Abstract
Oligonucleotide probes targeting the small-subunit rRNA are commonly used to detect and quantify bacteria in natural environments. We developed a PCR-based approach that allows synthesis of oligonucleotide probes targeting a variable region in the 16S rRNA without prior knowledge of the target sequence. Analysis of all 16S rRNA gene sequences in the Ribosomal Database Project database revealed two universal primer regions bracketing a variable, population-specific region. The probe synthesis is based on a two-step PCR amplification of this variable region in the 16S rRNA gene by using three universal bacterial primers. First, a double-stranded product is generated, which then serves as template in a linear amplification. After each of these steps, products are bound to magnetic beads and the primers are detached through hydrolysis of a ribonucleotide at the 3′ end of the primers. This ultimately produces a single-stranded oligonucleotide of about 30 bases corresponding to the target. As probes, the oligonucleotides are highly specific and could discriminate between nucleic acids from closely and distantly related bacterial strains, including different species of Vibrio. The method will facilitate rapid generation of oligonucleotide probes for large-scale hybridization assays such as screening of clone libraries or strain collections, ribotyping microarrays, and in situ hybridization. An additional advantage of the method is that fluorescently or radioactively labeled nucleotides can be incorporated during the second amplification, yielding intensely labeled probes.
The identification and quantification of microorganisms in environmental or clinical samples is a cornerstone of microbiology, as it is a crucial step in determining the roles of specific populations. This is increasingly achieved by the application of the rRNA phylogenetic framework, which consists of PCR-based retrieval of rRNA genes from samples, their characterization via sequencing, and the confirmation of the importance of specific organisms by rRNA-targeted oligonucleotide hybridization (5, 14). rRNA-targeted oligonucleotide hybridization takes advantage of extensive sequence variation in some regions of the ribosomal DNAs (rDNAs), allowing the design of specific probes that can be used to identify and quantify microbial populations in complex samples (1, 19). The 16S rRNAs have provided an especially useful tool, which is reflected in the rapidly growing number of sequences available in the databases for comparative purposes (10).
Despite this extensive sequencing effort, the diversity of microbial communities is still not sufficiently explored, a fact that is exacerbated by potentially significant variation in community structure even between physically and chemically similar environments (21). Thus, a major limiting step in the application of the rRNA framework remains the potentially large number of sequences to be determined and probes to be synthesized for a satisfactory correlation of specific environmental factors to the presence of specific populations. In this context, there is a need for an efficient means to generate large sets of oligonucleotides with high specificity.
Here, we present a novel method for the generation of oligonucleotide probes which give good phylogenetic resolution without prior knowledge of the target sequence. The method relies on two consecutive PCR amplifications with Bacteria-specific primers bracketing a highly variable region in the 16S rDNA, each followed by detachment of one primer by alkaline hydrolysis of a 3′ ribonucleotide. Ultimately, a single-stranded oligonucleotide of the variable region is obtained. Comparative analysis of sequences obtained from databases and empirical testing of representative isolates show good differentiation of targets by these PCR-generated probes. Because of the low cost and potential for production of the probes in microtiter plate format, anticipated applications include the rapid screening of clone libraries or of strain collections for potential abundance in specific environmental samples.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
A set of phylogenetically diverse isolates was used to develop and test the method. These included strains of Escherichia coli K-12, Bacillus subtilis JH 642, Roseobacter sp., Vibrio sp., Prochlorococcus sp., and Synechococcus sp., as well as marine isolates with representatives in Cytophaga-Bacteroides, gram-positive bacteria, and α-, β-, and γ-Proteobacteria (7) (Fig. 1). Synechococcus and Prochlorcoccus were grown in nutrient-enriched Sargasso seawater as previously described (12), whereas all other isolates were grown in Luria broth at 20°C.
