Abstract
The phytochrome family of red/far-red photoreceptors has been optimized to support photochemical isomerization of a bound bilin chromophore, a process that triggers a conformational change and modulates biochemical output from the surrounding protein scaffold. Recent studies have established that the efficiency of this photochemical process is profoundly altered by mutation of a conserved tyrosine residue (Tyr176) within the bilin-binding GAF domain of the cyanobacterial phytochrome Cph1 [Fischer, A. J., and Lagarias, J. C. (2004) Harnessing phytochrome’s glowing potential, Proc. Natl. Acad. Sci. U.S.A. 101, 17334–17339]. Here, we show that the equivalent mutation in plant phytochromes behaves similarly, indicating that the function of this tyrosine in the primary photochemical mechanism is conserved. Saturation mutagenesis of Tyr176 in Cph1 establishes that no other residue can support comparably efficient photoisomerization. The spectroscopic consequences of Tyr176 mutations also reveal that Tyr176 regulates the conversion of the porphyrin-like conformation of the bilin precursor to a more extended conformation. The porphyrin-binding ability of the Tyr176Arg mutant protein indicates that Tyr176 also regulates the ligand-binding specificity of apophytochrome. On the basis of the hydrogen-bonding ability of Tyr176 substitutions that support the nonphotochemical C15-Z,syn to C15-Z,anti interconversion, we propose that Tyr176 orients the carboxyl side chain of a conserved acidic residue to stabilize protonation of the bilin chromophore. A homology model of the GAF domain of Cph1 predicts a C5-Z,syn, C10-Z,syn, C15-Z,anti configuration for the chromophore and implicates Glu189 as the proposed acidic residue stabilizing the extended conformation, an interpretation consistent with site-directed mutagenesis of this conserved acidic residue.
Phytochromes are a widespread family of biliprotein photoreceptors specific for red and far-red light that were first identified in plants, where they modulate growth and development in response to changes in photosynthetically active radiation (1–6). Phytochromes and related photosensory proteins have also been identified in cyanobacteria, purple bacteria, nonphotosynthetic bacteria, and even fungi (7–15). Plant and microbial phytochromes incorporate linear tetrapyrrole molecules of different structures as their chromophores for sensing different shades of red and far-red light. Plant phytochromes (Phys)1 employ phytochromobilin (PΦB), while cyanobacterial phytochromes (Cphs) use phycocyanobilin (PCB), the same pigment that is used by phycobiliproteins for light harvesting (16–18). The bacteriophytochromes from non-oxygenic photosynthetic and heterotrophic eubacteria (BphPs) instead make use of biliverdin IXα (BV), the more oxidized bilin precursor from which plant and cyanobacterial phytochrome chromophores are derived (13, 19–21).
Absorption of a photon by the red-light-absorbing Pr form of phytochrome triggers the Z–E isomerization of the C15–C16 double bond of its bilin chromophore (22). This photoisomerization generates the spectrally distinct far-red-light-absorbing Pfr form, which can revert to Pr either upon absorption of a second photon or via a slow thermal process known as dark reversion. Numerous studies have established that the Pfr to Pr photoreversion reaction proceeds through spectrally distinct intermediates from the forward reaction (23, 24). The interconversion between Pr and Pfr ultimately triggers a conformational change that alters the biochemical activity of phytochrome’s C-terminal output region that is usually a protein kinase (8, 13, 18, 19, 25–28). For plant phytochromes, it is well-established that the N-terminal photosensory region transmits a second light signal via nuclear translocation and interaction with nuclear transcription factors (29–31). Phytochromes thereby transduce light signals via changes in gene expression, enabling long-term adaptation to changes in the light environment.
The photosensory input region of plant and microbial phytochromes typically consists of three evolutionarily related domains: a P2 domain that exhibits a weak homology to PAS domains, a P3 or bilin lyase domain recognized as a GAF domain, and a P4 or PHY domain [Figure 1A (32)]. Those phytochromes that employ BV, i.e., BphPs, possess a thioether linkage between the A-ring vinyl group of BV and a conserved Cys residue in the P2 domain (15, 20, 21, 33), while plant and cyanobacterial phytochromes utilize more reduced bilin chromophores, i.e., PCB and PΦB, that are instead linked to a conserved Cys in the P3 domain (16, 18). Interestingly, introduction of a Cys to the appropriate position in the P3 GAF domain of bacteriophytochromes permits covalent attachment of PCB (33, 34), indicating that plant and microbial phytochromes have considerable structural similarity in their bilin-binding pockets.
Figure 1.

Domain structure, chromophore configuration, and assembly of phytochromes. (A) Domain organization of the Cph1, BphP, and Phy subfamilies. The N-terminal photosensory region contains the P2 PAS domain, the P3 GAF bilin lyase domain, and the P4 PHY domain. Plant phytochromes (Phy) also possess a Ser/Thr-rich N-terminal P1 domain. The C-terminal regulatory region of these phytochromes contains a domain homologous to histidine kinase transmitter modules. Plant phytochrome C termini possess an additional inserted region that contains two additional PAS domains (labeled PAS1 and PAS2). Cys sites of covalent attachment to the bilin chromophore are indicated. (B) Conformations of the free chromophore precursor phycocyanobilin (PCB) and its protonated form during assembly. Rings are labeled in bold, and the carbons of the ring system are numbered. (Left) A cyclic, deprotonated C5 Z,syn, C10 Z,syn, C15 Z,syn configuration of the initially bound PCB is shown. (Center) Upon protonation of the ring system, PCB adopts a more extended conformation, here shown as C5 Z,syn, C10 Z,syn, C15 Z,anti. (Right) The covalent attachment of PCB to Cph1 results in a covalent thioether linkage to Cys259 with R stereochemistry (16). (C) Assembly reaction of PCB with wild-type phytochrome Cph1 is shown (38). After initially binding in a cyclic conformation, PCB becomes protonated and adopts a more extended conformation (“ext”). The thioether linkage is then formed (“ext, cov”). KD, initial binding constant for PCB and apoCph1; KPT, equilibrium constant for proton transfer and concomitant chromophore rearrangement; kchem, rate constant for covalent attachment.
It is generally accepted that the bilin prosthetic groups of phytochromes adopt semi-extended geometries in situ, but the overall conformation and protonation state of the Pr and Pfr chromophores has remained controversial until recently. Heteronuclear NMR spectroscopic analysis of the cyanobacterial phytochrome Cph1 from Synechocystis sp. PCC 6803 has established that the B/C-ring system of Pr and Pfr chromophores are both protonated (35), corroborating earlier vibrational spectroscopic investigations on plant phytochromes (36, 37). Recent holoprotein assembly studies on Cph1 using stopped-flow spectroscopy indicate that PCB adopts a cyclic or porphyrin-like C5-Z,syn, C10-Z,syn, C15-Z,syn conformation upon initial binding to apoCph1 (38). This species rapidly interconverts to a more strongly red-light-absorbing, red-shifted intermediate that slowly transforms to the more blue-shifted Pr form upon thioether bond formation, the overall rate-limiting step for assembly (Figure 1B). A recent theoretical study suggests that the red shift of the extended, noncovalent intermediate is due to protonation of the chromophore ring system, while the increased oscillator strength of the visible transition reflects a more extended conformation (39). The extended conformation could involve adoption of a Z,anti configuration about C5, C15, or both, a conclusion that has received support from density functional theory calculations coupled with experimental resonanced Raman spectroscopy (40). Very recent work with synthetic, sterically locked BV derivatives suggests that the Pr spectrum of an Agrobacterium tumefaciens BphP is best reproduced with a synthetic BV with a locked Z,anti configuration at C15, while a locked C15-E,anti BV analogue afforded an adduct resembling Pfr (41). When taken together, the simplest interpretation of these results and of the stopped-flow data is that the Pr chromophore possesses a C5-Z,syn, C10-Z,syn, C15-Z,anti configuration that arises from the initial porphyrin-like, all syn intermediate via a single, nonphotochemical syn–anti conformational interconversion (Figure 1).
Despite the current absence of a high-resolution phytochrome crystal structure, considerable indirect information is available for members of the phytochrome family in general and for the photosensory domains in particular. Truncation analysis has indicated that normal photochemistry does not require the C-terminal regulatory region (15, 42, 43). It is also possible to retain both covalent attachment of chromophore and photoconversion in the absence of the P4 or PHY domain, but such truncations exhibit reduced photoconversion efficiency, leading to the idea that this domain both stabilizes the core photosensory region (P2 and P3) and reduces futile de-excitation of the chromophore (15, 31, 44, 45). Point mutants also have been isolated in the PHY domain that shift the absorbance spectrum without significantly affecting photochemistry (46), again indicating that this domain serves an accessory role. Deletion mutagenesis of the cyanobacterial phytochrome 2 (Cph2) has revealed that a P3 GAF domain alone is sufficient for PCB attachment (45). In apparent contrast, however, a 23 amino acid motif within the P2 PAS domain of AphA, a Cph1 orthologue from the cyanobacterium Anabaena sp. PCC 7120, was shown to play a critical role in holoprotein assembly (47). When these results are taken together, they indicate that phytochrome P3 GAF domains contain most of the chromophore contact sites important for reversible photochemistry, while the P2 and P4 domains perform more indirect roles in assembly and spectral tuning.
Our laboratory recently reported that mutation of conserved Tyr176 in the P3 GAF domain of Cph1 significantly increases the fluorescence yield of the holoprotein while inhibiting the Pr to Pfr photoconversion process (46). In that study, we hypothesized that this Y176H mutation reduced the rate of Z–E photochemical relaxation of the Pr excited state via a steric gating mechanism, thereby enhancing its fluorescence yield. The present study was undertaken to more fully examine the structural and functional role of this tyrosine residue. To determine whether this conserved tyrosine plays a similar role throughout the phytochrome family, we have introduced the Tyr-to-His mutation into representative plant phytochromes, full-length Cph1, and a representative bacteriophytochrome. Saturation mutagenesis of Tyr176 in Cph1 demonstrated a critical function of this tyrosine in maintaining the protonated, extended conformation of its bilin chromophore. To identify the potential proton donor, homology modeling of the Cph1 P3 GAF domain was performed. These results implicated Glu189, a conserved acidic residue located near Tyr176, in the stabilization of the protonated form of the B- and C-ring nitrogens of the bilin prosthetic group. To test this hypothesis, site-directed mutations of Glu189 were characterized, revealing a critical role for this residue in adoption of the extended conformation of the Cph1 chromophore.
