Abstract
Current bacterial DNA-typing methods are typically based on gel-based fingerprinting methods. As such, they access a limited complement of genetic information and many independent restriction enzymes or probes are required to achieve statistical rigor and confidence in the resulting pattern of DNA fragments. Furthermore, statistical comparison of gel-based fingerprints is complex and nonstandardized. To overcome these limitations of gel-based microbial DNA fingerprinting, we developed a prototype, 47-probe microarray consisting of randomly selected nonamer oligonucleotides. Custom image analysis algorithms and statistical tools were developed to automatically extract fingerprint profiles from microarray images. The prototype array and new image analysis algorithms were used to analyze 14 closely related Xanthomonas pathovars. Of the 47 probes on the prototype array, 10 had diagnostic value (based on a chi-squared test) and were used to construct statistically robust microarray fingerprints. Analysis of the microarray fingerprints showed clear differences between the 14 test organisms, including the separation of X. oryzae strains 43836 and 49072, which could not be resolved by traditional gel electrophoresis of REP-PCR amplification products. The proof-of-application study described here represents an important first step to high-resolution bacterial DNA fingerprinting with microarrays. The universal nature of the nonamer fingerprinting microarray and data analysis methods developed here also forms a basis for method standardization and application to the forensic identification of other closely related bacteria.
The need to rapidly detect specific microorganisms is both varied and extensive, encompassing basic biochemical, genetic, and ecological research and numerous applications in the genetic identification and tracking of pathogenic microorganisms. Current epidemiological investigations of pathogenic microorganisms use fairly standard techniques for DNA fingerprinting or discriminating between closely related isolates. These include pulsed-field gel electrophoresis (2), variations on Southern hybridization (43), and PCR-based techniques such as randomly amplified polymorphic DNA PCR (39), repetitive element PCR (18, 24), analysis of restriction fragment length polymorphisms (20, 30), single-stranded conformation polymorphisms (26), denaturing gradient gel electrophoresis (29), and combinations thereof (40). In most cases, current DNA-typing methods access a limited complement of genetic information and the fingerprint is based on DNA fragment sizing technology (i.e., gels) that requires parallel processing with many independent restriction enzymes or probes to achieve statistical rigor and confidence in the resulting pattern of DNA fragments.
Despite the widespread acceptance of gel-based DNA fingerprinting techniques, they frequently fail to answer fundamental epidemiological questions. For example, Hancock et al. identified multiple sources of Escherichia coli O157:H7 in feedlots and dairy farms but were unable to discriminate between isolates by pulsed-field gel electrophoresis (16). Even the most advanced fluorescent amplified fragment length polymorphism techniques cannot unequivocally discriminate between near neighbors of Bacillus (39). In the absence of adequate resolving power, then, it is not possible to identify the source of disease outbreaks, determine how pathogens disseminate in the environment, or determine how they enter into and distribute between different vectors or hosts. Therefore, there is a continuing need to develop high-resolution DNA fingerprinting methods to discriminate between closely related microorganisms.
Nucleic acid microarrays are a relatively recent technology development that can overcome many of the limitations of gel-based, DNA fragment-sizing fingerprint methods. Microarrays typically contain hundreds to thousands of individual nucleic acid probes addressed at specific locations on a two- or three-dimensional support (4, 5, 8, 10, 31, 38, 44). Rather than relying solely on post-PCR size discrimination of the resulting DNA fragments (as with most gel-based fingerprinting systems), the microarray accesses information and interrogates the genome directly at the DNA sequence level. Thus, DNA microarrays (in general) offer tremendous potential for microbial detection, identification, and characterization in both basic and applied environmental science. Beattie (3) and coworkers were the first to use oligonucleotide microarrays for genomic fingerprinting, in a technique very similar to the nucleic acid scanning-by-hybridization membranes of Salazar and Caetano-Anollés (34). However, microarrays have not yet been developed for fingerprinting closely related microorganisms, nor have the quantitative analysis and statistical tools been developed to use microarrays for forensic analysis of microorganisms. In this study, we developed a random DNA fingerprinting microarray, automated image analysis tool, and requisite statistical algorithms for identifying and comparing microarray fingerprints, for the purpose of tracking pathogenic microorganisms in environmental systems and nonhuman vectors. The prototype array was applied to discriminate between pathogenic Xanthomonas species and subspecies that cannot be easily identified by gel-based fingerprinting methods.
MATERIALS AND METHODS
Bacterial isolates.
The bacterial isolates used in this study, their source, host plant, and associated disease are listed in Table 1. Xanthomonas and Pseudomonas isolates were cultured in TY-MOPS-buffered broth (11) and checked for purity and uniformity on TY-MOPS agar plates incubated at 30°C. Handling and long-term storage of the cultures were as described previously (12). Cultures of E. coli were grown at 37°C in Luria broth or Luria agar plates and were maintained and stored by standard methods (35).
TABLE 1.
Bacterial isolates used in this study
| Isolate | Source or origina | Disease or host | Reference(s) |
|---|---|---|---|
| X. axonopodis | |||
| pv. axonopodis | ATCC 19132T | Axonopus scoparius | |
| pv. malvacearum N | Cotton | 12 | |
| pv. citri 3213 | Florida | Citrus canker | 12 |
| pv. citri 62 | Japan | Citrus canker | 41c |
| pv. citri 100 | Pakistan | Citrus canker | 41 |
| pv. citri 166 | India | Citrus canker | 41 |
| pv. citri 169 | India | Citrus canker | 41 |
| pv. citrumelo | Florida | Citrus bacterial spot | 12, 41 |
| X. campestris | |||
| pv. campestris | ATCC 33913T | Cabbage | 12 |
| pv. campestris X3 | Cabbage | 22 | |
| pv. alfalfae KX-1 | Alfalfa | 12 | |
| X. oryzae | |||
| pv. oryzae | ATCC 43836b | Rice | |
| pv. oryzae | ATCC 43837b | Rice | |
| pv. oryzicola | ATCC 49072b | Rice | |
| E. coli strain B | Sigma | ||
| P. putida | ATCC 39169 |
T, type strain.
No longer sold by the American Type Culture Collection.
Isolates referred to in reference 41 were obtained from John Hartung, U.S. Department of Agriculture, Beltsville, Md.
Nucleic acid extraction.
High-molecular-weight chromosomal DNA was isolated from 5 ml of overnight culture using Genomic-Tip 100/G kits as specified by the manufacturer (Qiagen Inc., Valencia, Calif.). Plasmids from E. coli or Xanthomonas and chromosomal DNA from other (reference) bacteria were isolated by standard methods (35).
PCR amplification.
