Abstract
Campylobacter isolates from diverse samples within broiler production and processing environments were typed by using flaA short variable region DNA sequence analysis. Sixteen flocks from four different farms representing two broiler producers in Arkansas and California were analyzed. Fourteen of the flocks (87.5%) were Campylobacter-positive; two remained negative throughout the 6-week rearing period. In general, multiple clones were present within a flock. Additionally, clones found within a flock were also present on the final product, although the diversity of Campylobacter spp. on the final product appeared to be reduced relative to that observed within the flock. Comparison of clones between flocks on the same farm revealed that some clones of Campylobacter persisted in multiple flocks. Furthermore, some clones were identified across the two farms that were under the same management. In two sampling periods, environmental isolates were positive for Campylobacter prior to flock shedding. Environmental samples associated with five additional flocks were positive for Campylobacter concomitantly with recovery of Campylobacter from the birds. Analysis of the environmental isolates that were positive prior to flock shedding demonstrated that in some instances the environmental isolates possessed genotypes identical to those of isolates originating from the flock, while in other cases the environmental isolates possessed genotypes that were distantly related to isolates obtained from the flock. Analyses of environmental isolates that tested positive concurrently with the positive isolates from the flocks demonstrated varied results; in some instances the environmental isolates possessed genotypes identical to those of isolates originating from the flock, while in other cases the environmental isolates possessed genotypes that were distantly related to isolates obtained from the flock. These data suggest that the external environment may contribute to Campylobacter contamination during poultry production and processing. However, environmental contamination with Campylobacter does not appear to be the sole contributing factor.
Campylobacter jejuni, a gram-negative, microaerophilic bacterium, is currently believed to be the leading bacterial etiological agent of acute gastroenteritis in the human population; the total number of Campylobacter enteritis cases in the United States is estimated at 2.4 million per year, or approximately 1 to 2% of the population per year (3, 28, 33). The majority of C. jejuni cases are enteric. Most episodes are confined to local acute gastroenteritis characterized by nausea, abdominal cramps, diarrhea, and fatigue. Infections are generally self-limited and are resolved within several days after initial onset. Campylobacter infections have also been associated with unnecessary appendectomies, reactive arthritis, and development of Guillain-Barré syndrome, although these complications are infrequent (4-6, 35).
Handling and consumption of poultry or poultry-related products are considered to be a primary source for Campylobacter-induced disease in humans (5, 18, 24). Campylobacter has been cultured from as many as 75% of the live broiler population and from as much as 80% of processed poultry meat samples sold commercially (13, 14, 19, 27). The high colonization prevalence of poultry and the resultant clinical infections in humans have prompted a number of investigations focused upon identifying and subsequently eliminating sources of Campylobacter contamination in chickens. However, the pathways involved in Campylobacter contamination of poultry flocks remain unclear. Several suspected sources or vectors of contamination have been studied and include transmission from parent to progeny through the egg, exposure of birds to contaminated water, a previously contaminated rearing environment, hatchery pads, litter, feed, personnel, small animals on the farm, flies, and rodents (10, 11, 16, 17, 20, 25, 26, 29).
In an effort to further elucidate the means by which poultry flocks are contaminated with Campylobacter, 16 flocks from four different farms, representing two broiler producers in the United States, were studied. Samples from both poultry production and poultry processing environments were collected and cultured for the presence of Campylobacter. All Campylobacter isolates obtained were genotyped by using flaA short variable region (SVR) DNA sequence analysis to determine the relationships between the isolates (22). The identification of critical sources of Campylobacter contamination in poultry will allow for the development of intervention strategies that aggressively target specific locations. These interventions will facilitate the delivery of pathogen-free birds to the abattoir and consequently should reduce the incidence of human exposure.
MATERIALS AND METHODS
Broiler flocks.
Participating producers were located in Arkansas and California. Each producer provided access to two rearing houses located on two different farms; one rearing house was associated with high broiler growth performance, while the other rearing house had a history of low broiler growth performance. High and low broiler growth performance evaluations were based upon feed conversion ratios, weight gain, and overall performance. For all sites, one separate broiler flock was sampled over each of the four seasons, spring, summer, winter, and fall. Additional information regarding husbandry practices as they relate to the sampled flocks was previously described (32).
Sample collection.
Samples were collected in both the production environment and the processing environment for each of the flocks. Production samples were taken prior to flock placement, at delivery (delivery tray liners), and at 2-week intervals until slaughter (at 6 or 8 weeks of age). Production samples taken inside of the rearing facility at each sampling period included samples from broiler feces (n = 25), water lines (n = 6), water cups (n = 6), litter (n = 6), feed from the feed hopper (n = 2), feed from the feeder (n = 2), drag swabs (n = 3), wall swabs (n = 2), fan swabs (n = 2), mouse intestines (collected as trapped), insects (collected as observed), fly strips (n = 2), and cecal droppings (n = 5). Production-associated samples taken from the exterior of the rearing facility included samples from wild bird feces (collected as observed), animal feces (collected as observed), dirt (n = 1), standing water (n = 1), and boot swabs (n = 3).
Samples collected in the processing environment included samples from carcass rinses (n = 25), prechill water (n = 5), postchill water (n = 5), prescald water (n = 5), postscald water (n = 5), pretransport crate swabs (n = 10), and post-transport crate swabs (n = 10). In general, samples were collected into sterile plastic bags and placed in insulated boxes containing ice packs for overnight transport to the laboratory.
Bacterial isolates.
Fecal samples were weighed and diluted 1:3 (wt/vol) with Difco buffered peptone water (Becton Dickinson, Sparks, Md.). Serial dilutions were prepared and plated onto Campy-Cefex agar which was incubated at 42°C for 36 to 48 h in a microaerobic atmosphere (5% O2, 10% CO2, 85% N2) (30). Following incubation, a representative number of presumptive Campylobacter colonies were confirmed by observation of the typical cellular morphology by using phase-contrast microscopy and with a commercial latex agglutination kit (Integrated Diagnostics, Inc., Baltimore, Md.).
Carcass rinse samples were taken as previously described (9). Briefly, carcasses were sampled as they exited the final chill tank. Each carcass was placed into a clean bag with 100 ml of sterile tap water and vigorously shaken for 1 min. A 0.1-ml aliquot was plated onto duplicate Campy-Cefex agar and incubated as described for fecal samples. In addition, a 10-ml aliquot was used to inoculate 90 ml of Campylobacter enrichment broth (Acumedia Manufacturers, Inc., Baltimore, Md.) plus Campylobacter-selective supplement (Bolton's; Medox Diagnostics, Ogdensburg, N.Y.) which was enriched at 37°C for 4 h followed by a 40-h incubation at 42°C. Following a total of 44 h of incubation, the enriched samples were plated onto Campy-Cefex agar plates and incubated as previously described (32). Confirmation procedures for carcass rinse isolates were identical to those used for fecal isolates. All other samples were placed in Campylobacter enrichment broth plus Campylobacter-selective supplement and incubated as previously described (32).
Molecular subtype analysis.
One Campylobacter isolate originating from each positive sample type during each sampling period was chosen for subtype analysis. Isolated colonies of Campylobacter were resuspended in 300 μl of sterile H2O and placed at 100°C for 10 min. Ten microliters of each boiled cell suspension was used as the template for flaA SVR PCR with the primers FLA242FU (5′-CTA TGG ATG AGC AAT TWA AAA T-3′) and FLA625RU (5′-CAA GWC CTG TTC CWA CTG AAG-3′) (22). A 35-cycle reaction was used with 1 min of denaturing at 96°C, 1 min of annealing at 52°C, and a 1-min extension at 72°C. The resulting product was approximately 425 bp. Sequence data was generated by using either the FLA242FU primer or the FLA625RU primer with the Big Dye dye terminator cycle sequencing kit (Applied BioSystems Inc.-Perkin Elmer, Foster City, Calif.). Data were assembled with Sequencher version 4.1 (GeneCodes Corp., Ann Arbor, Mich.) and aligned by using ClustalX (34). The aligned sequences were compared, and dendrograms were generated by using the UPGMA algorithm with HKY85 distance measurements in version 4.0 of the Phylogenetic Analysis Using Parsimony (PAUP*) program (D. L. Swofford, Sinauer Associates, Sunderland, Mass., 1988).
