Abstract
Nineteen Listeria monocytogenes strains were characterized by automated ribotyping, pulsed-field gel electrophoresis, and plasmid profiling to determine the relationship between genotype and sanitizer resistance. Isolates within a ribogroup had a consistent sensitivity or resistance phenotype except for ribogroup C isolates. All isolates with resistance phenotypes harbored two plasmids. The sensitivity of L. monocytogenes strains to quaternary ammonium compounds (QACs) was correlated with sensitivity to sanitizers and antibiotics with other modes of action. All isolates tested contained the mdrL gene, which encodes an efflux pump that confers resistance to QACs and is both chromosome and plasmid borne.
Listeria monocytogenes is a pathogen of humans and animals which has been implicated in several outbreaks and sporadic cases of listeriosis, resulting in numerous food product recalls. The apparent increase in recalls due to L. monocytogenes contamination may be due to several factors, including more diligent surveillance since a large U.S. listeriosis outbreak in 1998 (4) and possible adaptation of L. monocytogenes to food production and food processing environments. One such adaptation may be the acquisition of resistance to sanitizers. Antimicrobial resistance can be either a natural property of an organism (intrinsic) or acquired by mutation or by means of plasmids or transposons (15).
Acquired antibiotic and sanitizer resistance in L. monocytogenes has been considered a very rare event, but during the last few years, resistance to different agents has been increasingly documented for this species (1, 9).
Nineteen isolates of L. monocytogenes were studied (Table 1). Five of the isolates (obtained from the Centers for Disease Control and Prevention, Atlanta, Ga.) were from the 1998 listeriosis outbreak. Five isolates were obtained in 1999 from a meat processing facility. The remaining nine isolates represent chronologically older isolates (designated LJH) dating from 1993 or earlier. Bacteria were grown at 37 and 30°C on tryptic soy agar (TSA; Oxoid). Prior to use, the cultures were subcultured on two consecutive days in tryptic soy broth (TSB; Difco) and incubated with shaking at 37 or 30°C.
TABLE 1.
Sources, ribogroups, pulsed-field types, serotypes, and plasmid profiles of the L. monocytogenes isolates used in this study
| Culture | Sourcea | Ribogroup | ApaI REDP | SmaI REDP | PFGE type | Serotype | Plasmid profile | Large plasmid | Small plasmid | Relative resistanceb |
|---|---|---|---|---|---|---|---|---|---|---|
| H7764 | Deli turkey, Sara Lee outbreak | A | I | I | A | 1 | A | + | + | R |
| H7596 | Turkey, Sara Lee outbreak | B | II | II | B | 4 | B | − | + | M |
| H7762 | Franks, Sara Lee outbreak | C | III | III | C | 4 | A | + | + | R |
| H7962 | Hot dogs, Sara Lee outbreak | C | II | II | B | 4 | A | + | + | R |
| N7969 | Franks, Sara Lee outbreak | C | III | III | C | 4 | B | − | + | M |
| C720 | Meat processing plant | D | IV | IV | D | 1 | A | + | + | M |
| C719 | Meat processing plant | D | IV | IV | D | 1 | A | + | + | S |
| C718 | Meat processing plant | D | IV | IV | D | 1 | A | + | + | M |
| C511 | Unknown CRIFS | D | IV | IV | D | 1 | A | + | + | S |
| C512 | Unknown CRIFS | D | IV | IV | D | 1 | B | − | + | M |
| C716 | Meat processing plant | E | V | V | E | 1 | C | + | + | R |
| C717 | Meat processing plant | F | VI | VI | F | 1 | D | + | + | R |
| LJH309 | Unknown CRIFS | G | VII | VII | G | 4 | B | − | + | M |
| LJH385 | Outbreak CRIFSG | G | XII | VII | H | 4b | B | − | + | M |
| LJH389 | Unknown CRIFS | 4b | B | − | + | S | ||||
| LJH375 | Rabbit CRIFS | I | VIII | VIII | I | 1/2a | B | − | + | S |
| LJH377 | Meat CRIFS | J | IX | IX | J | 1/2d | B | − | + | S |
| LJH381 | Health Canada CRIFS | K | X | X | K | 4b rough | B | − | + | S |
| C509 | Unknown CRIFS | L | XI | XI | L | 4 | E | + | + | M |
CRIFS, Canadian Research Institute for Food Safety.
