Abstract
Lactobacillus sakei is a lactic acid bacterium commonly used as a starter culture for dry sausage production and can utilize arginine via the arginine deiminase pathway. The arcABCTD cluster of L. sakei has been characterized, and transcriptional studies have shown that its expression is subject to carbon catabolite repression and induction by arginine. Downstream of arcD an additional gene has been found; this gene, arcR, codes for a putative regulatory protein of the Crp/Fnr family. Transcriptional studies have shown that arcR is coordinately transcribed with the remaining arc genes, and therefore, these genes constitute the arcABCTDR operon. Northern analysis also showed a complex pattern of transcripts, suggesting that processing and partial termination may play a role in regulation of the expression of individual genes of the operon. Inactivation of arcR led to arrest of transcription of the operon, indicating that the ArcR protein is essential for expression of the arc genes. The availability of this mutant made it possible to study whether the ability to utilize arginine affects the growth of L. sakei in meat fermentations. Under our experimental conditions, expression of arginine deiminase does not confer an obvious advantage to L. sakei, since the wild type and an arcR mutant strain displayed similar dynamics of growth.
The microbiota associated with dry fermented sausages encompasses a variety of bacteria, mostly lactic acid bacteria (LAB) and members of the Micrococcaceae, and undergoes marked changes during sausage production. LAB play an essential role in the production of dry fermented sausages. LAB, which are usually present at low levels in raw meat, grow rapidly in freshly prepared sausages, and the changes resulting from their metabolic activity make a major contribution to the final characteristics of the product (14). A number of different preparations and procedures are used for to manufacture dry fermented sausages; notwithstanding this, Lactobacillus sakei and Lactobacillus curvatus are the prevalent LAB in dry fermented sausages (5, 16, 25, 30, 33), and selected strains are normally used as starter cultures (15).
L. sakei is a facultatively heterofermentative LAB commonly found in plant material, meat, and fermented meat products (14). Unlike other facultatively heterofermentative lactobacilli, L. sakei can utilize arginine via the arginine deiminase (ADI) pathway (22). The ADI pathway comprises three reactions, which are catalyzed by ADI (EC 3.5.3.6), ornithine transcarbamoylase (EC 2.1.3.3), and carbamate kinase (EC 2.7.2.2), and converts arginine to ornithine, ammonia, and CO2, generating 1 mol of ATP per mol of arginine consumed. Previous studies have shown that genes encoding the proteins required for arginine catabolism in L. sakei are organized in a cluster (39). These studies showed that transcription of the pathway is repressed by glucose and is induced by arginine. Moreover, it has been shown that arginine enhances the survival of L. sakei in the stationary phase (4). Although arginine can be an important energy source in meat products, it has not been determined whether arginine metabolism has any effect on the evolution of the microbiota in meat fermentations.
Here a sixth gene in the ADI gene cluster of L. sakei, arcR, is described. This gene is essential for expression of the ADI pathway, as shown by transcriptional studies. The availability of an arcR mutant made it possible to study whether the ability to utilize arginine affects the dynamics of growth of L. sakei in a model dry sausage fermentation.
MATERIALS AND METHODS
Strains, media, growth conditions, and transformation.
The strains used in this study are listed in Table 1. L. sakei strains were routinely grown at 30°C in MRS medium (Oxoid Ltd., Basingstoke, United Kingdom), unless indicated otherwise. For determination of gene expression, MAM medium was used (39). Escherichia coli was grown in Luria-Bertani medium at 37°C with vigorous shaking, and 2% agar was added for solid media. Ampicillin (50 μg/ml), chloramphenicol (25 μg/ml), tetracycline (15 μg/ml), 5-bromo-4-chloro-3-indolyl-d-galactopyranoside (X-Gal) (80 μg/ml), and isopropyl-β-d-thiogalactopyranoside (1 mM) were added when required. Transformation of E. coli was performed by electroporation by using a Gene Pulser apparatus (Bio-Rad Laboratories, Richmond, Calif.) as recommended by the manufacturer. The electroporation and selection conditions used for L. sakei have been described previously (2, 17).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or vector | Genotype or description | Source or reference |
|---|---|---|
| E. coli XL1Blue | recA1 endA1 gyrA96 thi hsdR17 supE44 relA1 (F′ proAB lacIqlacZΔM15 Tn10) | Stratagene |
| L. sakei strains | ||
| 23K | Wild-type isolate | 2 |
| BL13 | Type strain | CECT 906 |
| BL213 | arcR variant of 23K | This study |
| BL214 | arcR variant of 23K | This study |
| S. xylosus BS108 | Isolate from a dry fermented sausage | 21 |
| Cloning vectors | ||
| pBluescriptII SK+ | Amr | Stratagene |
| pBSarcR | pBluescript containing the PCR-generated arcR gene cloned into BamHI-HindIII sites | This study |
| pBSH5B | pBluescript containing BamHI-HindIII fragment of pBSH5 spanning a stretch of arcA gene | 39 |
| pBSN2 | pBluescript containing an NheI genomic fragment of L. sakei spanning arcR and sequences upstream and downstream | 39 |
| pRV300 | Erir from pAβM1 | 17 |
| pDarcR | pRV300 containing the internal arcR fragment arcRF2R2 cloned into EcoRI-SalI sites | This study |
DNA techniques.
