Abstract
The carbazole 1,9a-dioxygenase (CARDO) system of Pseudomonas resinovorans strain CA10 consists of terminal oxygenase (CarAa), ferredoxin (CarAc), and ferredoxin reductase (CarAd). Each component of CARDO was expressed in Escherichia coli strain BL21(DE3) as a native form (CarAa) or a His-tagged form (CarAc and CarAd) and was purified to apparent homogeneity. CarAa was found to be trimeric and to have one Rieske type [2Fe-2S] cluster and one mononuclear iron center in each monomer. Both His-tagged proteins were found to be monomeric and to contain the prosthetic groups predicted from the deduced amino acid sequence (His-tagged CarAd, one FAD and one [2Fe-2S] cluster per monomer protein; His-tagged CarAc, one Rieske type [2Fe-2S] cluster per monomer protein). Both NADH and NADPH were effective as electron donors for His-tagged CarAd. However, since the kcat/Km for NADH is 22.3-fold higher than that for NADPH in the 2,6-dichlorophenolindophenol reductase assay, NADH was supposed to be the physiological electron donor of CarAd. In the presence of NADH, His-tagged CarAc was reduced by His-tagged CarAd. Similarly, CarAa was reduced by His-tagged CarAc, His-tagged CarAd, and NADH. The three purified proteins could reconstitute the CARDO activity in vitro. In the reconstituted CARDO system, His-tagged CarAc seemed to be indispensable for electron transport, while His-tagged CarAd could be replaced by some unrelated reductases.
Pseudomonas resinovorans strain CA10 is a bacterium that has the ability to utilize carbazole (CAR) as its sole source of carbon, nitrogen, and energy. As the initial degradation reaction, CAR is dioxygenated at the angular (C-9a) and adjacent (C-1) positions to yield the unstable cis-hydrodiol (25, 27). The resultant cis-hydrodiol is spontaneously converted to 2′-aminobiphenyl-2,3-diol, which is further converted to anthranilate via meta cleavage and hydrolysis. Such initial dioxygenation is called angular dioxygenation and contains the degradation pathways of CAR and its structural analogues (25).
Genes encoding CAR 1,9a-dioxygenase (CARDO) were cloned from P. resinovorans strain CA10, and CARDO was found to be a multicomponent enzyme system which consists of terminal oxygenase (CarAa), ferredoxin (CarAc), and ferredoxin reductase (CarAd) (32). Amino acid sequence homology and phylogenetic analysis revealed that CarAa is a rather unique type of oxygenase that shares low homology with other known dioxygenases (23, 32). Furthermore, from studies on substrate specificity, it was found that CARDO has a broad substrate range and can catalyze diverse oxygenations: angular dioxygenation, lateral dioxygenation, and monooxygenation (26). In addition, Habe et al. (11) have reported that CARDO can attack some chlorinated dioxin congeners in a manner similar to that of dibenzofuran 4,4a-dioxygenase from Terrabacter sp. strain DBF63 and dioxin dioxygenase from Sphingomonas wittichii strain RW1 (43), although some difference was observed in particular features. Compared to other degradative reactions for dioxin, angular dioxygenation is one of the attractive degradative mechanisms, because this reaction and subsequent spontaneous ring cleavage result in the destruction of the planar structure from which the toxicity of dioxin derives (27). In fact, strain CA10 expressing CARDO has been found to have the potential of decontaminating dioxin-contaminated soil (12). Therefore, the CARDO enzyme itself and bacteria with CARDO activity could be important tools in the bioremediation of dioxin contamination.
Considering these unique properties of the CARDO system, the oxygenation and recognition mechanisms of the substrate(s) are of great interest. For better understanding of the characteristics of CARDO, we report here the purification of the respective components of the CARDO system and describe some of their characteristics.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and culture conditions.
Escherichia coli strains BL21(DE3) (Novagen, Madison, Wis.) and JM109 (31) were used for protein expression and DNA propagation, respectively. Plasmids used in this study were pET-26b(+) (Novagen) and its derivatives, as well as pUCARA, pUCA152, and pUCA14 (32). The media used were Luria-Bertani (LB) medium (31) or SB medium (12 g of tryptone/liter, 24 g of yeast extract/liter, 5 ml of glycerol/liter, 12.5 g of K2HPO4/liter, 3.8 g of KH2PO4/liter) supplemented with kanamycin (50 μg/ml) or ampicilin (100 μg/ml). E. coli strains harboring appropriate plasmids were grown at 25 or 30°C for protein expression and at 37°C for DNA propagation. For protein purification, when the optical density at 600 nm reached approximately 0.5, 0.1 or 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added. After a 12-h incubation with IPTG, the E. coli cells were gathered from 2-liter cultures by centrifugation and used for extraction of the protein produced.
Construction of plasmids.
We constructed pET-26b(+)-based plasmids for overexpression of CarAc and CarAd whose C termini were tagged with six histidine residues. Artificial XbaI and XhoI sites were created on the respective end of the carAc- or carAd-containing DNA fragment by PCR using the appropriate primer sets (for carAc, 5′-TCTAGA-TCTAGA-TTTCTCGTCGTGCAGACATT-3′ and 5′-CTCGAG-CTCGAG-CTTCTTTTCTCCGGCGACAT-3′; for carAd, 5′-TCTAGA-TCTAGA-TAAGGTCTTTGGACTTGCTC-3′ and 5′-CTCGAG-CTCGAG-GAAAAATGCGTCAAAATGAA-3′ [XbaI and XhoI restriction sites are italicized]) and a template DNA, pUCARA (32). The resultant PCR amplicons were ligated into XbaI and XhoI sites of pET-26b(+) to produce pECAC1 and pECAD1, containing carAc and carAd, respectively.
To express CarAa as a native form in the E. coli expression system, the 5′ portion of the carAa gene was at first amplified by PCR from pUCARA by using primer set 5′-GAGATG-CATATG-GCGAACG-3′ and 5′-GAAC-GGATCC-ACCTTGAGAAC-3′ (artificial NdeI and BamHI sites are italicized). The resultant PCR product was ligated to pT7Blue T-vector (Clontech Laboratories Inc., Palo Alto, Calif.), to yield pTA2. An approximately 300 bp SmaI-EcoRI fragment containing the 3′ portion of the carAa gene from pUCA152 (32) was ligated into the corresponding sites of pTA2 to produce pTAA2. The NdeI-EcoRI fragment containing the entire carAa gene from pTAA2 was ligated into the corresponding site of pET-26b(+) to produce pES2.
For the activity measurement of CarAc, pUCA33 containing the carAaAaAd genes was constructed by XhoI digestion and subsequent self-ligation of pUCARA.
Purification of Car proteins.
All purification procedures were carried out at 4°C. E. coli BL21(DE3)(pECAC1) cells expressing His-tagged CarAc were harvested from a 2-liter culture and were then resuspended in TI buffer (20 mM Tris-HCl [pH 7.5] containing 10 mM imidazole and 0.5 M NaCl). The crude cell extract (CE) prepared by sonication and centrifugation was applied to a HiTrap Chelating HP column (column volume, 5 ml; Amersham Biosciences, Tokyo, Japan) equipped with the fast protein liquid chromatography instrument (Amersham Biosciences) according to the manufacturer's recommendation. After sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), the fractions containing His-tagged CarAc were pooled and concentrated by ultrafiltration using Centriprep-3 (Millipore, Bedford, Mass.). The resultant preparation was further purified by gel filtration chromatography (GFC) using a Sephacryl S-200 (26 by 600 mm; Amersham Biosciences) column and GFC buffer (20 mM Tris-HCl buffer [pH 7.5] containing 0.2 M NaCl and 10% glycerol). Purification of His-tagged CarAd was done similarly, except that plasmid pECAD1 and Centriprep-10 (Millipore) were used.