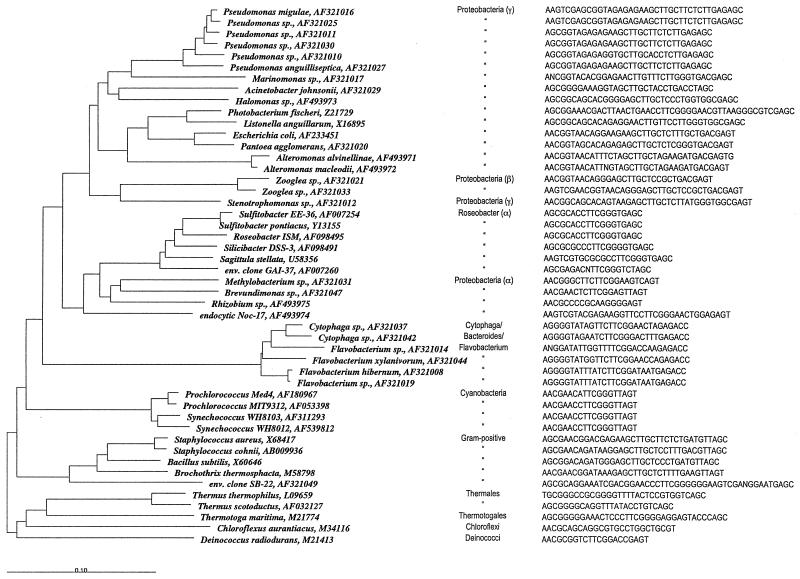
FIG. 1.
Neighbor-joining tree of 16S rRNAs for a collection of 44 bacterial isolates from a coastal marine environment, reference strains, and sequences retrieved from GenBank. The scale bar indicates 10% sequence difference. Organism names for the isolates are from the closest matches in GenBank. Group affiliations, GenBank accession numbers, and isolate-specific target sequences for PCR-generated 16S rRNA oligonucleotide probes are shown for each sequence.
Sequence manipulation and nucleotide sequence accession numbers.
All sequence manipulations were carried out by using the ARB software package (http://www.arb-home.de/). Sequences were initially aligned by using Fast Aligner version 1.03 and adjusted manually. The tree in Fig. 1 was constructed by using the neighbor-joining algorithm with the Jukes-Cantor correction. Tree topology was confirmed by maximum-likelihood analysis with FastDNAml. GenBank accession numbers for the complete or partial 16S rRNA sequences of the strains used in this study are given in Fig. 1.
Nucleic acid extractions.
DNA for PCR was extracted by bead beating of cell pellets in 2-ml tubes with 0.5 g of zirconium beads (0.2-mm diameter) in 650 μl of TE (10 mM Tris, 1 mM EDTA, pH 8) buffer for 30 s at 5,000 rpm. Lysates were incubated for 30 min at 37°C with 0.2 mg of proteinase K ml−1 and 0.06% sodium dodecyl sulfate (SDS), followed by inactivation of the enzyme at 65°C and addition of NaCl to a final concentration of 1 M. Nucleic acids were purified by standard phenol-chloroform extraction and by passing the final aqueous solution over Strataprep PCR purification columns (Stratagene, La Jolla, Calif.).
Total nucleic acids for membrane hybridizations were extracted by bead beating as described above except in the presence of 600 μl of RNase-free lysis buffer (5.25 M guanidine-HCl, 1.5% Triton X-100, 2.5 mM Tris, 0.25 mM EDTA, pH 7.5). Cell debris and beads were sedimented at 12,000 × g for 1 min, and 400 μl of the supernatant was transferred to a new tube containing 200 μl of isopropanol. Total nucleic acids were purified by using SNAP total RNA spin columns (Invitrogen, Carlsbad, Calif.).
Primer design.
The Bacteria-specific primers 64f (Table 1) and 104r (Table 2) were designed based on a complete alignment of the Ribosomal Database Project (RDP) in 1996 by using the following procedure. First, the alignment of representative 16S rRNA sequences published by the RDP was inspected for two stretches of high sequence conservation bracketing a region of high sequence variation (probe region) (Fig. 2). Second, a forward and a reverse PCR primer, each containing a single degeneracy, were designed matching the highly conserved regions. Third, additional degeneracies were added to the primer sequences based on comparison of the primers with the entire RDP alignment of several thousand 16S rRNA sequences to maximize matches of the primers against bacterial sequences. The resultant primers, 64f and 104r, were then tested for amplification success and properties against the 44 bacterial isolates and standard isolates listed in Fig. 1. In addition, the universal primer 519r (8) (Table 3) used in the first step of the probe synthesis was tested against the same alignment to test how well it matches bacterial sequences. Finally, all three primers were compared to the most recent alignment of 16,277 sequences in the RDP II (10) by using the Rose software (2). Archaeal sequences and database entries lacking sequence information in the target regions were manually excluded from the analysis.