MATERIALS AND METHODS
Plasmid Construction
The Synechocystis sp. PCC 6803 Cph1 plasmid pBAD-Cph1Δ and phycocyanobilin (PCB) biosynthetic plasmid pPL-PCB were previously described (48). The Pseudomonas aeruginosa PA 4117 BphP plasmids pASK-IBA2-PaBphP-ST (21) with and without the Tyr163His mutation were a gift from Nicole Frankenberg-Dinkel (University of Braunschweig, Germany). The pPL-PΦB plasmid that contains a synthetic operon consisting of the Synechocystis sp. PCC 6803 heme oxygenase ho1 (Cyanobase Locus SLL1184) and the Arabidopsis phytochromobilin synthase (HY2, NCBI Locus AB045112) fused to glutathione-S-transferase (GST) in the expression vector pPRO-LarA122 (Clontech) was constructed as follows. Using the plasmid DNA template pGEX-mHY2 (49) that contains the full-length HY2 cDNA lacking the transit peptide fused to GST the GST-HY2 chimera was PCR-amplified with the primers GST-mHY2-EcoRV, 5′-CGGATATCATGTCCCC-TATACTA-3′, and mHY2-NotI, 5′-GCGCGGCCGCTTAGC-CGATAAATTGTCC-3′. The resulting PCR product was subcloned into plasmid pCR2.1/HO1-RBS (48) using EcoRV and NotI sites to produce plasmid pCR2.1/HO1-RBS-GSTHY2. The entire HO1-GSTHY2 operon from this plasmid was then subcloned into pPROLarA122 using KpnI and NotI to produce pPL-PΦB. The full-length Cph1 plasmid pBAD-Cph1FL-myc/his was produced using pASK75B-Cph1-ST (8) as a PCR template with appropriate primers. The resulting PCR product was cloned into pBAD-myc/hisC (Invitrogen) with NcoI and HindIII. The Cph1-PAS/GAF truncation plasmid pBAD-Cph1(PAS/GAF)-myc/his was produced using pBAD-Cph1Δ (48) as a PCR template with the primers NcoIF-Cph1-P2, 5′-GGGCTAACAGGAGGAAT-TAACCATG-3,′ and PvuIIR-Cph1-P3, 5′-CAATAAAAC-CGCTTCATGCTCCGCCAG-3′. The PCR product, encoding the N-terminal 350 amino acids of Cph1, was cloned into pBAD-myc/hisC with NcoI and PvuII. pASK75B-AtPhyA(N599)-ST, encoding the N-terminal 599 amino acids of PHYA from Arabidopsis, was derived from the plasmid pA2a, a kind gift from Joanne Chory (Salk Institute, La Jolla, CA) by PCR using the primers 5′-CAGGATCCAGAAT-TCGAGCTCTC-3′ and 5′-GCGTCGACCGACCTTTGTAT-TCACATC-3′, and the PCR product was cloned into pASK75B-ST (8) with BamH1 and SalI. pBAD-AtPhyB-(N450)-6×his, encoding the N-terminal 450 amino acids of PHYB from Arabidopsis with a C-terminal His6 tag, was derived from pBS-AtPhyB-ST, which was generated from the XbaI–KpnI fragment of pYES2-AtPHYB-ST (50) and the similarly restricted pBluescript II KS+. The sequence corresponding to the N-terminal 450 amino acids was amplified using pBS-AtPhyB-ST and the primers AtBN450-(NcoI), 5′-CATGCCATGGCCGTTTCCGGAGTCGGGGG-TAG-3′, and AtBN450(EcoR1), 5′-GGAATTCCAGTGTCT-GCGTTCTCAAAACGCG-3′. The PCR product was cloned into the pBAD-myc/his-C vector using EcoRI and NcoI to produce pBAD-AtPhyB(N450)-6×his. QuikChange Site-Directed Mutagenesis (Stratagene) was used to generate all other site-specific mutations using 10 ng of dsDNA template with appropriate primers. All mutants and plasmid constructs derived from PCR reactions were confirmed by DNA sequencing (Davis Sequencing; http://www.davissequencing.com).
Recombinant Holophytochrome Expression and Purification
Phytochrome expressions for pASK-AtPhyA(N599)-ST and pASK-IBA2-PaBphP-ST were carried out as described (21). All other phytochrome expressions were performed in Escherichia coli strain LMG194 (Invitrogen) containing plasmids pPL-PCB or pPL-PΦB. Prior to expression, pPL-PCB- and pPL-PΦB-containing strains were maintained in minimal media (RM Media, Invitrogen) to repress expression of the bilin biosynthetic operon. Overnight precultures, in 3 mL of RM media containing 35 μg/mL kanamycin and 100 μg/mL ampicillin, were diluted 1:200 into 200 mL of the same medium and grown at 37 °C to an OD580 of ~0.5. These cultures were transferred into 600 mL of Luria–Bertani medium containing 100 μg/mL ampicillin, 35 μg/mL kanamycin, and 1 mM isopropyl-β-d-thiogalactoside (IPTG). After incubation for 1 h at 37 °C, l-arabinose was added to a final concentration of 0.002% (w/v) (48). The cultures were then incubated overnight (~20 h) at 20 °C, after which cells were collected by centrifugation and resuspended in 5 mL of cold lysis buffer [50 mM Tris-HCl at pH 7.0, 300 mM NaCl, 10% (v/v) glycerol, 0.05% (v/v) Tween 20, 1 mM 2-mercaptoethanol, and 20 mM imidazole]. Cell suspensions were passed though a French pressure cell (⅜″ D) 3 times at 10 000 psi. Cell lysates were incubated with 10 units of DNase I (RNase-free, Fermentas) at room temperature for 10 min with moderate agitation followed by ultracentrifugation at 200000g for 30 min. The resulting crude soluble protein extracts were applied to Talon metal-affinity spin columns (1 mL of bed volume, BD Biosciences) that had been pre-equilibrated with 3 mL of lysis buffer. Columns were then washed with 3 mL of lysis buffer, and bound protein was eluted with 2 mL of elution buffer (lysis buffer and 200 mM imidazole). Eluates were diluted 1:5 with protein buffer [25 mM Tes KOH at pH 7.5 and 10% (v/v) glycerol], passed through a 0.45 μm Acrodisc syringe filter (Gelman Sciences), and applied to a HiTrap Q HP anion-exchange column (1 mL of bed volume, Amersham Biosciences) pre-equilibrated with protein buffer. Columns were washed once with protein buffer containing 0.1 M NaCl (5 mL), and the recombinant protein was then eluted with protein buffer containing 0.5 M NaCl. Eluates were exchanged into protein buffer using an Amicon Ultra-4 Centrifugal Filter with a 30000 Da nominal molecular-weight limit, flash-frozen in liquid nitrogen, and stored at −80 °C.
Cyanogen Bromide Digestion and Chromopeptide Isolation
Approximately 100–200 μg of the purified Cph1Δ-PCB and Cph1Δ (Y176R)-PCB holoproteins were dialyzed into 50 mM ammonium acetate (pH 7.5) overnight at 4 °C. Protein solutions were then evaporated to ~25 μL in a Speedvac concentrator (Savant) and redissolved in 100 μL of deionized water. This solution was evaporated to dryness and dissolved in 60 μL of 70% aqueous TFA containing 2 M cyanogen bromide for protein digestion. Samples were incubated at room-temperature overnight in darkness to allow for complete digestion. These solutions were then dried under a stream of nitrogen gas, dissolved in 200 μL of 20% formic acid, and applied to a reversed-phase HPLC column (250 × 4.6 mm, Jupiter 4μ Proteo 90Å, Phenomenex). Peptides were eluted at 1 mL/min with a water/acetonitrile gradient in 20 mM formic acid. Absorbance was monitored at 220, 400, and 650 nm.
SDS–PAGE and Zinc-Blot Analysis
Protein samples were analyzed using 10% SDS–PAGE with the Laemmli buffer system (51). Zinc-blot analysis was carried out as described previously (52, 53).
Absorbance and Fluorescence Spectroscopy
Absorbance and difference spectra were obtained using a HP8453 UV–visible spectrophotometer (48). Red (650 ± 5 nm) and farred (720 ± 5 nm) light with fluence rates of >100 μmol m−2 s−1 were used for effecting phytochrome photoconversion, with sufficient duration of irradiation to ensure completeness. Fluorescence excitation and emission spectra were obtained with an SLM Aminco Bowman AB2 fluorimeter with both monochromators adjusted to a 4 nm bandpass. Fluorescence quantum yield measurements were determined with a Photon Technology International, Inc., SE 901M spectrofluorometer using purified cyanine dye (CY-5.18) as a standard as described previously (46, 54, 55).
Homology Modeling of Cph1 P3 Bilin Lyase Domain
An initial model for the P3 GAF region of Cph1, comprising amino acids 151–322 in the full-length sequence, was generated using 1MC0 (56) as a template with side-chain conformations generated by SCRWL (57). The region connecting the fourth and fifth strands (Asp226–Gly270) was poorly aligned to the template (see Supplemental Figure 1 in the Supporting Information). Therefore, the structure in this region was built manually. Inspection of the hydrophobic pattern suggested that the segment between Ser241 and His260 could be an amphipathic α helix. When this helix was docked to the rest of the template structure, the hydrophobic surface could be buried and isolated from the solvent. Furthermore, the helix also allowed Cys259 to point toward the conserved residue Tyr176. Two linker segments were then added to connect the helix to the main template. This structure was refined in explicit solvent for 200 ps using AMBER, during which a short helix between Asp226 and Val232 was formed. The resulting structure served as the initial model.
Supplemental Figure 1.

The Cph1 GAF domain and chromophore-binding pocket. (A) Sequence alignment of the Cph1 GAF domain and the two GAF domains of mouse phosphodiesterase 2A (PDB code 1MC0: (1)) with Tyr176 and Glu189 in red. (B) Residues within 3 Å of PCB (red) are continually colored by identity ranging from dark blue, absolutely conserved, to bright red, variable using an alignment of Cph1, the five phytochrome proteins in the A. thaliana genome, and BphP proteins from P. aeruginosa and Deinococcus radiodurans (2, 3).
To create a model incorporating chromophore, Merz–Kollman partial charges were calculated for PCB at the HF/6-31G level of theory using Gaussian 98 Revision A.9 (58) and assuming that the endo-vinyl side chain of the A ring had been reduced to mimic the situation after covalent attachment of the Cys residue. The resulting charges for the B/C-ring system were then scaled to a total of +1 for incorporation into the GROMOS96 43a1 and AMBER parameter sets (59, 60), and appropriate examples from these force fields were chosen for the remaining partial charges and other parameters. Parametrization of PCB in the 43a1 force field was sufficient to permit stable simulation of free PCB in explicit solvent using Gromacs 3.2 (61).