RESULTS
Analyses of cultures.
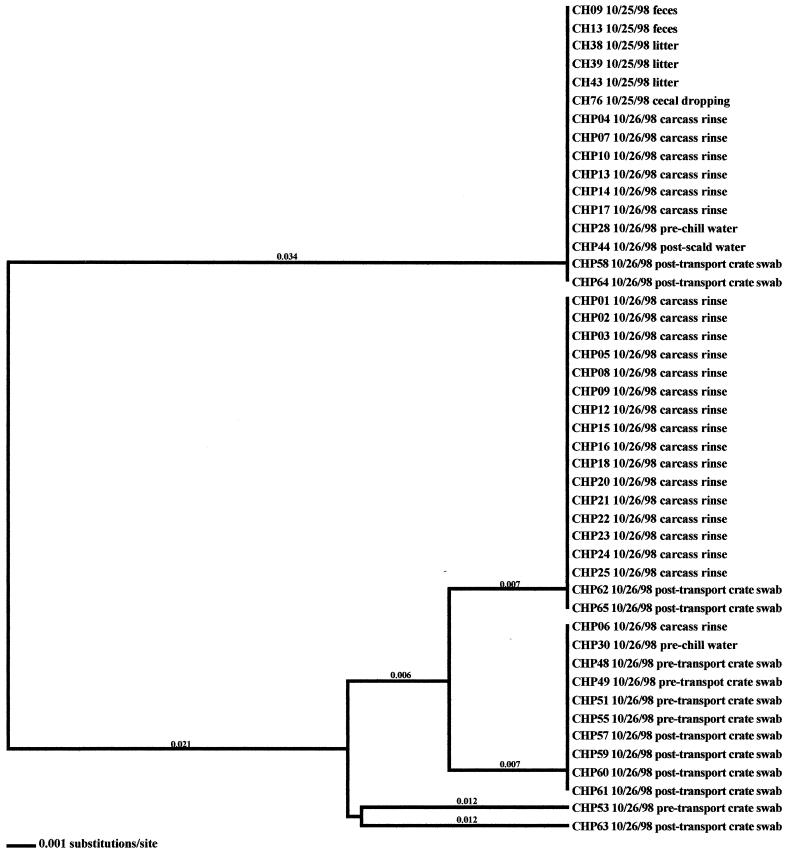
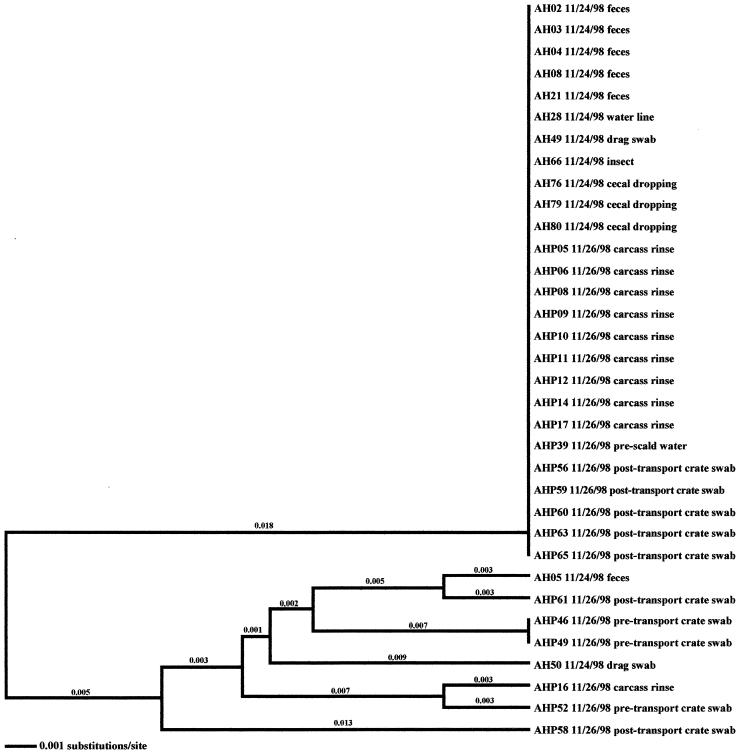
Sixteen flocks from four different farms, representing two U.S. broiler producers, were analyzed in this study. Flocks and isolates are designated such that the first letter indicates the producer code (A for Arkansas or C for California) and the second letter denotes whether the flock was reared on a high-performance (H) or low-performance (L) site. Isolates that were collected in the processing environment are further designated with a P as the third letter. All isolates analyzed in this investigation are presented in Tables 1 to 4. Fourteen (87.5%) of the flocks studied were Campylobacter positive; two flocks (CL-Summer and CL-Winter) remained negative throughout the 6-week rearing period. In 13 (92.9%) of the positive flocks, shedding of Campylobacter was detected at week 6; shedding was detected earlier, at week 4, in flock AL-Winter. In general, fecal samples, drag swabs from inside the house, cecal droppings, carcass rinses, pretransport crate swabs, and post-transport crate swabs comprised the majority of samples that were Campylobacter positive (Table 5). Samples from 12 of the 14 positive flocks included environmental samples that were Campylobacter positive; in 7 of these 12 flocks, samples taken from the external environment of the poultry house were Campylobacter positive. Only twice were environmental samples found to be positive prior to flock shedding (AL-Fall and CH-Winter) (Fig. 1 and 2).
TABLE 1.
Positive Campylobacter isolates from the producer A high-performance (AH) site
| Sample date (mo/day/yr) | Source | No. recovered |
|---|---|---|
| Spring | ||
| 04/06/98 | Feces from poultry | 13 |
| 04/07/98 | Carcass rinse from poultry | 1 |
| 04/07/98 | Prescald water | 3 |
| 04/07/98 | Postscald water | 3 |
| 04/07/98 | Pretransport crate swab | 5 |
| Summer | ||
| 07/13/98 | Feces from poultry | 3 |
| 07/13/98 | Drag swab from inside house | 3 |
| 07/13/98 | Cecal dropping | 5 |
| 07/23/98 | Carcass rinse from poultry | 7 |
| 07/23/98 | Pretransport crate swab | 1 |
| 07/23/98 | Post-transport crate swab | 7 |
| Fall | ||
| 11/24/98 | Feces from poultry | 6 |
| 11/24/98 | Water line | 1 |
| 11/24/98 | Drag swab from inside house | 2 |
| 11/24/98 | Insect | 1 |
| 11/24/98 | Cecal dropping | 3 |
| 11/26/98 | Carcass rinse from poultry | 10 |
| 11/26/98 | Prescald water | 1 |
| 11/26/98 | Pretransport crate swab | 3 |
| 11/26/98 | Post-transport crate swab | 7 |
| Winter | ||
| 01/25/99 | Feces from poultry | 4 |
| 01/25/99 | Drag swab from inside house | 1 |
| 01/25/99 | Cecal dropping | 1 |
| 01/25/99 | Postscald water | 1 |
| 01/25/99 | Post-transport crate swab | 7 |
TABLE 5.
