Abstract
The effect of dietary fat source (soy oil or a mixture of lard and tallow) and dietary supplementation with antibiotics (a combination of avilamycin at 10 mg kg of feed−1 and salinomycin at 40 mg kg of feed−1) on the bacterial community in the ileum of broiler chickens at different ages (7, 14, 21, and 35 days) was studied using PCR with denaturing gradient gel electrophoresis (DGGE) analysis and bacteriological culture. The bacterial origin of fragments in DGGE profiles was identified by sequencing. Bacterial enumeration results, together with PCR-DGGE profiles, showed that the composition of the microflora was age dependent and influenced by dietary fat source and antibiotic supplementation. An increased incidence of streptococci, enterobacteria, and Clostridium perfringens with age of the chickens was demonstrated. Lactobacilli and C. perfringens were the bacterial groups most strongly affected by the dietary treatments. Moreover, different strains (clonal variants of the alpha-toxin gene) of C. perfringens type A were detected in response to age, dietary fat source, and dietary supplementation with antibiotics.
Although the microbiology of the ceca of chickens has received considerable attention (2, 13, 16, 17), the small intestine has been less well studied (3, 23). This is surprising because the jejunum and ileum are the principal sites of nutrient absorption (10, 28). It is generally accepted that the gut microflora has an impact on avian growth and health and that the activities of the microflora can be manipulated by altering the diet (4, 15, 29). The effects of dietary carbohydrates have been studied (29, 33), but the role of dietary fat in relation to avian gut microbial ecology is unknown. The effects of antibiotic “growth promoters” added to the feed appear to be mediated through their impact on the microflora (4, 11). Gram-positive members of the microflora are the main target for antibiotic growth promoters in poultry feed. These bacteria putatively compete with the host for nutrients and, in the case of Clostridium perfringens, produce toxins (4, 7). Even more important may be the production of enzymes that deconjugate bile salts, resulting in impaired fat emulsification and lipid absorption (9, 31).
The majority of the studies investigating host-microflora-diet relationships in chickens have been based on traditional bacteriological culture and identification techniques. Analysis using only culture methods can bias results in favor of the more easily cultivated members of the community (22). These techniques can also be time-consuming, especially when a large number of samples must be processed. Culture-independent approaches, such as PCR with denaturing gradient gel electrophoresis (DGGE), are useful in studying microbial diversity (20, 21, 26, 27, 34). The aim of the present study was to use both bacteriological culture and PCR-DGGE to monitor the composition of the bacterial community in the ileal contents of 7-, 14-, 21-, and 35-day-old broiler chickens fed different dietary fat sources and subtherapeutic levels of the growth promoters salinomycin and avilamycin. We focused on the ileal microflora of broilers because of the importance of this intestinal region in nutrient absorption and because preliminary experiments showed that the microflora is the same in this site as in the jejunum (W. T. McBurney and G. W. Tannock, unpublished data).
MATERIALS AND METHODS
Animals, treatment, and sampling.
A total of 1,600 1-day-old male broiler chickens (Ross 208) were housed in 32 pens (50 birds per pen). The pens were assigned at random to one of four dietary treatments, where the random allocation of the treatments was balanced in relation to the position in the rearing house (eight replicate blocks). The diets used were wheat-based mash (Table 1), to which was added either 10% animal fat (A diet), which was a mixture of lard and tallow (1.5:1), or 10% soy oil (S diet). Each diet was left nonsupplemented (−) or was supplemented (+) with a combination of salinomycin (40 mg kg of feed−1) and avilamycin (10 mg kg of feed−1). Salinomycin and avilamycin were premixed in calcium carbonate before being added to the respective experimental diets. The animals had free access to feed and water and were subjected to continuous artificial illumination. From each pen, 16, 10, 8, and 6 animals were randomly selected at age 7, 14, 21, and 35 days, respectively, and killed by cervical dislocation. The number of birds removed at each sampling time was calculated to ensure that an adequate amount of intestinal material was available for analysis and to avoid major changes in stocking density (kilograms per square meter) during the experiment. The small intestine was rapidly excised at Meckel's diverticulum to the ileocecal junction (ileum). The ileal content was collected and pooled for the chickens within each pen at each sampling time. Samples were divided into aliquots for DNA-based and culture-based analysis. Samples for DNA analysis were stored at −20°C. The experiments complied with the guidelines of the Danish Ministry of Justice with respect to animal experimentation and care of animals under study.
TABLE 1.
Composition of the experimental diets
| Ingredient | Amt (g kg−1) in experimental dieta:
|
|||
|---|---|---|---|---|
| A− | A+ | S− | S+ | |
| Wheat | 501.3 | 501.3 | 501.3 | 501.3 |
| Soybean meal, toasted | 275.9 | 275.9 | 275.9 | 275.9 |
| Peas | 40.0 | 40.0 | 40.0 | 40.0 |
| Fishmeal | 40.0 | 40.0 | 40.0 | 40.0 |
| Lard | 60.0 | 60.0 | ||
| Tallow | 40.0 | 40.0 | ||
| Soy oil | 100.0 | 100.0 | ||
| Sodium chloride | 2.0 | 2.0 | 2.0 | 2.0 |
| Dicalcium phosphate | 19.5 | 19.5 | 19.5 | 19.5 |
| Calcium carbonate | 7.0 | 4.7 | 7.0 | 4.7 |
| Sodium bicarbonate | 1.5 | 1.5 | 1.5 | 1.5 |
| dl-Methionine (40%) | 5.0 | 5.0 | 5.0 | 5.0 |
| Vitamin and mineral mixb | 3.5 | 3.5 | 3.5 | 3.5 |
| Choline chloride (50%) | 0.4 | 0.4 | 0.4 | 0.4 |
| Chromoxid | 4.0 | 4.0 | 4.0 | 4.0 |
| Antibiotic premix in CaCO3c | 2.3 | 2.3 | ||
| Analyzed compositiond | ||||
| Dry matter (g/kg of feed) | 904.5 | 908.4 | 910.5 | 907.8 |
| Stoldt fat | 145.9 | 140.9 | 140.0 | 145.6 |
| Crude protein | 247.8 | 245.9 | 256.3 | 245.6 |
| Starch | 338.7 | 352.2 | 333.9 | 345.9 |
| Sugar | 61.6 | 62.8 | 61.3 | 59.7 |
| Fibre | 37.1 | 33.4 | 33.2 | 32.5 |
| Ash | 68.2 | 67.5 | 69.8 | 70.4 |
| Gross energy (MJ kg−1) | 20.8 | 20.7 | 20.9 | 20.6 |
The A diet consisted of 10% of a mixture of lard and tallow (1.5:1); the S diet consisted of 10% soy oil; − and + indicate a nonsupplemented diet and a diet supplemented with a combination of avilamycin and salinomycin, respectively.