For this study, we used repetitive extragenic palindromic (REP) consensus PCR primers (42) to sample microbial genomes and generate amplified genomic DNA fragments for subsequent analysis on the random oligonucleotide microarray. A minimum of two replicate PCR amplifications were performed for every isolate and test condition. Biotinylated PCR primers (REP1R-Dt, 5′-biotin-IIINCGNCGNCATCNGGC, and REP2-D, 5′-biotin-RCGYCTTATCVGGCCTAC, where I = inosine, Y = C or T, R = A or G, and V = G, A, or C) were obtained from Biosource International (Camarillo, Calif.). PCR reagents were from a HotStar Taq kit (Qiagen), except for the deoxynucleoside triphosphates (dNTPs) (Amersham Pharmacia Biotechnology, Piscataway, N.J.). PCR amplification was performed in a 50-μl total volume, using a Tetrad thermal cycler and 96-well plates (MJ Research, Watertown, Mass.). Final reaction conditions were (a minimum of) 150 ng of genomic DNA and 1× PCR buffer (Qiagen), 2.5 mM Mg2+, 200 μM each dNTP, 1 U of Taq polymerase, and 0.6 μM each REP primer. Reagent grade water was used as a negative control. Thermal cycling conditions were 1 cycle of 95°C for 15 min followed by 40 cycles of 95°C for 30 s, 40°C for 45 s, and 72°C for 3 min, and cooling to 4°C. PCR amplification was confirmed by analyzing 20-μl aliquots of the amplification reaction mixture on a 2% agarose gel in 1× Tris-acetate-EDTA (TAE) running buffer. The remaining, labeled amplification products were hybridized directly to microarrays without further manipulation, as described below. For gel-based fingerprinting, primer-labeled Xanthomonas REP-PCR amplification products were separated at 1 to 2 V cm−1 on 1.5% gels composed of a 50:50 mixture of SeaKem GTG:Metaphor agarose (FMC Bioproducts, Rockland, Maine) in 1× TAE running buffer, both containing 3 μg of ethidium bromide per ml.
Microarray probes.
A list of 2,000 nonamer microarray capture probes was generated by random computer selection based on the sequence of the E. coli K-12 genome (GenBank accession number U00096). Because the capture probes are only 9 nucleotides in length, any one probe is expected to occur (on average) once every 131,000 bases in any double-stranded genome (once every 49 bp = 262,000 bp; 131,000 bases in a double-stranded sequence). The computer program was written to perform the following screens: any repeated sequence was less than 4 nucleotides; there were no terminal, 3-nucleotide inverted repeats (hairpins); any probe containing a GGGCCC repeat was discarded; the G+C content was maintained between 44 and 55%; and any probe containing a palindrome was eliminated. From this analysis, we selected 47 nonamer probes (Table 2) that occur (on average) 35 times each within the E. coli genome, with nearly equal probability of hybridizing to each strand of the genome. In addition to the 47 nonamer capture probes, the prototype array contained a biotinylated quality control (QC) probe (5′-biotin-TTGTGGTGGTGGTGTGGTGGTGGGGTTGGG TGGTGG-3′) that served as a positional reference point and positive control for the array detection chemistry.
TABLE 2.
Sequences for the random nonamer capture probes
| Probe ID | Sequence | Tm (°C)a |
|---|---|---|
| 01 | 5′-GATTGCGGT-NH2-3′ | 34 |
| 02 | 5′-GTCATGGTG-NH2-3′ | 34 |
| 03 | 5′-GTCGCCATA-NH2-3′ | 34 |
| 04 | 5′-TCATCGCGT-NH2-3′ | 34 |
| 05 | 5′-TTGGTGGCT-NH2-3′ | 34 |
| 06 | 5′-CGGTATAAC-NH2-3′ | 32 |
| 07 | 5′-GAACAACGT-NH2-3′ | 32 |
| 08 | 5′-CGTTGAAGT-NH2-3′ | 32 |
| 09 | 5′-GTCAACAAC-NH2-3′ | 32 |
| 10 | 5′-AAGGCAAAC-NH2-3′ | 32 |
| 11 | 5′-ATTTCGGCA-NH2-3′ | 32 |
| 12 | 5′-GCTGTTTAC-NH2-3′ | 32 |
| 13 | 5′-TGTTTGTCG-NH2-3′ | 32 |
| 14 | 5′-AATCAGCTG-NH2-3′ | 32 |
| 15 | 5′-AATTGCTGC-NH2-3′ | 32 |
| 16 | 5′-ATGGCAATC-NH2-3′ | 32 |
| 17 | 5′-CAACTACAC-NH2-3′ | 32 |
| 18 | 5′-CAGATGATG-NH2-3′ | 32 |
| 19 | 5′-CGATGATGA-NH2-3′ | 32 |
| 20 | 5′-CTAACGACT-NH2-3′ | 32 |
| 21 | 5′-CTACGCTTA-NH2-3′ | 32 |
| 22 | 5′-GGACTTTCT-NH2-3′ | 32 |
| 23 | 5′-TATAGCCGT-NH2-3′ | 32 |
| 24 | 5′-TGGCATCAA-NH2-3′ | 32 |
| 25 | 5′-CGCTTTGGT-NH2-3′ | 34 |
| 26 | 5′-CGTTTGCAG-NH2-3′ | 34 |
| 27 | 5′-GGACAAACG-NH2-3′ | 34 |
| 28 | 5′-AACGCCATC-NH2-3′ | 34 |
| 29 | 5′-AAGTCAGCG-NH2-3′ | 34 |
| 30 | 5′-ACATCGGCA-NH2-3′ | 34 |
| 31 | 5′-ACCGTCTTC-NH2-3′ | 34 |
| 32 | 5′-ACGAACTGG-NH2-3′ | 34 |
| 33 | 5′-ACGACCAGA-NH2-3′ | 34 |
| 34 | 5′-ACGCTGAAG-NH2-3′ | 34 |
| 35 | 5′-AGCAGTTCC-NH2-3′ | 34 |
| 36 | 5′-AGTGGCAAC-NH2-3′ | 34 |
| 37 | 5′-ATACCGGTG-NH2-3′ | 34 |
| 38 | 5′-ATCTTCCGG-NH2-3′ | 34 |
| 39 | 5′-ATTCCGGCA-NH2-3′ | 34 |
| 42 | 5′-CCTGCATTG-NH2-3′ | 34 |
| 43 | 5′-CGGCAATCT-NH2-3′ | 34 |
| 44 | 5′-CGGTTAACC-NH2-3′ | 34 |
| 45 | 5′-GATGCCGTT-NH2-3′ | 34 |
| 46 | 5′-GCAGGTGAA-NH2-3′ | 34 |
| 47 | 5′-GTCCTTGAC-NH2-3′ | 34 |
| 48 | 5′-TCAGTGGCA-NH2-3′ | 34 |
| 49 | 5′-TGGCATTGC-NH2-3′ | 34 |
The approximate melting temperature was calculated using the nearest-neighbor method.
Microarray fabrication.
Amine modified oligonucleotides were printed on 12-well Teflon-masked slides (Erie Scientific, Portsmouth, N.H.) as previously described (7). Briefly, the slides were prepared for printing by being washed in 2% Microcleaner and rinsed with distilled water. The slides were submerged in a 3 N HCl bath for a minimum of 30 min and then given a 30-min wash in 3N H2SO4. They were thoroughly rinsed with distilled water and dried with compressed N2. They were then coated with 2% (vol/vol) epoxysilane (3-glycidoxypropyltrimethoxysilane [Aldrich, Milwaukee, Wis.]) in methanol for a minimum of 30 min, rinsed with 100% methanol, and immediately dried with compressed N2.
