Abstract
In the summer of 2000, we released genetically altered insect-pathogenic fungi onto a plot of cabbages at a field site on the Upper Marlboro Research Station, Md. The transformed derivatives of Metarhizium anisopliae ARSEF 1080, designated GPMa and GMa, carried the Aequorea victoria green fluorescent protein (gfp) gene alone (GMa) or with additional protease genes (Pr1) (GPMa). The study (i) confirmed the utility of gfp for monitoring pathogen strains in field populations over time, (ii) demonstrated little dissemination of transgenic strains and produced no evidence of transmission by nontarget insects, (iii) found that recombinant fungi were genetically stable over 1 year under field conditions, and (iv) determined that deployment of the transgenic strains did not depress the culturable indigenous fungal microflora. The major point of the study was to monitor the fate (survivorship) of transformants under field conditions. In nonrhizosphere soil, the amount of GMa decreased from 105 propagules/g at depths of 0 to 2 cm to 103 propagules/g after several months. However, the densities of GMa remained at 105 propagules/g in the inner rhizosphere, demonstrating that rhizospheric soils are a potential reservoir for M. anisopliae. These results place a sharp focus on the biology of the soil/root interphase as a site where plants, insects, and pathogens interact to determine fungal biocontrol efficacy, cycling, and survival. However, the rhizospheric effect was less marked for GPMa, and overall it showed reduced persistence in soils than did GMa.
Biocontrol experiments with fungi have often produced inconsistent results, and this has deterred commercial development (14). However, many fungi are amenable to genetic modification for purposes of enhancing utility for disease control, insect and plant pest management, or bioremediation. In such cases, genetically engineered fungi may provide environmentally preferred alternatives to current chemical-based control strategies. Much attention has focused on the ascomycete entomopathogen Metarhizium anisopliae. It is widely applied abroad, was recently registered for use in the United States and Europe (2), and offers particular promise as a suppressive agent for many soil insect pests that would otherwise provide a particular challenge to pest control specialists (6, 9). The addition and expression of pesticidal genes in M. anisopliae is quite straightforward and was used to genetically engineer a strain that overexpresses toxic proteases and kills insects faster than the wild type dose in laboratory tests (12).
This technology has potential for pest control (13), but there is an inherent uncertainty about the efficacy, survivability, and environmental risk posed by any introduced or engineered fungus because of our lack of knowledge about the fate of fungal genotypes at the population and ecosystem levels (1, 5). To achieve successful, reproducible, and safe (from the risk management point of view) biological control, we need to be able to study the ecology of the transformed genotype. After extensive laboratory analysis to test potential risks, including acquisition and evaluation of host range information, we were granted approval (38567-NMP-R) from the Biopesticides and Pollution Prevention Division of the EPA Office of Pesticide Program to conduct a planned release in a field of cabbage plants. The approval constrained us to establishing the technology required to monitor the fate of genetically enhanced M. anisopliae and to using this technology to determine the potential of engineered strains to establish and disperse over 1-year test period. This was achieved by combining conventional techniques used by soil microbiologists and ecologists with gfp (encoding green fluorescent protein) as a molecular marker. Since root exudates stimulate the growth of bacterial and fungal populations and the rhizosphere is of great importance to plant health and fertility (16), we focused on it as a potential refuge for transgenic fungi that could increase their persistence in the environment.
MATERIALS AND METHODS
Fungi and host.
The wild-type M. anisopliae strain ARSEF1080 was originally isolated from larvae of the cabbage looper (Trichoplusia ni: Noctuidae, Lepidoptera) in Florida. Allozyme analysis identified M. anisopliae strain 1080 as belonging to genotypic class 14, which is rare in North America (10). The recombinant strain gpd-Pr1-4 contains four copies of the Pr1a subtilisin gene under control of the constitutive gpd promoter from Aspergillus nidulans (12).
Transformation.