The CE containing CarAa was prepared from E. coli BL21(DE3)(pES2) cells suspended in TG buffer (20 mM Tris-HCl [pH 7.5] containing 10% glycerol) as described above and was then applied to a DEAE-Sepharose CL-6B column (25 by 150 mm; Amersham Biosciences). After a wash, the protein was eluted by incremental NaCl concentration under a linear gradient (400 ml) of 0.1 to 0.5 M NaCl. Fractions containing CarAa were concentrated and desalted by Centriprep-10 and were then applied to a POROS HQ/L column (7.5 by 100 mm; Perkin-Elmer Applied Biosystems). After a wash, elution was carried out using a gradient (150 ml) of 0.1 to 0.4 M NaCl in TG buffer. The resultant CarAa fraction was further purified by a second round of POROS HQ/L column chromatography using 20 mM Tris-H2SO4 buffer (pH 7.5) containing 10% glycerol as described above. Then the fraction was purified by GFC using a Sephacryl S-200 column as described above. Fractions containing CarAa were concentrated, and the buffer was replaced with TG buffer.
Protein manipulation.
Protein concentration was determined by using a protein assay kit (Bio-Rad Laboratories, Richmond, Calif.) with bovine serum albumin as a standard. Protein purity and the denatured molecular mass were determined by SDS-PAGE using a 10 or 12% polyacrylamide gel, or a 10-to-20% gradient gel (Bio-Rad Laboratories). N-terminal amino acid sequencing of proteins was determined with an Applied Biosystems model 492 Protein Sequencer according to the manufacturer's protocol.
Enzyme activity measurement.
The standard measurement of the NAD(P)H:cytochrome c oxidoreductase activity of His-tagged CarAd was performed as follows. The activity measurement system (1 ml) contained 50 mM Tris-HCl (pH 8.0), 500 μM NAD(P)H, 1 μM flavin adenine dinucleotide (FAD), 50 μM cytochrome c (Sigma Chemical Co., St. Louis, Mo.), and an appropriate amount of protein, unless otherwise stated. Reduction of cytochrome c was monitored as the increase in absorbance at 550 nm, and the molecular extinction coefficient at this wavelength was given as 21,000 M−1 cm−1 (41). Measurements were performed at 30°C by using a spectrophotometer (model DU-7400; Beckman Co., Fullerton, Calif.) equipped with a thermojacketed cuvette holder and a water circulation system. One unit of activity was defined as the amount of the enzyme required to reduce 1 μmol of electron acceptor per min. NAD(P)H-dependent oxidoreductase activity for other electron acceptors was assayed in the same reaction mixture with 88 μM 2,6-dichlorophenolindophenol (DCPIP) disodium salt, potassium hexacyanoferrate(III) (ferricyanide), or nitroblue tetrazolium (NBT). Reduction of DCPIP or ferricyanide was monitored by using the extinction coefficient 21,000 M−1 cm−1 at 600 nm (37) or 1,020 M−1 cm−1 at 420 nm (33), respectively. Reduction of NBT was monitored by the increase in absorbance at 550 nm, by using 28,600 M−1 cm−1 as the extinction coefficient (29). To monitor the effect of the flavin mononucleotide (FMN), 1 μM FMN was added to the reaction mixture instead of FAD. Kinetic constants (Km and Vmax) of His-tagged CarAd for NAD(P)H were calculated by fitting initial rate data in the NAD(P)H:DCPIP oxidoreductase assay to the Michaelis-Menten equation as described by Cleland (6) with SigmaPlot software. NADH and NADPH were added at final concentrations of 0.5 to 100 and 10 to 1,000 μM, respectively.
The CAR-oxygenating activity of the CARDO system reconstituted by the purified protein and/or CE is termed “CARDO system activity” in this study. CARDO system activity was routinely measured by using a reaction system (200 μl) containing 1.5 μg (11.4 pmol) of CarAa, 15 μg (1,154 pmol) of CarAc, 40 μg (1,081 pmol) of CarAd, 100 nmol of NAD(P)H, 100 nmol of [Fe(NH4)2(SO4)2 · 6H2O], and 200 pmol of FAD in 50 mM Tris-HCl (pH 7.5), unless otherwise stated. To avoid the possibility that the cofactors would be limiting for the reconstitution of the CARDO system in vitro, an excess amount of each cofactor was added. The reaction was started by addition of 2.5 to 50 nmol of [U-14C]CAR (7.9 mCi/mmol; Sigma Chemical Co.) dissolved in 5 μl of dimethyl sulfoxide. Because the transformation yield of CAR was linear within at least 30 min under the assay conditions employed (data not shown), the incubation time for determining CARDO system activity was routinely set for 10 min throughout this study. After a 10-min incubation at 30°C, the reaction was stopped by addition of 10 μl of 1 N HCl and holding on ice. A 5-μl aliquot of the reaction mixture was directly applied to a thin-layer chromatography (TLC) sheet (Chromatosheet; Wako Pure Chemical Industries Ltd., Osaka, Japan). After developing by use of an ethyl acetate-hexane (1:3, by volume) solvent system, the TLC sheet was cut into two parts at the region of 0.33 retardation factor (Rf). The radioactivity of the lower part containing the transformation product was counted by using a liquid scintillation system (model LSC-5100; Aloka Co., Ltd., Tokyo, Japan). Averages of duplicated determinations were calculated. One unit of activity was defined as the amount of the enzyme required to degrade 1 nmol of CAR per min at 30°C.
To assess the activities of CarAa during the purification process, the CARDO system activity reconstituted by 1.5 to 30 μg of a preparation containing CarAa and 180 μl of CE (2 mg of protein/ml of 50 mM Tris-HCl [pH 7.5] buffer) was determined as described above. The CEs were prepared from E. coli JM109 cells harboring pUCA14, which were grown in LB medium containing ampicillin and 0.1 mM IPTG at 37°C, as described above. His-tagged CarAc activity during the purification process was similarly assessed, except that 0.5 μg of a preparation containing His-tagged CarAc and CEs prepared from E. coli cells harboring pUCA33 were used.
To test the interchangeability of the components in the CARDO system, 100 μg of the ferredoxin component from spinach or Clostridium pasteurianum (both from Sigma Chemical Co.) was added to the reaction system instead of His-tagged CarAc. His-tagged CarAd was replaced with 600 μg of the ferredoxin reductase component from spinach (Sigma Chemical Co.).
The effect of reaction temperature on the reductase activity of His-tagged CarAd or on CARDO system activity was monitored between 5 and 65°C. The effects of pH on enzymatic activities were also determined by using 50 mM citrate buffer (pH 3.0 to 6.0), 50 mM morpholineethanesulfonic acid (MES)-NaOH (pH 5.5 to 7.0), 50 mM morpholinepropanesulfonic acid (MOPS)-NaOH (pH 6.5 to 8.0), 50 mM Tris-HCl (pH 7.0 to 9.0), or 50 mM glycine-NaOH (pH 8.6 to 10.6).
Other characterizations of Car proteins.