Total DNA was isolated from L. sakei as described by Posno et al. (27). Isolation of plasmid DNA from E. coli, restriction analysis, and ligation were performed by standard procedures (29). In order to sequence the region downstream of arcD, we used an inverse PCR-based strategy (24). DNA sequencing was carried out by using an ABI PRISM dRhodamine terminator cycle sequencing Ready Reaction kit with AmpliTaq DNA polymerase and an automatic ABI 310 DNA sequencer (Applied Biosystems, Foster City, Calif.).
The University of Wisconsin Genetics Computer Group (GCG) software package (version 8.0) was used for computer-assisted sequence analysis. Database searches were performed by using the BLAST server of the National Center for Biotechnology Information and FASTA and TFASTA of the EMBL-European BioInformatics Institute. Protein sequence alignments were constructed by using the Pileup program included in the GCG package.
Construction of plasmids.
An internal fragment of arcR was amplified by using primers arcRF2 (5′-TTTTGTCGACGATCAAACAGATGAGCGCAG-3′) and arcRR2 (5′-TTTTGAATTCGGATACGGCATATATGAGCG). This fragment was digested with EcoRI and SalI and ligated into the corresponding sites of pRV300, resulting in pDarcR. The complete arcR gene was amplified by using primers arcRF1 (5′-TTTTCCATGGGATCCATTGCCGCAGACGAATATCC-3′) and arcRR1 (5′-TTTTAAGCTTGTCGACCCTTAATTTTTAGG), digested with BamHI and HindIII, and ligated into the corresponding sites of pBluescript SK+, resulting in plasmid pBSarcR.
RNA isolation and Northern blot analysis.
Total RNA was isolated from L. sakei by using TRIZOL reagent (Gibco BRL, Grand Island, N.Y.). Cells were collected by centrifugation, washed with 50 mM EDTA (pH 8.0), resuspended in 1 ml of TRIZOL reagent, and then mechanically disrupted with glass beads in a cell disrupter (Savant Instruments, Holbrook, N.Y.). Additional steps were performed according to the instructions of the TRIZOL manufacturer. Sample preparation, denaturing agarose gel electrophoresis, and RNA transfer were performed by standard methods (29).
An RNA probe for arcA was synthesized from HindIII-digested plasmid pBSH5B with T3 RNA polymerase. An RNA probe for arcR was synthesized from BamHI-digested plasmid pBSarcR with T7 RNA polymerase. The reagents from a digoxigenin RNA labeling kit were used as instructed by the manufacturer (Roche Molecular Biochemicals, Indianapolis, Ind.).
Dry sausage model system.
Sausage manufacture was carried out as described previously (8), with the following modifications. The sausage mixture contained pork loin and raw pork bacon at a ratio of 75:25, 2.8% NaCl, 0.5% glucose, 0.015% NaNO2, 0.015% NaNO3, and 0.05% sodium ascorbate. The meat pieces were scalded, and their external layers were removed with a knife. The meat components were minced with an extruder with a 4-mm-diameter grid. Dry NaCl and solutions of glucose and curing salts were then added and mixed with a stainless steel bar, and the mixture was stored at 4°C for 24 h to allow diffusion of the salts. The batter was then divided into two portions and inoculated with Staphylococcus xylosus (105 CFU/g) and either L. sakei 23K or the arcR derivative BL213 (106 CFU/g). The L. sakei strains were grown in MRS fermentation broth (Reactivos Scharlau S.L., Barcelona, Spain) supplemented with 5 g of ribose per liter. Each mixture was thoroughly homogenized, stuffed into sterile dialysis tubing with a molecular mass cutoff of 12 to 14 kDa and a diameter of 23.8 mm (Medicell International Ltd., London, United Kingdom), and divided into pieces (approximately 35 g per sausage). The sausages were each hung from a wire frame and placed in a glass jar covered with a three-layer filter paper cap. All sausages were placed in a storeroom at 22°C with a relative humidity of 80% for 2 days (fermentation stage) and subsequently transferred to a storeroom at 15°C with a relative humidity of 75% for 14 days (ripening stage). In order to prevent exogenous contamination of the batter, all operations were carried out in a sterile airflow cabinet, the stainless steel tools used were autoclaved, the NaCl crystals were sterilized with dry heat, and the glucose and curing salts solutions were sterilized by filtration. The dialysis tubing was cleaned with 2% sodium bicarbonate and 1 mM EDTA at 80°C for 30 min and rinsed thoroughly in distilled water. The tubing was then immersed in 1 mM EDTA, autoclaved, and stored at 4°C until it was used.
Microbiological analysis and purification of total RNA from sausage samples.
For sampling, three sausages of each batch were removed after 2, 7, and 14 days for microbiological and molecular analyses and separately processed as follows, Each sausage was weighed, the pH was measured by inserting the pin electrode of a pH meter (micropH2001; Crison, Alella, Spain) into the center of the sausage, and the contents were transferred to a sterile stomacher bag. The sample was subsequently suspended in a 0.2% peptone solution at a sample/peptone solution ratio of 1:9, and the mixture was homogenized with a stomacher (Seward, London, United Kingdom). Aliquots were removed from the suspension to prepare serial dilutions, which were spread on MRS agar plates to count lactobacillus viable cells and on trypone soya agar (Difco, Detroit, Mich.) plates to count staphylococci. In order to verify that only the inoculated L. sakei strains grew during the fermentation, 40 colonies from each viable cell counting experiment were randomly selected from the MRS agar plates, and their randomly amplified polymorphic DNA profiles were determined as previously described (37). The remaining homogenate was filtered through filter paper, and the filtrate was centrifuged at 12,000 × g for 10 min at 4°C. The pellets were washed with 50 mM EDTA and stored at −80°C until they were used. RNA purification from the pellets was carried out by using TRIZOL reagent as described above.