The wild-type strain and gpd-Pr1-4 were transformed with pEGFP-CP (obtained from Don Nuss, Center of Agricultural Biotechnology, University of Maryland, College Park, Md. Plasmid EGFP-CP carries the gene for EGFP1 (a variant of the green fluorescent protein) under control of the glyceraldehyde 3-phosphate dehydrogenase (gpd) promoter from Cryphonectria parasitica (15).
Transformation was performed using a previously established protocol (12) with the modification that inoculated plates were incubated at 28°C for 30 h and transformants visible under a fluorescence microscope were rescued using a glass pipette. Transformants were purified by generating single-spore colonies, and these were subcultured on potato dextrose agar five times to confirm stability. CHEF (clamped homogeneous electric field) gel analysis employing a CHEF-DR-III apparatus (Bio-Rad) was used as described previously (11) to identify transformants carrying the pr1 and egfp1 genes at unlinked locations, i.e., on different chromosomes, so as to allow effective recovery of recombination events. Fluorescent transformant progeny of wild-type (GMa) and gpd-Pr1-4 (GPMa) chosen for the field trial had parent-type growth rate, colony morphology, level of conidial production, and relative virulence as determined by standard laboratory protocols (12).
Fungal release.
The field site was located in the University of Maryland Upper Marlboro research farm, Upper Marlboro, Md. It is a frequently cultivated (tilled) site, and the soil is a Monmouth fine sandy loam. The rectangular 0.2-ha field site was designed to allow for efficient maintenance and the detection of any dispersal of recombinant fungus outside the confines of the plot. The plot consisted of two 0.05-ha fungal application areas, each consisting of seven rows of cabbages separated by a five-row buffer. A barren, plant-free zone surrounded the subplots, and a low-maintenance fallow zone outside the plot was also monitored for marked (recombinant) fungus through the field tests. The cabbage plants (var. Early Flat Dutch) were sprayed on a low-wind day (14 June 2000) with a water-based application containing 0.01% Silwet L77 (Loveland Industries, Greeley, Colo.) at a rate of 1013 spores per ha. The ground and the plants in each row were sprayed with a backpack-mounted hydraulic sprayer (18-in. spray band). Application area 1 received GPMa; application area 2 received GMa. Because the purpose of this study was not quantitative, i.e., we were not attempting to compare virulence between GMa and GPMa, a single experimental plot for each was deemed sufficient.
Transfer of the fungus by mechanical means was minimized by using a field test design and field test protocol that included the buffer zone and tool and footwear disinfestation.
Collection of soil samples.
Before the start of the experiment and daily (first week), weekly (first 2 months), and at monthly intervals thereafter, soil samples were taken at defined depths using a 1-cm soil core sampler from 50, 20, and 15 evenly spaced locations within the application zones, buffer zone, and fallow zone, respectively. Soil samples from the innermost rows of the application areas were taken at 4 to 5 cm from the cabbage tap root as well as alongside the tap root (0 to 1 cm) to check for uneven distribution and persistence of spores close to the rhizosphere (vicinity of the root). Subsamples of soil were used for dry-weight determination.
Soil samples were stored for up to 3 days at 4°C before the propagules of M. anisopliae were quantitated by using Veens semiselective agar medium (3). Soil samples (1 g) were sonicated briefly in 0.05% Tween 80, serial dilutions were made, and 0.1-ml portions were spread on each of two to five plates of selective medium per dilution. The detection limit was less than 20 CFU per g of soil. After range finding experiments, only dilutions near those likely to produce countable numbers of CFU (up to 300 per plate) were plated. The medium was supplemented with hide protein azure to detect constitutive protease production by GPMa (12) and scanned with UV light to distinguish GFP-expressing recombinants from indigenous strains of Metarhizium spp. Proc MIXED was used to test for differences in rates of decline of spore titers between GMa and GPMa. Spore count data were transformed to the log scale before analyses. Means were compared using the Student-Newman-Keuls (SNK) method. All analyses were carried out using the SAS software package V8.2. (SAS Institute, Cary NC) (α = 0.05).