Oligonucleotide capture probes were resuspended in reagent grade water, and the concentration of each was measured in triplicate by spectrophotometry (Smartspec 3000; Bio-Rad, Hercules, Calif.). Subsequently, capture probes were diluted to 80 to 100 μM in 0.01% sodium dodecyl sulfate, 50 mM NaOH print buffer. Probes were printed with a 417 Pin and Ring arrayer (Affymetrix, Santa Clara, Calif.), with a complete 47 probe microarray contained within each well of the Teflon-masked slide (47 probes + 1 QC). After printing, the slides were baked for 30 min at 130°C and stored at room temperature.
Hybridization procedures.
Biotinylated REP-PCR products (20 μl) were diluted to 70 μl in hybridization buffer to achieve final concentrations of 4× SSC (1× SSC is 0.15 M NaCl plus 0.015 M trisodium citrate [pH 7.0]) and 5× Denhardt's solution (1 g of Ficoll 400 per liter, 1 g of polyvinylpyrrolidone per liter, 1 g of ultrapure bovine serum albumin per liter). Amplification products were heat denatured for 5 min at 95°C, snap cooled on ice, and divided evenly between two replicate arrays. Thus, the microarray fingerprint for each isolate was generated from four replicate hybridization reactions (two PCR amplifications × two microarrays per amplification). Denatured amplicons (in hybridization buffer) were hybridized overnight at 4°C and washed five times in an ice-cold solution of 4× SSC. Before the detection reagents were applied, the slides were incubated for 30 min in 35 μl of ice-cold reaction buffer (4× SSC, 5× Denhardt's solution). The reaction buffer was aspirated from each well and replaced with 35 μl of streptavidin alkaline phosphatase (SAAP) (Amersham) diluted 1:500 in reaction buffer. The slides were incubated for 1 h at 4°C, the SAAP was carefully aspirated from each well, and the slides were washed by immersion five times in 1× ELF-97 wash A (Molecular Probes, Eugene, Oreg.). Excess wash solution was aspirated from each slide, and a drop of ELF-97 wash A (20 to 35 μl) was applied to each well. Liquid from each well was carefully aspirated, and then 20 μl of ELF-97 substrate (diluted 1:100 in component C [developing buffer; Molecular Probes]) was applied to each well. Each slide was then incubated in the dark for 1 h at 4°C and washed five times in ELF final wash and three times each in a series of three distilled water tubes. The slides were air dried and imaged with a Fluor-S MultiImager (Bio-Rad) equipped with a 28- to 200-mm DL Hyperzoom macro lens (Sigma, Rödermark, Germany) that was fitted with a +1 close-up lens. Once dry, the ELF-97 crystals appeared to be stable and suitable for long-term storage and reimaging.
Image analysis and statistics.
Slide images were exported as 16-bit uncompressed TIFF files for data analysis and contained 12 × 48 spot arrays in a 6 × 8 offset grid pattern (see Fig. 1). A sample signature of estimated spot intensities for each array on each slide was extracted using Matlab (The Math Works, Inc., Natick, Mass.) and our toolbox of custom array-image analysis algorithms. The set of 64 sample signatures, four replicates each of 16 isolates, were then compared using multivariate statistical analysis techniques (37).
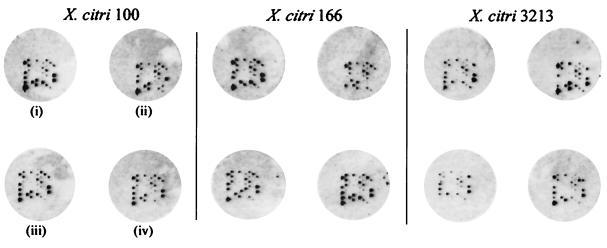
FIG. 1.
Variability in random nonamer microarray fingerprinting. Four replicate hybridizations are shown for each of three isolates. Each array contains one biotinylated QC probe in the lower left corner of the print pattern.
In this study, we used custom statistical models and algorithms, including a semiautomatic grid alignment procedure, to estimate isolate-specific signatures, one from each array on every slide. The statistical models and algorithms accommodate the uncontrollable variations in microarray geometry, probe spot location, and size due to the printing process and the variations in fluorescence intensity from nonspecific hybridization, local and global background noise, and stray light (36). First, the expected print layout is warped to the actual layout observed in a printed grid, resulting in an array template. Corresponding spots in the expected and printed grids are identified using a relatively small number of mouse clicks. Then, the general row and column spacing of a printed grid is estimated using a linear model and least-squares estimation (21). The warped grid is then fit automatically to each array in each slide image (64 arrays across six slides in this study) by using a second linear model that accounts for variation in displacement and orientation across arrays. Thus, the semiautomated grid algorithm and model allows for differences in spot location, orientation, and scale from array to array, within a slide and between slides, rather than extracting probe spot intensity (and background intensity) data from a fixed grid and spot size.
Once the expected locations of probe spots are identified within each array and slide image, we used the APEX (automated peak extraction) algorithm (19) to estimate the above-background pixels at each expected spot location and then the degree of reactivity (i.e. spot intensity). A key feature of the APEX algorithm is the automated identification of each spot's “above-background” pixels by using a stochastic model and a statistical hypothesis-testing framework that allows for variation in spot location, shape, and intensity on the array. Under the APEX model, all pixels in the neighborhood (including those of the expected spot) are hypothesized to be background pixels of nominally uniform intensity. The APEX test statistic tests the hypothesis of neighborhood pixel uniform intensity. A spot is called “on” if pixels in the expected spot location are more intense than adjacent pixels, so that the hypothesis of a uniform neighborhood is rejected; otherwise, the spot is deemed “off.” The estimated “on” spot intensity within the expected spot location is an indicator of the level of hybridization and can be deduced even in a highly variable local or regional background (see Results). The set of APEX-estimated spot intensities and “on/off” results is ordered by probe ID for each array and constitutes an estimated array signature.
For comparison, spot intensities were also estimated with Phoretix array software (version 1.00; Phoretix International, Newcastle, United Kingdom), a package that was initiated based on densitometry analysis of traditional dot blot data. In version 1.00, the user first defines a microarray grid according to a fixed mask or pattern, the geometry of the individual spots (square, circle, or rectangle), and the radius of each spot. The “appropriate” radius for each spot is defined as encompassing all of the spot area and enough of the surrounding background to be able to detect the difference between spot and background. Both the grid and individual spot size can be manually adjusted. For this study, we manually adjusted the grid to center each spot within a circle of fixed radius, with the radius being defined as the smallest circle required to isolate and detect the most intense spot. Phoretix results are reported as the raw volume value, which is a measure of [(spot intensity + background intensity) − total background].