Expression of the ADI operon was monitored by RNA dot blotting by using an RNA arcA probe. RNA was transferred to a nylon membrane (Hybond-N; Amersham) by using a Bio-Dot microfiltration apparatus (Bio-Rad) and the procedure described for use of digoxigenin-labeled probes (Roche Molecular Biochemicals).
Nucleotide sequence accession numbers.
The nucleotide sequence of arcR of L. sakei 23K has been deposited under accession number AF490616; the nucleotide sequence of L. sakei BL13 arcR and flanking regions has been deposited under accession number AJ001330 (updated record).
RESULTS
Sequence analysis of the arcR gene.
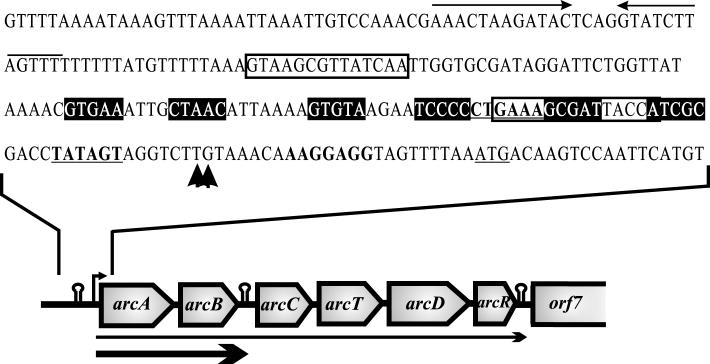
Sequencing of the region downstream of arcD revealed the presence of two additional open reading frames (ORFs) downstream of arcD on the same strand (Fig. 1). The first ORF is located 71 bp downstream of the stop codon of arcD, and it is preceded by a putative ribosome binding site (RBS) (5′-TGGGAGC-3′) located 8 bp upstream of the putative start codon, ATG (position 6219 in the sequence deposited under EMBL accession number AJ001330). A second possible start codon, TTG, is located 15 bp upstream of the ATG codon and is also preceded by a weak putative RBS (5′-ATTGAGA-3′) located 5 bp upstream. The intervening sequence between arcD and the first ORF does not contain either a potential rho-independent terminator or a promoter-like structure. The first possible start codon for the second ORF (orf7) is an ATG codon located 71 nucleotides downstream of the first ORF, but this putative start codon is not preceded by an RBS. Nevertheless, 60 nucleotides downstream of the first ATG (position 7055 in the sequence deposited under EMBL accession number AJ001330), there are two consecutive in-frame ATG codons. These codons are preceded by a putative RBS (5′-GGAGA-3′) four nucleotides upstream of the first ATG. The intervening sequence between the two ORFs does not contain any obvious promoter sequence; however, a putative rho-independent terminator is present (positions 6960 to 7001 in the sequence deposited under EMBL accession number AJ001330).
FIG. 1.
Genetic organization of the L. sakei arc operon and sequence of the ParcA promoter region. The ParcA promoter is indicated by a bent arrow. Putative rho-independent terminators are indicated by hairpins. The major RNA species observed are indicated by arrows under the operon structure. The sequence spanning the ParcA promoter region is shown in detail (positions 20 to 259 in the sequence deposited under EMBL accession no. AJ001330). The arrows at the top indicate a putative rho-independent terminator; the boxes contain putative cre sequences; the potential arcR binding sites are indicated by highlighting; −35 and −10 boxes are indicated by boldface type and underlining; the vertical arrows indicate transcription initiation points; the RBS is indicated by boldface type; and the translation start site of arcA is underlined.
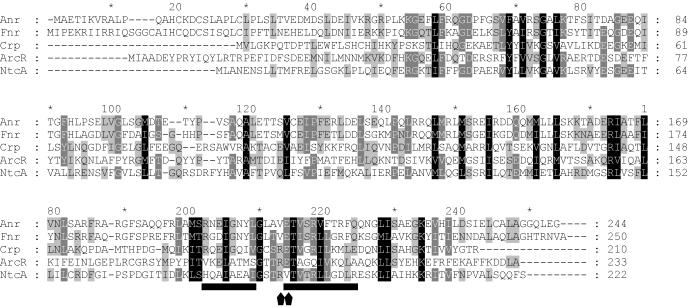
The ORF downstream of arcD encodes a predicted polypeptide containing 233 amino acids and has a calculated molecular mass of 27,461.3 Da. The results of sequence comparisons indicated that the putative polypeptide has features of a DNA binding protein of the Crp/Fnr family. Accordingly, this ORF was tentatively designated arcR (for arginine regulator). The most similar proteins included other putative members of the Crp/Fnr family associated with arc clusters in Enterococcus faecalis (arcE; accession no. CAC41344; 34% identical residues), Streptococcus pyogenes (SPY1548; accession no. AAK34340; 32% identical residues), Bacillus licheniformis (arcR; accession no. CAB95946; 30% identical residues), Oenococcus oeni (orf229; accession no. AF124851; 29% identical residues) (34), and Staphylococcus aureus (SA2424; accession no. BAB43729; 24% identical residues). A multiple-sequence alignment of arcR and selected representatives of the Crp/Fnr family showed that there are conserved residues throughout the complete sequence (Fig. 2). The similarity between ArcR and each of the proteins aligned is not high; however, the DNA-binding helix-turn-helix motif located in the C-terminal part is significantly more conserved. In particular, residues shown to be involved in recognition of the DNA binding site by Crp, such as Arg-213 and Glu-214 (numbering used in Fig. 2), are conserved, suggesting that ArcR must recognize a sequence similar to the sequence recognized by Crp.