Determinations are based on MIC results for BCl. R, resistant (MIC ≥ 5 μg/ml); S, sensitive (MIC ≤ 0.78 μg/ml); M, moderate sensitivity (MIC = 0.8 to 4.9 μg/ml).
Sanitizers used in the screening study included benzalkonium chloride (BCl; Sigma), Clinicide (a quaternary ammonium compound [QAC]; MTC Animal Health, Cambridge, Ontario, Canada), myristalkonium chloride (Mchl; Naschem Inc., Mississauga, Ontario, Canada), Iodophor (PHOR; Adage II, Diversey Lever Canada, Mississauga, Ontario, Canada), bleach, acetic acid (Sigma), hydrogen peroxide (H2O2, 30%), and ethidium bromide (EtBr; Sigma).
MICs of sanitizers were determined by the broth dilution method (20).
Isolates were serotyped with Bacto Listeria-O somatic antiserum types 1 and 4 (Difco Laboratories) by the slide agglutination test according to the manufacturer's instructions.
Pulsed-field gel electrophoresis (PFGE) was carried out according to a protocol modified from Brosch et al. (2). L. monocytogenes isolates were grown overnight at 37°C in TSB and harvested by centrifugation, and cells were washed twice in TE buffer (10 mM Tris-HCl-1 mM EDTA [pH 8]) before being resuspended in TE to a concentration of 109 cells/ml. The suspension was mixed with an equal concentration of 1.0% chromosomal agarose (Bio-Rad Laboratories, Hercules, Calif.) and dispensed into PFGE plug molds (Bio-Rad). Agarose plugs were incubated for 24 h at 37°C in a lysis solution containing 0.1 M EDTA, 1% sarcosyl (Sigma), 2 mg of deoxycholic acid (Sigma) per ml, and 2.5 mg of lysozyme (Boehringer Mannheim, Mannheim, Germany) per ml. Plugs were washed in TE (three 30-min washes) and deproteinized by incubation for 48 h at 50°C in a solution containing 0.5 M EDTA, 0.5% sarcosyl, and 2 mg of proteinase K (Gibco BRL, Burlington, Ontario, Canada) per ml. To remove the proteinase K, plugs were washed four times in TE buffer for 1 h each time. Plugs were stored in TE buffer at 4°C until restriction analysis.
The plugs were digested using the enzymes ApaI and SmaI (Roche Diagnostics, Laval, Quebec, Canada). Two-millimeter-thick sections of the plugs were digested in 150 μl of restriction buffer A (Roche) containing 0.15 mg of bovine serum albumin (New England Biolabs, Mississauga, Ontario, Canada) and 30 U of ApaI or SmaI. Plugs were incubated overnight in the restriction solution at 37°C for ApaI and at 25°C for SmaI.
The plug inserts were placed into a 1.1% PFGE agarose gel prepared with 0.5× TBE buffer (1× TBE buffer contains 89 mM Tris, 89 mM boric acid, and 2 mM EDTA [pH 8]). A lambda DNA PFGE ladder (Bio-Rad) was run alongside the samples. The gel was run in a Bio-Rad CHEF DR III electrophoresis system with the running buffer (0.5× TBE buffer) cooled to 14°C under the following electrophoresis conditions: for block 1, an initial switch time of 2 s, a final switch time of 20 s, and a run time of 12.5 h; and for block 2, an initial switch time of 15 s, a final switch time of 35 s, and a run time of 10 h. Both blocks were run at 6 V/cm with an included angle of 120°.