Percentage of Campylobacter-positive samples by sample type
| Sample type | No. (%) of positive samples
|
||||
|---|---|---|---|---|---|
| AH (n = 105)a | AL (n = 144) | CH (n = 103) | CL (n = 55) | Total (n = 407) | |
| Fecal | 26 (24.76) | 59 (40.97) | 38 (36.89) | 23 (41.82) | 146 (35.87) |
| Water line | 1 (0.95) | 0 (0) | 0 (0) | 0 (0) | 1 (0.25) |
| Water cup | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Litter | 0 (0) | 2 (1.39) | 6 (5.83) | 3 (5.45) | 11 (2.70) |
| Feed hopper | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Feeder | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Drag swab | 6 (5.71) | 9 (6.25) | 5 (4.85) | 5 (9.09) | 25 (6.14) |
| Wall swab | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Fan swab | 0 (0) | 0 (0) | 1 (0.97) | 0 (0) | 1 (0.25) |
| Mouse intestine | 0 (0) | 2 (1.39) | 1 (0.97) | 0 (0) | 3 (0.74) |
| Wild bird feces | 0 (0) | 3 (2.08) | 4 (3.88) | 0 (0) | 7 (1.72) |
| Animal feces | 0 (0) | 2 (1.39) | 0 (0) | 0 (0) | 2 (0.49) |
| Insects | 1 (0.95) | 3 (2.08) | 0 (0) | 0 (0) | 4 (0.98) |
| Dirt | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Standing water | 0 (0) | 0 (0) | 1 (0.97) | 0 (0) | 1 (0.25) |
| Boot swab | 0 (0) | 1 (0.69) | 0 (0) | 1 (1.82) | 2 (0.49) |
| Fly strip | 0 (0) | 1 (0.69) | 0 (0) | 0 (0) | 1 (0.25) |
| Cecal dropping | 9 (8.57) | 8 (5.56) | 6 (5.83) | 5 (9.09) | 28 (6.89) |
| Carcass rinse | 18 (17.14) | 22 (15.28) | 24 (23.30) | 8 (14.55) | 72 (17.69) |
| Prechill water | 0 (0) | 0 (0) | 2 (1.94) | 0 (0) | 2 (0.49) |
| Postchill water | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Prescald water | 7 (6.67) | 1 (0.69) | 0 (0) | 0 (0) | 8 (1.97) |
| Postscald water | 7 (6.67) | 4 (2.78) | 1 (0.97) | 0 (0) | 12 (2.95) |
| Pretransport crate swab | 9 (8.57) | 13 (9.03) | 5 (4.85) | 1 (1.82) | 28 (6.89) |
| Post-transport crate swab | 21 (20.00) | 14 (9.72) | 9 (8.74) | 9 (16.36) | 53 (13.02) |
AH, Arkansas high-production site; AL, Arkansas low-production site; CH, California high-production site; CL, California low-production site. n is the total number of samples.
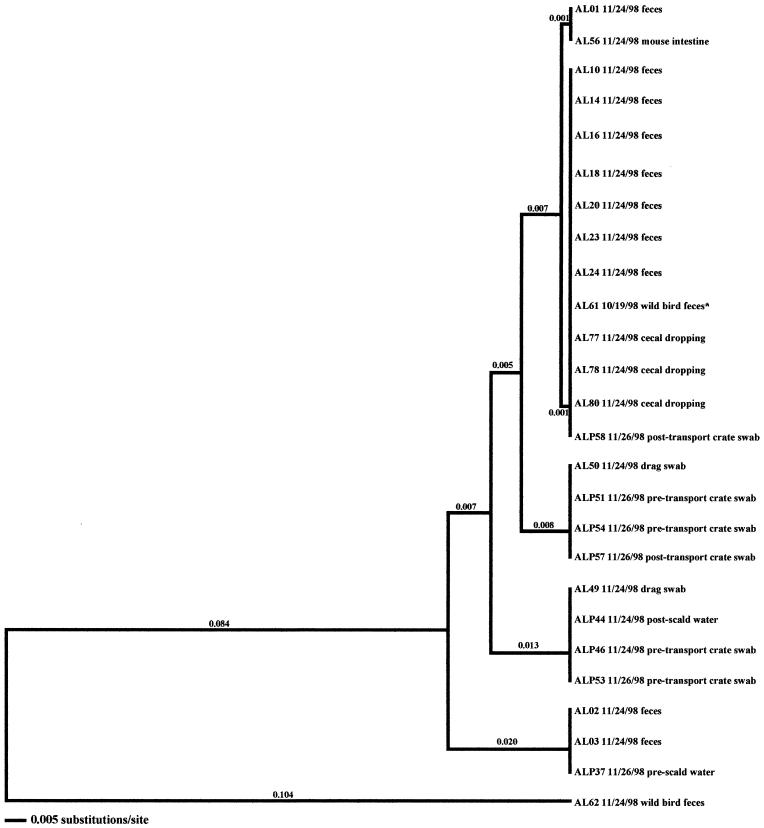
FIG. 1.
Relationships derived from comparison of the SVR DNA sequences of the flaA genes from Campylobacter organisms isolated from the Arkansas low-performance farm in the fall. The dendrogram was generated as described in Materials and Methods, and the isolates are labeled as described in Results. The dates of isolation and the sources of the isolates are also provided.
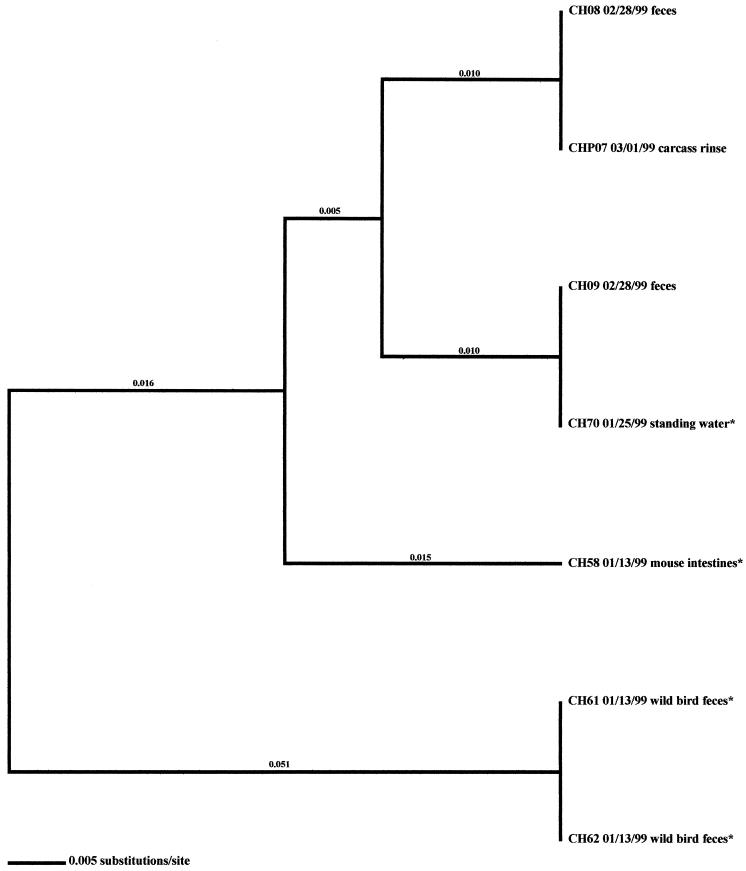
FIG. 2.
Relationships derived from comparison of the SVR DNA sequences of the flaA genes from Campylobacter organisms isolated from the California high-performance farm in the winter. The dendrogram was generated as described in Materials and Methods, and the isolates are labeled as described in Results. The dates of isolation and the sources of the isolates are also provided. Asterisks denote isolates that were obtained prior to flock shedding.
Molecular subtype analyses of production-associated isolates.