FIG. 2.
Alignment of the deduced amino acid sequences of ArcR and representative members of the Crp/Fnr superfamily. Anr from P. aeruginosa (accession no. P23926; 41% conserved and 20% identical residues in a 182-amino-acid overlap) and Fnr from E. coli (accession no. P03019; 41% conserved and 22% identical residues in a 208-amino-acid overlap) belong to the Fnr family, Crp from E. coli (accession no. P03020; 41% conserved and 22% identical residues in a 199-amino-acid overlap) belongs to the Crp family, and NtcA from Synechococcus sp. (accession no. P29283; 44% conserved and 22% identical residues in a 187-amino-acid overlap) belongs to the NtcA family. The alignment was performed with Pileup of the GCG software. The helix-turn-helix region is underlined. The positions of residues Arg-213 and Glu-214, involved in protein-DNA interactions, are indicated by arrowheads under the alignment.
Analysis of the partial sequence of the predicted product of orf7 indicated that this gene encodes a putative membrane protein. Interestingly, similarity searches showed that the product of orf7 is significantly similar to uncharacterized membrane proteins encoded by genes located in arc clusters in Borrelia afzelii (orf473; accession no. O51898; 31% identical residues), Borrelia burgdorferi (BB0843; accession no. O51783; 31% identical residues), Haemophilus influenzae (HI0594; accession no. P44023; 59% identical residues), Salmonella enterica (STY4802; accession no. Q8Z128; 35% identical residues), S. enterica serovar Typhimurium (STM4464; accession no. Q8ZK36; 35% identical residues), S. aureus (SA1014; accession no. Q99UT9; 73% identical residues), Streptococcus pneumoniae (SP2152; accession no. Q97NA1; 31% identical residues), and S. pyogenes (Spy1543; accession no. Q99YT7; 28% identical residues). Unfortunately, biochemical evidence regarding the functions of these proteins is lacking.
arcR is coordinately transcribed with the other arc genes.
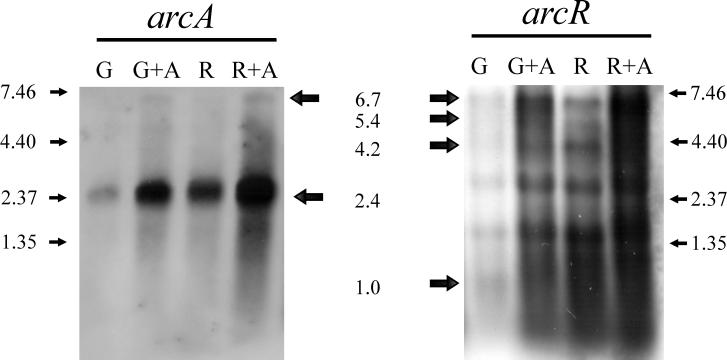
The transcription of arcR was studied by performing a Northern blotting analysis of samples of total RNA obtained from cultures of L. sakei grown in MAM media containing different combinations of sugars and arginine. Transcription of the arc cluster was monitored by using an RNA probe targeted to arcA. Transcription of arcR resulted in the same expression profile that was observed for the arc cluster (Fig. 3): expression was repressed by glucose and induced by arginine (39). Interestingly, the arcR transcription pattern was complex (Fig. 3). At least four different transcripts could be differentiated; a 6.7-kb transcript was the main mRNA species observed, but additional 5.4-, 4.2-, and 1-kb transcripts were also detected. The large 6.7-kb transcript was also detected with the arcA probe and could correspond to a transcript starting upstream of arcA and ending downstream of arcR. The additional transcripts observed may have been due to processing of the large transcript, since no significant differences were observed in the expression profiles.
FIG. 3.
Northern analysis of the arc operon with RNA probes targeted to arcA (left gel) and arcR (right gel). L. sakei 23K was grown in MAM medium supplemented as follows: lane G, 1 g of glucose per liter; lane G+A, 1 g of glucose per liter and 3 g of arginine per liter; lane R, 1 g of ribose per liter; lane R+A, 1 g of ribose per liter and 3 g of arginine per liter. The apparent sizes of the transcripts detected (in kilobases) are indicated between the gels. The sizes of the RNA size markers (in kilobases) are indicated on the right and left. The 2.5- and 1.6-kb signals probably correspond to 23S RNA and 16S RNA that were not efficiently denatured, which captured arc mRNA.
Transcription of the arc operon does not occur in arcR mutants.
In order to determine the role of arcR in the ADI pathway, arcR mutants were constructed by inserting the integrative vector pRV300 harboring an internal fragment of arcR (see Materials and Methods). The integrants were recovered by plating on MRS agar containing 5 μg of erythromycin per ml and were confirmed by Southern analysis by using digoxigenin-labeled pRV300 as a probe (results not shown). For transcriptional studies, total RNA was purified from L. sakei 23K and two integrants (BL213 and BL214) grown in MAM media containing different combinations of sugars and arginine. No transcript was detected in these integrants by Northern analysis under any of the growth conditions tested when either an arcA-targeted probe (Fig. 4) or an arcR-targeted probe (data not shown) was used, while transcription of the operon was readily detected in the wild-type strain.