Indigenous strains of M. anisopliae were characterized by allozyme analysis, which allows a large number of strains to be analyzed for recombination events, which we do by assigning a genetic basis to electrophoretic banding patterns (10). Cycloheximide was omitted from the Veens medium to study the abundance and composition of fungal populations other than M. anisopliae (total filamentous fungi).
Rhizosphere competence.
Samples taken alongside the root by using a 1-cm core borer may contain nonrhizospheric bulk soil that will cause the rhizospheric titer to be underestimated. Therefore, 4 months after planting, eight randomly selected cabbage plants from each application site were cut off above the soil and root samples with adhering soil (rhizosphere samples) were taken. Roots were sectioned into 2-cm segments, and the segments were shaken to collect soil adhering loosely to the roots (outer rhizosphere). To collect soil adhering after shaking but subject to removal by washing (inner rhizosphere), roots were weighed and ultrasonicated (15 s) in sterile water. Outer and inner rhizospheric suspensions were plated onto Veens medium for plate assays. Subsamples of the rhizospheric suspensions were used for dry-weight determinations. To sample rhizoplane M. anisopliae, root segments were further washed (10 times), air dried, weighed, and placed on Veens medium.
Monitoring strain stability.
Integrative transformants are very stable when grown for long periods in the absence of selection in pure culture under laboratory conditions (4, 11). However, stability may be different in a complex environment, in which case we reasoned that it would be unlikely for two unlinked markers (Pr1 and GFP) to be lost at once. There should usually be at least one marker remaining to positively distinguish a transformant from a native organism and detect recombination. To determine whether fungi retain the marker elements in their original form, we screened M. anisopliae isolates recovered from the application sites for any examples that have lost GFP but retained constitutive expression of Pr1 or for GFP-expressing strains demonstrating one or more phenotypic characteristics that differ significantly from those exhibited by the input transgenic strains. The growth rate, colony morphology, and level of conidial production were tested as described previously (12).
Monitoring nontarget arthropods.
During the course of the field tests, 50 pit fall traps embedded in the soil in and around the application sites were used to collect nontarget arthropods, particularly carabid beetles (important predators). These were maintained in the laboratory to determine if disease developed, and healthy as well as infected insects were analyzed for the presence of the marked fungus. Insects were placed in petri dishes containing Veens agar medium. Fungal growth over the medium was examined under UV illumination for GFP fluorescence. We anticipated that background levels would be high within the sprayed areas. Consequently, a representative portion (10%) of nontarget insects recovered from these areas were washed briefly in acetone followed by 95% ethanol to remove surface-associated fungal propagules. The individual (identified to species) insects were squashed and placed on Veens agar medium to detect internalized transgenic M. anisopliae spores and mycelia. These experiments were designed to determine the extent to which transgenic M. anisopliae strains can be recovered from insects (including nonhosts) within an intense deployment area in comparison to the extent found in the surrounding and remote sampling areas, i.e., to determine the potential of insect-mediated dispersal to nontargeted deployment areas.
RESULTS AND DISCUSSION
Effects on the indigenous culturable fungal microflora.
Before the application, soil samples from the top 3 cm were collected from locations within the application areas, buffer zone, and surrounding fallow zone. Veens medium minus cycloheximide was used to sample total fungal populations. Each fallow location contained at least 106 propagules/g comprising 30 or more fungal species. By contrast, the cultivated buffer and application areas were comparatively impoverished and contained only seven species with a total CFU of less than 104 propagules/g. Veens medium containing cycloheximide grew M. anisopliae from only three sites with a mean at these sites of 212 ± 24 propagules/g. Allozyme analysis of 20 colonies picked at random identified two genotypic classes based on electrophoretic phenotypes (10). Based on assigned genotypes (10), 14 of the colonies belonged to class 20 (field strain 1). The remaining 6 (field strain 2) differed only at the glutathione reductase locus, which demonstrated a mobility of 121 compared to the 100 shown by class 20. For several months after spraying with transformants, the indigenous strains of M. anisopliae were infrequently detected on plates. However, this would be accounted for by the large initial dilution (dilution factor 100) required to obtain countable numbers of GMa and GPMa. Sampling in the spring of the second year of bulk (nonrhizosphere) soil revealed that the original three locations still contained mixed populations of the two indigenous strains. Thus, there is no evidence of a detrimental effect on indigenous populations from introducing GMa and GPMa.