Based on the APEX on/off determination for each probe in each array, the frequency of positive probe hybridization (i.e., the number of “on” spots) was determined for all 47 probes across all isolates (0, 1, 2, 3, or 4 for four replicate arrays). The resulting frequency distributions of on/off determinations were then analyzed to identify the subset of probes containing discriminating information. For instance, a probe spot that was always “on” or “off” across all 64 arrays contained no discriminating information relative to this set of bacterial isolates. A probe spot whose on/off pattern is uniformly distributed across isolate types (i.e., independent of the isolate type) also contains no discriminating information. To identify the subset of discriminating probes, the observed distribution of on/off values was compared, via a chi-squared test of independence (P ≤ 0.5), to the expected uniform distribution obtained by assuming that the bacterial isolate type had no effect on a spot's being on or off (a Fisher exact test could be used instead of the chi-squared test). The on/off patterns of the rejected probes were visually reviewed as a check of the test's performance. Typically, rejected probes were called “on” in one or two of the four replicates for numerous isolates. Thus, the chi-squared test is a simple but effective measure for identifying noninformative probes and reducing the level of uncertainty associated with the estimated bacterial fingerprints.
After the frequency histogram is reduced to a presumptive fingerprint estimate of informative probes, a number of multivariate statistical analysis procedures (28) could be used to evaluate the relationships between fingerprint profiles (e.g. clustering, classification, discriminant analysis, and multivariate analysis of variance). For this study, we used principal-component analysis (PCA) to visualize and project (but not statistically evaluate) the distance between fingerprint profiles (25). Results from the PCA were displayed graphically using an isolate-specific microarray icon as the plotting symbol. The icon shows the fingerprint of an isolate within the context of the microarray print pattern. Thus, the PCA plots provide an estimate not only of distances between isolates but also of the specific details of the fingerprints used to calculate the distances.
RESULTS
Optimized hybridization conditions.
A thorough survey of salt conditions (1× to 4× SSC and 150 mM trisodium citrate, with 5× Denhardt's solution), incubation temperature (4°C and room temperature) and hybridization time (2 and 4 h and overnight [ca. 17 h]) was required to achieve any degree of hybridization to the fingerprinting array. In short, successful hybridization to the fingerprinting arrays was achieved only when labeled amplicons were hybridized at 4°C overnight. A 4°C hybridization temperature is counterintuitive to conditions based on a traditional understanding of solution- or membrane-phase hybridization kinetics (1). However, Drmanac et al. (9) articulate the theory and demonstrate the requirement for “cold” hybridizations when using hexa- or octamer probes. In particular, their analysis showed that the discriminatory ability of short-oligonucleotide hybridization either is temperature independent or decreases with increasing temperature. We think that the fingerprinting microarray described here is very similar to the short-oligonucleotide hybridization experiments described by Drmanac et al. and that “cold” hybridization temperatures are the preferred baseline condition for optimizing target hybridization to short-oligonucleotide microarrays.
APEX image analysis.
Figure 1 illustrates some of the inherent variability associated with capturing large DNA targets with nonamer probes immobilized on a two-dimensional support, in that both signal intensity and the visible hybridization pattern varied between replicate arrays. Variations in signal intensity and/or hybridization patterns result from all aspects of the analytical process (see, e.g., reference 36) but were captured, modeled, understood, and mitigated in this study through experimental replication of the entire fingerprinting method. However, microarray imaging itself is susceptible to a number of errors, including flat-field effects, overshine, bleeding, dark current, background noise, nonspecific hybridization, and autofluorescence. Furthermore, imaging (optical) artifacts can be easily confounded and exacerbated by the analysis software, resulting in erroneous spot identification, erroneous measures of spot intensities, and resulting errors in on/off declarations for each probe in the array signatures; hence, inaccurate fingerprint estimates are obtained.
Figure 2, a false-color image from one array and the attendant measures of signal intensity, illustrates these points and the rationale for developing the APEX algorithm for the DNA fingerprinting application. Variations in local and regional background intensity are obvious, some of which are due to ELF precipitate and others are due to autofluorescence, noise, or stray light. A flat-field effect can be seen in the bulk field, moving from the bottom to the top of the image. Overshine is visible in the bulk field between intensely hybridized probe spots. Localized background is much more intense in the upper right quadrant than in the lower left. These (and other) image effects can significantly affect measured signal intensities, depending on the algorithms and statistics used by the image analysis software. For example, region 1 encompasses two probe spots (spots 1 and 2) where there was no visible hybridization. The APEX algorithm (this study) correctly identified these as nonhybridized spots (Fig. 2B). However, not only did the commercial software indicate a positive hybridization in these areas, but also the estimated signal intensity for these two spots varied by an order of magnitude (172 and 1,999 relative light units). Region 2 shows two positive probes (probes 42 and 47) with obvious differences in signal intensity. In this case, the commercial software assigned a signal intensity of 30,020 to the more intense spot but a signal intensity of 31,686 to the obviously weaker spot. In contrast, the APEX algorithm assigned signal intensities of 3,483 and 2,753, respectively, values that are at least consistent with expectations based on the raw image (Fig. 2A). A similar situation is shown in regions 3 and 4 of Fig. 2.
FIG. 2.
False-color image for one fingerprinting array (A) and probe spot intensities (B) measured by Phoretix (top value) or APEX (bottom value). Regions 1 to 4 highlight different image analysis errors and erroneous intensity values deduced with the commercial software. Overshine is visible as a green haze in panel A. The asterisks in panel B indicate erroneous Phoretix on/off determinations. NS, no identifiable “on” spot using the APEX algorithm.
Image analysis with the APEX algorithms therefore showed that if we used the commercial software to analyze and quantify microarray signal intensities, we would erroneously declare six probes to be on and contributing to the overall fingerprint estimate when they were in fact off (probes 1, 2, 4, 19, 36, and 49). Consequently, we used the more conservative APEX algorithms to estimate spot intensities and determine whether nonamer probes were on or off and contributing to the microarray signature for each microorganism. Despite the demonstrated ability to more accurately define spot intensities with the APEX algorithm, only the binary on/off determinations (i.e., frequency histograms) were used in the chi-squared test for deducing microbial fingerprints.
Defining the microarray fingerprint.