Supplying (per kilogram of diet) retinol (retinyl acetate), 16,800 IU; cholecalciferol, 3,500 IU; vitamin E (dl-α-tocopheryl acetate), 42 IU; menadione, 3.5 mg; thiamine, 1.4 mg; riboflavin, 11.2 mg; pyridoxine, 4.2 mg; d-pantothenic acid, 14 mg; niacin, 56 mg; betaine anhydrate, 473 mg; folic acid, 2.1 mg; biotin, 140 μg; cyanocobalamin, 28 μg; BHT, 140 mg; FeSO4·7H2O, 112 mg; ZnO, 112 mg; MnO, 140 mg; CuSO4·5H2O, 21 mg; KI, 840 μg; Na2SeO3, 420 μg.
Supplying (per kilogram of diet) avilamycin, 10 mg; salinomycin, 40 mg.
Compositions are given in grams per kilogram of dry matter unless otherwise stated.
Enumeration of bacteria.
Ileal content (10 g) was immediately transferred under a flow of CO2 gas into sterile serum bottles containing 90 ml of a prereduced broth, composed of the following constituents in distilled water (grams per liter): peptone, 2.5; yeast extract, 2.5; NaCl, 0.9; KH2PO4, 0.45; CaCl2 · 2H2O, 0.027; MgCl2 · 6H2O, 0.02; MnSO4 · 4H2O, 0.012; CoCl2 · 6H2O, 0.01; (NH4)2SO4, 0.9; FeSO4 · 7H2O, 0.008; K2HPO4 · 3H2O, 0.45; hemin, 0.003; resazurin, 0.001; and 1.0 ml of Tween 80. The suspension was poured into a CO2-flushed plastic bag and homogenized in a stomacher laboratory blender (Seward Medical, London, United Kingdom) for 2 min. Serial 10-fold dilutions were performed by the technique of Miller and Wolin (19). The Lactobacillus count was determined using Rogosa agar (Merck 5413). This medium was incubated in an anaerobic cabinet (10:10:80 CO2-H2-N2 atmosphere) at 37°C for 48 h. Enterobacteria were enumerated on MacConkey agar (Merck 5465) after aerobic incubation at 37C° for 24 h. C. perfringens counts were made using tryptose sulfite agar (Merck 1972) supplemented with cycloserine (Oxoid SR088E). C. perfringens colonies were enumerated after 24 h of anaerobic incubation at 37°C.
DNA extraction.
Nucleic acids were extracted from the ileal content of broilers essentially as described by Walter et al. (34). For DNA extraction, 100 mg of ileal content was weighed into a sterile tube containing 300 mg of sterile zirconium beads (diameter, 0.1 mm) and suspended in 1 ml of TN150 buffer (10 mM Tris-HCl, 150 mM NaCl [pH 8.0]). The suspension was vortexed thoroughly and centrifuged at 14,600 × g for 5 min. The supernatant was discarded, and the pellet was washed twice with 1 ml of TN150 buffer. After the final wash, the pellet was resuspended in 1 ml of TN150 buffer. The cells were lysed by physical disruption in a mini-bead beater (Biospec Products, Bartlesville, Okla.) at 5,000 rpm for 3 min and placed on ice to cool. Subsequently, the sample was centrifuged at 14,600 × g for 5 min. A 500-μl volume of the supernatant was extracted three times with 500 μl of TE (10 mM Tris, 1 mM EDTA [pH 8.5])-saturated phenol, and the residual phenol was removed by extraction with an equal volume of chloroform-isoamyl alcohol (24:1). The nucleic acids were precipitated with 2 volumes of cold ethanol (−20°C) and 0.1 volume of 3 M sodium acetate and stored overnight at −20°C. The DNA was collected by centrifugation at 14,600 × g for 20 min at −4°C, and the pellet was dried at 37°C for 1 h. Finally, DNA was dissolved in 30 μl of TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 7.5]).
PCR amplification with HDA (total bacterial community) and species-specific primers.