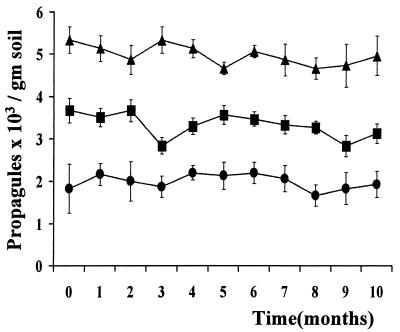
Total filamentous fungal populations in soil 0 to 1 cm from the roots was analyzed using repeated measurements analysis of variance (ANOVA) to determine whether the development of fungal populations is affected by the application of transgenic M. anisopliae (interaction between time and treatment). On the Veens medium minus cycloheximide, there was no significant difference in the total rhizospheric fungal populations or its composition in either GPMa- or GMa-treated plants compared to the untreated plants in the buffer zone. In fact, with (Fig. 1) or without (data not shown) application of GMA, the population levels of the three most frequently isolated genera, Paecilomyces, Penicillium, and Aureobasidium, did not change significantly over time.
FIG. 1.
Mean soil titers of propagules of Paecilomyces farinosus, (▴) Penicillium spp. (•), and Aureobasidium pullulans (▪) in the application area treated with M. anisopliae GMa. Soil samples were taken within 1 cm from cabbage plant roots at depths of 0 to 2 cm, and fungal propagules were quantified on Veens medium minus cycloheximide. Error bars indicate SD.
With the caveat that not all fungi are culturable, these results indicate that there is minimal risk of the engineered fungus displacing naturally occurring fungi. Given the impoverished fungal microflora observed in the cultivated compared to the noncultivated land the impact of introduced microorganisms in general is likely to be minor compared to that of common agricultural practices such as plowing or crop rotation.
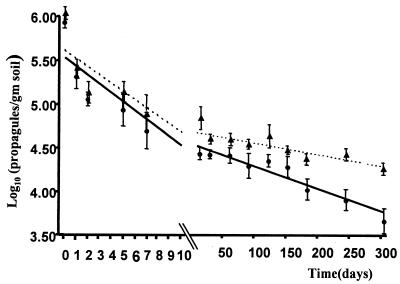
Soil persistence monitoring.
At 1 h following application, the differences in the titer of GMa (mean = 2.45 × 105, standard deviation [SD = 6.8 × 104] CFU/g) and GPMa (mean = 2.09 × 105 [SD = 5.2 × 104] CFU/g) were not significant. The data set for subsequent decline in spore numbers in the vicinity of cabbage roots (0 to 1 cm) behaved as piecewise regressions (Fig. 2). Thus, while soil titers of GMa and GPMa declined by 56 and 73%, respectively, in the first week, these strain differences were not significant (F = 2.9, P > 0.05). Differences between the strains became significant from day 10 (F = 7.8, P < 0.05). Thus, titers of GMa between 4 months (October) and 10 months (April of the second year) were reduced by 30% from 2.96 × 104, (SD = 1.00 × 103) CFU/g to 2.00 × 104 (SD = 5.29 × 103) CFU/g, respectively. During the same period, titers of GPMa declined by 70% from 1.53 × 104 (SD = 4.16 × 103) to 4.76 × 103 (SD = 1.15 × 103) CFU/g. Reduced fitness and survivability could reasonably be derived from deleterious effects of the additional genetic modifications, compared to the situation for transgenic fungi expressing GFP only. If so, then M. anisopliae may not just persist in the soil in a dormant state but characteristics for soil survival may include gene expression that can be interfered with by plasmid integration.
FIG. 2.