Following electrophoresis, the gel was stained for 1 h in 0.5 μg of EtBr per ml in distilled water and destained for 1 h in distilled water. The resulting DNA patterns were imaged with an Alpha Imager2200 documentation and analysis system (Alpha Innotech Corp., San Leandro, Calif.).
Automated ribotyping was carried out with a RiboPrinter microbial characterization system (Qualicon Inc., Wilmington, Del.) according to the manufacturer's instructions for a standard EcoRI batch (3). A dendrogram was generated from the resulting riboprint patterns with BioNumerics Cluster Analysis software (Applied Maths, Inc., Austin, Tex.).
Plasmids were isolated and tested with a Plasmid Midi isolation kit (Qiagen Inc., Mississauga, Ontario, Canada). Cells were harvested by centrifugation from 250 ml of exponential-growth-phase culture, and protoplasts were prepared with lysozyme (10 mg/ml) and incubated for 30 min at 37°C. DNA was examined on 0.7% agarose gels run at 70 V for 3 h in TAE buffer (40 mM Tris-acetate, 1 mM EDTA [pH 8.0]). The supercoiled DNA ladder (Sigma) was used for molecular size standards.
Colonies were picked from overnight cultures on TSA and cells were dispersed in 0.1 ml of sterile water in microcentrifuge tubes. After samples were boiled for 10 min, they were immediately placed on ice and left for 2 min before centrifugation at high speed (14,000 × g for 6 min).
Primers lltb1 (5′-AATGGATAACAGCGGCAG-3′) and lltb2 (5′-TGTAAGGTAAAATGTGCTGG-3′) were used to identify the mdrL gene (14). The PCR mixture consisted of a buffer of 10 mM Tris-HCl-50 mM KCl-2.5 mM MgCl2 (pH 8.3). Deoxyribonucleoside triphosphates (100 μM), 20 pmol of each of the two primers, and 0.5 U of AmpliTaq DNA polymerase were used in a total volume of 25 μl. The reaction procedure consisted of an initial denaturation step at 94°C for 120 s followed by 30 cycles of denaturation at 94°C for 1 min, primer annealing at 55°C for 1 min, and extension at 72°C for 90 s (10 min for the last extension). The expected size of the amplicons was 1,136 bp.
A second primer set, MT1 (5′-AAGAATTCGAGCTGGTTG-3′) and MT2 (5′-AACTGCAGTGTGATACTTT-3′), was also used for identification of the mdrL gene (11). For the amplification, one denaturation cycle of 8 min at 94°C, 25 elongation cycles (1 min at 94°C, 1 min at 48°C, and 1 min at 72°C), and an additional extension cycle of 5 min at 72°C were used. The expected size of the amplicons was 485 bp.
PCR amplification with primers orf1 (5′-AAATGATTGCTCGTGAAGCT-3′) and orf2 (5′-CGCACACCATTTTAATTCTG-3′) was used under the conditions described above to identify the orfA gene (expected fragment size, 467 bp) (14).
PCR products were detected by electrophoresis of the amplification mixture (10 μl) on a 1.3% agarose gel run in TAE buffer containing EtBr (0.2 μg/ml). Gels were visualized under UV light. A 100-bp DNA ladder (Pharmacia Biotech, Quebec, Canada) molecular weight marker was included in each gel.
Cultures of 19 L. monocytogenes isolates were screened for sensitivity to chlorine (bleach)- or iodine (PHOR)-based and QAC-type (Clinicide and Mchl) disinfectants, acetic acid, hydrogen peroxide (H2O2), BCl, and EtBr. The mechanisms of action for several of these sanitizers are known. Acetic acid disrupts intracellular pH homeostasis and membranes, while QACs cause generalized membrane damage and inactivate cellular enzymes (12). Hydrogen peroxide produces hydroxyl free radicals, which attack essential cell components. Chlorine and iodine sanitizers act as oxidizing agents on thiol groups (12), while EtBr inhibits DNA synthesis (18).