UV and visible absorption spectra were measured in cells of a 1-cm path length with a Beckman DU-7400 spectrophotometer as described above. Anaerobic samples were analyzed in anaerobic cuvettes fitted with rubber septa. The samples were rendered anaerobic by alternately evacuating the air and flushing the cuvette with argon which had been passed over a hot copper coil.
The flavin content of His-tagged CarAd was determined by high-performance liquid chromatography (HPLC) using a reverse-phase column (10 by 250 mm; ODS-4253-D; Senshu Scientific Co., Ltd., Tokyo, Japan) according to the method of Stuehr et al. (38). The iron content of protein was determined by using the iron-chelating reagent Feren S (Sigma Chemical Co.) as described by Zabinski et al. (46). A stock solution of Feren S reagent was prepared as described by Gottschall et al. (10). The amount of acid-labile sulfide was determined as described by Chen and Mortenson (5).
RESULTS
Purification and properties of His-tagged CarAd.
His-tagged CarAd was eluted at approximately 80 mM imidazole in metal chelation chromatography. After GFC, His-tagged CarAd was purified to near-homogeneity by SDS-PAGE (data not shown). His-tagged CarAd was purified 44-fold in a yield of 43%, as shown in Table 1. While the molecular mass of His-tagged CarAd detected by SDS-PAGE analysis was approximately 37 kDa, that of native His-tagged CarAd was approximately 37 kDa as calculated from GFC, suggesting that His-tagged CarAd is a monomeric protein. The amino acid sequence of the N terminus of the purified His-tagged CarAd, MYQLKIEGQAPGTXG (where X stands for any amino acid), was the same as the deduced amino acid sequence except for a cysteine residue that cannot be detected by the sequencer.
TABLE 1.
Purification of components of CARDO
| Component | Vol (ml) | Total activitya (U) | Total protein (mg) | Sp acta (U/mg) | Yield (%) | Purifi- cation (fold) |
|---|---|---|---|---|---|---|
| His-tagged CarAd | ||||||
| Crude extract | 87 | 28 | 510 | 0.055 | 100 | 1 |
| HiTrap Chelating | 9.1 | 16 | 12 | 1.3 | 57 | 24 |
| Sephacryl S-200 | 3.4 | 12 | 4.8 | 2.4 | 43 | 44 |
| His-tagged CarAc | ||||||
| Crude extract | 100 | 400 | 120 | 3.3 | 100 | 1 |
| HiTrap Chelating | 11 | 330 | 6.5 | 51 | 85 | 16 |
| Sephacryl S-200 | 3.2 | 220 | 1.5 | 150 | 56 | 45 |
| CarAa | ||||||
| Crude extract | 200 | 1,700 | 300 | 5.5 | 100 | 1 |
| DEAE | 11 | 440 | 20 | 21 | 26 | 4 |
| HQ/L (1)b | 12 | 330 | 3.8 | 86 | 20 | 16 |
| HQ/L (2)b | 1.7 | 310 | 2.3 | 130 | 19 | 24 |
| Sephacryl S-200 | 1.5 | 190 | 1.7 | 110 | 12 | 21 |
For the NADH:cytochrome c oxidoreductase activity of His-tagged CarAd, 1 U of activity was defined as the amount of the enzyme required to reduce 1 μmol of electron acceptor per min at 30°C. For CARDO system activities to determine CarAa and His-tagged CarAc activities, 1 U of activity was defined as the amount of the enzyme required to degrade 1 nmol of CAR per min at 30°C.
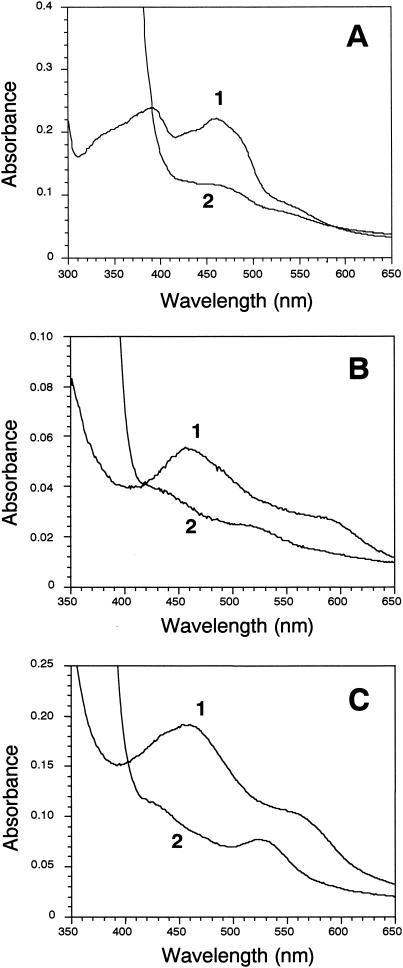
The UV and visible absorption spectra of air-oxidized His-tagged CarAd showed maxima at 278, 390, and 461 nm and shoulders at about 330, 430, and 550 nm (part of the absorption spectrum is shown in Fig. 1A, curve 1). These features are similar to those of reductases containing flavin and plant type [2Fe-2S] clusters (14, 30). Upon boiling, the yellow-brown His-tagged CarAd solution turned bright yellow. After precipitation of the white protein by centrifugation, the bright-yellow chromophore remaining in the supernatant was identified as FAD based on its absorption maxima at 265, 376, and 446 nm and its retention time (6.10 min) in HPLC analysis. Quantification by HPLC analysis showed that purified His-tagged CarAd contained 0.55 to 0.70 mol of FAD/mol of protein. This result indicated that 1 mol of His-tagged CarAd contains 1 mol of FAD, which is readily lost during purification. On the other hand, the iron and sulfur contents of His-tagged CarAd were determined as 1.8 to 2.0 and 1.8 to 1.9 mol/mol of protein, respectively, suggesting that 1 mol of CarAd protein has 1 mol of [2Fe-2S] cluster. The purified His-tagged CarAd was reduced and bleached by addition of NADH (Fig. 1A, curve 2).
FIG. 1.
UV-visible absorption spectra of His-tagged CarAd (A), His-tagged CarAc (B), and CarAa (C). In panel A, curve 1 shows the oxidized spectrum of His-tagged CarAd (10 nmol) in 1.0 ml of 50 mM Tris-HCl buffer (pH 8.0) under anaerobic conditions, and curve 2 shows the spectrum after the addition of NADH (0.5 μmol). In panel B, curve 1 shows the oxidized spectrum of His-tagged CarAc (10 nmol) in 1.0 ml of 50 mM Tris-HCl buffer (pH 7.0) under anaerobic conditions, and curve 2 shows the spectrum after the addition of NADH (0.5 μmol) and a catalytic amount of His-tagged CarAd (0.2 nmol). In panel C, curve 1 shows the oxidized spectrum of CarAa (10 nmol) in 1.0 ml of 50 mM Tris-HCl buffer (pH 7.0) under anaerobic conditions, and curve 2 shows the spectrum after 10 min from the addition of His-tagged CarAc (0.2 nmol), His-tagged CarAd (0.2 nmol), and NADH (0.5 μmol) (fully reduced CarAa).
Maximum cytochrome c reductase activity of His-tagged CarAd was observed at pH 7.5 to 8.5 and 35 to 45°C (data not shown). However, because the activity at 40°C was only 1.2 times higher than that at 30°C, further experiments were done at 30°C. His-tagged CarAd was almost 100% active for at least 10 h on ice and retained 93% activity after 100 h.