FIG. 4.
Autoradiograph of RNA isolated from L. sakei 23K and two arcR derivatives (BL213 and BL214) probed with an arcA-targeted probe. Cells were grown in MAM medium supplemented as follows: lane G, 1 g of glucose per liter; lane G+A, 1 g of glucose per liter and 3 g of arginine per liter; lane R, 1 g of ribose per liter; lane R+A, 1 g of ribose per liter and 3 g of arginine per liter.
Growth of L. sakei 23K and an arcR mutant derivative in a model sausage fermentation.
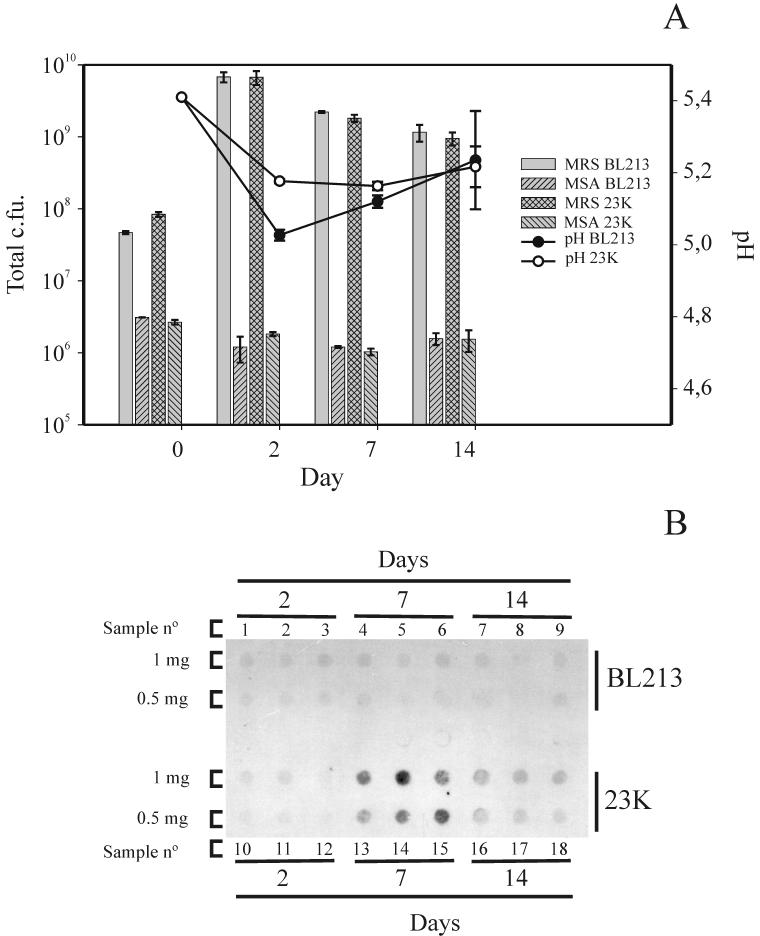
The dynamics of microbial populations in dry sausage fermentations performed with L. sakei 23K and BL213 were compared. Each sausage fermentation was characterized by a rapid increase in the L. sakei count during the fermentation stage and a slow decrease during the ripening stage (Fig. 5A). Randomly amplified polymorphic DNA analysis of colonies grown in MRS agar showed that only the inoculated L. sakei strains proliferated during the fermentation process (data not shown). The staphylococcal counts increased slightly during the fermentation stage and stayed at the same levels during the ripening stage. There were no significant differences in the dynamics of the microbial populations in the fermentations performed with the wild-type strain and the arcR mutant derivative. Nevertheless, the pH reached significantly lower values in fermentations performed with the arcR mutant derivative after the drying stage, but the differences became smaller during the ripening stage, so that after 14 days of fermentation the pH values were fairly similar (Fig. 5A). The decreases in water content were also similar in the two fermentations.
FIG. 5.
(A) Variation of total viable cell count (CFU expressed as [CFU per gram] × [weight of sausage] to account for the loss of water) and pH during the fermentation and ripening stages. The means of three independent samples are shown. The error bars indicate standard deviations. (B) RNA dot blot analysis of total RNA isolated from dry sausage samples as described in Materials and Methods and probed with an arcA-targeted probe. Samples were taken at the times indicated after inoculation. The numbers above and below the blot indicate independent samples. The amount of RNA loaded in each row is indicated on the left.
Transcription of arcA was monitored during fermentation as an indicator of expression of the ADI pathway. As previously observed in MAM medium, no induction of the pathway was observed for arcR strain BL213 (Fig. 5B). Strain 23K showed no induction after 2 days, transcription was maximal at 7 days, and after 14 days significant levels of transcription were still detected.
DISCUSSION
The role of ArcR in regulation of the ADI pathway in L. sakei was investigated in this study. The arcR gene is located downstream of arcD, the final member of the arcABCTD cluster encoding the ADI pathway. Inactivation of arcR stopped expression of the pathway, indicating that the protein encoded by arcR is essential for induction of expression of the pathway. Sequence similarities indicated that the product of arcR is a member of the Crp/Fnr family of transcriptional regulators. Although a low level of similarity was found in the N-terminal domain, the putative DNA binding domain is well conserved (Fig. 2). This domain contains the conserved residues Arg-213 and Glu-214 of Crp (numbering as in Fig. 2), which have been shown to make specific contacts with bases within the DNA site (26). Moreover, replacement of residues Val-213, Ser-217, and Gly-221 (numbering as in Fig. 2) in the Fnr helix-turn-helix motif with the Crp counterparts (Arg, Gly, and Lys, respectively) enables the mutant Fnr to recognize Crp binding sites (13). These three residues are also present in ArcR (Fig. 2). These data suggest that ArcR must recognize a binding site that is more similar to the binding site of Crp than to the binding site of Fnr.