Target DNA was amplified using Taq DNA polymerase (Roche, Mannheim, Germany) and a PCR-Express thermal cycler (Hybaid, Teddington, United Kingdom). PCR amplifications of total bacterial community DNA were carried out using primer pair HDA1-GC plus HDA2 and a PCR thermocycling program as described by Walter et al. (34). To specifically detect lactic acid bacteria and C. perfringens in ileal samples, amplifications with group-specific primers were conducted. Lactic acid bacteria-specific primers (Lac primers) Lac1 and Lac2-GC were used in the procedure reported by Walter et al. (35). A 324-bp fragment of the C. perfringens genome encoding the alpha-toxin gene (32) was amplified using the Cpa primers Cpa1-GC (CGC CCG GGG CGC GCC CCG GGC GGC CCG GGG GCA CCG GGG GGC TAA TGT TAC TGC CGT TGA-3′ [the GC clamp is in boldface]) and Cpa2 (5′-CCT CTG ATA CAT CGT GTA AG-3′) (14). The reaction mixture (50 μl) consisted of reaction buffer (final concentrations, 10 mM Tris-HCl, 2 mM MgCl2, and 50 mM KCl [pH 8.3]), each deoxynucleoside triphosphate at 120 μM, 0.5 μM each primer, 20 μg of bovine serum albumin, and 5.0 U of Taq DNA polymerase. The PCR program included 35 cycles of denaturation (1 min at 94°C), annealing (1 min at 55°C), and extension (1 min at 72°C). The integrity of the PCR products was checked by electrophoresis for 15 min in a 2% agarose gel stained with ethidium bromide (5 μg ml−1) and viewed by UV transillumination. To identify the PCR amplicons obtained using the Lac primers and to supplement identifications made by sequencing fragments eluted from gel slices, an identification ladder was prepared (34) that consisted of the following reference strains listed in order of migration distance in the DGGE gel: Lactobacillus plantarum ATCC 14917T, L. johnsonii ATCC 33200T, Weissella viridescens DSM 20410T, L. gasseri ATCC 33323T, L. acidophilus ATCC 4356T, L. crispatus ATCC 33820T, L. salivarius ATCC 11741T, L. ruminis ATCC 277780T, L. rhamnosus ATCC 7469T, and L. reuteri DSM 20016T. C. perfringens type A ATCC 13124T was included in the PCR-DGGE analysis as a reference in gels profiling amplicons obtained using Cpa primers.
DGGE analysis of PCR products.
The DGGE was performed as reported by Walter et al. (34), using a 16-cm by 16-cm by 1-mm-thick 6% polyacrylamide gel (ratio of acrylamide to bisacrylamide, 37.5:1), containing a 30 to 55% (HDA1-GC and HDA2 amplicons), a 30 to 45% (Lac amplicons), or a 20 to 35% (Cpa amplicons) gradient of urea and formamide. The similarity of bacterial community profiles within broiler groups (age and diet) was compared within each gel. Representative profiles of each age and diet group were then run in a single gel to obtain comparisons of bacterial communities in birds of different age groups. Therefore, interpretations of similarity and diversity of profiles between different electrophoretic gels did not occur. Profiles were compared using Quantity One version 4.2 software, which is part of the Discovery Series (Bio-Rad Laboratories, 2000) and were performed using Dice's similarity coefficient (Dsc) analysis.
Identification of bacteria by sequencing PCR-DGGE fragments.
DNA fragments of interest were excised aseptically from the polyacrylamide gel, placed in 100 μl of diffusion buffer, QIAEX II (0.5 M ammonium acetate, 10 mM magnesium acetate, 1 mM EDTA, 0.1% sodium dodecyl sulfate [pH 8.0]), and incubated overnight at 4°C to allow elution of the DNA. DNA was recovered using the QIAEX II kit (Qiagen, Hilden, Germany) as specified by the manufacturer. PCR amplification was accomplished using primers without the GC clamp. To avoid PCR artifacts with respect to heteroduplex formation and thereby obtain only the desired fragment, the number of cycles in the original thermocycling program was reduced to 15 or 20 cycles. The resulting PCR products were purified using the QIAquick PCR purification kit (Qiagen) and cloned into Escherichia coli DH5α by using the pGEM-T vector system (Promega, Madison, Wis.). To isolate plasmid DNA of selected transformants, 1.5 ml of the culture medium (24) was centrifuged for 5 min at 14,600 × g. The pellet was resuspended in 100 μl of GTE buffer (50 mM glucose, 25 mM Tris, 25 mM Tris-HCl, 10 mM EDTA [pH 8.0]) and subsequently in 200 μl of 0.2 M NaOH-1% sodium dodecyl sulfate-solution and placed on ice for 10 min. Then 150 μl of 5 M potassium acetate was added, and the sample was cooled on ice for 10 min. After two successive centrifugations in which the supernatant was retained, 900 μl of cold ethanol (−20°C) was added and the precipitated plasmid DNA was collected by centrifugation. Prior to sequencing, the plasmid DNA and the original template DNA, from which the band was excised, were subjected to 15 and 35 PCR thermal cycles, respectively, using the HDA1-GC and HDA2 primers and were checked by DGGE. Only products that migrated as a single band and to the same position with respect to the original sample were used for sequencing. For sequencing, the pGEM-T insert DNA was amplified using sequencing primers T7 and Sp6 (Promega). Sequencing was carried out by the Centre for Gene Research, University of Otago, Dunedin, New Zealand, as described by Walter et al. (34). The sequences retrieved were compared with the GenBank database using BLAST algorithm (1). Unless otherwise stated in Results, the BLAST search gave similarity values greater than 99% to database sequences.
Statistical analysis of culture results.
Values in the text are presented as least-square means ± standard error of the mean. The experiment can be regarded as a common 2 × 2 factorial design (30), and the statistical analysis was accomplished using the general-linear-models procedure of SAS (25). The statistical analysis was based on the following model:
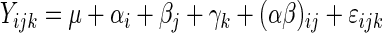 |
where Yijk is the observed response, μ the overall mean, and αi, βj, and γk denote the effect of fat, antibiotic, and block, respectively. The interaction between fat and antibiotic is designated (αβ)ij. Finally, ɛijk ≈ N(0,σ2) is the random error. The model was verified by plotting the residuals against the predicted values and by use of quantile plots of the residuals. In cases where the interaction was not significant (P ≥ 0.05), it was omitted from the model. If an overall effect was significant (P < 0.05), differences between means were compared pairwise (pdiff). For comparison of means of age effects, Student's t test was applied (30).