As shown in Table 2, the maximum His-tagged CarAd activity was observed when both NADPH and FAD were added to the reaction mixture. When NAD(P)H:DCPIP oxidoreductase activity was measured in the presence of FAD or FMN, activities 1.19 to 1.67 times higher than those without additional flavin were observed (Table 2). In the case of NAD(P)H:cytochrome c oxidoreductase activity, 1.93- to 6.71-times-higher activity was observed (Table 2), although the reason for such considerably higher activities is not known. Kinetic parameters of His-tagged CarAd were calculated for NADH and NADPH, with DCPIP as an electron acceptor. From the result, the Kms were calculated to be 3.37 (±0.20) and 182 (±20.6) μM for NADH and NADPH, respectively, and Vmaxs were calculated to be 16.0 (±0.24) and 36.9 (±1.45) U/mg for NADH and NADPH, respectively. The kcat/Km for NADH is 2.9 × 106 (±0.15) M−1 s−1, and that for NADPH is 1.3 × 105 (±0.93) M−1 s−1.
TABLE 2.
His-tagged CarAd activity with various cofactors and artificial electron acceptors
| Electron acceptor | Electron donor | Sp act (U/mg)a
|
||
|---|---|---|---|---|
| Without flavin | With FAD | With FMN | ||
| Cytochrome c | NADH | 0.80 ± 0.15 | 2.6 ± 0.24 | 2.7 ± 0.22 |
| NADPH | 1.4 ± 0.25 | 9.4 ± 0.51 | 2.7 ± 0.16 | |
| DCPIP | NADH | 18 ± 2.1 | 23 ± 1.4 | 22 ± 3.1 |
| NADPH | 27 ± 1.1 | 45 ± 3.4 | 32 ± 2.6 | |
| Ferricyanide | NADH | ND | 59 ± 4.1 | ND |
| NADPH | ND | 137 ± 8.7 | ND | |
| NBT | NADH | ND | 7.5 ± 0.33 | ND |
| NADPH | ND | 11.9 ± 0.73 | ND | |
Averages and standard deviations of four samples are shown. ND, not determined.
Purification and properties of His-tagged CarAc.
His-tagged CarAc was eluted at approximately 110 mM imidazole in metal chelation chromatography. The molecular masses of native and denatured His-tagged CarAc were similarly calculated to be approximately 13 kDa by GFC and SDS-PAGE analyses, respectively, suggesting that native His-tagged CarAc is a monomeric protein. After GFC, His-tagged CarAc was purified 45-fold in a yield of 56% (Table 1), resulting in near-homogeneity in SDS-PAGE (data not shown). The amino acid sequence of the N terminus of His-tagged CarAc, MNQIXLKVXAASDMQ, was the same as that expected from the nucleotide sequence except for cysteine and tryptophan residues.
A solution of purified His-tagged CarAc was brown. The air-oxidized protein had absorption maxima at 278, 325, and 457 nm, with a broad shoulder at about 570 nm, and the protein reduced by CarAd in the presence of NADH had absorption maxima at 432 and 515 nm (part of the absorption spectrum is shown in Fig. 1B). These are the characteristics of a class of ferredoxins containing a Rieske type [2Fe-2S] center (13, 15, 16, 39). In fact, the deduced amino acid sequence of CarAc predicted the binding of a [2Fe-2S] cluster (32). The result shown in Fig. 1B (curve 2) clearly indicates that purified His-tagged CarAc is reduced and bleached by catalytic amounts of His-tagged CarAd and NADH under anaerobic conditions. The iron and sulfur contents of His-tagged CarAc were determined as 1.7 to 1.9 and 1.3 to 1.6 mol/mol of protein, respectively, suggesting that 1 mol of CarAc has 1 mol of [2Fe-2S] cluster.
Purification and properties of CarAa.
Because the nucleotide sequence of the car locus indicated the presence of two putative initiation codons for the open reading frame (ORF) of carAa (32), we constructed two vectors to express CarAa proteins with different N termini. E. coli cells harboring the two respective plasmids showed similar transformation yields of CAR (data not shown). Considering that the shorter peptide was simultaneously synthesized from the downstream initiation codon on the expression vector containing the longer ORF, we used plasmid pES2, carrying the shorter ORF, in further experiments.
The typical purification process of CarAa is shown in Table 1. CarAa was purified 21-fold in a yield of 12%. The buffer system containing 10% glycerol stabilized the protein, and the final CarAa protein yield increased more than 8 times by use of a buffer system containing 10% glycerol in the purification procedure. In the presence of 10% glycerol, CarAa retained full activity at 4°C for 24 h, and more than 90% activity for 8 days on ice (data not shown). The protein yield was 1.7 mg from a 2-liter LB broth culture. By using SB medium instead of LB medium, the final CarAa yield was increased to approximately 10 mg from a 2-liter culture. The preparation of CarAa was nearly homogeneous on the gel in SDS-PAGE (data not shown). CarAa protein was detected as a 44-kDa protein in SDS-PAGE analysis, which is in agreement with the molecular mass calculated from the deduced amino acid sequence (43,782 Da). Because the native CarAa was detected as a 132-kDa protein in GFC, CarAa is found to be a trimer. Although the first methionine was not found in purified CarAa, the N-terminal amino acid sequence determined for purified CarAa (ANVDEAILKRVKGXA) was the same as that expected from the nucleotide sequence, except for tryptophan.
The solutions containing purified CarAa were brown. Like the reported spectra for several known oxygenase components (3, 30) and CarAc (Fig. 1B, curve 1), air-oxidized CarAa exhibited absorption maxima at 279, 322, and 459 nm, with a shoulder at about 560 nm (part of the absorption spectrum is shown in Fig. 1C, curve 1). When catalytic amounts of His-tagged CarAc, His-tagged CarAd, and NADH were added under anaerobic conditions, the purified CarAa was reduced within 10 min (Fig. 1C, curve 2). The iron and sulfur contents of CarAa were determined as 2.4 to 2.6 and 1.3 to 1.6 mol/mol of monomeric protein, respectively.
Properties of the CARDO system.
Maximum CARDO system activity was observed at 30°C, at pH 7.0 to 7.5, and at an ionic strength of 50 mM (Tris-HCl buffer). In the 40 to 80 mM ionic strength range, more than 90% activity was retained compared to that in 50 mM buffer (data not shown). When the three components were mixed and CARDO activity was measured after 24 h on ice, the mixture retained 90% activity.
When FAD and Fe2+ were included in the system with NADH, CARDO system activity increased approximately 1.9 times over that measured with NADH alone, suggesting that the FAD and Fe2+ added might supplement the cofactors lost during purification. The CARDO system can use NADPH as an electron donor, yielding 60% of the activity obtained with NADH.
When one of the CARDO system components was omitted from the reaction mixture without appropriate complementation by the other isofunctional protein, no activity was detected. When the ferredoxin from spinach or C. pasteurianum was added to the reaction system instead of His-tagged CarAc, the CARDO system activity detected was almost negligible (<1.3%) compared to the activity of the CARDO system reconstituted by purified CarAa, His-tagged CarAc, and His-tagged CarAd (125 U/mg). On the other hand, when His-tagged CarAd was replaced with the ferredoxin reductase from spinach, 96% activity was detected. Addition of CEs of E. coli JM109(pUC119), containing the unknown reductases, could also reconstitute CARDO system activity, at 4.5%.