Changes in the soil titer of GMa (▾) and GPMa (•), over 300 days, within 1 cm of cabbage plant roots at depths of 0 to 2 cm. Spore count data were transformed to the log scale. The lines represent the model outcome of the population decline of GMa (dashed line) and GPMA (solid line). The analysis was conducted using Proc MIXED, SAS. Error bars indicate SD.
Stability under field conditions.
Closely related, therefore, to the issue of population dynamics is the question whether an intensive deployment protocol into complex microbial communities will promote the generation and amplification of altered transgenic M. anisopliae strains. Growth rates of colonies isolated from application areas were similar to those of the input transgenic strains. In some cases the isolated colonies differed from their progenitors in producing fluffier, raised colonies and fewer conidia. In each case, the original phenotype returned after three serial propagations on Sabouraud dextrose agar, consistent with experimental and physiological variations rather than genetic differences. During the course of this study, more than 50,000 colonies were examined, and all fungi expressing GFP in application area 1 were also constitutive producers of Pr1 and vice versa. This indicates stability of plasmid DNA in the chromosomes and provides no evidence for recombination. At 10 months after spraying, allozyme analysis was performed on 100 colonies chosen at random from locations also containing field strains 1 and 2. No interisolate variability was detected with each of the eight enzymes possessing electromorphic forms characteristic of genotypic class 14, the class to which strain 1080 belongs (10). In particular, none of the eight loci displayed symmetrical three-banded phenotypes characteristic of heterozygotes or two banded electromorphs, which would be evidence of mixed alternatively homozygous cultures.
Spatial distribution of transgenic M. anisopliae.
Soil samples were taken at 4 to 5 cm as well as alongside cabbage tap roots (0 to 1 cm) to check for uneven distribution of spores. For GMa, the ratio of fungi alongside the root to 4 to 5 cm from the root increased from 1.4:1 (1 month postinoculation) to about 3:1 (4 months postinoculation) (Fig. 3). The value remained high after the cabbage plants were killed by frost (8 months), indicating that GMa was persisting on the decaying organic matter. The rhizosphere effect is also apparent in GPMa, although with a more rapid decline in spore titers, reflecting its reduced persistence.
FIG. 3.
Effect of proximity to cabbage roots on the persistence of M. anisopliae under field conditions. Mean soil titers of propagules of M. anisopliae GMa and GPMa at depths of 0 to 2 cm are shown. Error bars indicate SD.
Samples from the buffer and fallow zones contained no transgenic M. anisopliae or contained insufficient numbers to be detected using dilution plate counts.
Rhizosphere competence.
To further analyze rhizosphere competence, samples of soil were taken directly from roots 4 months after application of GMa. A four-fold-larger population of fungal propagules was observed in the inner rhizosphere soil than in the outer rhizosphere soil at the top 2 cm of the root base. This suggests that close proximity to the root and its exudates is involved in the rhizospheric effect. The titer of GMa at the root base (3.1 × 105 [SD = 2.5 × 104] CFU) was close to the original inoculum load. Most other studies using fungi known to be good root colonizers show a decline, perhaps because the initial population added is to large for the carrying capacity of the root (8). Evidently, soil in the vicinity of plant roots provides a refuge for M. anisopliae from factors in the environment that reduce fungal titer.
The colonization of roots by GMa in the outer and inner rhizosphere of roots formed a gradient, with the rhizosphere effect decreasing with increasing depth (Fig. 4). The presence of fungal propagules more than 10 cm from the stem in the inner but not the outer rhizosphere implies some degree of vertical movement along the roots through fungal growth or cracks in the soil or via percolating water. When unwashed root segments were placed on Veens medium, growth of fluorescent fungus was observed from roots up to 10 cm from the stem. In spite of this proximity, a lot of the fungus could be removed from the roots by serial washings (Fig 5). The patchy distribution of fungal colonies on washed roots compared with the total coverage of unwashed roots implies weak adhesion by most propagules.
FIG. 4.
Mean number of propagules of M. anisopliae GMa at different depths in the outer (▪) and inner (□) rhizosphere of cabbage roots, 4 months after application. Error bars indicate SD.