No significant difference in sensitivity to acetic acid, PHOR, bleach, and EtBr was observed among strains. About 4- to 10-fold differences in MICs of QACs and H2O2 were found among strains (Table 2). According to Emslie et al. (7), strains of Staphylococcus aureus were described as resistant if they were four to eight times more resistant than the most sensitive strains studied. Similar terminology seems appropriate for the L. monocytogenes strains used in this study, and groups were referred to as resistant, intermediate, or sensitive. Five strains (H7962, H7762, H7764, C717, and C716) showed a resistance to Clinicide (MIC, >3 μg/ml), Mchl (MIC, 4 μg/ml), and BCl (MIC, ≥5 μg/ml) whereas four strains (C719, C511, LJH381, and LJH389) were sensitive to these agents (MICs of all three agents, ≤1.25 μg/ml) and the rest exhibited intermediate resistance (MICs of at least two agents, ≥1.25 μg/ml) (Table 2). There were no differences in the MICs of QACs at 37, 30, or 20°C. Two strains (LJH375 and LJH381) were sensitive to QACs, bleach, and H2O2, and the MICs for all the agents tested except PHOR seemed to be low for LJH381 (Table 2). Three (H7962, H7762, and H7764) of the five strains most resistant to BCl were also most resistant to H2O2, and the MICs of Mchl were high for them. All five resistance phenotypes were found in recent isolates. This fact may reflect the general increase in sanitizer resistance among listeriae, but the phenotypes of older strains may have changed during repeated subculture.
TABLE 2.
Sensitivities of 19 L. monocytogenes isolates to several sanitizers
| Culture | MICa of:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Clinicideb | BCl | Mch1 | Bleach | PHOR | H2O2 | Acetic acid | EtBr | |
| H7962 | 3.08 | 6.25 | 4 | 1.64 | >6 | 75 | 2.5 | 25 |
| N7969 | 1.54 | 1.25 | 1 | 1.64 | >6 | 18.75 | 2.5 | 25 |
| H7762 | 3.08 | 5.0 | 4 | 1.64 | >6 | 75 | 2.5 | 25 |
| H7764 | 3.08 | 6.25 | 4 | 1.64 | >6 | 75 | 2.5 | 25 |
| H7596 | 1.54 | 1.25 | 1 | 1.64 | >6 | 18.75 | 2.5 | 25 |
| C720 | 3.08 | 1.25 | 2 | 1.64 | >6 | 37.5 | 2.5 | 25 |
| C719 | 0.77 | 0.78 | 1 | 1.64 | >6 | 18.75 | 2.5 | 25 |
| C718 | 1.54 | 1.25 | 2 | 1.64 | >6 | 18.75 | 2.5 | 25 |
| C717 | 3.08 | 5 | 4 | 1.64 | >6 | 18.75 | 2.5 | 25 |
| C716 | 3.08 | 5 | 4 | 1.64 | >6 | 18.75 | 2.5 | 25 |
| C509 | 1.54 | 1.25 | 2 | 1.64 | >6 | 18.75 | 2.5 | 12.5 |
| C511 | 0.77 | 0.78 | 1 | 1.64 | >6 | 37.5 | 2.5 | 25 |
| C512 | 1.54 | 1.25 | 1 | 0.82 | >6 | 18.75 | 2.5 | 25 |
| LJH375 | 1.54 | 0.62 | 0.5 | 0.82 | >6 | 9.4 | 2.5 | 25 |
| LJH377 | 1.54 | 0.62 | 2 | 1.64 | >6 | 18.75 | 2.5 | 25 |
| LJH381 | 0.77 | 0.78 | 1 | 0.82 | >6 | 9.4 | 0.63 | 6.25 |
| LJH385 | 1.54 | 1.25 | 1 | 1.64 | >6 | 37.5 | 2.5 | 25 |
| LJH389 | 0.77 | 1.25 | 1 | 0.82 | >6 | 18.75 | 2.5 | 25 |
| LJH309 | 3.08 | 1.25 | 2 | 1.64 | >6 | 37.5 | 2.5 | 12.5 |
The MICs of bleach and acetic acid are in milligrams per milliliter; all other MICs are in micrograms per milliliter.