Crp interacts with a 22-bp symmetric DNA site (consensus sequence, 5′-AAATGTGATCTAGATCACATTT-3′). Inspection of the complete sequence of the L. sakei arc cluster by using a broad criterion made it possible to locate three putative Crp-like binding sequences in the promoter region upstream of arcA (Fig. 1). A few additional putative binding sites meet this criterion, but they are located within ORFs and were not considered. Nevertheless, the actual DNA sequence recognized by ArcR has not been determined, and the DNA binding activity of ArcR remains to be investigated.
Regulation of the ADI pathway varies widely among microorganisms in relation to their different metabolic potentials (6). Biochemical studies have shown that in general, the ADI pathway is induced by arginine and anaerobiosis in microorganisms possessing a functional respiratory metabolism. Regulation at a molecular level has been studied in P. aeruginosa and B. licheniformis, both of which are facultatively anaerobic microorganisms. In P. aeruginosa Anr mediates anaerobic induction of the pathway and ArgR mediates induction by exogenous arginine (12, 18). In B. licheniformis, anaerobic induction is mediated by ArcR, a protein encoded by arcR, which is located downstream of the ADI operon, while ArgR mediates arginine induction (19, 20). Therefore, there is functional parallelism between these two organisms; both use ArgR to regulate arginine induction and a homolog of Crp/Fnr for anaerobic induction. Nevertheless, there is evidence which indicates that under anaerobic conditions activation by ArcR in B. licheniformis must be mediated by another system (19). Regulation of the pathway is less well known in other bacteria. In obligately fermentative organisms arginine and energy depletion are the primary signals for induction. It has been shown that the pathway is repressed by glucose in Clostridium sporogenes (35), L. sakei (39), and several enterococci and streptococci (6, 7). Moreover, there is evidence which indicates that ADI activity is enhanced under anaerobic conditions in L. sakei (4). On the basis of the role of ArcR homologs in P. aeruginosa and B. licheniformis, it may be suggested that ArcR plays the same role in L. sakei.
Analysis of the transcription of arcR revealed that arcR forms part of an operon together with arcA, arcB, arcC, arcT, and arcD. The arcR gene is transcribed mainly in a long messenger that is also detected with an arcA-targeted probe, although additional transcripts could be detected, indicating that the transcriptional profile of this operon is complex. Nevertheless, the results obtained suggest that transcription of the operon starts from the ParcA promoter exclusively. First, no significant differences between the levels of the transcripts were observed when total RNA isolated from cells grown under different conditions was analyzed (Fig. 3). Second, inactivation of arcR eliminates transcription of every transcript observed. Results reported previously (39) and results presented here suggest that partial termination and processing may account for the complex profile observed. Interestingly, complex transcriptional patterns have also been described for the arc operons of P. aeruginosa (11) and Halobacterium salinarium (28). In P. aeruginosa, partial termination and processing modulate the level of expression of the different genes of the operon.
Isogenic mutants of L. sakei 23K that were unable to express the ADI pathway were used to study whether the ability to use arginine confers a growth advantage in a meat environment. The dynamics of growth of L. sakei and S. xylosus in our dry sausage model system were similar to the dynamics reported by other authors (3, 33). Hence, L. sakei strains grew rapidly during the first stages of the fermentation, and the populations slowly declined during the ripening stage, while the S. xylosus population initially increased slightly and stayed at the same level during the ripening stage. After the fermentation stage, sausages containing the wild-type strain had a significantly higher pH than sausages containing the ArcR mutant strain. Although no induction of the pathway was detected at this stage (Fig. 5B), possibly due to the presence of glucose in the mixture, it should be noted that the cells used to inoculate the batter had been grown in media containing ribose and thus actively expressed the ADI pathway (Fig. 3). Therefore, it is conceivable that the cells still retained significant ADI activity that could account for the observed differences. Unfortunately, accurate measurements of ADI activity in cells growing in the batter were not obtained.
During the ripening stage, ADI was induced in strain 23K, showing a peak of expression at 7 days. This peak can be explained by activation of expression of the ADI pathway after glucose depletion. However, the pH slowly increased in all sausages fermented with either strain 23K or strain BL213. Clearly, the change in the pH during the ripening stage cannot be explained by ADI activity. The increase in pH can be explained by proteolytic activity that led to an increase in the concentration of free amino acids (reference 36 and references therein). The initial hydrolysis of muscle proteins has been attributed mainly to endogenous cathepsins, while the role of bacterial enzymes may be degradation of oligopeptides into small peptides and free amino acids (36). The proteolytic system of L. sakei has been partially characterized (23, 31, 32), yet regulation of the expression of the enzymes has received little attention.