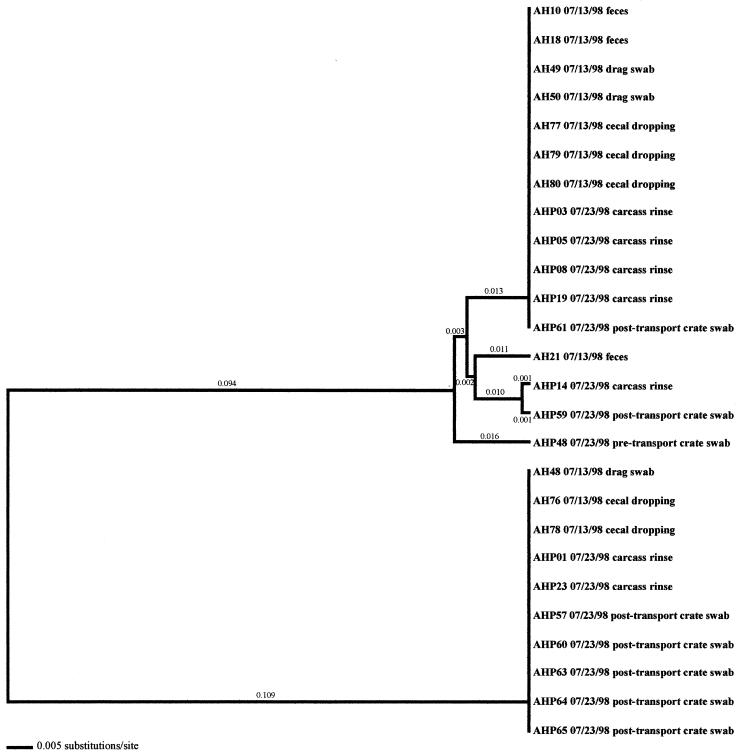
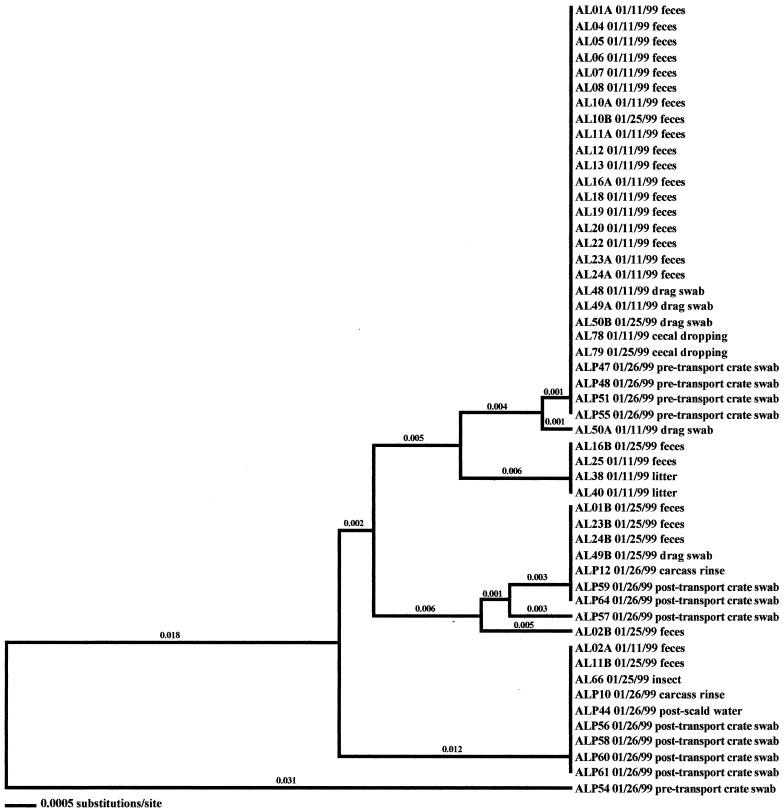
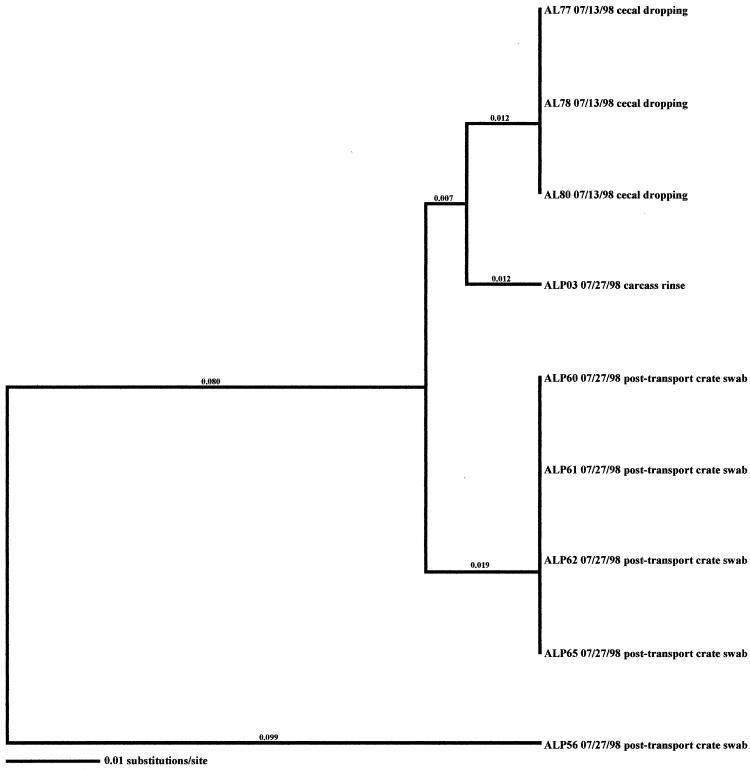
Molecular subtype analyses of Campylobacter isolated from individual flocks revealed that overall, multiple clones of Campylobacter were present within a single flock. As many as six distinct clones were isolated within a flock (AH-Spring) from samples representative of the broiler, fecal, and cecal samples (Fig. 3). The three exceptions observed in this investigation were the two positive flocks from the low-performance farm from producer C (CL-Spring and CL-Fall) (data not shown) and the samples obtained from producer A (AL-Spring) (Fig. 4). Only one predominant clone was observed for all related poultry samples collected during each of these specific investigations. Ten environmental samples were positive for Campylobacter during the AL-Spring analyses. Six of these isolates (two from drag swabs, AL48 and AL50; two from animal feces, AL63 and AL64; one from a fly strip, AL74; and one from an insect sample, AL68) possessed flaA SVR DNA sequences identical to those from poultry, while three isolates (one from a mouse sample, AL56; one from an insect sample, AL66; and one from a boot swab, AL71) demonstrated 2.4% variability. The remaining isolate, AL62, from a wild bird, demonstrated 21.3% variability from poultry isolates. With respect to CL-Spring and CL-Fall, the same clone was detected for all environmental and processing samples collected for each specific investigation. Interestingly, a pretransport crate swab sample (CLP48) collected during the CL-Spring investigation was the same clone that was seen within the corresponding flock and on the carcasses. Further subtype analyses demonstrated that all Campylobacter isolates obtained from the producer C low-performance farm, regardless of season, had identical flaA SVR DNA sequences and were therefore considered to be clonal.
FIG. 3.
Relationships derived from comparison of the SVR DNA sequences of the flaA genes from Campylobacter organisms isolated from the Arkansas high-performance farm in the spring. The dendrogram was generated as described in Materials and Methods, and the isolates are labeled as described in Results. The dates of isolation and the sources of the isolates are also provided.
FIG. 4.
Relationships derived from comparison of the SVR DNA sequences of the flaA genes from Campylobacter organisms isolated from the Arkansas low-performance farm in the spring. The dendrogram was generated as described in Materials and Methods, and the isolates are labeled as described in Results. The dates of isolation and the sources of the isolates are also provided.
As stated previously, environmental samples that were positive for Campylobacter prior to flock shedding were obtained from two flocks (AL-Fall and CH-Winter) in this investigation (Fig. 1 and 2). An isolate originating from a wild bird, AL61 10/19/98, was collected prior to the placement of flock AL-Fall. Subtype analysis demonstrated that this isolate was very closely related to 12 isolates representative of the broilers (fecal and cecal isolates, namely, AL01, AL10, AL14, AL16, AL18, AL20, AL23, AL24, AL77, AL78, AL79, and AL80). Four additional environmental isolates were collected from this farm once the flock was positive for Campylobacter. Only one of the isolates (AL56), from a mouse, was related to the Campylobacter that was observed in the broilers. Two of the three remaining isolates (drag swab isolates AL49 and AL50) were distinct from the broiler isolates; however, they matched isolates obtained from processing samples (pretransport crate swabs and post-transport crate swabs). The remaining isolate (AL62), from a wild bird, was distantly related (21.4% variability) to all other isolates obtained from the sampling of this flock.
Four environmental samples associated with the CH-Winter sampling were Campylobacter positive. Of these samples, three were obtained prior to flock placement (one mouse sample, CH58, and two samples from wild bird feces, CH61 and CH62). The remaining environmental isolate (a sample from standing water, CH70) was positive 2 weeks later; however, flock shedding had not yet begun. Subtype analyses demonstrated that the three isolates obtained prior to flock placement were distinct from broiler-associated isolates (2.7 to 6.6% variability). The isolate from standing water, however, was identical to a fecal isolate.