PCB was then manually docked into the protein model using VMD (62) with the following four assumptions: one, the propionate side chains of PCB were more likely to be exposed to the solvent than the more hydrophobic PCB ring system; two, the register of the first and second β strands was chosen to place highly conserved residues (including Tyr176) facing the presumptive chromophore-binding pocket; three, the stereocenter generated by the Cys–PCB bond should be in the R configuration, by analogy to plant phytochrome (16); and four, the Pr form of PCB should be protonated on both B and C rings with the C15–C16 double bond in the Z configuration. Brief (0.1 ps) molecular dynamics simulations in Gromacs or AMBER were used to relieve strain introduced during docking. After initial docking, rotation of either the A or D ring was considered as alternative means of generating the extended Pr conformation of PCB. Rotation of the A ring while retaining proper R stereochemistry implied either burial of the PCB carboxylates in an unfavorable hydrophobic environment or adoption of a strained A-ring geometry that was not maintained during simulation (data not shown). In contrast, rotation of the D ring readily permitted retention of stereochemistry with burial of the more hydrophobic ring system and exposure of the propionate side chains to the solvent; therefore, this configuration was chosen as the basis for further refinement. The resulting holoprotein models were subjected to longer molecular dynamics simulations in AMBER or Gromacs to generate two configurations each having favorable and unfavorable aspects.
On the basis of sequence homology and the expectation that the chromophore-ring system should be largely excluded from the solvent to favor photochemistry, these models were combined into a single apoprotein model using MODELLER version 8.0 (63). The resulting model was then further refined by shifting the register of the sixth β strand with MODELLER to place a conserved Trp residue in the interior to account for observed energy transfer between chromophore and a tryptophan (64). This was followed by subsequent 0.1 ps simulation in Gromacs and finally by generation of several possible configurations for the loop between the second and third β strands with MODELLER. One of these loops was chosen as offering the best burial of PCB, and this model was then subjected to 1.2 ns of simulation time in Gromacs, with the last 400 ps giving little change in the model structure. The final model conforms to the four assumptions stated above and predicts that the Pr configuration of Cph1 should adopt a C5 Z,syn, C10 Z,syn, C15 Z,anti conformation.
To model a cyclic conformation as observed during the Cph1–PCB assembly reaction (38), partial charges were calculated for PCB as described above with deprotonation of either the B or C ring. The resulting partial charges were then slightly adjusted (≤ 10%) such that the total charge on the B/C-ring system was approximately 0 and were then incorporated into the 43a1 force field to give two deprotonated tautomers. The final model for protonated chromophore was manually modified in VMD to restore the porphyrin-like configuration observed upon initial binding of PCB to Cph1 (i.e., C5 Z,syn, C10 Z,syn, C15 Z,syn, Figure 1). The complexes between Cph1 and both deprotonated PCB tautomers were modeled as covalent holoproteins, because the present study demonstrates that the cyclic chromophore configuration is compatible with covalent attachment. The resulting complexes were subjected to 1 ns simulations in Gromacs with explicit protonation of Glu189 as the most obvious candidate proton donor in our model. Apoprotein was similarly modeled by editing the holoprotein PDB file to remove PCB and then carrying out 1 ns of simulation with Gromacs. Figures were prepared using VMD (62), Tachyon (65), STRIDE (66), and Adobe Photoshop CS (Adobe Systems, Inc.). Preparation of Supplemental Figure 1 in the Supporting Information also used STAMP (67), CLUSTALW (68), and homolmapper (N. C. Rockwell and J. C. Lagarias, unpublished results).
Theoretical pKa Calculations
pKa calculations were performed using MEAD and Redti (69) on six snapshots taken at 20 ps intervals across the last 100 ps of the trajectories to compensate for slight variations in the structure during molecular dynamics. For MEAD calculations, changes in the ionization state of the PCB-ring system (and hence site–site interactions between the PCB-ring system and titrating groups on the protein) were neglected because the pKa value for the ring system in isolated PCB is itself ill-defined and because the errors incorporated in the current homology model seem likely to be at least as significant for this application. pKa calculations were performed on apoprotein (mean pKa for Glu189, 4.4 ± 0.7), deprotonated PCB (8.1 ± 1.0 and 7.4 ± 1.2 for the tautomers with B and C ring deprotonated, respectively) and protonated PCB (−1.4 ± 1.2) with explicit consideration of ionization for all Asp, Glu, His, Lys, Arg, Cys, and Tyr residues except for Cys259, which was covalently attached to PCB and thus was nontitrating. The ionization of the PCB propionate moieties was also explicitly included in the calculation. The non-physiological pKa value for protonated chromophore is likely to indicate that protonation of Glu189 in the context of protonated chromophore will cause structural change (70).
RESULTS
Tyr176 and Its Equivalents Perform a Conserved Photochemical Function in Plant and Cyanobacterial Phytochromes
The demonstration that a single amino acid substitution at a conserved position in the GAF domain (Y176H) was sufficient to evolve Cph1 into a fluorescent protein (46) led us to ask whether this was a phenomenon unique to Cph1 or reflected an inherent property of phytochromes in general. To examine this question, a construct expressing the N-terminal 450 amino acids of PhyB from Arabidopsis [equivalent to the P1–P2–P3 domains (31)], with or without the equivalent Y276H mutation, was expressed and purified from PCB-producing E. coli cells (48). Like the Y176H mutant of Cph1Δ, the corresponding AtPhyB-(N450) mutant exhibited enhanced fluorescence and reduced efficiency of photoconversion (<10% of the wild type, Figure 2). Similar results were observed for an Arabidopsis phytochrome A Y242H mutant holoprotein containing the entire photosensory domain (data not shown). Because plant phytochromes use PΦB instead of PCB, we examined the influence of chromophore substitution in the YH mutant. PΦB adducts of both Cph1Δ and AtPhyB(N450) YH mutant proteins yielded fluorescent proteins with reduced photoconversion efficiencies, indicating that the enhanced fluorescence is not specific for the PCB chromophore (Supplemental Table 1 in the Supporting Information and data not shown). We also examined the corresponding Y163H mutation in the Pseudomonas aeruginosa bacterial phytochrome PaBphP, which unusually exhibits a Pfr spectrum at the thermal ground state (21). In this case, the wild-type holoprotein BV adduct exhibited considerable fluorescence, as has also recently been reported for multiple bacteriophytochromes from Rhodopseudomonas palustris (28), but the Y163H mutant PaBphP protein exhibited no fluorescence enhancement (data not shown). When these results are taken together, they indicate that the conserved P3 GAF domain tyrosine residue performs a similar photochemical gating role in plant (Phy) and cyanobacterial (Cph1) phytochromes but plays a distinct role in bacteriophytochromes.
Figure 2.

Spectroscopic and fluorescent properties of PCB adducts of recombinant Arabidopsis PhyB(N450) wild-type (- - -) and Y276H (—) proteins. (A) Absorbance difference spectra were normalized for equal absorbance at the blue band, demonstrating that the amount of photoconversion is highly reduced for the Y276H mutant compared to the wild-type. (B) Absorbance spectra normalized to the blue band, showing that there is little difference in the bilin configuration or λmax between the wild-type and Tyr276 mutant. (C) Fluorescence excitation (thin line) and emission (thick line) spectra of the wild type and mutant (samples were adjusted to have equal absorbance at 648 nm).
Supplemental Table 1.
Spectral properties of phytochrome constructsa
| Protein | Construct | Bilin | λmax | %ΔΔA | Φf | λex | λem |
|---|---|---|---|---|---|---|---|
| Cph1Δ | wildtype | PCB | 365, 663 | 100 | 0.005 | 356, 647 | 676 |
| “ | Y176H | PCB | 365, 635 | 1.9 | 0.145 | 356, 647 | 671 |
| “ | Y176W | PCB | 355, 642 | 0.3 | 0.077 | 356, 647 | 671 |
| “ | Y176Q | PCB | 357, 651 | 1.6 | 0.121 | 355, 649 | 678 |
| “ | Y176E | PCB | 365, 660 | 7.5 | 0.086 | 359, 654 | 681 |
| “ | wildtype | PΦB | 380, 675 | 100 | 0.002 | 383, 647 | 688 |
| “ | Y176H | PΦB | 376, 655 | 2.0 | 0.067 | 387, 650 | 683 |
| “ | Y176W | PΦB | 382, 656 | 0 | 0.080 | 385, 649 | 682 |
| “ | Y176Q | PΦB | 380, 661 | 0.63 | 0.071 | 388, 650 | 685 |
| “ | Y176E | PΦB | 377, 672 | 0.3 | 0.042 | 390, 668 | 692 |
Spectral properties of various phytochrome constructs were determined as described in Table 1. Units for wavelengths are in nm. Fluorescence quantum yield is a dimensionless quantity.
n.d., not determined.
The Y176H mutation in Cph1 was also analyzed both in full-length Cph1 and in a more truncated Cph1 consisting of only the N-terminal P2 PAS and P3 GAF domains (approximately equivalent to AtPhyBN450), to assess the possibility that the novel spectroscopic properties were due to altered protein folding of the specific truncation constructs tested. In both cases, the Y176H mutation resulted in reduced photoconversion and increased fluorescence compared with its WT counterpart (data not shown). The Y176H mutant derived from the Cph1 PAS/GAF construct had a reduced fluorescence quantum yield (0.083) compared with the Cph1Δ mutant (0.145). This suggests that more energy is lost through futile modes of de-excitation upon removal of the PHY domain, a result consistent with earlier studies (45).
Tyr176 Is Essential for Normal Cph1 Photochemistry
To assess the range of possible phenotypes associated with the mutation of Tyr176, saturation mutagenesis of this residue was performed. All amino acid substitutions at this position resulted in soluble covalently bound biliproteins except for the proline mutant, which was poorly expressed (Supplemental Figure 2 in the Supporting Information). These results indicate that Tyr176 is not required for proper apoprotein folding or chromophore attachment. All of the purified Tyr176 mutant proteins exhibited reduced photoconversion efficiencies as measured by difference spectroscopy with the next most photoactive substitution mutants, i.e., Y176S and Y176T, exhibiting 34 and 13% of the wild-type photoconversion efficiency, respectively (Table 1). Tyr176 is thus necessary for optimal photochemistry. The reduced difference spectra could indicate that the forward reaction (Pr to Pfr) is specifically affected by mutation of Tyr176, which seems plausible because the reverse reaction proceeds through different intermediates and is thus a distinct pathway (23, 24), that the Pr and Pfr forms of the mutant proteins have indistinguishable spectra or that the conversion to Pfr has become so energetically disfavorable that the contribution of the reverse reaction to the measured difference spectra is negligible. We have not attempted to discriminate between these possibilities.