Clinicide contains two QACs as active ingredients: 60% didecyl dimethyl ammonium chloride and 40% benzalkonium chloride.
Mereghetti et al. (14) reported a correlation between serotype and resistance to BCl. No such correlation was found in the present study.
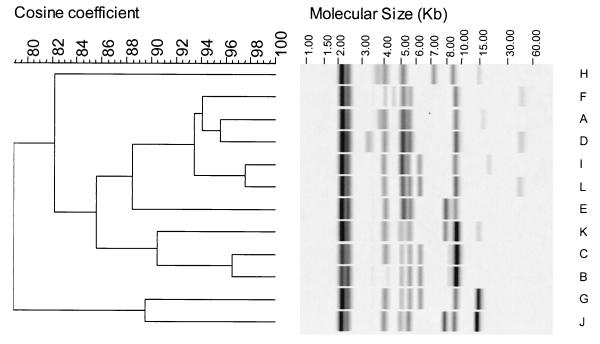
EcoRI has been shown to be highly discriminatory for L. monocytogenes isolates (5). The 19 isolates used in the present study were grouped into 11 ribogroups (Table 1). Three isolates from the meat processing facility (C718, C719, and C720) had identical ribotypes, suggesting that the isolates were from a common source and were disseminated within the packaging area. A dendrogram showing the relationship among groups is shown in Fig. 1. A given ribogroup had a consistent sensitivity or resistance phenotype with one exception. Ribogroup C consisted of two resistant isolates (H7762 and H7962) and one sensitive isolate (N7969). Interestingly, the plasmid profile of N7969 differed from that of the other two in the group and was the same as that of ribogroup B isolate H7596, another sensitive isolate from the Sara Lee group of strains whose ribopattern differed only very slightly from that of ribogroup C (Table 1).
FIG. 1.
Dendrogram showing relationships among L. monocytogenes ribogroups. Ribogroups are assigned letter designations. Refer to Table 1 for strain numbers assigned to each group.
A total of 12 ApaI restriction endonuclease digestion profiles (REDP) and 11 SmaI REDP were identified among the 19 isolates (Table 1). SmaI was slightly less discriminatory than ApaI, which has been reported elsewhere (2, 19). REDP I and II were very similar for enzymes SmaI and ApaI, differing by only one band, indicating that they are closely related. REDP VII and XII of ApaI differed by three bands, suggesting that a single genetic event may be responsible for the difference in patterns (21). The remaining patterns differed by more than five to eight bands, indicating distant relationships for these isolates (21). The REDP patterns resulted in the 19 isolates being grouped into 12 PFGE types. The groupings were generally consistent with the ribogroup patterns, with differences being seen within ribogroups G and C. Sensitive or resistant isolates do not appear clustered on the dendrogram.
Some reports suggest that sanitizer resistance is plasmid mediated (16, 17, 18,). Lemaitre et al. (9) reported extrachromosomal DNA in all BCl-resistant strains of L. monocytogenes, which was transferable with EtBr resistance. This resistance seemed to be associated with the ability to resist an accumulation of EtBr, and results indicate that these properties might be plasmid mediated. All our isolates accumulated EtBr, as determined by the presence of pink fluorescing colonies when the isolates were grown on TSA plates containing 0.25 μg of EtBr per ml, and this agrees with MIC results that indicate sensitivity to EtBr (Table 2). The MICs of EtBr observed in this study ranged from 6.25 to 25 μg/ml, in comparison with the 256 μg/ml reported by Lemaitre et al. (9) for resistant L. monocytogenes.