RESULTS
Bacterial enumeration by culture.
The bacterial populations in the ileal contents of broilers aged 7, 14, 21, and 35 days, fed the experimental diets, are shown in Table 2. From days 7 to 35, the counts of lactose-fermentative and nonfermentative enterobacteria increased (P < 0.001). The C. perfringens population also increased (approximately 3 log units) from days 7 to 35 (P < 0.001). On average, lactobacilli were abundant in all age groups, and the viable counts were unaffected by the dietary lipid source. The number of lactobacilli was reduced in the ileum of 14- and 21-day old broilers fed the diets supplemented with antibiotics (Table 2). In general, the number of enterobacteria was unaffected by lipid source and antibiotic supplementation during the entire growth period. In contrast, the viable count of C. perfringens was influenced by both lipid source and antibiotic supplementation. The number of C. perfringens was smaller in the soy oil-fed broilers than in the broilers fed animal fat (Table 2). Reduced counts of C. perfringens were also observed in broilers fed antibiotic-supplemented feed compared to those in broilers fed the nonsupplemented diets (Table 2). The effects of antibiotics and fat source were additive: statistical analysis did not show a significant interaction between the two factors.
TABLE 2.
Effect of dietary fat source and antibiotic supplementation on counts of bacteria in the ileal content from broilers
| Bacterial group | Bacterial counts (least-squares mean log10 CFU) after following treatmenta:
|
SEM | Effects
|
|||||
|---|---|---|---|---|---|---|---|---|
| A− | A+ | S− | S+ | Fat P value | Antibiotic P value | Fat × antibiotic P value | ||
| Day 7 | ||||||||
| Lactobacilli | 8.52 | 8.50 | 8.62 | 8.46 | 0.065 | 0.6556 | 0.1991 | 0.3315 |
| Fermentative enterobacteria | 5.40 | 5.83 | 5.06 | 5.22 | 0.281 | 0.0990 | 0.2911 | 0.6367 |
| Nonfermentative enterobacteria | 4.29 | 4.65 | 4.39 | 4.55 | 0.240 | 0.9866 | 0.2945 | 0.6851 |
| C. perfringens | 5.04 | 3.32 | 4.50 | 3.20 | 0.204 | 0.1109 | 0.0001 | 0.3202 |
| Day 14 | ||||||||
| Lactobacilli | 8.12 | 7.51 | 7.99 | 7.45 | 0.114 | 0.4216 | 0.0001 | 0.7780 |
| Fermentative enterobacteria | 5.49 | 5.90 | 5.82 | 5.58 | 0.271 | 0.9742 | 0.7515 | 0.2330 |
| Nonfermentative enterobacteria | 4.18 | 4.48 | 4.04 | 4.26 | 0.239 | 0.4617 | 0.2834 | 0.8756 |
| C. perfringens | 6.35 | 3.03 | 5.50 | 2.81 | 0.199 | 0.0087 | 0.0001 | 0.1132 |
| Day 21 | ||||||||
| Lactobacilli | 7.76 | 7.14 | 7.73 | 7.41 | 0.134 | 0.3686 | 0.0008 | 0.2670 |
| Fermentative enterobacteria | 6.09 | 6.09 | 6.38 | 6.35 | 0.218 | 0.2259 | 0.9464 | 0.9474 |
| Nonfermentative enterobacteria | 3.85 | 4.44 | 4.09 | 4.53 | 0.203 | 0.4127 | 0.0124 | 0.7131 |
| C. perfringens | 6.62 | 3.93 | 6.46 | 3.13 | 0.199 | 0.0176 | 0.0001 | 0.1101 |
| Day 35 | ||||||||
| Lactobacilli | 8.88 | 8.73 | 8.59 | 8.68 | 0.107 | 0.1339 | 0.7923 | 0.2485 |
| Fermentative enterobacteria | 7.25 | 7.16 | 7.31 | 7.60 | 0.187 | 0.1829 | 0.6063 | 0.3227 |
| Nonfermentative enterobacteria | 5.96 | 5.99 | 6.34 | 6.36 | 0.190 | 0.0489 | 0.9065 | 0.9865 |
| C. perfringens | 8.04 | 6.08 | 7.24 | 5.44 | 0.188 | 0.0003 | 0.0001 | 0.6633 |
Diet A consisted of 10% of a mixture of lard and tallow (1.5:1); diet S consisted of 10% soy oil; and − and + indicate a nonsupplemented diet or a diet supplemented with a combination of avilamycin (10 mg/kg of feed) and salinomycin (40 mg/kg of feed), respectively.
DGGE profiles generated with HDA (total-microflora) primers.