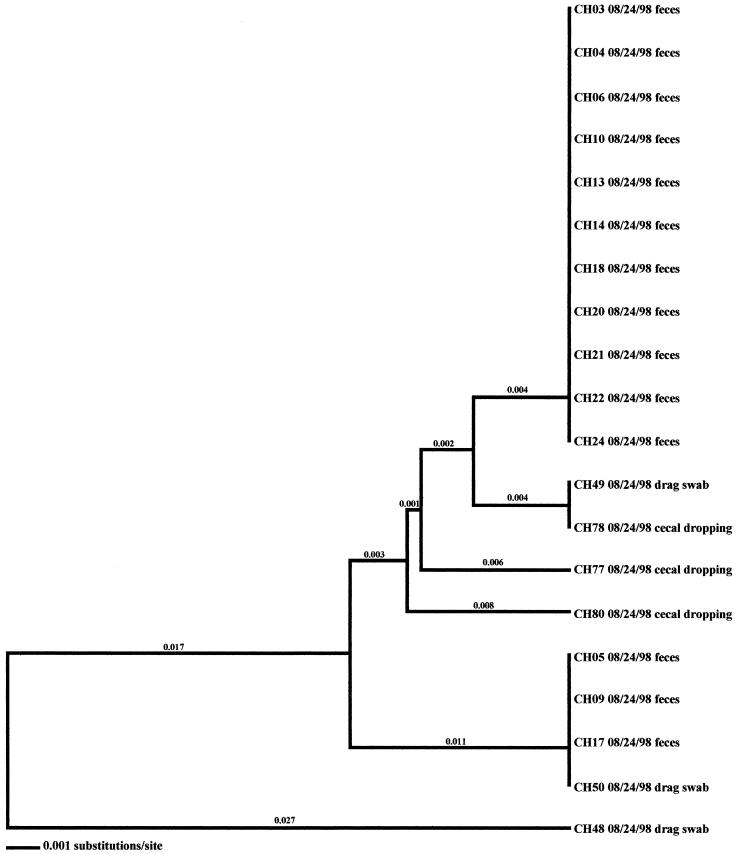
In the seven remaining flocks where environmental samples were positive, three flocks were from the high-performance site of producer C (CH-Spring, CH-Summer, and CH-Fall) (Fig. 5, 6, and 7), three were from the producer A high-performance site (AH-Summer, AH-Fall, and AH-Winter) (Fig. 8, 9, and 10), and one was from the producer A low-performance site (AL-Winter) (Fig. 11). A total of four distinct Campylobacter clones were observed from the broiler-associated samples from the AL-Winter investigation (Fig. 11). One of the subtypes, comprising isolates AL01B, AL23B, and AL24B, was present only during week 6 of sampling; all other subtypes were present at both week 4 and week 6 of sampling. Eight Campylobacter-positive environmental samples were isolated from this flock; five were isolated in week 4, when flock shedding began (two are litter isolates, AL38 01/11/99 and AL40 01/11/99, and three are drag swab isolates, AL48 01/11/99, AL49A 01/11/99, and AL49B 01/11/99), while three were isolated in week 6 (two are drag swab isolates, AL49B 01/25/99 and AL50B 01/25/99, and one is an insect isolate, AL66 01/25/99). All eight of the environmental Campylobacter isolates matched broiler-associated isolates. Additionally, all environmental isolates were comprised of the four distinct subtypes associated with the broilers.
FIG. 5.
Relationships derived from comparison of the SVR DNA sequences of the flaA genes from Campylobacter organisms isolated from the California high-performance farm in the spring. The dendrogram was generated as described in Materials and Methods, and the isolates are labeled as described in Results. The dates of isolation and the sources of the isolates are also provided.
FIG. 6.
Relationships derived from comparison of the SVR DNA sequences of the flaA genes from Campylobacter organisms isolated from the California high-performance farm in the summer. The dendrogram was generated as described in Materials and Methods, and the isolates are labeled as described in Results. The dates of isolation and the sources of the isolates are also provided.
FIG. 7.
Relationships derived from comparison of the SVR DNA sequences of the flaA genes from Campylobacter organisms isolated from the California high-performance farm in the fall. The dendrogram was generated as described in Materials and Methods, and the isolates are labeled as described in Results. The dates of isolation and the sources of the isolates are also provided.
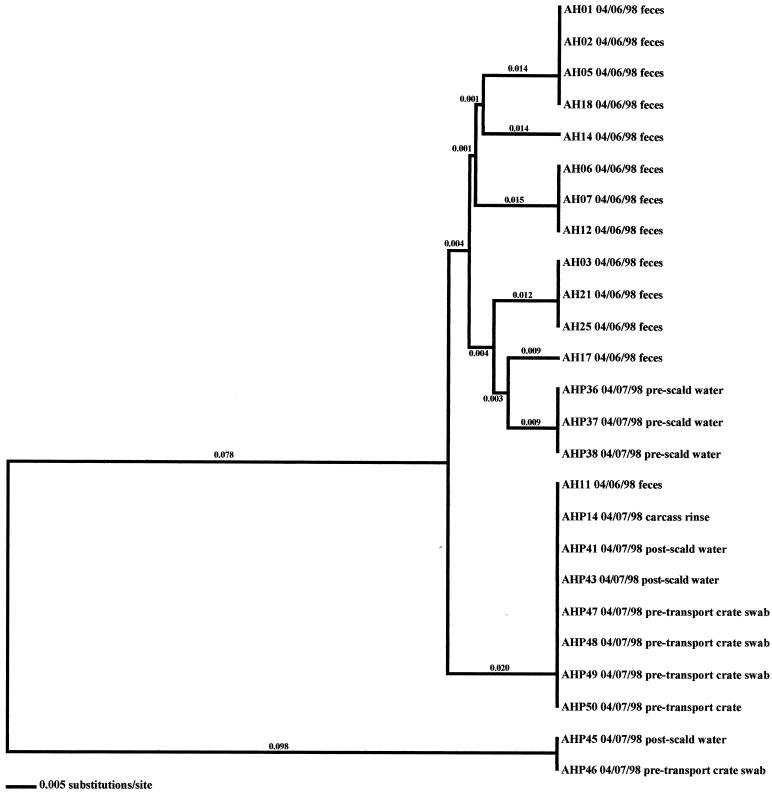
FIG. 8.
Relationships derived from comparison of the SVR DNA sequences of the flaA genes from Campylobacter organisms isolated from the Arkansas high-performance farm in the summer. The dendrogram was generated as described in Materials and Methods, and the isolates are labeled as described in Results. The dates of isolation and the sources of the isolates are also provided.
FIG. 11.
Relationships derived from comparison of the SVR DNA sequences of the flaA genes from Campylobacter organisms isolated from the Arkansas low-performance farm in the winter. The dendrogram was generated as described in Materials and Methods, and the isolates are labeled as described in Results. The dates of isolation and the sources of the isolates are also provided.
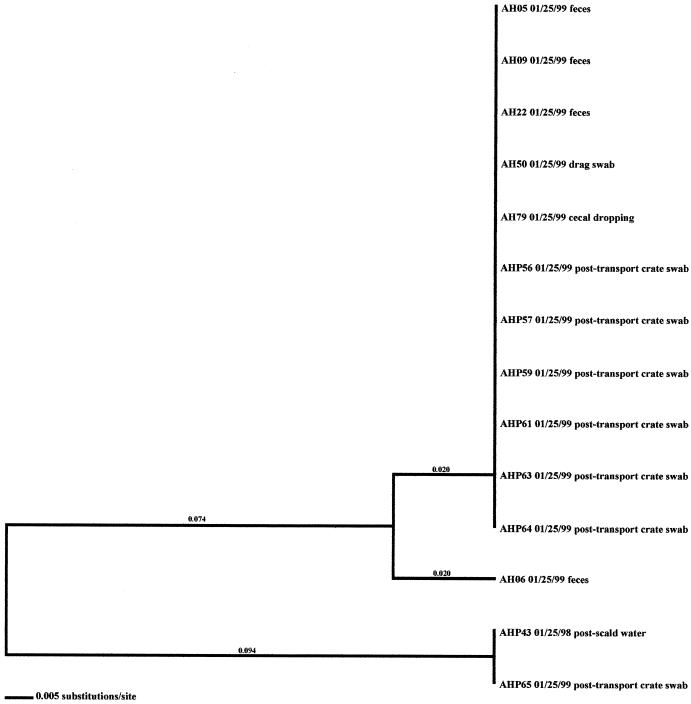
In the six other flocks where environmental isolates were Campylobacter positive, all environmental-associated isolates were obtained during the same week as the broiler-associated samples. Four of these flocks possessed environmental isolates that all had flaA SVR DNA sequences identical to those of the respective fecal and/or cecal isolates. These environmental samples included three drag swabs (AH 48, AH49, and AH50) from AH-Summer (Fig. 8), one drag swab (AH50) from AH-Winter (Fig. 10), three litter samples (CH38, CH39, and CH43) from CH-Fall (Fig. 7), and a variety of samples from CH-Spring that included three litter samples (CH39, CH42, and CH43), two drag swabs (CH49 and CH50), one fan swab (CH53), one sample from wild bird feces (CH62), and one sample from animal feces (CH63) (Fig. 5). Subtype analysis of environmental isolates from AH-Fall (Fig. 9) and CH-Summer (Fig. 6) revealed that some isolates possessed subtypes that are identical to the respective fecal and/or cecal isolates, while other environmental isolates did not. Three isolates from AH-Fall (one water line isolate, AH28, one drag swab isolate, AH49, and one insect isolate, AH66) had flaA SVR DNA sequences that were identical to each other as well as to those of broiler-associated isolates; one isolate, a drag swab isolate (AH50), was distantly related to all other isolates obtained from this flock. Three drag swab isolates were obtained from CH-Summer; two isolates (CH49 and CH50) had subtypes identical to those of broiler-associated isolates, while the third isolate (CH48) was distantly related to all other isolates (5.5% variability).