FIG. 5.
UV micrographs of 2- to 5-cm-deep cabbage roots from application area 2 (4 months after spraying with GMa) placed on Veens medium for 48 h and showing growth by fluorescent M. anisopliae. The roots were shaken free of rhizosphere soil (A) or, in addition, ultrasonicated and subjected to a series of 10 water washes (B), which removed the inner rhizosphere and most but not all fungal propagules (indicated by arrows).
If rhizosphere competence is a general phenomenon among insect pathogens, its impact on plant ecology could be considerable and it possesses implicit coevolutionary implications. In addition, most efforts employing M. anisopliae for biocontrol have ignored habitat preferences and survival outside the host. Evidently, factors associated with soil dwelling may be even more critical in the selection of an isolate than virulence per se (1). However, rhizosphere competence also imposes potential risks since it might increase the difficulty of eliminating a pathogen following unanticipated and deleterious environmental effects.
Monitoring of naturally occurring insects.
A natural infestation of Pieris rapae occurred on the cabbages. Eight of 20 and 6 of 20 third-instar or older P. rapae larvae collected from application areas 1 and 2, respectively, within 10 days of the application died from infections with GPMa and GMa, respectively. A majority of 43 P. rapae larvae collected from the test sites 1 month following application died of bacterial infections and parasitoids and further analysis of these was not possible. However, three larvae died from fungal infections and produced spores of GMa. Flea beetles (Alticinae) collected from cabbages up to 1 week after spraying also had spores of GMa and GPMa on their surfaces, but none of 30 beetles maintained for 10 days in the laboratory on cabbage seedlings succumbed to overt fungal infection. More than 3,000 arthropods were collected from pit fall traps in and around the application sites in the summer of the first year. These included four species of carabids (Amara and Stenolophus spp.), other beetles including predatory rove beetles (Staphylinidae), five species of ants, and many types of aphids, springtails, spiders, and mites. About 5% of the arthropods monitored under laboratory conditions died of a variety of overt fungal infections and 27% died of septicemia or other unidentified causes, but we did not detect external or internalized M. anisopliae in healthy, sick, or dead insects. These results suggest the potential of insect-mediated dispersal to nontargeted deployment areas is low.
Conclusion.
The results of this trial do not suggest any safety concerns to using GMa or GPMa that would detract from their being environmentally preferred alternatives to current chemical-based control strategies. However, their survival into the second year is significant, since time increases the possibility of adaptation for increased fitness (7). It cannot be assumed, therefore, that either strain will die out because of current reduced fitness. There was no evidence for phenotypic instability of the introduced fungi, but this might not be expected if genetic changes with clear phenotypes follow the punctuated- equilibrium model of evolution, with long periods of apparent stability punctuated by large infrequent changes. Given that the long-term fitness of a genetically engineered pathogen that persists in nature is difficult to predict, it is all the more essential to establish technologies such as the use of gfp that will permit informed risk assessment through monitoring the fate of marked strains.
Acknowledgments
This work was supported by the USDA risk assessment program (CSREES-99331208284).
We thank Mark Spiknall and the agricultural technicians at the Upper Marlboro Research Station for their comprehensive technical assistance in planting and maintaining the field site. Several undergraduate students from the 2000 and 2001 BSCI105 course at the University of Maryland, particularly Franklin Johnson, Jason Lloyd, and Jordan Newmark, participated in collecting and analyzing samples. We thank Mary Christman, Department of Agriculture, University of Maryland, for help with statistical analyses.
REFERENCES
- 1.Bidochka, M. J. 2001. Monitoring the fate of biocontrol fungi, p. 193-218. In T. M. Butt, C. Jackson, and N. Morgan, (eds.), Fungal biocontrol agents: progress, problems and potential. CAB International, Wallingford, United Kingdom.
- 2.Butt, T. M., C. Jackson, and N. Magan. 2001. Introduction-fungal biological control agents: progress, problems and potential, p. 1-8. In T. M. Butt, C. Jackson, and N. Morgan (ed.), Fungal biocontrol agents: progress, problems and potential. CAB International, Wallingford, United Kingdom.