Supplemental Figure 2.
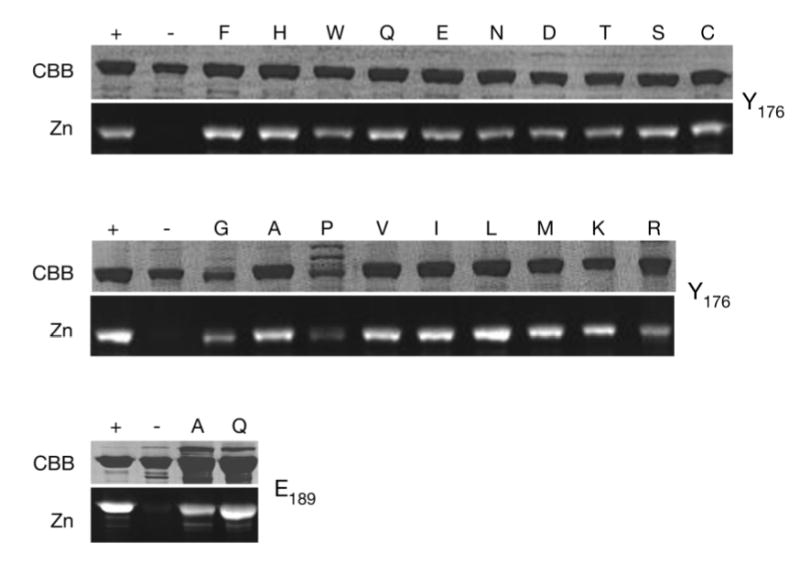
SDS/PAGE (CBB) and zinc-blot (Zn) analysis of Cph1Δ mutant proteins as PCB adducts. 3 μg of total protein was run per lane except for Y176P (30 μg). Wildtype holoCph1Δ (+) and apoCph1Δ (−) are shown for each gel. The remaining lanes are labeled with the amino acid substituted for Tyr at position 176 (top two panels) or for Glu at position 189 (bottom panel). Y176P is largely insoluble; therefore a ten-fold excess of total protein was loaded in order to demonstrate its presence. The Cph1Δ proteins all migrate with a relative molecular mass of 60 kDa.
Table 1.
Spectral Properties of PCB–Cph1Δ Holoproteinsa
| Cph1 | λmaxb | R/Bc | % ΔΔAd | Φfe | λexf | λemf |
|---|---|---|---|---|---|---|
| wild type | 365, 663 | 3.33 | 100 | 0.005 | 356, 647 | 676 |
| Y176S | 359, 650 | 2.0 | 34 | ndg | ndg | ndg |
| Y176T | 364, 655 | 0.9 | 13 | ndg | ndg | ndg |
| Y176W | 365, 635 | 0.71 | 0.3 | 0.077 | 356, 647 | 671 |
| Y176H | 355, 645 | 2.5 | 1.9 | 0.145 | 356, 647 | 671 |
| Y176Q | 357, 651 | 2.5 | 1.6 | 0.121 | 355, 649 | 678 |
| Y176E | 365, 660 | 2.0 | 7.5 | 0.086 | 359, 664 | 681 |
| Y176K | 356, 629 | 0.5 | 0.5 | ndg | ndg | ndg |
| Y176I | 364, 610 | 0.5 | 0.2 | ndg | ndg | ndg |
| Y176F | 364, 605 | 0.52 | 3.0 | ndg | ndg | ndg |
| Y176L | 362, 609 | 0.55 | 0.2 | ndg | ndg | ndg |
| Y176V | 362, 601 | 0.55 | 0.9 | ndg | ndg | ndg |
| Y176A | 364, 645 | 1.0 | 9.5 | ndg | ndg | ndg |
| Y176C | 364, 648 | 1.0 | 5.0 | ndg | ndg | ndg |
| Y176N | 357, 646 | 1.11 | 4.7 | ndg | ndg | ndg |
| Y176D | 364, 655 | 1.25 | 7.9 | ndg | ndg | ndg |
| Y176M | 364, 645 | 1.25 | 5.6 | ndg | ndg | ndg |
| Y176G | 365, 644 | 1.25 | 3.7 | ndg | ndg | ndg |
| E189A | 353, 635 | 0.5 | 4.0 | ndg | ndg | ndg |
| E189Q | 366, 650 | 0.88 | 10 | ndg | ndg | ndg |
Wild-type and mutant Cph1Δ holoproteins were purified as described in the Materials and Methods after expression in E. coli cells synthesizing PCB (48). Reported values are for proteins in the Pr form. Y176P and Y176R are not included in the table. Y176P was poorly expressed and was not characterized in detail. As shown in Figure 4, Y176R primarily bound to a porphyrin, with absorbance maxima at 403, 503, 539, 568, and 623 nm, fluorescence excitation maxima at 402, 502, 537, and 567 nm, and fluorescence emission maxima at 622, 650, and 690 nm.
Two values are presented. The first is the wavelength of maximal absorbance for the blue band (350–400 nm), while the second is the wavelength of maximal absorbance for the red band (600–700 nm). All values are in nanometers.
R/B ratio is defined as the ratio of the peak absorbance value of the red band divided by that of the blue band.
Percent photoconversion efficiency was determined by subtracting the Pfr spectrum from the Pr spectrum, subtracting the resulting difference minimum from the difference maximum, and normalizing to the wild type as 100%. Spectra were normalized to the blue band absorbance prior to the calculation.
Fluorescence quantum yield.
Maximal fluorescence excitation (λex) and emission (λem) wavelengths are presented as instrument-corrected values in nanometers.
nd = not determined.
In addition to the Y176H substitution, three other amino acid substitutions conferred enhanced fluorescence upon Cph1Δ, i.e., Gln, Glu, and Trp (Table 1). These four mutant proteins exhibited fluorescence emission maxima ranging from 671 to 681 nm as PCB adducts; all four mutants were poorly photoactive. Y176H was the most fluorescent as determined by quantum yield measurements (Table 1). Enhanced fluorescence was also observed for their PΦB adducts; however, in comparison with the corresponding PCB adducts, all such mutants had reduced fluorescence quantum yields and red-shifted fluorescence emission maxima except for Y176W, which resulted in a PΦB adduct with a similar fluorescence quantum yield to the PCB adduct (Supplemental Table 1 in the Supporting Information).
Tyr176 Is Required for the Protonated, Extended Chromophore Conformation
All substitutions at position 176 led to a reduction in the red/blue absorbance ratio, indicating that the bilin prosthetic group is bound to the mutant proteins in a more cyclic conformation [Figure 3A and Table 1 (39, 71)]. Additionally, all Tyr176 mutant proteins displayed blue-shifted absorbance maxima for the long wavelength transition (Table 1). A plot of the blue shift versus the R/B ratio demonstrated a correlation between a decreasing R/B ratio and an increasing blue shift, with the R/B ratio decreasing from that of wild-type Cph1Δ (R/B ratio > 3) to an apparent minimum ratio of approximately 0.5 (Figure 3B). Interestingly, mutants exhibiting enhanced fluorescence or relatively efficient photoconversion had the largest R/B ratios, with the exceptions being Y176W and Y176T (Table 1). Extended chromophore conformations therefore also correlate with de-excitation of the excited state via either photoconversion or fluorescence, reflecting minimized excited-state decay through nonproductive, radiationless pathways. Intermediate cases may reflect the presence of two populations within the purified preparation: a population with extended chromophore, which is competent for photoconversion and has a high R/B ratio, and a population with cyclic chromophore, which is incompetent for photoconversion and has a low R/B ratio.
Figure 3.

Substitution mutants at Tyr176 and Glu189 alter chromophore configuration. (A) Absorbance spectra were normalized to the blue band. Tyr176 substitutions exhibiting various ranges of R/B ratios are shown with representative absorbance spectra. The spectra shown are wild type, Y176H, Y176A, and Y176I. (B) A plot of the R/B ratio versus the maximal absorbance wavelength for the long wavelength band is shown for the wild-type and mutant proteins with substitutions at Tyr176 (○) or Glu189 (□). Proteins whose spectra are shown in A are indicated in one-letter code. Substitutions at Tyr176 which resulted in fluorescence, are shown in •, while the wild type and other substitutions are shown as ○ and were fitted to a smooth hyperbolic function (—).
Previous stopped-flow studies of apoCph1 assembly demonstrated that the chromophore of the Cph1·PCB complex converts from a porphyrin-like conformation to a more extended conformation prior to covalent attachment [Figure 1 (38)]. The authors of this work proposed that this conformational interconversion reflects protonation of both B/C-ring system pyrrole nitrogens of the bilin chromophore, by analogy to the pH dependence of free bilin spectra (71). This interpretation is also consistent with recent NMR data showing that all four PCB nitrogens are protonated in the Pr state (35). Protonation and isomerization of the chromophore thus seem to be coupled processes both in solution and in the phytochrome-binding pocket, with covalent attachment occurring more slowly than chromophore isomerization. Because most of the Tyr176 mutant proteins have covalently attached chromophores with R/B absorbance ratios consistent with the cyclic deprotonated conformation, our results suggest that chromophore protonation is not a prerequisite for covalent bond formation. It should be noted that these mutant proteins are assembled into holoproteins over a 20 h period. For this reason, we cannot rule out that deprotonation occurs following chromophore attachment. When these data are taken together, they demonstrate that Tyr176 is required for maintenance of the extended, protonated conformation of the chromophore.
The Y176R Mutant Binds an Endogenous Porphyrin
In contrast to wild-type Cph1 and all other Tyr176 mutant proteins, the Y176R mutant surprisingly exhibited an absorbance spectrum with features of a nonbilin pigment. On the basis of an intense 400 nm absorbance maximum and the fluorescence excitation/emission spectra (Figure 4), it was clear that a porphyrin was associated with this mutant. However, the broader absorbance maximum at longer wavelengths (~623 nm, Figure 4) suggested that the purified protein also contained a bilin chromophore, indicating that the purified preparation consisted of two populations with different chromophores. To assess whether the porphyrin was covalently attached, wild-type and Y176R proteins were digested with CNBr and the resulting peptide mixture was separated by reverse-phase HPLC. Two chromopeptides were observed, one eluting at the same time as the single wild-type Cph1Δ chromopeptide and a second eluting 1.5 min later (Supplemental Figure 3 in the Supporting Information). The absorbance spectrum of the former was similar to the PCB-bound chromopeptide derived from wild-type Cph1Δ, while the absorbance spectrum of the second chromopeptide clearly demonstrated a bound porphyrin. These results indicated that the Y176R mutation altered the chromophore specificity of Cph1Δ, apparently enabling an endogenous porphyrin to covalently bind to the apoprotein. The chemical structure and linkage of this bound porphyrin is the subject of an ongoing investigation.