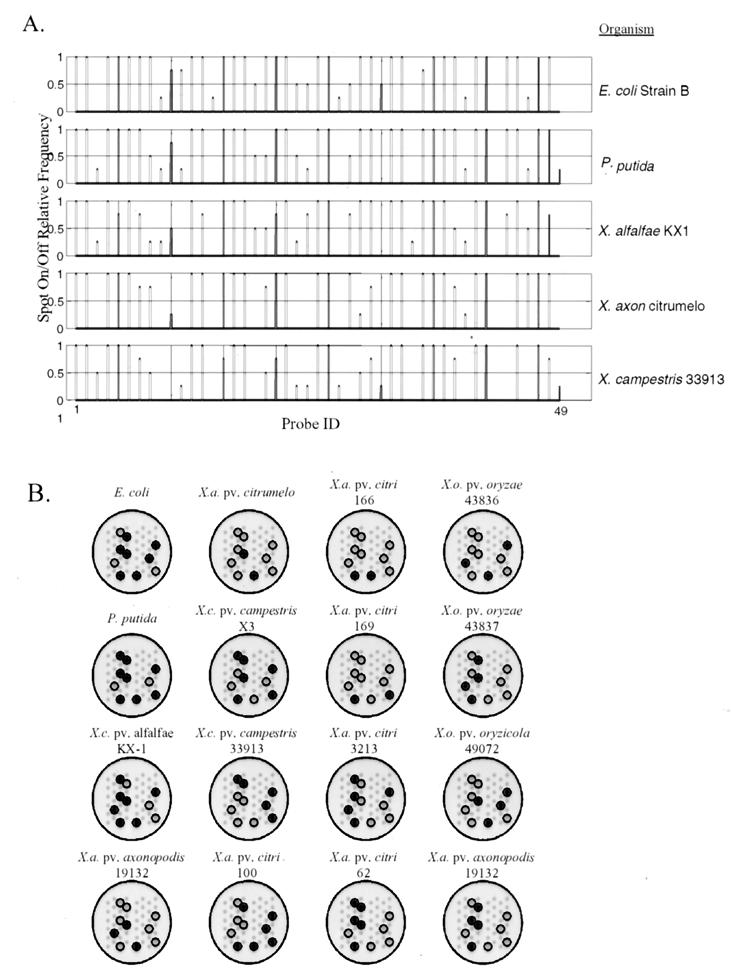
From replicate amplification reactions and replicate arrays, we generated a probe frequency histogram for each isolate; a subset of these are shown in Fig. 3A. The frequency histogram is a quantitative, multivariate vector indexed by probe ID and is analogous to a traditional gel fingerprint where the profile is indexed by DNA size. As multivariate vectors, the frequency histograms could be analyzed, clustered, and compared with each other by using any number of multivariate statistical approaches. In contrast to a gel image, however, the frequency histogram is a quantitative summary that captures experimental replication and the true variability. Hence, the microarray frequency histogram is a more realistic fingerprint estimate than a gel image or single microarray image. To extract a robust fingerprint from the replicates, we performed the chi-squared independence test to identify nonamer probes with discriminatory value, given the set of isolates under consideration in the study. In this study, 10 of the 47 nonamer probes had diagnostic value for the 16 organisms and are displayed in Fig. 3B as microarray icons. In practice, adding new isolates to the study for the purpose of creating a library of fingerprints may result in a different subset of informative probes and different fingerprints. Thus, the informative probes (resulting from the chi-squared test) and deduced fingerprints depend on the context of the experiment and the extent of the library to which a fingerprint is being compared.
FIG. 3.
Relative-frequency histogram for selected Xanthomonas isolates (A) and the corresponding microarray icon with informative probes given the set of isolates used in this study (B).
Discriminating between isolates.
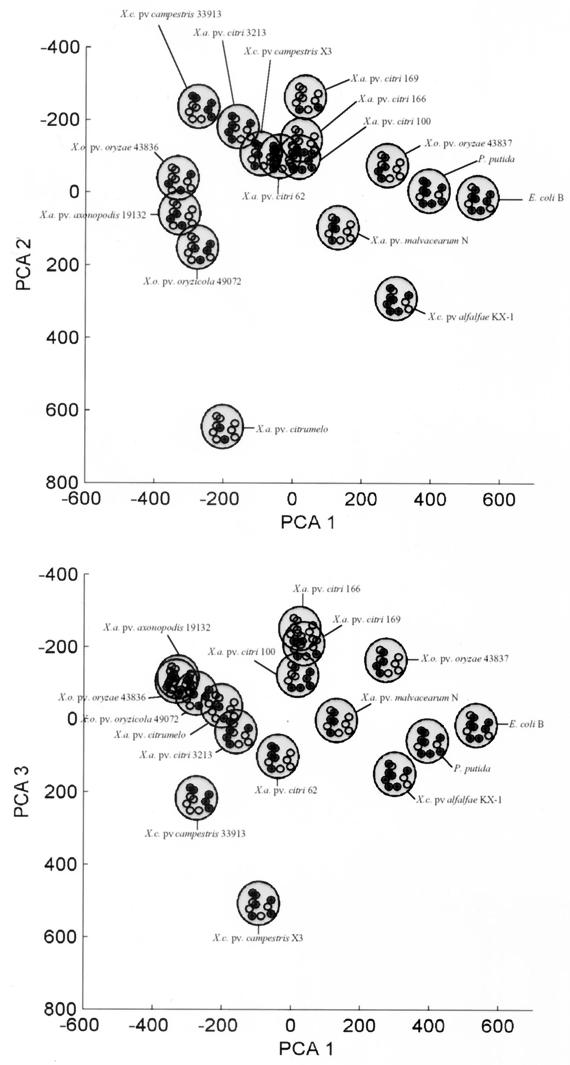
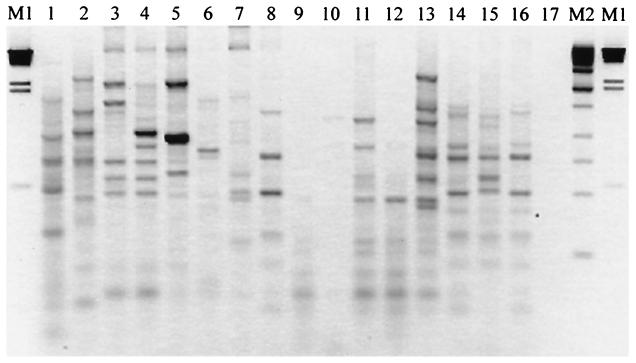
In contrast to the frequency histograms in Fig. 3, the set of informative spots identified by the chi-squared test constitute a statistically robust “bar code” and 10-dimensional signature vector for each isolate that can be confidently analyzed by multivariate statistics. We used PCA to project and visualize the differences between microarray fingerprints (i.e., microorganisms); the results are illustrated in Fig. 4. The first three principal components accounted for 70% of the variability in the data set, and the first seven principal components accounted for 95% of the variability. The first three principal components were plotted against each other to visualize the separation between isolate signatures. The separation between signatures in PCA plots is indicative of the discrimination between the 14 test organisms but is not a statistical test of the genetic distance between organisms. Using PCA to visualize the microarray fingerprints, however, we could clearly separate X. oryzae strains 43836 and 49072 that could not be separated by traditional gel electrophoresis of REP-PCR amplification products (Fig. 5).
FIG. 4.
PCA of microarray fingerprint profiles. (Top) principal component 1 versus principal component 2; (bottom) principal component 1 versus principal component 3.
FIG. 5.
Fingerprinting Xanthomonas isolates by REP-PCR and agarose gel electrophoresis. Lanes: M1, λ × HindIII (Life Technologies, Gaithersburg, Md.); M2 = kilobase DNA ladder (Stratagene, La Jolla, Calif.); 1, E. coli strain B; 2, P. putida 39169; 3, X. campestris pv. campestris 33913; 4, X. campestris pv. campestris X3; 5, X. campestris pv. campestris KX-1; 6, X. axonopodis pv. citrumelo 3048; 7, X. axonopodis pv. malvacearum N; 8, X. axonopodis pv. axonopodis 19132; 9, X. axonopodis pv. citri 62; 10, X. axonopodis pv. citri 3213; 11, X. axonopodis pv. citri 100; 12, X. axonopodis pv. citri 166; 13, X. axonopodis pv. citri 169; 14, X. oryzae pv. oryzae 43836; 15, X. oryzae pv. oryzae 43837; 16, X. oryzae pv. oryzicola 49072; 17, no-template control.