Plasmids were observed in all 19 strains (Table 1). Ten isolates possessed two plasmids, whereas the rest had only one. Most of the isolates harbored a small plasmid of approximately 20 kb. The sizes of the large plasmids varied among the isolates (40 to 120 kb). Studies have indicated that the percentage of plasmid-positive strains in Listeria spp. is higher in strains of food and environmental origins than in clinical isolates, as was also the case with our isolates (6, 10).
McLauchlin et al. (13) reported that the resistance to cadmium and arsenic and the presence of plasmid DNA varied markedly with the serotype and that plasmid DNA was strongly associated with cadmium resistance in serogroup 1 and 2 cultures but not within those of serogroup 4. However, Lemaitre et al. (9) have shown that resistant strains of L. monocytogenes harboring plasmids belonged both to serotype 1 and serotype 4, as was observed in this study.
In the Sara Lee isolates, there was a correlation between the presence of the two plasmids and resistance to QACs (Table 1). The two isolates (N7969 and H7596) that did not contain the large plasmid were susceptible to QACs. In the other Listeria isolates, however, the presence of a large plasmid was not correlated with resistance to QACs, although all of the resistant Listeria isolates tested contained two plasmids.
As an adaptation to adverse environments, bacteria can use proton motive force-driven multidrug efflux pumps to eliminate and avoid the effects of an accumulation of chemicals. The most well-studied species for the association of efflux mechanism and resistance to BCl is the QAC group of genes in S. aureus. The same resistance mechanism appears to be present in some strains of L. monocytogenes (1). The mdrL gene of L. monocytogenes can form MdrL, a protein that is homologous to the Bmr and Blt proteins of the B. subtilis multidrug resistance efflux pump family (8). Acquired QAC resistance may be directly related to the efficiency of such efflux pump systems. Mereghetti et al. (14) reported that overexpression of the MdrL protein may be required for a resistance phenotype. Conditions for the activation of the genes and the resulting expression of efflux pumps are required for resistance.
Two different pairs of primers were used to detect the presence of the mdrL gene in chromosomal and plasmid DNA. The results indicate that the mdrL gene is present in all our chromosomal and plasmid isolates. Thus, contrary to a previous report (14), it appears that the mdrL gene can be both chromosome and plasmid borne.
A second putative protein, OrfA, which was identified at the same time as MdrL, may be a transcriptional repressor of mdrL gene expression (8). The orfA gene was also found in all the isolates.
REFERENCES
- 1.Aase, B., G. Sundheim, S. Langsrud, and L. V. Rørvik. 2000. Occurrence of and a possible mechanism for resistance to a quaternary ammonium compound in Listeria monocytogenes. Int. J. Food Microbiol. 62:57-63. [DOI] [PubMed] [Google Scholar]
- 2.Brosch, B., C. Buchrieser, and J. Rocourt. 1991. Subtyping of Listeria monocytogenes serovar 4b by use of low-frequency-cleavage restriction endonuclease and pulsed-field gel electrophoresis. Res. Microbiol. 142:667-675. [DOI] [PubMed] [Google Scholar]
- 3.Bruce, J. 1996. Automated system rapidly identifies and characterizes microorganisms in food. Food Technol. 50:77-81. [Google Scholar]
- 4.Centers for Disease Control and Prevention. 1998. Multistate outbreak of listeriosis—United States, 1998. Morb. Mortal. Wkly. Rep. 47:1085-1086. [PubMed] [Google Scholar]