FIG. 10.
Relationships derived from comparison of the SVR DNA sequences of the flaA genes from Campylobacter organisms isolated from the Arkansas high-performance farm in the winter. The dendrogram was generated as described in Materials and Methods, and the isolates are labeled as described in Results. The dates of isolation and the sources of the isolates are also provided.
FIG. 9.
Relationships derived from comparison of the SVR DNA sequences of the flaA genes from Campylobacter organisms isolated from the Arkansas high-performance farm in the fall. The dendrogram was generated as described in Materials and Methods, and the isolates are labeled as described in Results. The dates of isolation and the sources of the isolates are also provided.
Molecular subtype analyses of processing-associated isolates.
Comparison of Campylobacter subtypes between production- and processing-associated isolates of the same flock demonstrated that in general, clones found within the flock during production were the same clones that were recovered from the carcasses, although the number of distinct clones was reduced on the carcasses. Four of the 14 Campylobacter-positive flocks were Campylobacter negative with respect to carcass rinse samples and therefore no comparison could be made. In 9 of the remaining 10 Campylobacter-positive flocks sampled, at least one subtype recovered from production-associated samples was also recovered from the carcass rinse samples. The last flock, AL-Summer (Fig. 12), had one carcass rinse isolate with no apparent matches to production isolates; however, the number of isolates recovered from this flock sampling was low.
FIG. 12.
Relationships derived from comparison of the SVR DNA sequences of the flaA genes from Campylobacter organisms isolated from the Arkansas low-performance farm in the summer. The dendrogram was generated as described in Materials and Methods, and the isolates are labeled as described in Results. The dates of isolation and the sources of the isolates are also provided.
Post-transport crate swab isolates were recovered from 9 of the 14 Campylobacter-positive flocks. In eight of these flocks, the post-transport crate isolates matched the isolates from the broiler production samples. Furthermore, the subtypes of post-transport crate isolates also matched the subtypes found in the majority of the respective carcass rinse isolates. Pretransport crate swab isolates were recovered from eight positive flocks. Five of these eight flocks (AH-Spring, AL-Spring, AL-Fall, AL-Winter, and CL-Spring) (Fig. 3, 4, 9, 11, and data not shown, respectively), demonstrated that the subtypes of the pretransport crate swab isolates were identical to the subtypes that were found in the production sample isolates from the respective flock. In two of the eight flocks, CH-Fall and AH-Fall (Fig. 7 and 9), two flaA SVR DNA sequence subtypes were observed. In each flock, one subtype was unique, while the second subtype matched isolates from carcass rinse samples only. Pretransport crate isolates from flock AH-Summer (Fig. 8) possessed subtypes that were unique and matched no other isolates.
Analysis of the remaining processing samples (from prescald water, postscald water, prechill water, and postchill water) revealed that a greater number of Campylobacter-positive samples were obtained from producer A than from producer C. Additionally, more Campylobacter isolates were collected from pre- and postscald water (14 isolates) than from pre- and postchill water (2 isolates). Molecular subtype analysis demonstrated that the prescald water isolates were comprised of both unique subtypes and subtypes that matched fecal isolates and carcass rinse isolates from the respective flocks. The postscald water isolates possessed flaA SVR DNA sequence subtypes that matched those of the respective production-associated isolates. The two prechill water isolates (CHP28 and CHP30) from CH-Fall had subtypes that matched carcass rinse isolates and pretransport crate isolates.
Molecular subtype analyses of Campylobacter isolates between flocks.
As stated previously, molecular subtype analysis of all Campylobacter isolates obtained from the producer C low-performance farm, regardless of season, had identical flaA SVR DNA sequences and were therefore considered to be clonal. Further analysis demonstrated that this subtype was also present in all flocks sampled from the high-performance farm of producer C; however, additional subtypes were also present. Analysis of all Campylobacter isolates obtained from the producer C high-performance farm during all seasons revealed that several of the same subtypes were present in different flocks. A similar observation, namely, that particular subtypes persist throughout the year on different farms, was made for producer A. Subtype analysis of all Campylobacter isolates obtained in this study from both producer A and producer C revealed that closely related subtypes were present within the operations of the two individual producers over all of the seasons investigated.
DISCUSSION
The Centers for Disease Control and Prevention estimates that Campylobacter enteritis is a multibillion-dollar disease and that the consumption of poultry is a primary source for the corresponding clinical infections in humans. Therefore, an understanding of the pathways involved in Campylobacter contamination and transmission in poultry flocks is critical for the development of intervention strategies and for the subsequent reduction of Campylobacter in poultry. These data demonstrate that, in general, multiple clones of Campylobacter are present within a flock and that the clones that are present within a flock are also present on the final product, although the number of distinct clones is reduced. These findings concur with what has been observed in the United Kingdom and elsewhere (1, 2, 21). One explanation for the decrease in the diversity of Campylobacter clones recovered from production relative to processing is that during processing, exposure to adverse conditions (chlorination, heat treatment, cold treatment, exposure to oxygen, etc.) reduces the total number of Campylobacter organisms, thus reducing the diversity (23). A second possible explanation is that the processing-associated isolates were recovered by using an enrichment methodology, while the fecal isolates were recovered by using direct plating. It has been previously demonstrated that the use of enrichment media for the recovery of Campylobacter may result in the preferential selection of certain subtypes (23).
Our findings also demonstrate that closely related clones are present throughout the poultry industry, regardless of the producer and location. It is not believed that this observation is a result of a media preference, because direct plating, rather than enrichment, was used for the recovery of Campylobacter from broiler fecal samples. However, it should be noted that all culture methodologies possess an inherent bias in that only a finite number of colonies can be picked and subsequently typed. Consequently, this limitation allows for only the most predominant clones to be detected. A second explanation is that some subtypes of Campylobacter may be better adapted for survival in different niches than are others. It may well be that the environmental stresses associated with having a chicken as a niche have led to the preferential survival of specific Campylobacter clones. A third, but related, explanation may be the transmission of Campylobacter from breeder hens to the broiler offspring (7-9, 12, 15, 20). A limited number of Campylobacter clones may be present within the population of breeder grandparents as well as the subsequent breeder hens and roosters. These clones may be passed on to the broiler offspring, where they proliferate and become the predominant clones. Should either of the two scenarios occur, new intervention strategies directed against the predominant clones or focused on the breeder hens and roosters will need to be developed.
Production-associated environmental samples were positive in 12 of the 16 flocks sampled. In 10 of these 12 flocks, environmental samples turned positive during the same week in which flock shedding began. In general, environmental isolates obtained from within the rearing facility (samples from drag swabs, litter, water line swabs, and fan swabs) possessed subtypes identical to those of the fecal and cecal isolates from the respective flocks. Considering that most of these samples were negative for Campylobacter prior to flock shedding, it is likely that shedding of Campylobacter from the broilers led to the contamination of the rearing-facility samples rather than the reverse. Analyses of external environmental isolates demonstrated varied results. In four of the five flocks where samples from the external environment of the rearing facility turned positive with flock shedding, environmental isolates had flaA SVR DNA sequences identical to those of the fecal and cecal isolates of the broilers. The exception, AL-Spring, revealed that some environmental isolates possessed genotypes identical to those of isolates originating from the flock, while other environmental isolates possessed genotypes that were distantly related to isolates obtained from the flock. Considering the free access that some vectors (mice, insects, and wild birds) have inside and outside of the rearing facility, coupled with the fact that positive isolates were obtained concurrently from all samples, it is difficult to determine which vector, poultry or environment, served as the source of the observed Campylobacter contamination.