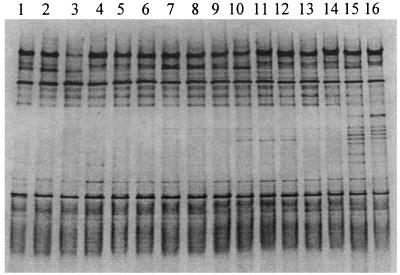
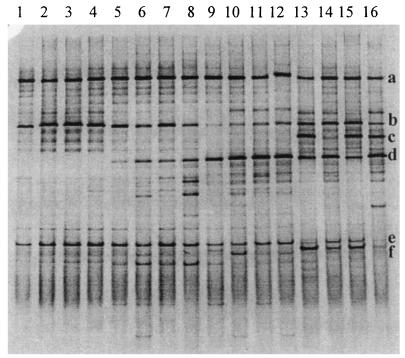
Although some minor differences were noted in position, staining intensity, and number of DNA fragments present, the ileal bacterial community profiles were consistent within replicate pools of each age group. An example is provided in Fig. 1, in which profiles from pools of ileal contents of 7-day-old birds are shown. The Dsc values indicated that a large proportion of the fragment profile (average, 73.8%; standard deviation, 8.6) in each of these birds was shared, regardless of the dietary group. Comparison of representative DGGE profiles from all groups (Fig. 2) showed an effect of age and dietary supplementation on the composition of the microflora in the ileum of broilers. Average Dsc values showed increasing disparity between the profiles of different age groups (7 days old compared to 14 days old, 71.8%; 7 days old compared to 21 days old, 50.7%; 7 days old compared to 35 days old, 47.9%). Fragment a was identified by DNA sequencing as L. johnsonii and was detected in all age groups. Fragment b corresponded to L. crispatus and was also present in all of the birds. Fragment c represented L. salivarius and was present only in profiles from 35-day-old birds. It was not detected in ileal contents from chickens fed antibiotics and animal fat (Fig. 2, lane 14). Fragment d was identified as a member of the genus Streptococcus. Definitive identification of this bacterium at species level was not obtained because the BLAST search showed an equally high similarity to several species such as Streptococcus alactolyticus, S. bovis, and S. equinus. The Streptococcus organism was not detectable in the youngest chickens, but its DNA fragment was present in profiles of 14-day-old and older chickens. Fragment e was detected throughout the growth period. Sequencing of fragment e showed 98% (196 of 199 bp) and 97% (194 of 199 bp) similarity to a yet uncultured bacterium (GenBank accession number AJ400274) and L. reuteri, respectively. C. perfringens (fragment f) was detected only in samples from 35-day-old broilers. DNA fragments representing C. perfringens were faintly stained in samples from birds given antibiotics (lanes 14 and 16).
FIG. 1.
PCR-DGGE profiles (30 to 55% denaturing gradient gel) generated from ileal template DNA from 7-day-old chickens, using primer pair HDA1-GC plus HDA2. Lanes 1 to 8 are from replicate pooled samples from 16 chickens fed the S− diet, and lanes 9 to 16 are from replicate pooled samples from 16 chickens fed the S+ diet. Note the consistency of profiles detected in birds of the same age.
FIG. 2.
PCR-DGGE profiles (30 to 55% denaturing gradient gel) generated from representative ileal content pools, using primer pair HDA1-GC plus HDA2. Lanes: 1 to 4, samples from 7-day-old chickens fed the A− diet (lane 1), the A+ diet (lane 2), the S− diet (lane 3), and the S+ diet (lane 4); 5 to 8, samples from 14-day-old chickens fed the A− diet (lane 5), the A+ diet (lane 6), the S− diet (lane 7), and the S+ diet (lane 8); 9 to 12, samples from 21-day-old chickens fed the A− diet (lane 9), the A+ diet (lane 10), the S− diet (lane 11), and the S+ diet (lane 12); 13 to 16, samples from 35-day-old chickens fed the A− diet (lane 13), the A+ diet (lane 14), the S− diet (lane 15), and the S+ diet (lane 16). The fragments were allotted by sequence analysis to the following genera or species: fragment a, L. johnsonii; fragment b, L. crispatus; fragment c, L. salivarius; fragment d, Streptococcus spp.; fragment e, sequence similarity of 98% to an uncultured bacterium and 97% to L. reuteri; fragment f, C. perfringens.
DGGE profiles generated with Lac primers.
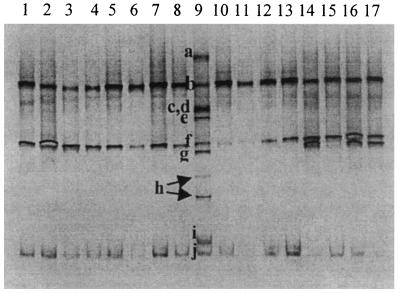
All of the fragments detected by PCR with Lac primers plus DGGE could be identified by reference to the Lactobacillus species ladder. The presence of the four different Lactobacillus species detected by PCR with HDA primers plus DGGE and DNA sequencing was confirmed using the lactic acid bacteria-specific primers (Fig. 3; L. johnsonii, L. crispatus, L. salivarius, and L. reuteri). L. salivarius was not detected in birds fed the antibiotic-supplemented diet containing animal fat.
FIG. 3.
Detection of lactobacilli in representative ileal content pools, using primer pair Lac1 plus Lac2-GC in a 30 to 45% gradient gel. Lanes: 1 to 4, samples from 7-day-old chickens fed the A− diet (lane 1), the A+ diet (lane 2), the S− diet (lane 3), and the S+ diet (lane 4); 5 to 8, samples from 14-day-old chickens fed the A− diet (lane 5), the A+ diet (lane 6), the S− diet (lane 7), and the S+ diet (lane 8); 9, identification ladder composed of sequences from reference strains of Lactobacillus species (a, L. plantarum; b, L. johnsonii; c, Weissella viridescens; d, L. gasseri; e, L. acidophilus; f, L. crispatus; g, L. salivarius; h, L. ruminis; i, L. rhamnosus; j, L. reuteri); 10 to 13, samples from 21-day-old chickens fed the A− diet (lane 10), the A+ diet (lane 11), the S− diet (lane 12), and the S+ diet (lane 13); 14 to 17, samples from 35-day-old chickens fed the A− diet (lane 14), the A+ diet (lane 15), the S− diet (lane 16), and the S+ diet (lane 17).