DISCUSSION
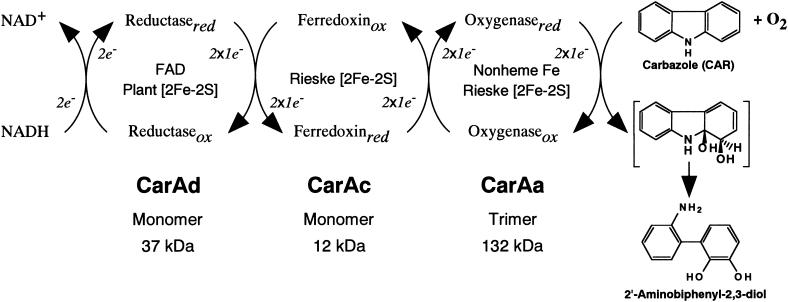
Considering the spectroscopic characteristics and activities of the three purified components of the CARDO system, the functions of each component are suggested to be as shown in Fig. 2. A 13-kDa monomer, CarAc, and a 37-kDa monomer, CarAd, function as a ferredoxin and a ferredoxin reductase, respectively, to transport electrons from NADH to terminal oxygenase. When a 132-kDa homotrimeric terminal oxygenase made up of 44-kDa monomeric CarAa coexisted with the ferredoxin and reductase proteins, CARDO system activity was observed. These are in agreement with the functions suggested by their deduced amino acid sequences (32).
FIG. 2.
Components and functions of the CARDO system. The proposed electron transfer reactions and the conversion of CAR to 2′-aminobiphenyl-2,3-diol are illustrated.
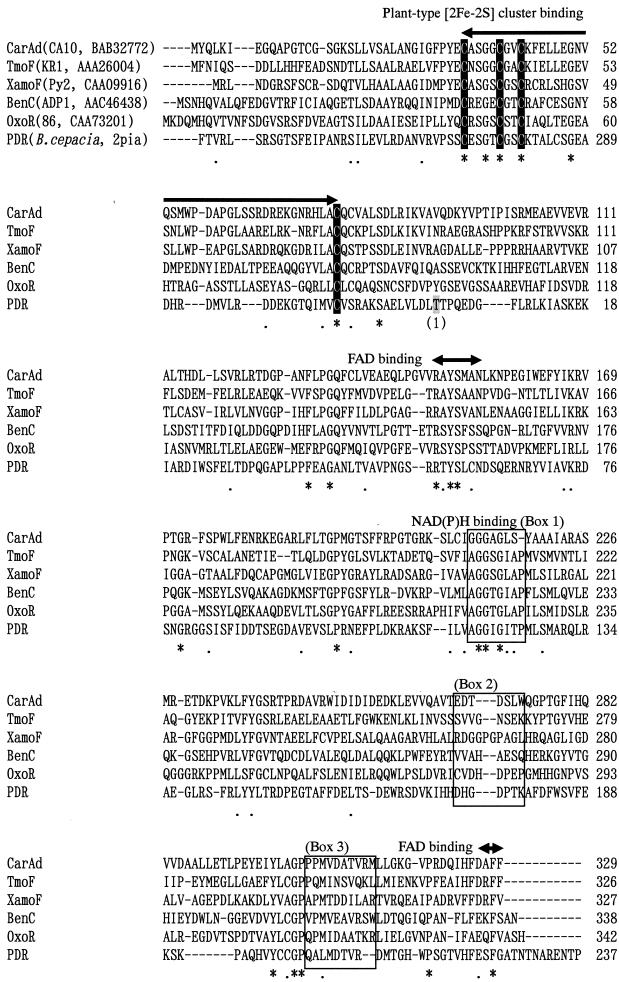
Compared to the data previously reported (14, 40), the FAD content in purified His-tagged CarAd, 0.55 to 0.7 mol/mol of protein, is relatively high. Also, the iron and sulfur contents were around 2 mol/mol of protein for both His-tagged CarAc and His-tagged CarAd. These relatively high contents of FAD, iron, and sulfur supposedly resulted from the small number of manipulations (only two steps). Taking these results into account, it can be concluded that fusion of the histidine tags to the C termini of CarAc and CarAd did not significantly affect their activities. As for CarAc, this consideration is supported by the three-dimensional structure of the biphenyl dioxygenase ferredoxin component (BphF), whose amino acid sequence shares 34% identity with that of CarAc (7). The structure of BphF revealed that both the N and C termini are extruded to the outside of the protein. We are now performing X-ray crystallography of His-tagged CarAc, and it has been found that both termini of His-tagged CarAc are also extruded to the outside of the protein (J.-W. Nam,, Z. Fujimoto, H. Mizuno, K. Iwata, T. Yoshida, H. Habe, H. Nojiri, and T. Omori, unpublished data). Therefore, it is highly possible that the six-histidine tag had little effect on the inside folding of CarAc. As for the reductase, when we started this experiment, the structures of reductases from phthalate dioxygenase (PDR) (8) and biphenyl dioxygenase (BphA4) (35) were available. Based on the classification system proposed by Batie et al. (1), CarAd is a class III reductase containing FAD and the [2Fe-2S] cluster, although PDR is a class IA reductase containing FMN and a plant type [2Fe-2S] cluster. Amino acid sequence comparison suggested that the location of the ferredoxin domain containing the [2Fe-2S] cluster in CarAd is different from that in PDR (Fig. 3). In BphA4, classified as a class IIB reductase in Batie's system, the [2Fe-2S] cluster is not present. Therefore, the structure of CarAd could not be predicted. Since the C-terminal His-tagged CarAd protein showed significant activity, as shown in Table 2, we used this modified CarAd protein for convenience in rapid purification. Very recently, the X-ray crystal structure of benzoate 1,2-dioxygenase reductase, BenC, from Acinetobacter sp. strain ADP1 was reported (17). Benzoate 1,2-dioxygenase (BenABC) has been assigned to Batie's class IB, and the reductase BenC contains plant type [2Fe-2S] and FAD. As shown in Fig. 3, the amino acid sequence of CarAd could be well aligned with that of BenC, suggesting that the structure of CarAd might be similar to that of BenC. Karlsson et al. (17) indicated that the C-terminal region of BenC is involved in FAD binding. Especially, Phe335 of BenC, which is the 4th residue from the C terminus, was found to interact with the isoalloxazine ring of flavin by the stacking of an aromatic side chain. Based on the alignment shown in Fig. 3, this residue corresponds to Phe328 of CarAd. In addition, the C terminus of CarAd showed significant similarity to those of TmoF (44) and XamoF (47) (Fig. 3). These similarities predict the possible involvement of the C-terminal region of CarAd in FAD binding. However, considering the fact that significant reductase activity was observed in His-tagged CarAd (Table 2), the addition of His residues at the C terminus might have little effect on the inside folding and electron transport function of CarAd.
FIG. 3.
Alignment of some NAD(P)H-dependent reductase components. The shaded Thr residue is the first residue of PDR and is marked by the numeral 1 in parentheses. Identical and similar amino acid residues among the aligned sequences are marked by asterisks and dots, respectively. Conserved residues for plant type [2Fe-2S] cluster binding and flavin binding are indicated by arrows, and the Cys residues involved in the binding of the [2Fe-2S] cluster are shown against a solid background. Conserved residues for NAD(P)H binding are boxed.