- 3.Goettel, M. S., and G. D. Inglis. 1997. Fungi: hyphomycetes, p. 367-394. In L. Lacey, (ed.), Manual of techniques in insect pathology. Academic Press, Inc., San Diego, Calif.
- 4.Goettel, M. S., R. J. St. Leger, S. Bhairi, M. K. Jung, B. R. Oakley, D. W. Roberts, and R. C. Staples. 1990. Pathogenicity and growth of Metarhizium anisopliae stably transformed to benomyl resistance. Curr. Genet. 17:129-132. [Google Scholar]
- 5.Hajek, A. E., I. Delalibera, and M. L. McManus. 2000. Introduction of exotic pathogens and documentation of their establishment and impact, p. 339-370. In L. A. Lacey and H. K. Kaya (ed.), Field manual of techniques in invertebrate pathology. Kluwer Academic Press, Dordrecht, The Netherlands.
- 6.Milner, R. J. 1992. Selection and characterization of strains of Metarhizium anisopliae for control of soil insects in Australia, p. 200-207. In C. I. Lomer and C. Prior (ed.), Biological control of locusts and grasshoppers. CAB International, Wallingford, United Kingdom.
- 7.Mundt, C. C. 1995. Models from plant pathology on the movement and fate of new genotypes of microorganisms in the environment. Annu. Rev. Phytopathol. 33:467-488. [DOI] [PubMed] [Google Scholar]
- 8.Parke, J. L. 1991. Root colonization by indigenous and introduced microorganisms, p. 33-42. In D. L. Keister and P. B. Cregan (ed.), The rhizosphere and plant growth. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 9.Roberts, D. W., and A. E. Hajek. 1992. Entomopathogenic fungi as bioinsecticides, p. 144-159. In G. F. Leatham (ed.), Frontiers in industrial mycology. Chapman & Hall, New York, N.Y.
- 10.St. Leger, R. J., B. May, L. L. Allee, R. C. Frank, R. C. Staples, and D. W. Roberts. 1992. Genetic differences in allozymes and in formation of infection structures among isolates of the entomopathogenic fungus Metarhizium anisopilae. J. Invertebr. Pathol. 60:89-101. [Google Scholar]
- 11.St. Leger, R. J., M. J. Bidochka, and D. W. Roberts. 1995. Co-transformation of Metarhizium anisopliae: by electroporation or using the gene gun to produce stable GUS transformants. Curr. Genet. 131:289-294. [Google Scholar]
- 12.St. Leger, R. J., L. Joshi, M. J. Bidochka, and D. W. Roberts. 1996. Construction of an improved mycoinsecticide over-expressing a toxic protease. Proc. Natl. Acad. Sci. USA 93:6349-6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.St. Leger, R. J. 2001. Development and testing of genetically improved mycoinsecticides, p. 229-239. In J. Gressel, T. Butts, G. Harman, A. Pilgeram, R. St. Leger, and D. Nuss (ed.), Enhancing biocontrol agents and handling risks. Proceedings of a NATO Advanced Research Workshop, 9-15 June, 2001. IOS Press, Amsterdam, The Netherlands.
- 14.St. Leger, R. J., and S. Screen. 2001. Prospects for strain improvement of fungal pathogens of insects and weeds, p. 219-238. In T. M. Butt, C. Jackson, and N. Morgan (ed.), Fungal biocontrol agents: progress, problems and potential. CAB International, Wallingford, United Kingdom.
- 15.Suzuki, N., L. M. Geletka, and D. L. Nuss. 2000. Essential and dispensable virus-encoded replication elements revealed by efforts to develop hypoviruses as gene expression vectors. J. Virol. 74:7568-7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whipps, J. M. 2001. Microbial interactions and biocontrol in the rhizosphere. J. Exp.Bot. 52:487-511. [DOI] [PubMed] [Google Scholar]