DGGE profiles generated with Cpa primers.
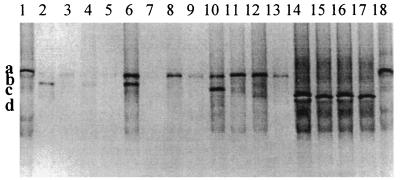
The DGGE profiles generated with Cpa primers (Fig. 4) revealed two fragments in Cpa amplicons from 14- and 21-day-old chickens fed the A− diet (fragments a and b in lane 6 and fragments a and c in lane 10), while only one fragment was present in the amplicons from chickens fed the S− diet (fragment a in lanes 8 and 12). An antibiotic-associated effect in 14- and 21-day-old chickens was detected, which was more noticeable in the youngest birds. Thus, two fragments were detected in chickens fed the A− diet (fragments a and b in lane 6 and fragments a and c in lane 10), but none or one fragment was detected in 14- and 21-day old chickens fed the A+ diet, respectively (lane 7 and fragment a in lane 11). One fragment was present in 35-day-old chickens irrespective of the dietary treatment (fragment d in lanes 14 to 17). Fragment a had a migration distance similar to that of the reference strain (lanes 1 and 18). Sequencing of the fragments and subsequent nucleotide alignment showed that the alpha-toxin genes differed from each other by a few nucleotides (fragment a, GenBank accession number AF477010; fragment b, AF477008; fragment c, AF475144; fragment d, AF477009), which explained the different migration distances in the denaturing gel. Amino acid alignment of the alpha-toxins (Table 3), showed that fragment a was different from fragments b, c, and d at positions 174 (Ala174 to Asp174) and 177 (Thr177 to Ala177). Fragments b, c, and d had the same amino acid sequence.
FIG. 4.
Detection of C. perfringens in ileal content pools, using primer pair Cpa1-GC plus Cpa2 in a 20 to 35% gradient gel. Lanes: 1, C. perfringens ATCC 13124T; 2 to 5, samples from 7-day-old chickens fed the A− diet (lane 2), the A+ diet (lane 3), the S− diet (lane 4), and the S+ diet (lane 5); 6 to 9, samples from 14-day-old chickens fed the A− diet (lane 6), the A+ diet (lane 7), the S− diet (lane 8), and the S+ diet (lane 9); 10 to 13, samples from 21-day-old chickens fed the A− diet (lane 10), the A+ diet (lane 11), the S− diet (lane 12), and the S+ diet (lane 13); 14 to 17, samples from 35-day-old chickens fed the A− diet (lane 14), the A+ diet (lane 15), the S− diet (lane 16), and the S+ diet (lane 17); 18, C. perfringens ATCC 13124T. Fragments a (AF477010), b (AF477008), c (AF475144), and d (AF477009) represent four different strains of C. perfringens type A.
TABLE 3.
Amino acid sequence differences between alpha-toxins of C. perfringens strains detected in ileal content of chickens
| DGGE fragment | Amino acid sequencea |
|---|---|
| a | 171TNEAFYTD178 |
| b, c, d | 171TNEDFYAD178 |
Amino acid differences between the peptides are shown in bold type.
DISCUSSION
The study showed that the ileal contents of chickens of different ages that were fed different dietary supplements harbored a bacterial community of relatively simple composition in which lactobacilli, streptococci, and C. perfringens were commonly detected. Other bacterial populations were present, as demonstrated by faintly staining fragments in DGGE gels, but the center of our focus was on the fragments that were affected by dietary modification. Both PCR-DGGE and bacteriological culture showed that the composition of the ileal microflora changed in relation to age of the birds. Streptococci were not detected by PCR-DGGE in the youngest chickens but were present in older birds. A culture medium selective for streptococci was not included in the study, so that quantitative information about these bacteria was not obtained. An identification of the streptococci to species level was not accomplished by the BLAST search, but S. alactolyticus has been isolated in a previous culture-based study of broiler chickens (A. Knarreborg, unpublished data). Enterobacteria and C. perfringens became more numerous as the chickens grew older.
Regardless of age and dietary treatment, the total counts of lactobacilli were >107 CFU g of ileal content−1. Lactobacillus counts were lowered in 14- and 21-day-old chickens fed antibiotics. An insight into the composition of the Lactobacillus population was provided by DGGE using HDA as well as Lac primers. Four Lactobacillus species were detected by PCR-DGGE, of which L. salivarius was detected only in 35-day-old birds. L. salivarius was not detected in birds fed the animal fat-supplemented diet containing antibiotics. Thus, PCR-DGGE detected a modification in the composition of the bacterial community that was not demonstrated by observation of total Lactobacillus numbers. Other Lactobacillus species must have proliferated in the absence of L. salivarius such that the total number of lactobacilli remained constant.