Analysis of Campylobacter isolates from the two investigations where positive samples were obtained prior to flock shedding, namely, the AL-Fall and CH-Winter investigations, demonstrated that particular Campylobacter subtypes from these previously isolated environmental samples were identical to some of the Campylobacter subtypes observed within the flocks once shedding began. An isolate originating from a wild bird, AL61, had a flaA SVR DNA sequence identical to that of a subset of isolates collected from the broiler-associated samples. Additional subtypes were also present within this flock. A standing-water isolate, CH70, obtained from the CH-Winter investigation, had a subtype identical to that of a subsequently isolated broiler isolate. It should be noted that the number of Campylobacter-positive isolates obtained from this particular investigation was quite low. The above observations suggest that the environment can be a contributing factor to Campylobacter contamination of broiler flocks. Certain husbandry practices, such as opening the rearing facility to the outside environment for cooling, may lead to this contamination. Conversely, practices such as the reuse of litter and limited disinfection of the rearing facility between flocks appeared to have little effect on contamination of flocks. The presence of additional subtypes within broiler flocks, coupled with the previous observation that environmental samples are rarely positive prior to flock shedding, indicates, however, that the environment may not be the sole source for Campylobacter contamination of broiler flocks. Alternatively, Campylobacter organisms that were sublethally injured from environmental exposure may not have been adequately recovered in this investigation. Improved methods of culture and molecular detection would greatly facilitate epidemiologic investigations.
Analysis of Campylobacter isolates recovered from the processing environment revealed that contamination of the final product (carcass) originated primarily from the intestinal contents of the broiler flock. This may have occurred during processing or, more likely, transpired during transport of the birds (31). However, additional sources within the processing environment, such as pretransport crates and residual Campylobacter from previously processed flocks, also contributed to contamination of the final product. These observations suggest that more robust cleaning of crates and of the processing facilities may result in a further decline of Campylobacter contamination of the final market product. Improved detection methods and additional epidemiologic investigations are needed to further elucidate the means by which broiler flocks become contaminated with Campylobacter.
TABLE 2.
Positive Campylobacter isolates from the producer A low-performance (AL) site
| Sample date (mo/day/yr) | Source | No. recovered |
|---|---|---|
| Spring | ||
| 04/06/98 | Feces from poultry | 23 |
| 04/06/98 | Drag swab from inside house | 2 |
| 04/06/98 | Mouse intestine | 1 |
| 04/06/98 | Wild bird feces | 1 |
| 04/06/98 | Animal feces | 2 |
| 04/06/98 | Insects | 2 |
| 04/06/98 | Boot swab | 1 |
| 04/06/98 | Fly strip | 1 |
| 04/08/98 | Carcass rinse from poultry | 19 |
| 04/08/98 | Postscald water | 2 |
| 04/08/98 | Pretransport crate swab | 4 |
| Summer | ||
| 07/13/98 | Cecal dropping | 3 |
| 07/27/98 | Carcass rinse from poultry | 1 |
| 07/27/98 | Post-transport crate swab | 5 |
| Fall | ||
| 10/19/98 | Wild bird feces | 1 |
| 11/24/98 | Feces from poultry | 10 |
| 11/24/98 | Drag swab from inside house | 2 |
| 11/24/98 | Mouse intestine | 1 |
| 11/24/98 | Wild bird feces | 1 |
| 11/24/98 | Cecal dropping | 3 |
| 11/26/98 | Prescald water | 1 |
| 11/26/98 | Postscald water | 1 |
| 11/26/98 | Pretransport crate swab | 4 |
| 11/26/98 | Post-transport crate swab | 2 |
| Winter | ||
| 01/11/99 | Feces from poultry | 19 |
| 01/11/99 | Litter from house | 2 |
| 01/11/99 | Drag swab from inside house | 3 |
| 01/11/99 | Cecal dropping | 1 |
| 01/25/99 | Feces from poultry | 7 |
| 01/25/99 | Drag swab from inside house | 2 |
| 01/25/99 | Insects | 1 |
| 01/25/99 | Cecal dropping | 1 |
| 01/26/99 | Carcass rinse from poultry | 2 |
| 01/26/99 | Postscald water | 1 |
| 01/26/99 | Pretransport crate swab | 5 |
| 01/26/99 | Post-transport crate swab | 7 |
TABLE 3.
Positive Campylobacter isolates from the producer C high-performance (CH) site
| Sample date (mo/day/yr) | Source | No. recovered |
|---|---|---|
| Summer | ||
| 08/24/98 | Feces from poultry | 14 |
| 08/24/98 | Drag swab from inside house | 3 |
| 08/24/98 | Cecal dropping | 3 |
| Fall | ||
| 10/25/98 | Feces from poultry | 2 |
| 10/25/98 | Litter | 3 |
| 10/25/98 | Cecal dropping | 1 |
| 10/26/98 | Carcass rinse from poultry | 23 |
| 10/26/98 | Prechill water | 2 |
| 10/26/98 | Postscald water | 1 |
| 10/26/98 | Pretransport crate swab | 5 |
| 10/26/98 | Post-transport crate swab | 9 |
| Winter | ||
| 01/13/99 | Mouse intestine | 1 |
| 01/13/99 | Wild bird feces | 2 |
| 01/25/99 | Standing water | 1 |
| 02/28/99 | Feces from poultry | 2 |
| 03/01/99 | Carcass rinse from poultry | 1 |
| Spring | ||
| 04/28/99 | Feces from poultry | 20 |
| 04/28/99 | Litter pretransport crate swab | 3 |
| 04/28/99 | Drag swab from inside house | 2 |
| 04/28/99 | Fan swab | 1 |
| 04/28/99 | Wild bird feces | 1 |
| 04/28/99 | Animal feces | 1 |
| 04/28/99 | Cecal dropping | 2 |
TABLE 4.
Positive Campylobacter isolates from the producer C low-performance (CL) site
| Sample date (mo/day/yr) | Source | No. recovered |
|---|---|---|
| Fall | ||
| 10/25/98 | Feces from poultry | 2 |
| 10/25/98 | Litter | 1 |
| 10/25/98 | Drag swab from inside house | 3 |
| 10/25/98 | Boot swab | 1 |
| 10/25/98 | Cecal dropping | 3 |
| 10/25/98 | Carcass rinse from poultry | 1 |
| 10/25/98 | Post-transport crate swab | 4 |
| Spring | ||
| 04/28/99 | Feces from poultry | 21 |
| 04/28/99 | Litter | 2 |
| 04/28/99 | Drag swab from inside house | 2 |
| 04/28/99 | Cecal dropping | 2 |
| 04/28/99 | Carcass rinse from poultry | 7 |
| 04/28/99 | Pretransport crate swab | 1 |
| 04/28/99 | Post-transport crate swab | 5 |
Acknowledgments
This work was supported by funding from FSIS contract number 53-3A94-97-10.
We also acknowledge the participating companies and the technical support of Aphrodite Douris, Susan Brooks, Latoya Wiggins, and Debbie Posey.
REFERENCES
- 1.Ayling, R. D., M. J. Woodward, S. Evans, and D. G. Newell. 1996. Restriction fragment length polymorphism of polymerase chain reaction products applied to the differentiation of poultry campylobacters for epidemiological investigations. Res. Vet. Sci. 60:168-172. [DOI] [PubMed] [Google Scholar]
- 2.Berndtson, E., M. Tivemo, and A. Engvall. 1992. Distribution and numbers of Campylobacter in newly slaughtered broiler chickens and hens. Int. J. Food Microbiol. 15:45-50. [DOI] [PubMed] [Google Scholar]
- 3.Blaser, M. J., and L. B. Reller. 1981. Campylobacter enteritis. N. Engl. J. Med. 305:1444-1452. [DOI] [PubMed] [Google Scholar]
- 4.Bokkenheuser, V. D., and V. L. Sutter. 1981. Campylobacter infections, p. 301-310. In A. Balows and W. J. Hausler (ed.), Bacterial, mycotic and parasitic infections, 6th ed. American Public Health Association, Washington, D.C.