Both NADH and NADPH are able to transfer electrons to other artificial electron acceptors such as cytochrome c, DCPIP, ferricyanide, and NBT via His-tagged CarAd. When NADPH was supplied as an electron donor, the reductase activities for artificial electron acceptors were higher than those with NADH (Table 2). However, since the kcat/Km for NADH is 22.3-fold higher than that for NADPH, the physiological electron donor of CarAd seems to be NADH. In many of the ring-hydroxylating oxygenases, the preferred reductant is NADH (20). For example, PDR (2) and the reductases of toluene dioxygenase of Pseudomonas putida (40) are specific for NADH. Although several dioxygenase systems showed reductase activity in the presence of either NADH or NADPH as an electron donor, NADH seemed to be the physiological electron donor based on the higher activity and the kcat/Km values (14, 16, 30, 36). On the other hand, in the toluene 4-monooxygenase system of Pseudomonas mendocina KR1, NADH and NADPH were equally effective as electron donors (42). The residues involved in the coenzyme specificity for the reductases of iron-sulfur flavoproteins have been studied extensively from the structure of PDR (8). In the sequence alignment of CarAd with those of some reductases, the amino acid sequence of PDR from residue 238 to the end was placed upstream of the residues from 1 to 237 (Fig. 3). Conserved motifs have previously been proposed to be involved in the binding of [2Fe-2S], flavin cofactor, and NAD(P)H (24), and the amino acid residues that are possibly involved in NAD(P)H coenzyme specificity are boxed in Fig. 3. The binding site for NADH in PDR is known: the adenine ring is positioned between a loop formed by residues 173 to 179 (Fig. 3, box 2) and 202 to 211 (Fig. 3, box 3), by van der Waals contacts with Asp173 and Thr207. The pyrophosphate is adjacent to Ile121. Hydrogen bonding between hydroxyl groups in the adenosine ribose and an acidic side chain is a hallmark of many NAD(H) binding proteins, and the corresponding acidic residue in PDR is Asp173. This structural and sequence similarity study (8) assigned PDR to a distinct family of flavoprotein reductases, all related to ferredoxin-NADP+ reductase (FNR). Medina et al. (21) studied coenzyme specificity in FNR and found the amino acid residues involved. None of those residues are found in the reductase components shown in Fig. 3. Another region that might account for NAD(P)H specificity is the GGXGXTP region for NAD+/H-dependent enzymes (residues 119 to 125 in PDR [see Fig. 3, box 1]). As shown in Fig. 3, the reductases except for PDR have the sequence GGXGXA(S)P(Y) at this region, and some of these reductases have been found to be able to use NADPH as an electron donor (30, 36, 42). Because this amino acid sequence is similar to those of NADP+/H-dependent enzymes, T(P)GTGXAP (21), the acceptance of NAD(P)H as an electron donor seems to be reasonable. Phylogenetic analysis of the reductase components showed that the CarAd of strain CA10 is clustered together with TmoF of strain KR1 (42, 44) and XamoF of strain Py2 (36) (data not shown), suggesting that these reductases originated from the same ancestor, which can function not only with NADH but also with NADPH. Since the comparative analyses cannot fully explain the high activity of CarAd toward NADPH, we are now crystallizing His-tagged CarAd, aiming at the determination of the three-dimensional structure.
In the multicomponent oxygenase systems, two types of reductases, FNR type and glutathione reductase (GR) type reductases, are involved. The FNR type includes Batie's class I and III reductases, and the GR type includes Batie's class II reductases. They have different structures involved in the hydride transfer reaction, related to the stacking or contact between the nicotinamide group and the flavin ring (8), and the hydride transfer mechanism is pro-R or pro-S mode for FNR type or GR type reductases, respectively (34). The reductases coupled with the multicomponent nonheme iron oxygenases might be recruited from at least two different sources (34). While the amino acid sequence of CarAd has homology with those of Batie's class IB or III reductases, the amino acid sequence of CarAc has homology with those of Batie's class IIB or III ferredoxins. It has also been found that the three-dimensional structure of CarAc (Nam et al., unpublished) is similar to that of the Batie class IIB ferredoxin BphF. It is quite interesting that the hydride retrieved by different mechanisms from NAD(P)H in CarAd (probably by the pro-R mode) or BphG of biphenyl dioxygenase (probably by the pro-S mode) is transferred to structurally homologous ferredoxins, CarAc or BphF. From this observation, we postulate that the electron transfer from reductase to ferredoxin occurs by similar mechanisms in these two different types of reductases. Since no experimental evidence has been obtained for the binding site of ferredoxin in the reductase molecule, the pathway of the electron flow from reduced FAD to ferredoxin remains unclear.
Concerning the possible coevolution of ferredoxin and reductase, the relationship between CARDO and phenanthrene dioxygenase of Alcaligenes faecalis strain AFK2 (accession numbers: BAA76321 for reductase PhnAa, BAA76320 for ferredoxin PhnAb) is noteworthy. PhnAb and PhnAa have 39% homologies with CarAc and CarAd, respectively. Based on phylogenetic analyses (data not shown), PhnAa and PhnAb are the proteins most closely related to CarAc and CarAd, respectively, suggesting the possibility that these sets of genes have evolved from a common ancestor. However, the genetic organization of phn gene clusters (accession number AB024945) is not similar to that of car gene clusters. In fact, a 318-bp ORF7 resides between carAc and carAd in the car gene cluster (32), while phnAa and phnAc are flanked by 24 bases. In addition, the oxygenase component of the phenanthrene dioxygenase system of strain AFK2 consists of two proteins, the catalytic α subunit PhnAc and the β subunit PhnAd, and PhnAc shows only 9% identity with CarAa. Because the mosaic structure of the car locus has been built by several genetic rearrangements (28), it is possible that the flanking region of reductase and ferredoxin genes may have been under dynamic reorganization.
There are many reports that E. coli cells carrying genes encoding only part of an oxygenase system showed oxygenating activity as a result of complementation with a reductase component or with both ferredoxin and reductase components from the host cell (9, 18, 45). Also, dioxygenase systems for naphthalene (14), toluene (40), and biphenyl (16) have been reported to retain activities with the spinach FNR. On the other hand, phthalate oxygenase is reported to be specific for PDR as its reductant (1). In the CARDO system, the reductase component could be replaced by some isofunctional proteins, although ferredoxin could not be replaced. This indicates possible differences between the electron transport mechanisms of the oxygenase systems discussed above. It is likely that in the CARDO system there is a rather strict recognition mechanism, at least between CarAa and CarAc. Of course, from the present data, we cannot exclude the possibility that a similar ferredoxin (for example, PhnAb of strain AFK2) would work well in the CARDO system. Therefore, more-detailed investigation on the interaction between the components of the CARDO system is necessary for better understanding of the electron transport mechanism in CARDO.
The difference in the interactions among components is supposedly related to the origins of the respective components. This again supports the suggestion of Nam et al. (23) to establish a new classification system for nonheme iron-containing oxygenases in which classification of oxygenase systems would be complemented by phylogenetic classification of the terminal oxygenase components. We now suggest further that classification of reductase components and ferredoxin should be added to the oxygenase system classification as well. Butler and Mason (4) have made a similar suggestion, that each component may be classified better as an enzyme with a separate EC number.