DISCUSSION
Xanthomonas pathovars.
Members of the bacterial genus Xanthomonas cause disease in many important crops and are a potential threat agent for agricultural terrorism. For these reasons, the U.S. Department of Agriculture Animal and Plant Health Inspection Service and the European Plant Protection Organization regulate the national and international shipment of seed, fruit, and plants. The legal statutes governing the treatment of pathogen outbreaks can be extreme. For example, Asiatic bacterial canker of citrus (X. axonopodis pv. citri A) recently reemerged in Florida and had a significant impact on the citrus industry. Regulatory statutes, in this case, mandated that infected fields (and nearby trees) be cleared and burned to eradicate the disease (15) and prevent the continued spread of Xanthomonas (canker). The need for forensic Xanthomonas identification and tracking programs was also evident in a 1984 disease outbreak in Florida. In this episode, a number of lawsuits were filed as a result of a “canker” outbreak in nursery stock of Single citrumelo (Citrus paradisi × C. trifoliata). This outbreak, while not affecting mature fruit-bearing trees, nonetheless resulted in the imposition of quarantine on all Florida citrus and the destruction, by burning, of acres of nursery stock. Gabriel et al. (12) proposed the name Xanthomonas campestris pv. citrumelo for the pathogen causing one form of the nursery leaf-spotting disease. The current proposed designation for this particular pathogen is X. axonopodis pv. citrumelo, the causative agent of citrus bacterial spot (41). Under existing regulatory statutes, citrus bacterial spot is of little economic importance and falls outside of the statutory requirements for eradication, yet the economic damage was already inflicted on the industry, with the current Asiatic canker outbreak costing the industry millions of dollars (15).
Limitations of gel-based fingerprinting methods.
The need to accurately identify closely related microorganisms is therefore obvious; it includes many applications in source attribution, epidemiology, and public health. State-of-the science microbial DNA fingerprinting relies primarily on repetitive-element PCR techniques, methods that are used extensively to identify, geolocate, and track the dissemination of xanthomonads (and other microorganisms) in the environment (11, 13, 23, 24, 27, 32, 33). The PCR and repetitive DNA primers (e.g., REP, ERIC, and BOX) are used to sample a microbial genome, with the resulting PCR amplicons being separated and indexed based on size. However, fragment-sizing techniques are not unambiguous, as illustrated in several samples in Fig. 3B and in even a cursory review of published gel fingerprinting images (see, e.g., reference 6). For example, objective identification or definition of a “band” continues to be problematic, especially with smeared backgrounds (Fig. 5, lane 2) or low- and high-intensity bands (Fig. 5, lane 5). Criteria for including or excluding data (bands) above or below a given size are arbitrary, and a single gel cannot simultaneously resolve low- and high-molecular-weight bands. There is also no guarantee of genetic identity between two bands of the same size unless they are verified by Southern blot analysis. Gels are also susceptible to warps, bubbles, distortions, and other anomalies (lane 13) that cannot be objectively corrected within or between gels, even with internal standards and advanced software (39). For these reasons, gel electrophoresis (fragment sizing) frequently cannot resolve near neighbors, as illustrated for X. oryzae strains 43836 and 49072, and is not conducive to automated, objective scoring across gels. The inability to objectively score gel fingerprints and/or resolve near neighbors therefore led us to explore microarrays as an alternative detection technology for microbial fingerprinting.
Microarrays as an alternative detection technology.
The random-fingerprinting microarray described here is predicated on the nucleic acid scanning-by-hybridization technique of Salazar and Caetoano-Anollés (34) for separating clonal isolates of E. coli O157:H7. A similar membrane-based hybridization approach was used to detect rifampin-resistant strains of tuberculous mycobacteria in lieu of culture-based tests (14), and Beattie (3) showed how the random fingerprinting concept could be configured in microarray format. The power of microarrays for fingerprinting microorganisms is embodied in several subtle but important ways. First, oligonucleotide probes are physically immobilized in space and therefore remove the positional variation inherent in detecting and defining shared bands or comparing gel fingerprints across multiple gels or experiments (Fig. 3B) (39). Second, even though we used a simple 47-probe microarray for this study, many thousands of capture probes can be immobilized on a microarray. Thus, microarrays provide a multiplicative increase in resolving power over agarose or polyacrylamide gels, which are typically limited to <50 identifiable bands in any one lane. Third, microarray capture probes interrogate target DNA based on the DNA sequence, ensuring that common elements in two microarray fingerprints are genetically informative and identical. Therefore, the microarray profile embodies the added genetic information of a Southern blot without any additional effort. Finally, microarray technology is more conducive to automation, replication, and standardization than are sieving media (gels).
Because nonamer probes occur once every 131 kbp (on average) in double-stranded DNA, a priori knowledge of target DNA sequences is not required and the same chip can be used to generate fingerprints from any microorganism. Indeed, we have obtained similar results using the same chip in preliminary tests of E. coli O157:H7 isolates amplified with REP and ERIC primers (data not shown). The universal nature of a random fingerprinting microarray is a compelling feature for method standardization, but it is also a potential weakness. That is, nonamer probes may be complementary to repetitive DNA PCR primers that are used (or required) to sample genomes of interest. The REP 1R primer used in this study, for example, is so degenerate at the 5′ end (5′-IIINGCNGC) that it is partially homologous to several of the capture probes (although perfectly matched with none). Fortunately, positive hybridization signals that result from DNA sequence contained within the repetitive DNA PCR primer are shared by all profiles under consideration. Thus, primer homology to the capture probes results, at worst, in an uninformative signal (probe spot) that is discarded from the individual fingerprint by using the statistical methods developed here; at best, it serves as an internal positive control for successful hybridization and signal development. As described, then, the microarray method is conceptually similar to standard gel fingerprinting techniques without the drawbacks of gels: probes spots are akin to bands on a gel, hybridization intensity is similar to band intensity (e.g., accumulation of detectable amplicon), and the binary microarray profile is equivalent to the binary profile of gel bands. However, while the linkage between microarray and gel fingerprinting is obvious and a natural extension of prior work (3), the statistical foundation for image analysis, quantitatively defining a microarray DNA fingerprint, and statistically comparing fingerprint profiles is not.
Image analysis, statistics, and microarray fingerprints.
There are numerous software products for image analysis, many of which are being applied to the analysis of microarray images. A majority of available microarray software, however, is designed for two-color expression profiling studies, such that the underlying assumptions, statistical models, and computational algorithms are not designed to address alternate biological questions or microarray formats, such as the work described here. For single-color fingerprinting applications, the principal information used to construct a fingerprint is whether specific probes are on or off. Because the true hybridization intensity for any given probe is always unknown, any signal above background is, in principle, a significant data point contributing to the overall estimate of a microbial fingerprint. Therefore, the microarray software must be able to accurately discriminate between true signal and background and must be capable of modeling the major sources of microarray variability (described in references 17 and 36).