Figure 4.

Y176R mutant protein affects chromophore-binding specificity. (A) Absorbance (thick solid line), fluorescence excitation (thin solid line), and fluorescence emission (dashed line) spectra of the Y176R mutant Cph1Δ protein. The multiple fluorescence excitation peaks and sharp fluorescence emission peak at approximately 620 nm are characteristic of a porphyrin (71), while the broader absorbance around 620 nm is indicative of a second population with bound bilin. (B) Absorbance and fluorescence excitation and emission spectra of the Y176H mutant Cph1Δ protein for comparison. Spectra are the same as in A.
Supplemental Figure 3.
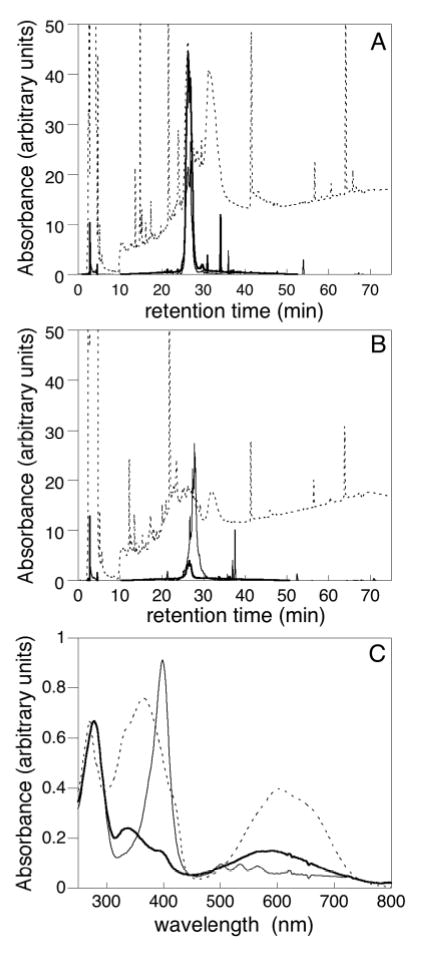
Analysis of chromopeptides. Peptides derived from wildtype (A) and Y176R (B) Cph1 were analyzed by HPLC: 220 nm (dashed), 400 nm (porphyrin, thin line), and 650 nm (bilin, thick line). (C) Absorbance spectra for wildtype (dashed) and Y176R (peak 1, thick line; peak 2, thin line) chromopeptides.
Homology Modeling Predicts Close Association of Tyr176 and Glu189 in the P3 Domain of Cph1
To develop hypotheses concerning the role of Tyr176 in chromophore protonation, we generated homology models of the Cph1 P3 bilin lyase domain using published crystal structures of GAF domains as templates (56, 72, 73). Using a multiple sequence alignment with both GAF domains of mouse phosphodiesterase 2A (PDB ID 1MC0, Supplemental Figure 1 in the Supporting Information), a homology model of the Cph1 P3 domain predicted a six-stranded, antiparallel β-sheet scaffold with helices on either side (Figure 5A). Because there is little sequence identity within the phytochrome family in the large insert between the fourth and fifth β strands, substantial refinement of this region was not undertaken. The resulting model placed several residues conserved among phytochromes about a central cavity that is likely to constitute the chromophore-binding pocket.
Figure 5.

Stereoviews of the Cph1 bilin lyase domain homology model. (A) Proposed Cph1 bilin lyase fold shows a central twisted β sheet, with Tyr176 and Glu189 (blue) on adjacent strands (β1 and β2, respectively). Both side chains point up into the proposed chromophore-binding pocket, with the nucleophilic Cys259 (blue) above Tyr176 and Glu189. β strands are numbered on the basis of their position in the sequence starting from the N terminus. (B) PCB is shown attached to Cys259, with Tyr176 and Glu189 lying below the chromophore (colors: C, gray; H, white; N, blue; O, red; and S, yellow). For Tyr176 and Glu189, atoms within 3.4 Å of PCB are also shown as solvent-accessible surfaces (Tyr176, yellow; and Glu189, orange). PCB atoms within 3.4 Å of Tyr176 are shown as a solvent-accessible surface (light blue). (C) Interactions between Glu189 and PCB. The Glu189 side chain is hydrogen-bonded to the B/C-ring system of the protonated chromophore, stabilizing the positive charge. The rings of PCB are labeled for clarity.
We then manually docked PCB chromophore into this pocket and carried out additional refinement as described in the Materials and Methods. The final model was subjected to 1.2 ns of molecular dynamics simulation in explicit solvent in Gromacs 3.2.1 (61). The resulting model stably incorporates PCB into the proposed chromophore-binding pocket; PCB is predicted to assume the C5-Z,syn, C10-Z,syn, C15- Z,anti configuration in the Pr form (Figure 1), while also maintaining the stereochemistry of the Cys–PCB linkage based on that of plant phytochromes (16). This model also exhibited a number of favorable interactions between PCB and highly conserved protein residues (Figure 5 and Supplemental Figure 1 in the Supporting Information). In particular, the Tyr176 side chain lies within 3.5 Å of both the C15 methine bridge and the D-ring methyl group (Figure 5B). This model also predicts that Tyr176 is hydrogen-bonded to the carboxyl side chain of Glu189 (Figure 5B). Because Glu189 can form multiple hydrogen bonds with the positively charged B/Cring system of the protonated chromophore in this model (Figure 5C), its involvement in stabilizing the charge of the protonated chromophore appeared reasonable.
Glu189 Is Required for Chromophore Protonation
To test the hypothesis that Glu189 is required to stabilize the protonated, extended chromophore conformation, this residue was mutated to Gln and Ala. A previous study utilizing a short in vitro assembly reaction had shown this residue to be essential for holoprotein formation, but we hoped that in vivo assembly would permit assembly of Glu189 mutant holophytochromes. Indeed, the two Glu189 mutations yielded soluble holophytochromes with bound chromophore (Supplemental Figure 2 in the Supporting Information), which exhibited highly reduced photoconversion efficiencies (4% for E189A and 10% for E189Q, Table 1). Both Glu189 mutants displayed significantly reduced red/blue ratios, consistent with their chromophores adopting cyclic conformations (Table 1). The long wavelength absorbance band was also blue-shifted with both mutant proteins (Figure 3B, □), indicating that the mutation of Glu189 resulted in comparable effects to the mutation of Tyr176.
DISCUSSION
The data that we present here demonstrate that the conserved Tyr176 in the P3 domain of Cph1 plays a key role in the structure and function of holophytochromes. First, the gain-of-function fluorescence observed in the Y176H mutant of Cph1 (46) is also observed in at least two plant phytochromes upon introduction of the equivalent mutation. This gain-of-function fluorescence is observed with both PΦB and PCB chromophores, indicating that the spectral gating function of the P3 GAF domain Tyr is similar for both bilin chromophores. Moreover, we show that Tyr176 is essential for optimal photoconversion; no other residue supports even 50% of wild-type levels of photoconversion to Pfr. Indeed, the most conservative substitution (i.e., Y176F) is among the least photoconvertible, i.e., 3% of the wild type (Table 1). Our studies show that Tyr176 is critical for adoption of the extended, protonated Pr chromophore conformation, because many mutant proteins instead covalently bind chromophore in a cyclic, deprotonated conformation reminiscent of a porphyrin (Figure 3). In the most extreme example of this altered conformational specificity, substitution of Arg for Tyr at this position resulted in a mutant phytochrome that preferentially bound a porphyrin (Figure 4 and Supplemental Figure 3 in the Supporting Information).
The increased fluorescence associated with mutation of Tyr176 in both Cph1 and plant phytochromes indicates that this residue has a conserved role in the photochemical interconversion of Pr to the primary intermediate Lumi-R (46). Both Cph1 and plant phytochromes are known to exhibit de-excitation on a subnanosecond time scale with their native chromophores (74, 75). Use of the alternative chromophore phycoerythrobilin, which has a saturated C15 methine bridge, extends the excited-state lifetime into the nanosecond time scale, leading to enhanced fluorescence (75, 76). By analogy, the mutation of Tyr176 or the equivalent residue in plant phytochromes could prevent the normal path of photoreaction and prolong the lifetime of the excited state, resulting in increased fluorescence.
Interestingly, the introduction of the equivalent Y163H mutation into the bacteriophytochrome PaBphP did not result in a comparable enhancement of fluorescence. This indicates that there must be differences in the chromophore–protein interactions and/or photochemical reaction pathways of the BphP subfamily relative to plant and cyanobacterial phytochromes. In contrast to the very low fluorescence observed with wild-type Cph1 or plant phytochrome (Table 1), wild-type bacteriophytochromes RpBphP2 and RpBphP3 from R. palustris both exhibit substantial fluorescence in the Pr state (28), as did wild-type PaBphP in our hands (data not shown). These results suggest that members of the BphP subfamily may have an intrinsically higher Pr fluorescence and hence a longer excited-state lifetime for photoconversion from Pr to Pfr compared with plant (Phy) and cyanobacterial (Cph1) phytochromes. Mutation of this conserved Tyr residue apparently does not have a comparable effect on the BphP excited-state lifetime.
The current study does not fully elucidate the role(s) of Tyr176 in the photochemistry of Cph1. On the basis of current data, the proposal that this residue is involved in sterically gating the Pr to Pfr transition (46) is still plausible. Indeed, the contacts between PCB and Tyr176 in the current homology model are consistent with such a role, although this model is unlikely to be accurate in atomic detail. In the present study, we show that four residues resulted in a significant increase in fluorescence when substituted for Tyr at position 176: His, Gln, Glu, and Trp (Table 1). All of these residues possess potential hydrogen-bonding donor/acceptor side chains that are positioned four atoms removed from the peptide backbone. As we proposed earlier (46), a fortuitous hydrogen bond with the bilin chromophore, another residue in the PCB-binding pocket, or even water could account for an increased activation barrier for photochemical de-excitation of the Pr excited state. The molecular basis for the fluorescence enhancement may differ between the four mutants. Introduction of the large Trp residue at position 176 might inhibit rotation of the D ring on steric grounds, while the other mutations may enhance D-ring rigidity via a different mechanism(s). The reduced R/B ratio seen with Y176W relative to the other fluorescent mutant proteins (Figure 3B) is consistent with this interpretation. Further studies, especially those that generate experimental structural information, are needed to more fully understand the structural basis of the fluorescence enhancement.