C. perfringens was present in small numbers in 7-day-old chickens, but its numbers increased gradually thereafter in birds that had not been given antibiotics. C. perfringens numbers were even smaller when the diet contained soy oil. Both PCR-DGGE profiles of total bacterial community and counts of C. perfringens showed an additive effect between fat source and antibiotics, particularly in 35-day-old birds. A dietary effect was not demonstrated in this age group when Cpa primers were used to detect the clostridia. This showed that primers with different PCR detection limits can be useful when screening for changes in the composition of the microflora where the species of interest are present in all animals but of different population sizes. A possible explanation for the additive effect of soy oil and antibiotics on clostridial numbers could be that both of the antibiotics have lipophilic properties (18). Hence, the activities of these substances may be influenced by the dietary fat source. Due to a higher content of unsaturated fatty acids, soy oil exhibits a higher solubility in the micellar phase (10), which may facilitate an enhanced dispersion of antibiotics in the small intestine. The type of fat source is also likely to indirectly influence the intestinal microflora through its impact on viscosity of the digesta, intestinal transit time, and digestion in the small intestine (6). Our observations concerning the impact of dietary fat source and antibiotics on the ileal clostridial population were particularly important because it may be possible to relate clostridial numbers to bile salt hydrolase activity in the small intestine and growth performance in broilers (5, 9). Further, a lower incidence of C. perfringens in the small intestine of chickens may reduce the likelihood that the birds will develop necrotic enteritis (7). Salinomycin, which is widely used as an anticoccidial drug, inhibits the growth of C. perfringens (7, 8). The continued use of salinomycin in broiler feed may be responsible for the fact that broiler producers have not noticed an increase in acute or subacute necrotic enteritis following the mandatory withdrawal of other antibiotic feed additives from farm use in Europe (7).
The DGGE profile generated with Cpa primers showed that different strains of C. perfringens type A, based on clonal variation of the alpha-toxin gene, were present in the ileum of chickens in the different experimental groups. Ginter et al. (12) demonstrated that the amino acid sequence of the alpha-toxin produced by the type culture of C. perfringens type A differed from the sequences of the alpha-toxins produced by isolates of C. perfringens from the small intestine of calves and that these amino acid changes were responsible for the different immunological properties of the toxins (12). There is thus clonal variation of the alpha-toxin gene among strains of C. perfringens inhabiting the gut of farm animals. The impact of this clonal variation on chicken welfare, if any, should be further investigated by inoculating birds with variant strains and determining the effect on broiler growth.
In conclusion, enumeration of bacteria by culture demonstrated an effect of antibiotics on the total Lactobacillus population and an effect of fat source and antibiotics on the C. perfringens population in ileal contents of broilers. Both PCR-DGGE and bacterial counts showed that the composition of the microflora changed in relation to the age of the chickens. PCR-DGGE analysis revealed details of the species composition of the Lactobacillus population and of the impact of antibiotic administration and dietary fat source on L. salivarius and C. perfringens and detected clonal variation within the C. perfringens population. Thus, the culture-dependent and independent methods were complementary and permitted a detailed analysis of the ileal microflora in relation to age and dietary treatments.
Acknowledgments
We express our gratitude to the technicians at Research Centre Foulum for their excellent care of the animals and their skillful technical assistance in collecting data. We thank the staff of G. W. Tannock's laboratory for an excellent introduction to the DGGE technique.
Financial support from the Danish Ministry of Food, Agriculture and Fisheries is greatly appreciated.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Apajalahti, J. H. A., A. Kettunen, M. R. Bedford, and W. E. Holben. 2001. Percent G+C profiling accurately reveals diet-related differences in the gastrointestinal microbial community of broiler chickens. Appl. Environ. Microbiol. 67:5656-5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnes, E. M., G. C. Mead, and D. A. Barnum. 1972. The intestinal flora of the chicken in the period 2 to 6 weeks of age, with particular reference to the anaerobic bacteria. Br. Poult. Sci. 13:311-326. [DOI] [PubMed] [Google Scholar]
- 4.Coates, M. E., R. Fuller, G. F. Harrison, M. Lev, and S. F. Suffolk. 1963. A comparison of the growth of chicks in the Gustafsson germ-free apparatus and in a conventional environment with and without dietary supplement of penicillin. Br. J. Nutr. 17:141-150. [DOI] [PubMed] [Google Scholar]
- 5.Cole, C. B., and R. Fuller. 1984. Bile acid deconjugation and attachment of chicken gut bacteria: their possible role in growth depression. Br. Poult. Sci. 25:227-231. [DOI] [PubMed] [Google Scholar]
- 6.Dänicke, S., W. Vahjen, O. Simon, and H. Jeroch. 1999. Effects of dietary fat source and xylanase supplementation to rye-based broiler diets on selected bacterial groups adhering to the intestinal epithelium, on transit time of feed, and on nutrient digestibility. Poult. Sci. 78:1292-1299. [DOI] [PubMed] [Google Scholar]
- 7.Elwinger, K., E. Berndtson, B. Engström, O. Fossum, and L. Waldenstedt. 1998. Effect of antibiotic growth promoters and anticoccidials on growth of Clostridium perfringens in the caeca and on performance of broiler chickens. Acta Vet. Scand. 39:433-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engberg, R. M., M. S. Hedemann, T. D. Leser, and B. B. Jensen. 2000. Effect of zinc bacitracin and salinomycin on intestinal microflora and performance of broilers. Poult. Sci. 79:1311-1319. [DOI] [PubMed] [Google Scholar]
- 9.Feighner, S. D., and M. P. Dashkevicz. 1987. Subtherapeutic levels of antibiotics in poultry feeds and their effects on weight gain, feed efficiency, and bacterial cholyltaurine hydrolase activity. Appl. Environ. Microbiol. 53:331-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freeman, C. P. 1976. Digestion and absorption of fat, p. 117-142. In K. N. Boorman and B. M. Freeman (ed.), Digestion in the fowl. British Poultry Science, Edinburgh, United Kingdom.
- 11.Fuller, R., C. B. Cole, and M. E. Coates. 1984. The role of Streptococcus faecium in antibiotic-relieved growth depression of chickens, p. 395-403. In M. Woodbine (ed.), Antimicrobials and agriculture. Butterworths, London, United Kingdom.