Because the background signal varies in time and space and every signal is potentially informative, we do not think that it is sufficient to normalize against simple measures of “global” background intensity, a fixed window or spot size, or the intensity of unhybridized (negative control) spots. Thus, results presented in Fig. 2 illustrate the interdependence of statistics with biology when using microarrays and the need to understand the image analysis challenge and software. The Phoretix software package used for this study was version 1.0 and probably originated from a dot blot densitometry algorithm, wherein the hybridized (macro)spots are all of uniform size and shape. The latest version of Phoretix software (v3.1), for example, contains more sophisticated spot-finding tools and probably produces a more accurate measure of raw volume data. Many new commercial software packages also contain similar (and more advanced) features and algorithms, so that the results here are not an indictment of commercial products (in general) or Phoretix software (in particular). The simple result shown in Fig. 2, however, illustrates why we developed the APEX algorithm for image analysis rather than relying exclusively on closed-source software or inaccessible (i.e., commercially developed and proprietary) algorithms to find and extract our microarray images. Equally important to the fingerprinting method described here, however, is the statistical framework (chi-squared test, linear model, and experimental replication) that allowed us to establish a fingerprint estimate that can be quantitatively compared across slides, days, etc., by using well-established multivariate statistics. The inability to quantitatively compare fingerprints continues to be a significant (forensic) limitation to many gel-based microbial fingerprinting methods, a limitation that has been overcome in the microarray approach described here.
Summary.
The proof-of-application study described here represents an important first step to high-resolution bacterial DNA fingerprinting with microarrays, even though we demonstrated the efficacy of the method with a very simple, 47-probe chip. Increasing the number of capture probes will provide an additive increase in useful (discriminatory) data. Amplifying genomic DNA with additional repetitive or arbitrary PCR primers will provide a multiplicative increase in the number of effective fingerprint probes for each isolate without adding any more capture probes to the array. Finally, performing quantitative comparisons between microarray fingerprints will require additional statistical and computational effort and development. Regardless, the power of the microarray fingerprinting technique described here ultimately lies in understanding, modeling, and capturing the variability in the entire experimental process. Forensic applications of microarray fingerprinting will therefore require a more precise definition for experimental replication to ensure that the fingerprint is representative of microbial sample profiles collected on different days with slides printed from different batches, etc. Without the proper replication, observed differences between true “unknowns” and a fingerprint library will be confounded with sources of variability inherent to the experimental process and a reliable identification of microbial presence in the sample will not be possible. Application of the fingerprinting array to microbial forensics will also require a more thorough understanding of natural (microbial) variability in microarray fingerprints, improved analytical methods to improve sample quality and signal-to-noise ratios, and enhanced open-source image analysis software for unambiguous feature extraction and generation of microarray fingerprints that can be broadly disseminated to the user community.
Acknowledgments
This work was supported by the U.S. Department of Energy (DOE) under the Pacific Northwest National Laboratory Scientist VI LDRD program. Pacific Northwest National Laboratory is operated by Battelle Memorial Institute for the U.S. DOE under contract DE-AC06-76RL0 1830.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1995. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 2.Baggesen, D. L., D. Sandvang, and F. M. Aarestrup. 2000. Characterization of Salmonella enterica serovar typhimurium DT104 isolated from Denmark and comparison with isolates from Europe and the United States. J. Clin. Microbiol. 38:1581-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beattie, K. L. 1997. Genomic fingerprinting using oligonucleotide arrays, p. 213-224. In G. Caetano-Anolles and P. M. Gresshoff (ed.), DNA markers: protocols, applications, and overviews. John Wiley and Sons, New York, N.Y.
- 4.Bertucci, F., K. Bernard, B. Loriod, Y.-C. Chang, S. Granjeaud, D. Birnhaum, C. Nguyen, K. Peck, and B. R. Jordan. 1999. Sensitivity issues in DNA array-based expression measurements and performance of nylon microarrays for small samples. Hum. Mol. Genet. 8:1715-1722. [DOI] [PubMed] [Google Scholar]
- 5.Brown, P. O., and D. Botstein. 1999. Exploring the new world of the genome with DNA microarrays. Nat. Genet. 21:33-37. [DOI] [PubMed] [Google Scholar]
- 6.Brumlik, M. J., U. Szymajda, D. Zakowska, X. Liang, R. J. Redkar, G. Patra, and V. G. Del Vecchio. 2001. Use of long-range repetitive polymorphism-PCR to differentiate Bacillus anthracis strains. Appl. Environ. Microbiol. 67:3021-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Call, D. R., D. P. Chandler, and F. J. Brockman. 2001. Fabrication of DNA microarrays using unmodified oligomer probes. BioTechniques 30:368-379. [DOI] [PubMed] [Google Scholar]
- 8.Cheung, V. G., M. Morley, F. Aguilar, A. Massimi, R. Kucherlapati, and G. Childs. 1999. Making and reading microarrays. Nat. Genet. 21:15-19. [DOI] [PubMed] [Google Scholar]
- 9.Drmanac, R., Z. Strezoska, I. Labat, S. Drmanac, and R. Crkvenjakov. 1990. Reliable hybridization of oligonucleotides as short as six nucleotides. DNA Cell Biol. 9:527-534. [DOI] [PubMed] [Google Scholar]
- 10.Edman, C. F., D. E. Raymond, D. J. Wu, E. Tu, R. G. Sosnowski, W. F. Butler, M. Nerenberg, and M. J. Heller. 1997. Electric field directed nucleic acid hybridization on microchips. Nucleic Acids Res. 25:4907-4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gabriel, D. W., J. E. Hunter, M. T. Kingsley, J. W. Miller, and G. R. Lazo. 1988. Clonal population structure of Xanthomonas campestris and genetic diversity among citrus canker strains. Mol. Plant-Microbe Interact. 1:59-65. [Google Scholar]
- 12.Gabriel, D. W., M. T. Kingsley, J. E. Hunter, and T. Gottwald. 1989. Reinstatement of Xanthomonas citri (ex Hasse) and X. phaseoli (ex Smith) to species and reclassification of all X. campestris pv. citri strains. Int. J. Sys. Bacteriol. 39:14-22. [Google Scholar]
- 13.George, M. L. C., M. Bustamam, W. T. Cruz, J. E. Leach, and R. J. Nelson. 1997. Movement of Xanthomonas oryzae pv oryzae in southeast Asia detected using PCR-based DNA fingerprinting. Phytophathology 87:302-309. [DOI] [PubMed] [Google Scholar]
- 14.Gingeras, T. R., G. Ghandour, E. Wang, A. Berno, P. M. Small, F. Drobniewski, D. Alland, E. Desmond, M. Holdniy, and J. Drenkow. 1998. Simultaneous genotyping and species identification using hybridization pattern recognition analysis of generic Mycobacterium DNA arrays. Genome Res. 8:435-448. [DOI] [PubMed] [Google Scholar]
- 15.Gottwald, T. R., G. Hughes, J. H. Graham, X. Sun, and T. Riley. 2001. The citrus canker epidemic in Florida: the scientific basis of regulatory eradication policy for an invasive species. Phytopathology 91:30-34. [DOI] [PubMed] [Google Scholar]
- 16.Hancock, D. D., T. E. Besser, D. H. Rice, E. D. Ebel, D. E. Herriott, and L. V. Carpenter. 1998. Multiple sources of Escherichia coli O157 in feedlots and dairy farms in the northwestern USA. Prev. Vet. Med. 35:11-19. [DOI] [PubMed] [Google Scholar]
- 17.Herzel, H., D. Beule, S. Kielbasa, J. Korbel, C. Sers, A. Malik, H. Eickhoff, H. Lehrach, and J. Schuchhardt. 2001. Extracting information from cDNA arrays. CHAOS 11:98-107. [DOI] [PubMed] [Google Scholar]
- 18.Jackson, P. J., E. A. Walthers, A. S. Kalif, K. L. Richmond, D. M. Adair, K. K. Hill, C. R. Kuske, G. L. Andersen, K. H. Wilson, M. E. Hugh-Jones, and P. Keim. 1997. Characterization of the variable-number tandem repeats in vrrA from different Bacillus anthracis isolates. Appl. Environ. Microbiol. 63:1400-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jarman, K. H., D. S. Daly, K. K. Anderson, and K. L. Wahl. June2001. U.S. patent 6253162 B1.