Our data also show that Tyr176 is important for the adoption of the normal Pr chromophore conformation. Many substitutions at this position result in a holoprotein in which the chromophore exhibits a slight blue shift and a substantially lower R/B ratio, indicative of a chromophore in a cyclic configuration (71). Gas-phase calculations suggest that the red enhancement observed during formation of wild-type Pr holoprotein is the direct consequence of rotation about the C5 or C15 methine bridges, while the red shift of the absorbance maximum is the consequence of protonation of the B/C-ring system pyrrole nitrogens (39). Our mutant proteins show a rough correlation between these two spectral trends (Figure 3B). While the R/B ratio reaches an apparent minimum value that would presumably be expected for the cyclic conformation, it is unclear whether the value observed for the wild type reflects a maximum. Moreover, the mutants that have a minimal R/B ratio exhibit a range of absorbance maxima. It thus seems likely that our current spectral data reflect the presence of two species: a cyclic, deprotonated form that predominates in most of the mutant proteins and an extended, protonated form that predominates in wild-type Cph1. Our data thus show that Tyr176 is an important residue both for the adoption of the proper chromophore configuration and for photoconversion, perhaps indicating overlap between the structural changes involved in assembly of holoprotein and those associated with the photochemical cycle.
The simplest explanation for the reduced R/B ratios of the different Tyr176 mutant proteins is that they reflect varying degrees of chromophore protonation, because protonation strongly favors formation of the extended C15 Z,anti conformation of the bilin chromophore (71). This implicates Tyr176 as critical for stabilization of the protonated state of PCB either via direct interaction with the proton donor itself or via its ability to optimally position an anionic residue near the B/C-ring positive charge. By analogy with the role of aspartate residues in stabilizing the protonated state of bilin chromophores in phycocyanin (77, 78), we reason that a residue with a carboxyl side chain would best fulfill this role in phytochrome. On the basis of previous studies indicating that the invariant acidic residue Glu189 is critical for chromophore attachment (45) along with homology modeling in the present study, this residue is a plausible candidate for maintaining the extended, protonated conformation of the Cph1 chromophore. Indeed, this hypothesis is consistent with the spectroscopic properties of the E189A and E189Q mutants, whose chromophores adopt porphyrin-like conformations. In view of the many assumptions made in developing our homology model, however, it remains possible that another conserved acidic residue within the GAF domain, e.g., Asp207 or Asp226, could perform this role.
While the identity of the actual proton donor responsible for triggering the C15,syn to C15,anti conformational rearrangement is conceivably the same acidic residue that stabilizes the protonated chromophore, we cannot conclude this with certainty. Our homology model implicates His260 or Glu189 as the most likely candidates to fulfill this role. His260 is conserved, and the participation of His residues in proton-transfer reactions is well-documented in many protein systems. In our model, however, His260 lies closer to the A ring and thus appears better suited to mediate thioether attachment. In contrast, Tyr176 and Glu189 are more closely associated with the bilin B and C rings. Because the pKa for a tyrosine phenol side chain is normally too high to play a significant role in such a proton transfer, Glu189 is the more reasonable candidate. However, the pKa values for isolated Glu side chains are typically <5, indicating that the pKa of Glu189 would have to be significantly increased for this residue to be a viable proton donor. Such increases for Glu pKa values are not unknown; for instance, Glu35 in lysozyme has a pKa value >6 (79–81). Initial attempts to estimate the Glu189 pKa using the MEAD software suite (69) and the current model structure indicate that this residue has a normal pKa in the apoprotein, a substantially elevated pKa in the proposed apoprotein/PCB intermediate complex (protonated Glu189 with one proton on either the B or C ring of PCB), and a substantially decreased pKa in the holoprotein complex depicted in Figure 5 (data not shown). While these calculations are likely to incorporate significant systematic error, they do suggest that Glu189 is a viable candidate for proton transfer to PCB during holoprotein assembly.
This hypothesis led us to examine the role of Glu189 in Cph1’s assembly and photochemistry. This residue had previously been shown to be required for formation of holoprotein in a short in vitro assembly assay (45), but we reasoned that the in vivo expression system used in the present study might permit successful synthesis of the holoprotein. Indeed, both E189Q and E189A Cph1 were isolated as holoproteins, and both exhibited spectra consistent with cyclic, deprotonated chromophore configurations (Table 1). Mutation of Glu189 also ablated normal Pr to Pfr photoconversion. It thus appears that Glu189, like Tyr176, is required for the formation of the extended, protonated Pr chromophore conformation. In the absence of a crystal structure for any phytochrome, it is difficult to determine the precise mechanistic basis for the observed roles of these two residues in the photochemistry of Cph1 and plant phytochromes. However, the data presented here establish that both Tyr176 and Glu189 are required for the chromophore to adopt the extended, protonated C15, Z,anti conformation found in wild-type Cph1. Both residues are also required for efficient photoconversion. It will be interesting to see whether this correlation reflects actual similarities between the initial assembly pathway of holoprotein assembly and the phytochrome photocycle. During the wild-type photocycle, we hypothesize that the orientation of these two residues is likely to change, a process that is likely to underlie the transmission of the light signal.
SUPPORTING INFORMATION AVAILABLE
(Supplemental Figure 1) Additional information about the sequence alignment used in homology modeling and the chromophore–protein interactions predicted by the resulting model. (Supplemental Figure 2) Purified mutant proteins reported in this paper. (Supplemental Figure 3) Chromopeptide analysis of the wild-type and Y176R Cph1. (Supplemental Table 1) Additional spectral information for those Tyr176 mutations that resulted in fluorescent holoproteins. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
We thank former Lagarias lab members Beronda Montgomery, John Murphy, Jennifer Santos, and Tedd Elich for construction of the plasmids pPL-PΦB, pASK75B-AtPhyA-(N599), pBAD:Cph1fl, and pA2a, respectively. The original phyA and phyB plasmids were kind gifts from J. Chory. The paBphP plasmids were a gift from Nicole Frankenberg-Dinkel (University of Braunschweig, Germany).
Footnotes
This work was supported in part by grants from the National Institutes of Health GM068552 (to J.C.L.), GM64458 (to Y.D.), and GM67168 (Y.D.), from the National Science Foundation EIA0330135 (to A.S.W.), and from the National Science Foundation Center for Biophotonics Science and Technology PHY-0120999.
Abbreviations: BphP, bacteriophytochrome; BV, biliverdin IXα; CNBr, cyanogen bromide; Cph1, cyanobacterial phytochrome 1 from Synechocystis sp. PCC 6803; Cph1Δ, truncation of the cyanobacterial phytochrome 1 consisting of the N-terminal 514 amino acids; GAF, domain acronym derived from vertebrate cGMP-specific phosphodiesterases, cyanobacterial adenylate cyclases, and formate hydrogen lyase transcription activator FhlA; P2, phytochrome PAS domain; P3, phytochrome GAF or bilin lyase domain; P4, phytochrome PHY, GAF-related domain; PAS, domain acronym derived from period clock (PER) protein, aromatic hydrocarbon receptor nuclear translocator (ARNT), and single minded (SIM); PΦB, phytochromobilin; PCB, phycocyanobilin; PCR, polymerase chain reaction; Phy, plant phytochrome; Pr, red absorbing form of phytochrome; Pfr, far-red absorbing form of phytochrome; R/B, red/blue ratio measured by absorbance.
References
- 1.Smith H. Phytochromes and light signal perception by plants—An emerging synthesis. Nature. 2000;407:585–591. doi: 10.1038/35036500. [DOI] [PubMed] [Google Scholar]
- 2.Schäfer, E., and Nagy, F., Eds. (2005) Photomorphogenesis in Plants and Bacteria: Function and Signal Tranduction Mechanisms, 3rd ed., pp 667, Springer Publishers, Dordrecht, The Netherlands.
- 3.Nagy F, Schäfer E. Phytochromes control photomorphogenesis by differentially regulated, interacting signaling pathways in higher plants. Annu Rev Plant Biol. 2002;53:329–355. doi: 10.1146/annurev.arplant.53.100301.135302. [DOI] [PubMed] [Google Scholar]
- 4.Quail PH. Phytochrome photosensory signalling networks. Nat Rev Mol Cell Biol. 2002;3:85–93. doi: 10.1038/nrm728. [DOI] [PubMed] [Google Scholar]
- 5.Chen M, Chory J, Fankhauser C. Light signal transduction in higher plants. Annu Rev Gen. 2004;38:87–117. doi: 10.1146/annurev.genet.38.072902.092259. [DOI] [PubMed] [Google Scholar]
- 6.Franklin KA, Whitelam GC. Light signals, phytochromes, and cross-talk with other environmental cues. J Exp Bot. 2004;55:271–276. doi: 10.1093/jxb/erh026. [DOI] [PubMed] [Google Scholar]
- 7.Kehoe DM, Grossman AR. Similarity of a chromatic adaptation sensor to phytochrome and ethylene receptors. Science. 1996;273:1409–1412. doi: 10.1126/science.273.5280.1409. [DOI] [PubMed] [Google Scholar]
- 8.Yeh KC, Wu SH, Murphy JT, Lagarias JC. A cyanobacterial phytochrome two-component light sensory system. Science. 1997;277:1505–1508. doi: 10.1126/science.277.5331.1505. [DOI] [PubMed] [Google Scholar]
- 9.Hughes J, Lamparter T, Mittmann F, Hartmann E, Gärtner W, Wilde A, Börner T. A prokaryotic phytochrome. Nature. 1997;386:663. doi: 10.1038/386663a0. [DOI] [PubMed] [Google Scholar]
- 10.Jiang ZY, Swem LR, Rushing BG, Devanathan S, Tollin G, Bauer CE. Bacterial photoreceptor with similarity to photoactive yellow protein and plant phytochromes. Science. 1999;285:406–409. doi: 10.1126/science.285.5426.406. [DOI] [PubMed] [Google Scholar]
- 11.Davis SJ, Vener AV, Vierstra RD. Bacteriophytochromes: Phytochrome-like photoreceptors from nonphotosynthetic eubacteria. Science. 1999;286:2517–2520. doi: 10.1126/science.286.5449.2517. [DOI] [PubMed] [Google Scholar]
- 12.Herdman M, Coursin T, Rippka R, Houmard J, de Marsac NT. A new appraisal of the prokaryotic origin of eukaryotic phytochromes. J Mol Evol. 2000;51:205–213. doi: 10.1007/s002390010082. [DOI] [PubMed] [Google Scholar]
- 13.Giraud E, Fardoux J, Fourrier N, Hannibal L, Genty B, Bouyer P, Dreyfus B, Verméglio A. Bacteriophytochrome controls photosystem synthesis in anoxygenic bacteria. Nature. 2002;417:202–205. doi: 10.1038/417202a. [DOI] [PubMed] [Google Scholar]
- 14.Idnurm A, Heitman J. Light controls growth and development via a conserved pathway in the fungal kingdom. PLoS Biol. 2005;3:e95. doi: 10.1371/journal.pbio.0030095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karniol, B., and Vierstra, R. D. (2005) Structure, function, and evolution of microbial phytochromes, in Photomorphogenesis in Plants (Schafer, E., Ed.) Martinus Nijhoff Publishers, Dordrecht, The Netherlands.