- 5.De Cesare, A., J. L. Bruce, T. R. Dambaugh, M. E. Guerzoni, and M. Weidmann. 2001. Automated ribotyping using different enzymes to improve discrimination of Listeria monocytogenes isolates, with a particular focus on serotype 4B strains. J. Clin. Microbiol. 39:3002-3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Earnshaw, A. M., and L. M. Lawrence. 1998. Sensitivity to commercial disinfectants and the occurrence of plasmids within various Listeria monocytogenes genotypes isolated from poultry products and the poultry-processing environment. J. Appl. Microbiol. 84:642-648. [DOI] [PubMed] [Google Scholar]
- 7.Emslie, K. R., D. E. Townsend, and W. B. Grubb. 1986. Isolation and characterisation of a family of small plasmids encoding resistance to nucleic acid-binding compounds in Staphylococcus aureus. J. Med. Microbiol. 22:9-15. [DOI] [PubMed] [Google Scholar]
- 8.Huillet, E., S. Larin, P. Pardon, and P. Berche. 1999. Identification of a new locus in Listeria monocytogenes involved in cellobiose-dependent repression of hly expression. FEMS Microbiol. Lett. 174:265-272. [DOI] [PubMed] [Google Scholar]
- 9.Lemaitre, J. P., H. Echchannaoui, G. Michaut, C. Divies, and A. Rousset. 1998. Plasmid-mediated resistance to antimicrobial agents among listeriae. J. Food Prot. 61:1459-1464. [DOI] [PubMed] [Google Scholar]
- 10.Margolles, A., and C. C. de los Reyes-Gavilan. 1998. Characterization of plasmids from Listeria monocytogenes and Listeria innocua strains isolated from short-ripened cheeses. Int. J. Food Microbiol. 39:231-236. [DOI] [PubMed] [Google Scholar]
- 11.Mata, M. T., F. Baquero, and J. C. Perez-Diaz. 2000. A multidrug efflux transporter in Listeria monocytogenes. FEMS Microbiol. Lett. 187:185-188. [DOI] [PubMed] [Google Scholar]
- 12.McDonnell, A. G., and A. D. Russell. 1999. Antiseptics and disinfectants: activity, action, and resistance. Clin. Microbiol. Rev. 12:147-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McLauchlin, J., M. Hampton, S. Shah, E. Threlfall, A. Wieneke, and G. Curtis. 1997. Subtyping of Listeria monocytogenes on a basis of plasmid profiles and arsenic and cadmium susceptibility. J. Appl. Microbiol. 83:381-388. [DOI] [PubMed] [Google Scholar]
- 14.Mereghetti, L., R. Quentin, N. Marquet-Van Der Mee, and A. Audurier. 2000. Low sensitivity of Listeria monocytogenes to quaternary ammonium compounds. Appl. Environ. Microbiol. 66:5083-5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Russell, A. D., and I. Chopra. 1996. Understanding antibacterial action and resistance, 2nd ed. Prentice-Hall, London, United Kingdom.
- 16.Russell, A. D. 1997. Plasmid and bacterial resistance to biocides. J. Appl. Microbiol. 82:155-165. [DOI] [PubMed] [Google Scholar]
- 17.Russell, A. D., and M. J. Day. 1996. Antibiotic and biocide resistance in bacteria. Microbios 85:45-65. [PubMed] [Google Scholar]
- 18.Sasatsu, M., Y. Shibata, N. Noguchi, and M. Kono. 1992. High-level resistance to ethidium bromide and antiseptics in Staphylococcus aureus. FEMS Microbiol. Lett. 93:109-114. [DOI] [PubMed] [Google Scholar]
- 19.Senczek, D., R. Stephan, and F. Untermann. 2000. Pulsed-field gel electrophoresis (PFGE) typing of Listeria strains isolated from a meat processing plant over a two-year period. Int. J. Food Microbiol. 62:155-159. [DOI] [PubMed] [Google Scholar]
- 20.Sundheim, G., T. Hagtvedt, and R. Dainty. 1992. Resistance of meat associated to a quaternary ammonium compound. Food Microbiol. 9:161-167. [Google Scholar]
- 21.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]