- 20.Johansson, M. L., G. Molin, B. Pettersson, M. Uhlen, and S. Ahrne. 1995. Characterization and species recognition of Lactobacillus plantarum strains by restriction fragment length polymorphism (RFLP) of the 16S rRNA gene. J. Appl. Bacteriol. 79:536-541. [Google Scholar]
- 21.Kutner, M. H., C. J. Nachtschiem, W. Wasserman, and J. Neter. 1996. Applied linear statistical models. Irwin/McGraw-Hill Co., New York, N.Y.
- 22.Lazo, G. R., R. Roffey, and D. W. Gabriel. 1987. Pathovars of Xanthomonas campestris are distinguishable by restriction fragment length polymorphisms. Int. J. Syst. Bacteriol. 37:214-221. [Google Scholar]
- 23.Leach, J. E., M. L. Rhoads, C. M. Vera Cruz, F. F. White, T. W. Mew, and H. Leung. 1992. Assessment of genetic diversity and population structure of Xanthomonas oryzae pv. oryzae with a repetitive DNA element. Appl. Environ. Microbiol. 58:2188-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Louws, F. J., D. W. Fulbright, C. T. Stephens, and F. J. de Bruijn. 1994. Specific genomic fingerprints of phytopathogenic Xanthomonas and Pseudomonas pathovars and strains generated with repetitive sequences and PCR. Appl. Environ. Microbiol. 60:2286-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malinowski, E. R. 2002. Factor analysis in chemistry. John Wiley & Sons, Inc., New York, N.Y.
- 26.McClelland, M., H. Arensdorf, R. Cheng, and J. Welsh. 1994. Arbitrarily primed PCR fingerprints resolved on SSCP gels. Nucleic Acids Res. 22:1770-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McDonald, J. G., and E. Wong. 2001. Use of a monoclonal antibody and genomic fingerprinting by repetitive-sequence-based polymerase chain reaction to identify Xanthomonas populi pathovars. Can. J. Plant Pathol. 23:47-51. [Google Scholar]
- 28.Morrison, D. F. 1976. Multivariate statistical methods. McGraw Hill Book Co., New York, N.Y.
- 29.Muyzer, G., E. C. De Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Navarro, E., P. Simonet, P. Normand, and R. Bardin. 1992. Characterization of natural populations of Nitrobacter spp. using PCR/RFLP analysis of the ribosomal intergenic spacer. Arch. Microbiol. 157:107-115. [DOI] [PubMed] [Google Scholar]
- 31.Pease, A. C., D. Solas, E. J. Sullivan, M. T. Cronin, C. P. Holmes, and S. P. A. Fodor. 1994. Light-generated oligonucleotide arrays for rapid DNA sequence analysis. Proc. Natl. Acad. Sci. USA 91:5022-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rademaker, J. L. W., B. Hoste, F. J. Louws, K. Kersters, J. Swings, L. Vauterin, P. Vauterin, and F. J. de Bruijn. 2000. Comparison of AFLP and rep-PCR genomic fingerprinting with DNA-DNA homology studies: Xanthomonas as a model system. Int. J. Syst. E vol. Microbiol. 50:665-677. [DOI] [PubMed] [Google Scholar]
- 33.Restrepo, S., M. T. Duque, J., and V. Verdier. 1999. AFLP fingerprinting: an efficient technique for detecting genetic variation of Xanthomonas axonopodis pv. manihotis. Microbiology 145:107-114. [DOI] [PubMed] [Google Scholar]
- 34.Salazar, N. M., and G. Caetano-Anollés. 1996. Nucleic acid scanning-by-hybridization of enterohemorrhagic Escherichia coli isolates using oligodeoxynucleotide arrays. Nucleic Acids Res. 24:5056-5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Schuschhardt, J., D. Beule, A. Malik, E. Wolski, H. Eickhoff, H. Lehrach, and H. Herzel. 2000. Normalization strategies for cDNA microarrays. Nucleic Acids Res. 28:e47i-e47v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seber, G. A. F. 1984. Multivariate observations. John Wiley & Sons, Inc., New York, N.Y.
- 38.Southern, E. M., U. Maskos, and J. K. Elder. 1992. Analyzing and comparing nucleic acid sequences by hybridization to arrays of oligonucleotides: evaluation using experimental models. Genomics 13:1008-1017. [DOI] [PubMed] [Google Scholar]
- 39.Ticknor, L. O., A.-B. Kolstφ, K. K. Hill, P. Keim, M. T. Laker, M. Tonks, and P. J. Jackson. 2001. Fluorescent amplified fragment length polymorphism analysis of Norwegian Bacillus cereus and Bacillus thuringiensis soil isolates. Appl. Environ. Microbiol. 67:4863-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van der Wurff, A. W. G., Y. L. Chan, N. M. van Straalen, and J. Schouten. 2000. TE-AFLP: combining rapidity and robustness in DNA fingerprinting. Nucleic Acids Res. 28:e105.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verniere, C., J. S. Hartung, O. P. Pruvost, E. L. Civerolo, A. M. Alvarez, P. Maestri, and J. Luisetti. 1998. Characterization of phenotypically distinct strains of Xanthomonas axonopodis pv. citri from Southwest Asia. Eur. J. Plant Pathol. 104:477-487. [Google Scholar]
- 42.Versalovic, J., T. Koeuth, and J. R. Lupski. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waterhouse, R. N., and L. A. Glover. 1993. Identification of procaryotic repetitive DNA suitable for use as fingerprinting probes. Appl. Environ. Microbiol. 59:1391-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yershov, G., V. Barsky, A. Belgovskiy, E. Kirillov, E. Kreindlin, I. Ivanov, S. Parinov, D. Guschin, A. Drobishev, S. Dubiley, and A. Mirzabekov. 1996. DNA analysis and diagnostics on oligonucleotide microchips. Proc. Natl. Acad. Sci. USA 93:4913-4918. [DOI] [PMC free article] [PubMed] [Google Scholar]