- 16.Lagarias JC, Rapoport H. Chromopeptides from phytochrome. The structure and linkage of the Pr form of the phytochrome chromophore. J Am Chem Soc. 1980;102:4821–4828. [Google Scholar]
- 17.Wu SH, McDowell MT, Lagarias JC. Phycocyanobilin is the natural precursor of the phytochrome chromophore in the green alga Mesotaenium caldariorum. J Biol Chem. 1997;272:25700–25705. doi: 10.1074/jbc.272.41.25700. [DOI] [PubMed] [Google Scholar]
- 18.Hübschmann T, Börner T, Hartmann E, Lamparter T. Characterization of the Cph1 holo-phytochrome from Synechocystis sp. PCC 6803. Eur J Biochem. 2001;268:2055–2063. doi: 10.1046/j.1432-1327.2001.02083.x. [DOI] [PubMed] [Google Scholar]
- 19.Bhoo SH, Davis SJ, Walker J, Karniol B, Vierstra RD. Bacteriophytochromes are photochromic histidine kinases using a biliverdin chromophore. Nature. 2001;414:776–779. doi: 10.1038/414776a. [DOI] [PubMed] [Google Scholar]
- 20.Lamparter T, Carrascal M, Michael N, Martinez E, Rottwinkel G, Abian J. The biliverdin chromophore binds covalently to a conserved cysteine residue in the N-terminus of Agrobacterium phytochrome Agp1. Biochemistry. 2004;43:3659–3669. doi: 10.1021/bi035693l. [DOI] [PubMed] [Google Scholar]
- 21.Tasler R, Moises T, Frankenberg-Dinkel N. Biochemical and spectroscopic characterization of the bacterial phytochrome of Pseudomonas aeruginosa. FEBS J. 2005;272:1927–1936. doi: 10.1111/j.1742-4658.2005.04623.x. [DOI] [PubMed] [Google Scholar]
- 22.Rüdiger W, Thümmler F, Cmiel E, Schneider S. Chromophore structure of the physiologically active form (Pfr) of phytochrome. Proc Natl Acad Sci USA. 1983;80:6244–6248. doi: 10.1073/pnas.80.20.6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sineshchekov VA. Photobiophysics and photobiochemistry of the heterogeneous phytochrome system. Biochim Biophys Acta Bioenerg. 1995;1228:125–164. [Google Scholar]
- 24.Braslavsky, S. E. (2003) Phytochrome, in Photochromism: Molecules and Systems (Dürr, H., and Bouas-Laurent, H., Eds.) pp 738–755, Elsevier Science BV, Amsterdam, The Netherlands.
- 25.Yeh KC, Lagarias JC. Eukaryotic phytochromes: Light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc Natl Acad Sci USA. 1998;95:13976–13981. doi: 10.1073/pnas.95.23.13976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamparter T, Esteban B, Hughes J. Phytochrome Cph1 from the cyanobacterium Synechocystis PCC6803—Purification, assembly, and quaternary structure. Eur J Biochem. 2001;268:4720–4730. doi: 10.1046/j.1432-1327.2001.02395.x. [DOI] [PubMed] [Google Scholar]
- 27.Karniol B, Vierstra RD. The pair of bacteriophytochromes from Agrobacterium tumefaciens are histidine kinases with opposing photobiological properties. Proc Natl Acad Sci USA. 2003;100:2807–2812. doi: 10.1073/pnas.0437914100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giraud E, Zappa S, Vuillet L, Adriano JM, Hannibal L, Fardoux J, Berthomieu C, Bouyer P, Pignol D, Vermeglio A. A new type of bacteriophytochrome acts in tandem with a classical bacteriophytochrome to control the antennae synthesis in Rhodopseudomonas palustris. J Biol Chem. 2005;280:32389–32397. doi: 10.1074/jbc.M506890200. [DOI] [PubMed] [Google Scholar]
- 29.Martinez-Garcia JF, Huq E, Quail PH. Direct targeting of light signals to a promoter element-bound transcription factor. Science. 2000;288:859–863. doi: 10.1126/science.288.5467.859. [DOI] [PubMed] [Google Scholar]
- 30.Matsushita T, Mochizuki N, Nagatani A. Dimers of the N-terminal domain of phytochrome B are functional in the nucleus. Nature. 2003;424:571–574. doi: 10.1038/nature01837. [DOI] [PubMed] [Google Scholar]
- 31.Oka Y, Matsushita T, Mochizuki N, Suzuki T, Tokutomi S, Nagatani A. Functional analysis of a 450-amino acid N-terminal fragment of phytochrome B in Arabidopsis. Plant Cell. 2004;16:2104–2116. doi: 10.1105/tpc.104.022350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Montgomery BL, Lagarias JC. Phytochrome ancestry. Sensors of bilins and light. Trends Plant Sci. 2002;7:357–366. doi: 10.1016/s1360-1385(02)02304-x. [DOI] [PubMed] [Google Scholar]
- 33.Lamparter T, Michael N, Caspani O, Miyata T, Shirai K, Inomata K. Biliverdin binds covalently to Agrobacterium phytochrome Agp1 via its ring A vinyl side chain. J Biol Chem. 2003;278:33786–33792. doi: 10.1074/jbc.M305563200. [DOI] [PubMed] [Google Scholar]
- 34.Quest B, Gärtner W. Chromophore selectivity in bacterial phytochromes: Dissecting the process of chromophore attachment. Eur J Biochem. 2004;271:1117–1126. doi: 10.1111/j.1432-1033.2004.04015.x. [DOI] [PubMed] [Google Scholar]
- 35.Strauss HM, Hughes J, Schmieder P. Heteronuclear solution-state NMR studies of the chromophore in cyanobacterial phytochrome Cph1. Biochemistry. 2005;44:8244–8250. doi: 10.1021/bi050457r. [DOI] [PubMed] [Google Scholar]
- 36.Kneip C, Hildebrandt P, Schlamann W, Braslavsky SE, Mark F, Schaffner K. Protonation state and structural changes of the tetrapyrrole chromophore during the Pr → Pfr phototransformation of phytochrome: A resonance Raman spectroscopic study. Biochemistry. 1999;38:15185–15192. doi: 10.1021/bi990688w. [DOI] [PubMed] [Google Scholar]
- 37.Foerstendorf H, Benda C, Gartner W, Storf M, Scheer H, Siebert F. FTIR studies of phytochrome photoreactions reveal the C=O bands of the chromophore: Consequences for its protonation states, conformation, and protein interaction. Biochemistry. 2001;40:14952–14959. doi: 10.1021/bi0156916. [DOI] [PubMed] [Google Scholar]
- 38.Borucki B, Otto H, Rottwinkel G, Hughes J, Heyn MP, Lamparter T. Mechanism of Cph1 phytochrome assembly from stopped-flow kinetics and circular dichroism. Biochemistry. 2003;42:13684–13697. doi: 10.1021/bi035511n. [DOI] [PubMed] [Google Scholar]
- 39.Göller AH, Strehlow D, Hermann G. The excited-state chemistry of phycocyanobilin: A semiempirical study. ChemPhysChem. 2005;6:1259–1268. doi: 10.1002/cphc.200400667. [DOI] [PubMed] [Google Scholar]
- 40.Mroginski MA, Murgida DH, von Stetten D, Kneip C, Mark F, Hildebrandt P. Determination of the chromophore structures in the photoinduced reaction cycle of phytochrome. J Am Chem Soc. 2004;126:16734–16735. doi: 10.1021/ja043959l. [DOI] [PubMed] [Google Scholar]
- 41.Inomata K, Hammam MA, Kinoshita H, Murata Y, Khawn H, Noack S, Michael N, Lamparter T. Sterically locked synthetic bilin derivatives and phytochrome Agp1 from Agrobacterium tumefaciens form photoinsensitive Pr- and Pfr-like adducts. J Biol Chem. 2005;280:24491–24497. doi: 10.1074/jbc.M504710200. [DOI] [PubMed] [Google Scholar]
- 42.Tu, S.-L., and Lagarias, J. C. (2005) The phytochromes, in Handbook of Photosensory Receptors (Briggs, W. R., and Spudich, J. A., Eds.) pp 121–149, Wiley VCH, Weinheim, Germany.
