells in coeliac disease. Lancet. 2003;362(9377):30–7. doi: 10.1016/S0140-6736(03)13803-2. [DOI] [PubMed] [Google Scholar]
- 35.Meresse B, Chen Z, Ciszewski C, Tretiakova M, Bhagat G, Krausz TN, Raulet DH, Lanier LL, Groh V, Spies T, Ebert EC, Green PH, Jabri B. Coordinated induction by IL15 of a TCR-independent NKG2D signaling pathway converts CTL into lymphokine-activated killer cells in celiac disease. Immunity. 2004;21(3):357–66. doi: 10.1016/j.immuni.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 36.Hue S, Mention JJ, Monteiro RC, Zhang S, Cellier C, Schmitz J, Verkarre V, Fodil N, Bahram S, Cerf-Bensussan N, Caillat-Zucman S. A direct role for NKG2D/MICA interaction in villous atrophy during celiac disease. Immunity. 2004;21(3):367–77. doi: 10.1016/j.immuni.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 37.Tuckova L, Novotna J, Novak P, Flegelova Z, Kveton T, Jelinkova L, Zidek Z, Man P, Tlaskalova-Hogenova H. Activation of macrophages by gliadin fragments: isolation and characterization of active peptide. J Leukoc Biol. 2002;71(4):625–31. [PubMed] [Google Scholar]
- 38.Hadjivassiliou M, Williamson CA, Woodroofe N. The immunology of gluten sensitivity: beyond the gut. Trends Immunol. 2004;25(11):578–82. doi: 10.1016/j.it.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 39.Vader LW, Stepniak DT, Bunnik EM, Kooy YM, de Haan W, Drijfhout JW, Van Veelen PA, Koning F. Characterization of cereal toxicity for celiac disease patients based on protein homology in grains. Gastroenterology. 2003;125(4):1105–13. doi: 10.1016/s0016-5085(03)01204-6. [DOI] [PubMed] [Google Scholar]
- 40.Shan L, Mathews II, Khosla C. Structural and mechanistic analysis of two prolyl endopeptidases: role of interdomain dynamics in catalysis and specificity. Proc Natl Acad Sci U S A. 2005;102(10):3599–604. doi: 10.1073/pnas.0408286102. [DOI] [PMC free article] [PubMed] [Google Scholar]