- 12.Ginter, A., E. D. Williamson, F. Dessy, P. Coppe, H. Bullifent, A. Howells, and R. W. Titball. 1996. Molecular variation between the α-toxins from the type strain (NCTC8237) and clinical isolates of Clostridium perfringens associated with disease in man and animals. Microbiology 142:191-198. [DOI] [PubMed] [Google Scholar]
- 13.Gong, J., R. J. Forster, H. Yu, J. R. Chambers, P. M. Sabour, R. Wheatcroft, and S. Chen. 2002. Diversity and phylogenetic analysis of bacteria in the mucosa of chicken ceca and comparison with bacteria in the cecal lumen. FEMS Microbiol. Lett. 208:1-7. [DOI] [PubMed] [Google Scholar]
- 14.Kanakaraj, R., D. L. Harris, J. G. Songer, and B. Bosworth. 1998. Multiplex PCR assay for detection of Clostridium perfringens in feces and intestinal contents of pigs and in swine feed. Vet. Microbiol. 63:29-38. [DOI] [PubMed] [Google Scholar]
- 15.Langhout, D. J. 1998. The role of the intestinal flora as affected by non-starch polysaccharides in broiler chickens. Ph.D. thesis, Wageningen Agricultural University. Department of Animal Nutrition and Physiology, TNO Nutrition and Food Research Institute, Wageningen, The Netherlands.
- 16.Lev, M., and C. A. E. Briggs. 1956. The gut flora of the chick. II. The establishment of the flora. J. Appl. Bacteriol. 19:224-230. [Google Scholar]
- 17.Mead, G. C. 1989. Microbes of the avian cecum: types present and substrates utilised. J. Exp. Zool. Suppl. 3:48-54. [DOI] [PubMed] [Google Scholar]
- 18.Merck & Co. 1996. The merck index, 12th ed. Merck & Co., Inc., Whitehouse Station, N.J.
- 19.Miller, T. L., and M. J. Wolin. 1974. A serum bottle modification of the Hungate technique for cultivating obligate anaerobes. Appl. Microbiol. 27:985-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muyzer, G., and K. Smalla. 1998. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Leeuwenhoek Int. J. Gen. Microbiol. 73:127-141. [DOI] [PubMed] [Google Scholar]
- 21.Nielsen, E. M., J. Engberg, V. Fussing, L. Petersen, C. H. Brogren, and S. L. W. On. 2000. Evaluation of phenotypic and genotypic methods for subtyping Campylobacter jejuni isolates from humans, poultry, and cattle. J. Clin. Microbiol. 38:3800-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Sullivan, D. J. 1999. Methods for analysis of the intestinal microflora, p. 23-44. In G. W. Tannock (ed.), Probiotics: a critical review. Horizon Scientific Press, Wymondham, United Kingdom.
- 23.Salanitro, J. P., I. G. Blake, P. A. Muirhead, M. Maglio, and J. R. Goodman. 1978. Bacteria isolated from the duodenum, ileum, and cecum of young chicks. Appl. Environ. Microbiol. 35:782-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.SAS Institute Inc. 1988. SAS user's guide: statistics. SAS Institute Inc., Cary, N.C.
- 26.Satokari, R. M., E. E. Vaughan, A. D. L. Akkermans, M. Saarela, and W. M. de Vos. 2001. Bifidobacterial diversity in human feces detected by genus-specific PCR and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 67:504-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simpson, J. M., V. J. McCracken, H. R. Gaskins, and R. I. Mackie. 2000. Denaturing gradient gel electrophoresis analysis of 16S ribosomal DNA amplicons to monitor changes in fecal bacterial populations of weaning pigs after introduction of Lactobacillus reuteri MM53. Appl. Environ. Microbiol. 66:4705-4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sklan, D. 1980. Site of digestion and absorption of lipids and bile acids in the rat and turkey. Comp. Biochem. Physiol. Ser. 65:91-95. [Google Scholar]
- 29.Smits, C. H. M., A. Veldman, H. J. Verkade, and A. C. Beynen. 1998. The inhibitory effect of carboxymethylcellulose with high viscosity on lipid absorption in broiler chickens coincides with reduced bile salt concentration and raised microbial numbers in the small intestine. Poult. Sci. 77:1534-1539. [DOI] [PubMed] [Google Scholar]
- 30.Snedecor, G. W., and W. G. Cochran. 1979. Statistical methods, 6th ed. Iowa State University Press, Ames.
- 31.Tannock, G. W., M. P. Dashkevicz, and S. D. Feighner. 1989. Lactobacilli and bile salt hydrolase in the murine intestinal tract. Appl. Environ. Microbiol. 55:1848-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Titball, R. W., S. E. C. Hunter, K. L. Martin, B. C. Morris, A. D. Shuttleworth, T. Rubidge, D. W. Anderson, and D. C. Kelly. 1989. Molecular cloning and nucleotide sequence of the alpha-toxin (phospholipase C) of Clostridium perfringens. Infect. Immun. 57:367-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vahjen, W., K. Gläser, K. Schäfer, and O. Simon. 1998. Influence of xylanase-supplemented feed on the development of selected bacterial groups in the intestinal tract of broiler chicks. J. Agric. Sci. 130:489-500. [Google Scholar]
- 34.Walter, J., G. W. Tannock, A. Tilsala-Timisjarvi, S. Rodtong, D. M. Loach, K. Munro, and T. Alatossava. 2000. Detection and identification of gastrointestinal Lactobacillus species by using denaturing gradient gel electrophoresis and species-specific PCR primers. Appl. Environ. Microbiol. 66:297-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walter, J., C. Hertel, G. W. Tannock, C. M. Lis, K. Munro, and W. P. Hammes. 2001. Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella species in human feces by using group-specific PCR primers and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 67:2578-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]