- 43.Vierstra RD. Illuminating phytochrome functions. Plant Physiol. 1993;103:679–684. doi: 10.1104/pp.103.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park CM, Shim JY, Yang SS, Kang JG, Kim JI, Luka Z, Song PS. Chromophore–apoprotein interactions in Synechocystis sp. PCC6803 phytochrome Cph1. Biochemistry. 2000;39:6349–6356. doi: 10.1021/bi992916s. [DOI] [PubMed] [Google Scholar]
- 45.Wu SH, Lagarias JC. Defining the bilin lyase domain: Lessons from the extended phytochrome superfamily. Biochemistry. 2000;39:13487–13495. doi: 10.1021/bi001123z. [DOI] [PubMed] [Google Scholar]
- 46.Fischer AJ, Lagarias JC. Harnessing phytochrome’s glowing potential. Proc Natl Acad Sci USA. 2004;101:17334–17339. doi: 10.1073/pnas.0407645101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao KH, Ran Y, Li M, Sun YN, Zhou M, Storf M, Kupka M, Böhm S, Bubenzer C, Scheer H. Photochromic biliproteins from the cyanobacterium Anabaena sp. PCC 7120: Lyase activities, chromophore exchange, and photochromism in phytochrome AphA. Biochemistry. 2004;43:11576–11588. doi: 10.1021/bi0491548. [DOI] [PubMed] [Google Scholar]
- 48.Gambetta GA, Lagarias JC. Genetic engineering of phytochrome biosynthesis in bacteria. Proc Natl Acad Sci USA. 2001;98:10566–10571. doi: 10.1073/pnas.191375198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frankenberg N, Mukougawa K, Kohchi T, Lagarias JC. Functional genomic analysis of the HY2 family of ferredoxin-dependent bilin reductases from oxygenic photosynthetic organisms. Plant Cell. 2001;13:965–978. doi: 10.1105/tpc.13.4.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elich TD, Chory J. Biochemical characterization of Arabidopsis wild-type and mutant phytochrome B holoproteins. Plant Cell. 1997;9:2271–2280. doi: 10.1105/tpc.9.12.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Laemmli UK. Cleavage of structural proteins during the assembly of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 52.Berkelman TR, Lagarias JC. Visualization of bilin-linked peptides and proteins in polyacrylamide gels. Anal Biochem. 1986;156:194–201. doi: 10.1016/0003-2697(86)90173-9. [DOI] [PubMed] [Google Scholar]
- 53.Li L, Lagarias JC. Phytochrome assembly—Defining chromophore structural requirements for covalent attachment and photoreversibility. J Biol Chem. 1992;267:19204–19210. [PubMed] [Google Scholar]
- 54.Murphy JT, Lagarias JC. Purification and characterization of recombinant affinity peptide-tagged oat phytochrome A. Photochem Photobiol. 1997;65:750–758. doi: 10.1111/j.1751-1097.1997.tb01920.x. [DOI] [PubMed] [Google Scholar]
- 55.Southwick PL, Ernst LA, Tauriello EW, Parker SR, Mujumdar RB, Mujumdar SR, Clever HA, Waggoner AS. Cyanine dye labeling reagents—Carboxymethylin-docyanine succinimidyl esters. Cytometry. 1990;11:418–430. doi: 10.1002/cyto.990110313. [DOI] [PubMed] [Google Scholar]
- 56.Martinez SE, Wu AY, Glavas NA, Tang XB, Turley S, Hol WG, Beavo JA. The two GAF domains in phosphodiesterase 2A have distinct roles in dimerization and in cGMP binding. Proc Natl Acad Sci USA. 2002;99:13260–13265. doi: 10.1073/pnas.192374899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bower MJ, Cohen FE, Dunbrack RL., Jr Prediction of protein side-chain rotamers from a backbone-dependent rotamer library: A new homology modeling tool. J Mol Biol. 1997;267:1268–1282. doi: 10.1006/jmbi.1997.0926. [DOI] [PubMed] [Google Scholar]
- 58.Frisch, M. J., Trucks, G. W., Schlegel, H. B., Scuseria, G. E., Robb, M. A., Cheeseman, J. R., Zakrzewski, V. G., Montgomery, J. A., Jr., Stratmann, R. E., Burant, J. C., Dapprich, S., Millam, J. M., Daniels, A. D., Kudin, K. N., Strain, M. C., Farkas, O., Tomasi, J., Barone, V., Cossi, M., Cammi, R., Mennucci, B., Pomelli, C., Adamo, C., Clifford, S., Ochterski, J., Petersson, G. A., Ayala, P. Y., Cui, Q., Morokuma, K., Malick, D. K., Rabuck, A. D., Raghavachari, K., Foresman, J. B., Cioslowski, J., Ortiz, J. V., Baboul, A. G., Stefanov, B. B., Liu, G., Liashenko, A., Piskorz, P., Komaromi, I., Gomperts, R., Martin, R. L., Fox, D. J., Keith, T., Al-Laham, M. A., Peng, C. Y., Nanayakkara, A., Gonzalez, C., Challacombe, M., Gill, P. M. W., Johnson, B. G., Chen, W., Wong, M. W., Andres, J. L., Head-Gordon, M., Replogle, E. S., and Pople, J. A. (1998) Gaussian 98 (Revision A.9), Gaussian, Inc., Pittsburgh, PA.
- 59.Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang J, Kollman P. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 60.van Gunsteren, W. F., Billeter, S. R., Eising, A. A., Hünenberger, P. H., Krüger, P., Mark, A. E., Scott, W. R. P., and Tironi, I. G. (1996) The GROMOS96 Manual and User Guide, VdF Hochsculverlag AG an der ETH Zürich, Zürich, Switzerland.
- 61.Lindahl E, Hess B, van der Spoel D. GROMACS 3.0: A package for molecular simulation and trajectory analysis. J Mol Model. 2001;7:306–317. [Google Scholar]
- 62.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graphics. 1996;14:27–28. doi: 10.1016/0263-7855(96)00018-5. 33–38. [DOI] [PubMed] [Google Scholar]
- 63.Fiser A, Sali A. Modeller: Generation and refinement of homology-based protein structure models. Methods Enzymol. 2003;374:461–491. doi: 10.1016/S0076-6879(03)74020-8. [DOI] [PubMed] [Google Scholar]
- 64.Sarkar HK, Song PS. Nature of phototransformation of phytochrome As probed by intrinsic tryptophan residues. Biochemistry. 1982;21:1967–1972. doi: 10.1021/bi00537a041. [DOI] [PubMed] [Google Scholar]
- 65.Stone, J. (1998) Master’s thesis in Computer Science Department, University of Missouri—Rolla.
- 66.Frishman D, Argos P. Knowledge-based protein secondary structure assignment. Proteins. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- 67.Russell RB, Barton GJ. Multiple protein sequence alignment from tertiary structure comparison: Assignment of global and residue confidence levels. Proteins. 1992;14:309–323. doi: 10.1002/prot.340140216. [DOI] [PubMed] [Google Scholar]
- 68.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bashford D, Gerwert K. Electrostatic calculations of the pKa values of ionizable groups in bacteriorhodopsin. J Mol Biol. 1992;224:473–486. doi: 10.1016/0022-2836(92)91009-e. [DOI] [PubMed] [Google Scholar]
- 70.Spassov VZ, Luecke H, Gerwert K, Bashford D. pKa calculations suggest storage of an excess proton in a hydrogen-bonded water network in bacteriorhodopsin. J Mol Biol. 2001;312:203–219. doi: 10.1006/jmbi.2001.4902. [DOI] [PubMed] [Google Scholar]
- 71.Falk, H. (1989) The Chemistry of Linear Oligopyrroles and Bile Pigments, pp 621, Springer-Verlag, Vienna, Austria.
- 72.Ho YS, Burden LM, Hurley JH. Structure of the GAF domain, a ubiquitous signaling motif, and a new class of cyclic GMP receptor. EMBO J. 2000;19:5288–5299. doi: 10.1093/emboj/19.20.5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Martinez SE, Bruder S, Schultz A, Zheng N, Schultz JE, Beavo JA, Linder JU. Crystal structure of the tandem GAF domains from a cyanobacterial adenylyl cyclase: Modes of ligand binding and dimerization. Proc Natl Acad Sci USA. 2005;102:3082–3087. doi: 10.1073/pnas.0409913102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bischoff M, Hermann G, Rentsch S, Strehlow D. First steps in the phytochrome phototransformation: A comparative femtosecond study on the forward (Pr → Pfr) and back reaction (Pfr → Pr) Biochemistry. 2001;40:181–186. doi: 10.1021/bi0011734. [DOI] [PubMed] [Google Scholar]
- 75.Heyne K, Herbst J, Stehlik D, Esteban B, Lamparter T, Hughes J, Diller R. Ultrafast dynamics of phytochrome from the cyanobacterium Synechocystis, reconstituted with phycocyanobilin and phycoerythrobilin. Biophys J. 2002;82:1004–1016. doi: 10.1016/S0006-3495(02)75460-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Murphy JT, Lagarias JC. The phytofluors: A new class of fluorescent protein probes. Curr Biol. 1997;7:870–876. doi: 10.1016/s0960-9822(06)00375-7. [DOI] [PubMed] [Google Scholar]
- 77.Schirmer T, Bode W, Huber R. Refined three-dimensional structures of two cyanobacterial C-phycocyanins at 2.1 and 2.5 Å resolution. A common principle of phycobilin–protein interaction. J Mol Biol. 1987;196:677–695. doi: 10.1016/0022-2836(87)90040-4. [DOI] [PubMed] [Google Scholar]
- 78.Duerring M, Schmidt GB, Huber R. Isolation, crystallization, crystal structure analysis, and refinement of constitutive C-phycocyanin from the chromatically adapting cyanobacterium Fremyella diplosiphon at 1.66 Å resolution. J Mol Biol. 1991;217:577–592. doi: 10.1016/0022-2836(91)90759-y. [DOI] [PubMed] [Google Scholar]
- 79.Kuramitsu S, Hamaguchi K, Miwa S, Nakashima K. Ionization of the catalytic groups and tyrosyl residues in human lysozyme. J Biochem. 1980;87:771–778. doi: 10.1093/oxfordjournals.jbchem.a132806. [DOI] [PubMed] [Google Scholar]
- 80.Bashford D, Karplus M. pKa’s of ionizable groups in proteins: Atomic detail from a continuum electrostatic model. Biochemistry. 1990;29:10219–10225. doi: 10.1021/bi00496a010. [DOI] [PubMed] [Google Scholar]
- 81.Inoue M, Yamada H, Hashimoto Y, Yasukochi T, Hamaguchi K, Miki T, Horiuchi T, Imoto T. Stabilization of a protein by removal of unfavorable abnormal pKa: Substitution of undissociable residue for glutamic acid-35 in chicken lysozyme. Biochemistry. 1992;31:8816–8821. doi: 10.1021/bi00152a018. [DOI] [PubMed] [Google Scholar]
- 1.Martinez SE, Wu AY, Glavas NA, Tang XB, Turley S, Hol WG, Beavo JA. The two GAF domains in phosphodiesterase 2A have distinct roles in dimerization and in cGMP binding. Proc Nat Acad Sci USA. 2002;99:13260–5. doi: 10.1073/pnas.192374899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tasler R, Moises T, Frankenberg-Dinkel N. Biochemical and spectroscopic characterization of the bacterial phytochrome of Pseudomonas aeruginosa. FEBS J. 2005;272:1927–36. doi: 10.1111/j.1742-4658.2005.04623.x. [DOI] [PubMed] [Google Scholar]
- 3.Davis SJ, Vener AV, Vierstra RD. Bacteriophytochromes: phytochrome-like photoreceptors from nonphotosynthetic eubacteria. Science. 1999;286:2517–20. doi: 10.1126/science.286.5449.2517. [DOI] [PubMed] [Google Scholar]