- 5.Bryan, F., and M. Doyle. 1995. Health risks and consequences of Salmonella and Campylobacter jejuni in raw poultry. J. Food Prot. 58:326-344. [DOI] [PubMed] [Google Scholar]
- 6.Butzler, J. P., and M. B. Skirrow. 1979. Campylobacter enteritis. Clin. Gastroenterol. 8:737-765. [PubMed] [Google Scholar]
- 7.Chuma, T., T. Yamada, K. Yano, K. Okomoto, and H. Yugi. 1994. A survey of Campylobacter jejuni in broilers from assignment to slaughter using DNA-DNA hybridization. J. Vet. Med. Sci. 56:697-700. [DOI] [PubMed] [Google Scholar]
- 8.Clark, A., and D. Bueschkens. 1985. Laboratory infection of chicken eggs with Campylobacter using pressure differential. Appl. Environ. Microbiol. 49:1467-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox, N. A., J. E. Thomson, and J. S. Bailey. 1981. Sampling of broiler carcasses for Salmonella with low volume water rinse. Poultry Sci. 60:768-770. [DOI] [PubMed] [Google Scholar]
- 10.Cox, N. A., N. J. Stern, K. L. Hiett, and M. E. Berrang. 2002. Identification of a new source of Campylobacter contamination in poultry: transmission from breeder hens to broiler chickens. Avian Dis. 46:535-541. [DOI] [PubMed] [Google Scholar]
- 11.Genigeorgis, C. A., M. Hassuney, and P. Collins. 1986. Campylobacter jejuni infection on poultry farms and its effect on poultry meat contamination during slaughtering. J. Food Prot. 49:895-903. [DOI] [PubMed] [Google Scholar]
- 12.Hiett, K. L., N. A. Cox, and N. J. Stern. 2002. Direct polymerase chain reaction detection of Campylobacter spp. in poultry hatchery samples. Avian Dis. 46:219-223. [DOI] [PubMed] [Google Scholar]
- 13.Hood, A. M., A. D. Pearson, and M. Shalamat. 1988. The extent of surface contamination of retailed chickens with Campylobacter jejuni serogroups. Epidemiol. Infect. 100:17-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Izat, A. L., and F. A. Gardner. 1986. Marketing and products: incidence of Campylobacter jejuni in processed egg products. Poultry Sci. 67:1431-1435. [DOI] [PubMed] [Google Scholar]
- 15.Jacobs-Reitsma, W. F. 1997. Aspects of epidemiology of Campylobacter in poultry. Vet. Q. 19:113-117. [DOI] [PubMed] [Google Scholar]
- 16.Jacobs-Reitsma, W. F. 1998. Experimental horizontal spread of Campylobacter amongst one-day-old broilers, p. 377-378. In A. J. Lastovica, D. G. Newell, and E. E. Lastovica (ed.), Campylobacter, Helicobacter and related organisms, 1st ed. University of Cape Town, Cape Town, South Africa.
- 17.Kazwala, R. R., J. D. Collins, J. Hannin, A. P. Crinion, and H. O'Mahoney. 1990. Factors responsible for the introduction and spread of Campylobacter jejuni infection in commercial poultry production. Vet. Rec. 126:305-306. [PubMed] [Google Scholar]
- 18.Kinde, H., C. A. Genigeorgis, and M. Pappaioanou. 1983. Prevalence of Campylobacter jejuni in chicken wings. Appl. Environ. Microbiol. 45:1116-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lam, K. M., A. J. DaMassa, T. Y. Morishita, H. L. Shivaprasad, and A. A. Bickford. 1992. Pathogenicity of Campylobacter jejuni for turkeys and chickens. Avian Dis. 36:359-363. [PubMed] [Google Scholar]
- 20.Lindblom, G. B., E. Sjogren, and B. Kaijser. 1986. Natural Campylobacter colonization in chickens raised under different environmental conditions. J. Hyg. 96:385-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mead, G. C., W. R. Hudson, and M. H. Hinton. 1995. Effect of changes in processing to improve hygiene control on contamination of poultry carcasses with Campylobacter. Epidemiol. Infect. 115:495-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meinersmann, R. J., L. O. Helsel, P. I. Fields, and K. L. Hiett. 1997. Discrimination of Campylobacter jejuni isolates by fla gene sequencing. J. Clin. Microbiol. 35:2810-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newell, D. G., J. E. Shreeve, M. Toszeghy, G. Domingue, S. Bull, T. Humphrey, and G. C. Mead. 2001. Changes in the carriage of Campylobacter strains by poultry carcasses during processing in abattoirs. Appl. Environ. Microbiol. 67:2636-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park, C. E., Z. K. Stankiewicz, J. Lovett, and J. Hunt. 1981. Incidence of Campylobacter jejuni in fresh eviscerated whole market chickens. Can. J. Microbiol. 27:841-842. [DOI] [PubMed] [Google Scholar]
- 25.Pearson, A. D., M. Greenwood, and T. D. Healing. 1993. Colonization of broiler chickens by waterborne Campylobacter jejuni. Appl. Environ. Microbiol. 59:987-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pearson, A. D., M. H. Greenwood, R. K. Feltham, T. D. Healing, J. Donaldson, D. M. Jones, and R. R. Colwell. 1996. Microbial ecology of Campylobacter jejuni in a United Kingdom chicken supply chain: intermittent common source, vertical transmission, and amplification by flock propagation. Appl. Environ. Microbiol. 62:4614-4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosef, O., B. Gondroson, and G. Kapperud. 1984. Campylobacter jejuni and Campylobacter coli as surface contaminants of fresh and frozen poultry carcasses. Int. J. Food Microbiol. 1:205-215. [Google Scholar]
- 28.Slutsker, L., S. F. Altekruse, and D. L. Swerdlow. 1998. Foodborne diseases. Emerging pathogens and trends. Infect. Dis. Clin. North Am. 12:199-216. [DOI] [PubMed] [Google Scholar]
- 29.Stern, N. J. 1992. Reservoires for Campylobacter jejuni and approaches for intervention in poultry, p. 49-60. In I. Nachamkin, M. J. Blaser, and L. S. Thompkins (ed.), Campylobacter jejuni: current status and future trends, 1st ed. American Society for Microbiology, Washington, D.C.
- 30.Stern, N., B. Wojton, and K. Kwiatek. 1992. A differential-selective medium and dry ice-generated atmosphere for recovery of Campylobacter jejuni. J. Food Prot. 55:514-517. [DOI] [PubMed] [Google Scholar]
- 31.Stern, N. J., M. R. S. Clavero, J. S. Bailey, N. A. Cox, and M. C. Robach. 1995. Campylobacter spp. in broilers on the farm and after transport. Poultry Sci. 74:937-941. [DOI] [PubMed] [Google Scholar]
- 32.Stern, N. J., P. Fedorka-Cray, J. S. Bailey, N. A. Cox, S. E. Craven, K. L. Hiett, M. T. Musgrove, S. Ladely, D. E. Cosby, and G. C. Mead. 2001. Distribution of Campylobacter spp. in selected U.S. poultry production and processing operations. J. Food. Prot. 64:1705-1710. [DOI] [PubMed] [Google Scholar]
- 33.Tauxe, R. V. 1992. Epidemiology of Campylobacter jejuni infections in the United States and other industrialized nations, p. 9-19. In I. Nachamkin, M. J. Blaser, and L. S. Thompkins (ed.), Campylobacter jejuni: current status and future trends, 1st ed. American Society for Microbiology, Washington, D.C.
- 34.Taylor, D. N. 1992. Campylobacter infections in developing countries, p. 20-30. In I. Nachamkin, M. J. Blaser, and L. S. Thompkins, (ed.), Campylobacter jejuni: current status and future trends, 1st ed. American Society for Microbiology, Washington, D.C.
- 35.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walker, R. I., M. B. Caldwell, E. C. Lee, P. Guerry, T. J. Trust, and G. M. Ruiz-Palacios. 1986. Pathophysiology of Campylobacter enteritis. Microbiol. Rev. 50:81-94. [DOI] [PMC free article] [PubMed] [Google Scholar]