Among known multicomponent oxygenase systems, the structures of PDR (8), the naphthalene dioxygenase system oxygenase component (19), the biphenyl dioxygenase system ferredoxin component (7), the biphenyl dioxygenase system reductase component (35), and the benzoate 1,2-dioxygenase system reductase component (17) have been analyzed. The availability of highly purified components of the CARDO system from P. resinovorans strain CA10 has allowed us to start crystallization of each component. We have already determined the structure of His-tagged CarAc by X-ray crystallography (Nam et al., unpublished) and have obtained crystals of CarAa (22). Determination of the three dimensional structure of CarAa is now under way. Information on the structures of CARDO components will provide insights not only into substrate specificity and the substrate oxygenation mechanism but also into component interactions and the mechanism of electron transfer. These will provide us with important cues for designing an engineered CARDO system which will have enhanced activity for CAR or dioxin degradation.
Acknowledgments
We thank N. Kinoshita and Y. Ohnishi of The University of Tokyo and Z. Fujimoto of the National Institute of Agrobiological Sciences for experimental assistance, and we thank M. Nishiyama and N. Kobashi of The University of Tokyo for helpful discussions.
This work was supported by grants from the Program for Promotion of Basic Research Activities for Innovative Bioscience (PROBRAIN) in Japan.
REFERENCES
- 1.Batie, C. J., D. P. Ballou, and C. C. Correll. 1991. Phthalate dioxygenase reductase and related flavin-iron-sulfur containing electron transferases. Chem. Biochem. Flavoenzymes 3:543-556. [Google Scholar]
- 2.Batie, C. J., E. LaHaie, and D. P. Ballou. 1987. Purification and characterization of phthalate oxygenase and phthalate oxygenase reductase from Pseudomonas cepacia. J. Biol. Chem. 262:1510-1518. [PubMed] [Google Scholar]
- 3.Bünz, P. V., and A. M. Cook. 1993. Dibenzofuran 4,4a-dioxygenase from Sphingomonas sp. strain RW1: angular dioxygenation by a three-component enzyme system. J. Bacteriol. 175:6467-6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butler, C. S., and J. R. Mason. 1997. Structure-function analysis of the bacterial aromatic ring-hydroxylating dioxygenases. Adv. Microb. Physiol. 38:47-84. [DOI] [PubMed] [Google Scholar]
- 5.Chen, J.-S., and L. E. Mortenson. 1977. Inhibition of methylene blue formation during determination of the acid-labile sulfide of iron-sulfur protein samples containing dithionite. Anal. Biochem. 79:157-165. [DOI] [PubMed] [Google Scholar]
- 6.Cleland, W. W. 1979. Statistical analysis of enzyme kinetic data. Methods Enzymol. 63:103-138. [DOI] [PubMed] [Google Scholar]
- 7.Colbert, C. L., M. M.-J. Couture, L. D. Eltis, and J. T. Bolin. 2000. A cluster exposed: structure of the Rieske ferredoxin from biphenyl dioxygenase and the redox properties of Rieske Fe-S proteins. Structure 8:1267-1278. [DOI] [PubMed] [Google Scholar]
- 8.Correll, C. C., C. J. Batie, D. P. Ballou, and M. L. Ludwig. 1992. Phthalate dioxygenase reductase: a modular structure for electron transfer from pyridine nucleotides to [2Fe-2S]. Science 258:1604-1610. [DOI] [PubMed] [Google Scholar]
- 9.Eaton, R. W., O. V. Selifonova, and R. M. Gedney. 1998. Isopropylbenzene catabolic pathway in Pseudomonas putida RE204: nucleotide sequence analysis of the ipb operon and neighboring DNA from pRE4. Biodegradation 9:119-132. [DOI] [PubMed] [Google Scholar]
- 10.Gottschall, D. W., R. F. Dietrich, S. J. Benkovic, and R. Shiman. 1982. Phenylalanine hydroxylase. Correlation of the iron content with activity and preparation and reconstitution of the apoenzyme. J. Biol. Chem. 257:845-849. [PubMed] [Google Scholar]
- 11.Habe, H., J.-S. Chung, J.-H. Lee, K. Kasuga, T. Yoshida, H. Nojiri, and T. Omori. 2001. Degradation of chlorinated dibenzofurans and dibenzo-p-dioxins by two types of bacteria having angular dioxygenases with different features. Appl. Environ. Microbiol. 67:3610-3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Habe, H., K. Ide, M. Yotsumoto, H. Tsuji, H. Hirano, J. Widada, T. Yoshida, H. Nojiri, and T. Omori. 2001. Preliminary examinations for applying a carbazole-degrader, Pseudomonas sp. strain CA10, to dioxin-contaminated soil remediation. Appl. Microbiol. Biotechnol. 56:788-795. [DOI] [PubMed] [Google Scholar]
- 13.Haddock, J. D., D. A. Pelletier, and D. T. Gibson. 1997. Purification and properties of ferredoxinBPH, a component of biphenyl 2,3-dioxygenase of Pseudomonas sp. strain LB400. J. Ind. Microbiol. Biotechnol. 19:355-359. [DOI] [PubMed] [Google Scholar]
- 14.Haigler, B. E., and D. T. Gibson. 1990. Purification and properties of NADH-ferredoxinNAP reductase, a component of naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816. J. Bacteriol. 172:457-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haigler, B. E., and D. T. Gibson. 1990. Purification and properties of ferredoxinNAP, a component of naphthalene dioxygenase from Pseudomonas sp. strain NCIB-9816. J. Bacteriol. 172:465-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hurtubise, Y., D. Barriault, J. Polowski, and M. Sylvestre. 1995. Purification and characterization of the Comamonas testosteroni B-356 biphenyl dioxygenase components. J. Bacteriol. 177:6610-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karlsson, A., Z. M. Beharry, D. M. Eby, E. D. Coulter, E. L. Niedle, D. M. Kurtz, Jr., H. Eklund, and S. Ramaswamy. 2002. X-ray crystal structure of benzoate 1,2-dioxygenase reductase from Acinetobacter sp. strain ADP1. J. Mol. Biol. 318:261-272. [DOI] [PubMed] [Google Scholar]
- 18.Kasuga, K., H. Habe, J. S. Chung, T. Yoshida, H. Nojiri, H. Yamane, and T. Omori. 2001. Isolation and characterization of the genes encoding a novel oxygenase component of angular dioxygenase from the Gram-positive dibenzofuran-degrader Terrabacter sp. strain DBF63. Biochem. Biophys. Res. Commun. 283:195-204. [DOI] [PubMed] [Google Scholar]
- 19.Kauppi, B., K. Lee, E. Carredano, R. E. Parales, D. T. Gibson, H. Eklund, and S. Ramaswamy. 1998. Structure of an aromatic ring-hydroxylating dioxygenase-naphthalene 1,2-dioxygenase. Structure 6:571-586. [DOI] [PubMed] [Google Scholar]
- 20.Mason, J. R., and R. Cammack. 1992. The electron-transport proteins of hydroxylating bacterial dioxygenases. Annu. Rev. Microbiol. 46:277-305. [DOI] [PubMed] [Google Scholar]