Interestingly, under our experimental conditions, the ability to utilize arginine does not confer an obvious advantage to L. sakei, since the dynamics of growth of the wild-type strain and the arcR mutant were fairly similar. Similarly, no differences were observed in the dynamics of growth of S. xylosus. The arginine levels in raw meat are low (9); therefore, proteolytic activity is required to increase the levels of arginine. Peptidases can either be endogenous or have a microbial origin, but their activity can be compromised by the composition of the medium. In particular, muscle arginyl-aminopeptidase is severely inhibited at a low pH and a low temperature (10). In L. sakei, two arginyl-aminopeptidases have been reported and are probably active during sausage fermentation (31). Nevertheless, the differences in reported arginine levels in dry fermented sausages (1, 38) suggest that the effect of arginine degradation can vary considerably.
In summary, our results demonstrate that the arcR gene is essential for activation of transcription of the arcABCTDR operon. Control of expression of the ADI pathway is exerted mainly on ParcA, although other secondary control elements cannot be ruled out. The ability to utilize arginine did not confer any obvious selective advantage to L. sakei 23K in a dry sausage model system compared to an arcR mutant derivative. Nevertheless, due to the heterogeneity of the dry sausage preparations, this result cannot be extrapolated to other preparations.
Acknowledgments
This work was financed by the Spanish Government (project ALI98-0714). M. Zúñiga was supported by a European Union Marie Curie return fellowship.
REFERENCES
- 1.Beriain, M. J., G. Lizaso, and J. Chasco. 2000. Free amino acids and proteolysis involved in "salchichon' processing. Food Control 11:41-47. [Google Scholar]
- 2.Berthier, F., M. Zagorec, M. C. Champomier-Verg[grave]es, and F. Morel-Deville. 1996. High frequency transformation of Lactobacillus sake by electroporation. Microbiology 142:1273-1279. [DOI] [PubMed] [Google Scholar]
- 3.Bover-Cid, S., M. Izquierdo-Pulido, and M. C. Vidal-Carou. 2001. Effect of the interaction between a low tyramine-producing Lactobacillus and proteolytic staphylococci on biogenic amine production during ripening and storage of dry sausages. Int. J. Food Microbiol. 65:113-123. [DOI] [PubMed] [Google Scholar]
- 4.Champomier-Verg[grave]es, M.-C., M. Zuñiga, F. Morel-Deville, G. Pérez-Martínez, M. Zagorec, and S. D. Ehrlich. 1999. Relationships between arginine degradation, pH and survival in Lactobacillus sakei. FEMS Microbiol. Lett. 180:297-304. [DOI] [PubMed] [Google Scholar]
- 5.Cocolin, L., M. Manzano, C. Cantoni, and G. Comi. 2001. Denaturing gradient gel electrophoresis analysis of the 16S rRNA gene V1 region to monitor dynamic changes in the bacterial population during fermentation of Italian sausages. Appl. Environ. Microbiol. 67:5113-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cunin, R., N. Glansdorff, A. Piérard, and V. Stalon. 1986. Biosynthesis and metabolism of arginine in bacteria. Microbiol. Rev. 50:314-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curran, T. M., Y. Ma, G. C. Rutherford, and R. E. Marquis. 1998. Turning on and turning off the arginine deiminase system in oral streptococci. Can. J. Microbiol. 44:1078-1085. [DOI] [PubMed] [Google Scholar]
- 8.Dauneau, P. 1997. Production de composes aromatiques par des bacteries lactiques. Ph.D. thesis. Institut National Agronomique Paris-Grignon, Paris, France.
- 9.Dierick, N., P. Vandekerckhove, and D. Demeyer. 1974. Changes in nonprotein nitrogen compounds during dry sausage ripening. J. Food Sci. 39:301-304. [Google Scholar]
- 10.Flores, M., Y. Sanz, and F. Toldrá. 1998. Curing agents as regulators of muscle and microbial aminopeptidase activity in dry fermented sausages, p. 862-863. In Proceedings of the 44th ICoMST: Barcelona, Spain. Estrategias Alimentarias S.L.-Eurocarne, Madrid, Spain.
- 11.Gamper, M., B. Ganter, M. Polito, and D. Haas. 1992. RNA processing modulates the expression of the arcDABC operon in Pseudomonas aeruginosa. J. Mol. Biol. 226:943-957. [DOI] [PubMed] [Google Scholar]
- 12.Gamper, M., A. Zimmermann, and D. Haas. 1991. Anaerobic regulation of transcription initiation in the arcDABC operon of Pseudomonas aeruginosa. J. Bacteriol. 173:4742-4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Green, J., A. S. Irvine, W. Meng, and J. R. Guest. 1996. Fnr-DNA interactions at natural and semisynthetic promoters. Mol. Microbiol. 19:125-137. [DOI] [PubMed] [Google Scholar]
- 14.Hammes, W. P., A. Bantleon, and S. Min. 1990. Lactic acid bacteria in meat fermentation. FEMS Microbiol. Rev. 87:165-174. [Google Scholar]
- 15.Hammes, W. P., and H. J. Knauf. 1994. Starters in processing of meat products. Meat Sci. 36:155-168. [DOI] [PubMed] [Google Scholar]
- 16.Hugas, M., M. Garriga, T. Aymerich, and J. M. Monfort. 1993. Biochemical characterization of lactobacilli isolated from dry fermented sausages. Int. J. Food Microbiol. 18:107-113. [DOI] [PubMed] [Google Scholar]