- 21.Medina, M., A. Luquita, J. Tejero, J. Hermoso, T. Mayoral, J. Sanz-Aparicio, K. Grever, and C. Gomez-Moreno. 2001. Probing the determinants of coenzyme specificity in ferredoxin-NADP+ reductase by site-directed mutagenesis. J. Biol. Chem. 276:11902-11912. [DOI] [PubMed] [Google Scholar]
- 22.Nam, J.-W., Z. Fujimoto, H. Mizuno, H. Yamane, T. Yoshida, H. Habe, H. Nojiri, and T. Omori. 2002. Crystallization and preliminary crystallographic analysis of the terminal oxygenase component of carbazole 1,9a-dioxygenase of Pseudomonas resinovorans strain CA10. Acta Crystallogr. Sect. D 58:1350-1352. [DOI] [PubMed] [Google Scholar]
- 23.Nam, J.-W., H. Nojiri, T. Yoshida, H. Habe, H. Yamane, and T. Omori. 2001. New classification system for oxygenase components involved in ring-hydroxylating oxygenations. Biosci. Biotechnol. Biochem. 65:254-263. [DOI] [PubMed] [Google Scholar]
- 24.Neidle, E. L., C. Hartnett, L. N. Ornston, A. Bairoch, M. Rekik, and S. Harayama. 1991. Nucleotide sequences of the Acinetobacter calcoaceticus benABC genes for benzoate 1,2-dioxygenase reveal evolutionary relationships among multicomponent oxygenases. J. Bacteriol. 173:5385-5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nojiri, H., H. Habe, and T. Omori 2001. Bacterial degradation of aromatic compounds via angular dioxygenation. J. Gen. Appl. Microbiol. 47:279-305. [DOI] [PubMed] [Google Scholar]
- 26.Nojiri, H., J.-W. Nam, M. Kosaka, K. Morii, T. Takemura, K. Furihata, H. Yamane, and T. Omori. 1999. Diverse oxygenations catalyzed by carbazole 1,9a-dioxygenase from Pseudomonas sp. strain CA10. J. Bacteriol. 181:3105-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nojiri, H., and T. Omori. 2002. Molecular bases of aerobic bacterial degradation of dioxins: involvement of angular dioxygenation. Biosci. Biotechnol. Biochem. 66:2001-2016. [DOI] [PubMed] [Google Scholar]
- 28.Nojiri, H., H. Sekiguchi, K. Maeda, M. Urata, S. Nakai, T. Yoshida, H. Habe, and T. Omori. 2001. Genetic characterization and evolutionary implications of a car gene cluster in the carbazole degrader Pseudomonas sp. strain CA10. J. Bacteriol. 183:3663-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roerig, D. L., L. Mascaro, Jr., and S. D. Aust. 1972. Microsomal electron transport: tetrazolium reduction by rat liver microsomal NADPH-cytochrome c reduction. Arch. Biochem. Biophys. 153:475-479. [DOI] [PubMed] [Google Scholar]
- 30.Rosche, B., B. Tshisuaka, S. Fetzner, and F. Lingens. 1995. 2-Oxo-1,2-dihydroquinoline 8-monooxygenase, a two-component enzyme system from Pseudonomas putida 86. J. Biol. Chem. 270:17836-17842. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Sato, S., J.-W. Nam, K. Kasuga, H. Nojiri, H. Yamane, and T. Omori. 1997. Identification and characterization of genes encoding carbazole 1,9a-dioxygenase in Pseudomonas sp. strain CA10. J. Bacteriol. 179:4850-4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schellenberg, K. A., and L. Hellerman. 1958. Oxidation of reduced diphosphopyridine nucleotide. J. Biol. Chem. 231:547-556. [PubMed] [Google Scholar]
- 34.Schlafli, H. R., D. P. Baker, T. Leisinger, and A. M. Cook. 1995. Stereospecificity of hydride removal from NADH by reductases of multicomponent nonheme iron oxygenase systems. J. Bacteriol. 177:831-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Senda, T., T. Yamada, N. Sakurai, M. Kubota, T. Nishizaki, E. Masai, M. Fukuda, and Y. Mitsui. 2000. Crystal structure of NADH-dependent ferredoxin reductase component in biphenyl dioxygenase. J. Mol. Biol. 304:397-410. [DOI] [PubMed] [Google Scholar]
- 36.Small, F. J., and S. A. Ensign. 1997. Alkene monooxygenase from Xanthobacter strain Py2. Purification and characterization of a four-component system central to the bacterial metabolism of aliphatic alkenes. J. Biol. Chem. 272:24913-24920. [DOI] [PubMed] [Google Scholar]
- 37.Steyn-Parve, E. P., and H. Beinert. 1958. On the mechanism of dehydrogenation of fatty acyl derivatives of coenzyme A. J. Biol. Chem. 233:843-852. [PubMed] [Google Scholar]
- 38.Stuehr, D. J., H. J. Cho, H. S. Kwon, M. F. Weise, and C. F. Nathan. 1991. Purification and characterization of the cytokine-induced macrophage nitric oxide synthase: an FAD- and FMN-containing flavoprotein. Proc. Natl. Acad. Sci. USA 88:7773-7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Subramanian, V., T.-N. Liu, W. K. Yeh, C. M. Serdar, L. P. Wackett, and D. T. Gibson. 1985. Purification and properties of ferredoxinTOL. A component of toluene dioxygenase from Pseudomonas putida F1. J. Biol. Chem. 260:2355-2363. [PubMed] [Google Scholar]
- 40.Subramanian, V., T.-N. Liu, W. K. Yeh, M. Narro, and D. T. Gibson. 1981. Purification and properties of NADH-ferredoxinTOL reductase. A component of toluene dioxygenase from Pseudomonas putida. J. Biol. Chem. 256:2723-2730. [PubMed] [Google Scholar]
- 41.Ueda, T., E. T. Lode, and M. J. Coon. 1972. Enzymatic ω-oxidation. VI. Isolation of homogeneous reduced diphosphopyridine nucleotide-rubredoxin reductase. J. Biol. Chem. 247:2109-2116. [PubMed] [Google Scholar]
- 42.Whited, G. M., and D. T. Gibson. 1991. Toluene-4-monooxygenase, a three-component enzyme system that catalyzes the oxidation of toluene to p-cresol in Pseudomonas mendocina KR1. J. Bacteriol. 173:3010-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilkes, H., R.-M. Wittich, K. N. Timmis, P. Fortnagel, and W. Francke. 1996. Degradation of chlorinated dibenzofurans and dibenzo-p-dioxins by Sphingomonas sp. strain RW1. Appl. Environ. Microbiol. 62:367-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yen, K. M., and M. R. Karl. 1992. Identification of a new gene, tmoF, in the Pseudomonas mendocina KR1 gene cluster encoding toluene-4-monooxygenase. J. Bacteriol. 174:7253-7261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yen, K. M., M. R. Karl, L. M. Blatt, M. J. Simon, R. B. Winter, P. R. Fausset, H. S. Lu, A. A. Harcourt, and K. K. Chen. 1991. Cloning and characterization of a Pseudomonas mendocina KR1 gene cluster encoding toluene-4-monooxygenase. J. Bacteriol. 173:5315-5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zabinski, R., E. Munck, P. M. Champion, and J. M. Wood. 1972. Kinetic and Mossbaur studies on the mechanism of protocatechuic acid 4,5-oxygenase. Biochemistry 11:3213-3219. [DOI] [PubMed] [Google Scholar]
- 47.Zhou, N. Y., A. Jenkins, C. K. Chan Kwo Chion, and D. J. Leak. 1999. The alkene monooxygenase from Xanthobacter strain Py2 is closely related to aromatic monooxygenases and catalyzes aromatic monohydroxylation of benzene, toluene, and phenol. Appl. Environ. Microbiol. 65:1589-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]