- 17.Leloup, L., S. D. Ehrlich, M. Zagorec, and F. Morel-Deville. 1997. Single-crossover integration in the Lactobacillus sake chromosome and insertional inactivation of the ptsI and lacL genes. Appl. Environ. Microbiol. 63:2117-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu, C. D., H. Winteler, A. Abdelal, and D. Haas. 1999. The ArgR regulatory protein, a helper to the anaerobic regulator ANR during transcriptional activation of the arcD promoter in Pseudomonas aeruginosa. J. Bacteriol. 181:2459-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maghnouj, A., A. A. W. Abu-Bakr, S. Baumberg, V. Stalon, and C. Vander Wauven. 2000. Regulation of anaerobic arginine catabolism in Bacillus licheniformis by a protein of the Crp/Fnr family. FEMS Microbiol. Lett. 191:227-234. [DOI] [PubMed] [Google Scholar]
- 20.Maghnouj, A., T. F. de Sousa Cabral, V. Stalon, and C. Vander Wauven. 1998. The arcABDC gene cluster, encoding the arginine deiminase pathway of Bacillus licheniformis, and its activation by the arginine repressor argR. J. Bacteriol. 180:6468-6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miralles, M. C., J. Flores, and G. Pérez-Martínez. 1996. Biochemical tests for the selection of Staphylococcus strains as potential meat starter cultures. Food Microbiol. 13:227-236. [Google Scholar]
- 22.Montel, M. C., and M. C. Champomier. 1987. Arginine catabolism in Lactobacillus sake isolated from meat. Appl. Environ. Microbiol. 53:2683-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montel, M.-C., M.-P. Seronine, R. Talon, and M. Hébraud. 1995. Purification and characterization of a dipeptidase from Lactobacillus sakei. Appl. Environ. Microbiol. 61:837-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ochman, H., A. S. Gerber, and D. L. Hartl. 1988. Genetic applications of an inverse polymerase chain reaction. Genetics 120:621-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parente, E., S. Grieco, and M. A. Crudele. 2001. Phenotypic diversity of lactic acid bacteria isolated from fermented sausages produced in Basilicata (southern Italy). J. Appl. Microbiol. 90:943-952. [DOI] [PubMed] [Google Scholar]
- 26.Parkinson, G., C. Wilson, A. Gunasekera, Y. Ebright, R. Ebright, and H. Berman. 1996. Structure of the CAP-DNA complex at 2.5 [angst]A resolution: a complete picture of the protein-DNA interface. J. Mol. Biol. 260:395-408. [DOI] [PubMed] [Google Scholar]
- 27.Posno, M., R. J. Leer, N. van Luijk, M. J. F. van Giezen, P. T. H. M. Heuvelmans, B. C. Lokman, and P. H. Pouwels. 1991. Incompatibility of Lactobacillus vectors with replicons derived from small cryptic Lactobacillus plasmids and segregational instability of the introduced vectors. Appl. Environ. Microbiol. 57:1822-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruepp, A., and J. Soppa. 1996. Fermentative arginine degradation in Halobacterium salinarium (formerly Halobacterium halobium): genes, gene products, and transcripts of the arcRACB gene cluster. J. Bacteriol. 178:4942-4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 30.Samelis, J., S. Stavropoulos, A. Kakouri, and J. Metaxopoulos. 1994. Quantification and characterization of microbial populations associated with naturally fermented Greek dry salami. Food Microbiol. 11:447-460. [DOI] [PubMed] [Google Scholar]
- 31.Sanz, Y., and F. Toldrá. 1997. Aminopeptidase activities from Lactobacillus sake in models of curing ingredients and processing conditions for dry sausage. J. Food Sci. 62:1211-1213, 1234. [Google Scholar]
- 32.Sanz, Y., and F. Toldra. 2001. Purification and characterization of an X-prolyl-dipeptidyl peptidase from Lactobacillus sakei. Appl. Environ. Microbiol. 67:1815-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schillinger, U., and F. K. L[uml]ucke. 1987. Identification of lactobacilli from meat and meat products. Food Microbiol. 4:199-208. [Google Scholar]
- 34.Tonon, T., J. P. Bourdineaud, and A. Lonvaud-Funel. 2001. The arcABC gene cluster encoding the arginine deiminase pathway of Oenococcus oeni, and arginine induction of a Crp-like gene. Res. Microbiol. 152:653-661. [DOI] [PubMed] [Google Scholar]
- 35.Venugopal, V., and G. B. Nadkarni. 1977. Regulation of the arginine dihydrolase pathway in Clostridium sporogenes. J. Bacteriol. 131:693-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verplaetse, A. 1994. Influence of raw meat properties and processing technology on aroma quality of raw fermented meat products, p. 45-65. In Proceedings of the 40th International Congress of Meat Science and Technology, The Hague, The Netherlands. TNO Nutrition and Food Research, Zeist, The Netherlands.
- 37.Veyrat, A., M. C. Miralles, and G. Pérez-Martínez. 1999. A fast method for monitoring the colonization rate of lactobacilli in a meat model system. J. Appl. Microbiol. 87:49-61. [DOI] [PubMed] [Google Scholar]
- 38.Waade, C., and L. H. Stahnke. 1997. Dried sausages fermented with Staphylococcus xylosus at different temperatures and with different ingredient levels. Part IV. Amino acid profile. Meat Sci. 46:101-114. [DOI] [PubMed] [Google Scholar]
- 39.Zúñiga, M., M.-C. Champomier-Verg[grave]es, M. Zagorec, and G. Pérez-Martínez. 1998. Structural and functional analysis of the gene cluster encoding the enzymes of the arginine deiminase pathway of Lactobacillus sake. J. Bacteriol. 180:4154-4159. [DOI] [PMC free article] [PubMed] [Google Scholar]