TABLE 1.
Design of primer 64f and similarity to 16S rRNA sequences in the RDP II databasea
| Nucleotide | Mismatchesb for the following nucleotide in the 64f primer target consensus (sense, 5′→3′):
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| U | A | A | Nc | A | C | A | U | G | C | A | A | G | U | C | G | |
| A | 5/0.2 | 2/0.2 | 0/0 | 1/0.0 | 0/0 | 9/0.4 | 0/0 | 6/0.3 | 38/1.8 | |||||||
| T | 215/10 | 4/0.2 | 0/0 | 0/0 | 2/0.1 | 3/0.1 | 3/0.1 | 2/0.1 | 26/1.2 | 0/0 | 31/1.5 | 7/0.3 | ||||
| C | 1/0 | 5/0.2 | 1/0.0 | 6/0.3 | 0/0 | 0/0 | 9/0.4 | 0/0 | 0/0 | 4/0.2 | 1/0.0 | 3/0.1 | ||||
| G | 1/0 | 5/0.2 | 0/0 | 83/3.4 | 0/0 | 0/0 | 0/0 | 1/0.0 | 1/0.0 | 0/0 | 0/0 | 7/0.3 | ||||
| Primer 64, forward (5′→3′) | T | A/T | A | N | G/A | C | A | T | G | C | A | A | G | T | C | G |
The forward primer 64f targets positions 49 to 64 in E. coli 16S rRNA.
Absolute number/percentage of nucleotide-specific mismatches when the primer target consensus sequence was compared against 2,179 16S rRNA sequences retrieved from the RDP II database. Degenerate positions introduced in primers in order to eliminate mismatches are in boldface. 0/0 denotes <0.05.
N, degenerate nucleotide position (A, T, G, or C).
TABLE 2.
Design of primer 104r and similarity to 16S rRNA sequences in the RDP II databasea
| Nucleotide | Mismatchesb for the following nucleotide in the 104r primer target consensus (sense, 3′→5′):
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | A | U | G | A | G | U | G | G | G | C | A | Nc | G | C | G | G | |
| A | 1/0.0 | 0/0 | 3/0.1 | 4/0.2 | 0/0 | 0/0 | 1/0.0 | 18/0.8 | 267/12 | 1/0.0 | 2/0.1 | 19/0.9 | |||||
| T | 9/0.4 | 2/0.1 | 2/0.1 | 3/0.1 | 8/0.4 | 1/0.0 | 1/0.0 | 20/0.9 | 67/3 | 1/0.0 | 0/0 | 10/0.4 | 1/0.0 | 3/0.1 | |||
| C | 1/0.0 | 0/0 | 4/0.2 | 0/0 | 142/6.4 | 0/0 | 4/0.2 | 11/0.5 | 1/0 | 1/0.0 | 5/0.2 | 0/0 | 3/0.1 | 3/0.1 | |||
| G | 4/0.2 | 4/0.2 | 21/0.9 | 3/0.0 | 8/0.4 | 0/0 | 1/0.0 | 0/0 | |||||||||
| Primer 104, reverse (5′→3′) | T | T | A | C | G/T | C | A | C | C | C | G | T | N | T/C | G | C | C |
The reverse primer 104r targets position 104 to 119 in E. coli 16S rRNA.
Absolute number/percentage of nucleotide-specific mismatches when the primer target consensus sequence was compared against 2,233 16S rRNA sequences retrieved from the RDP II database. Degenerate positions introduced in primers in order to eliminate mismatches are in boldface. (See Table 1.)
N, degenerate nucleotide position (A, T, G, or C).
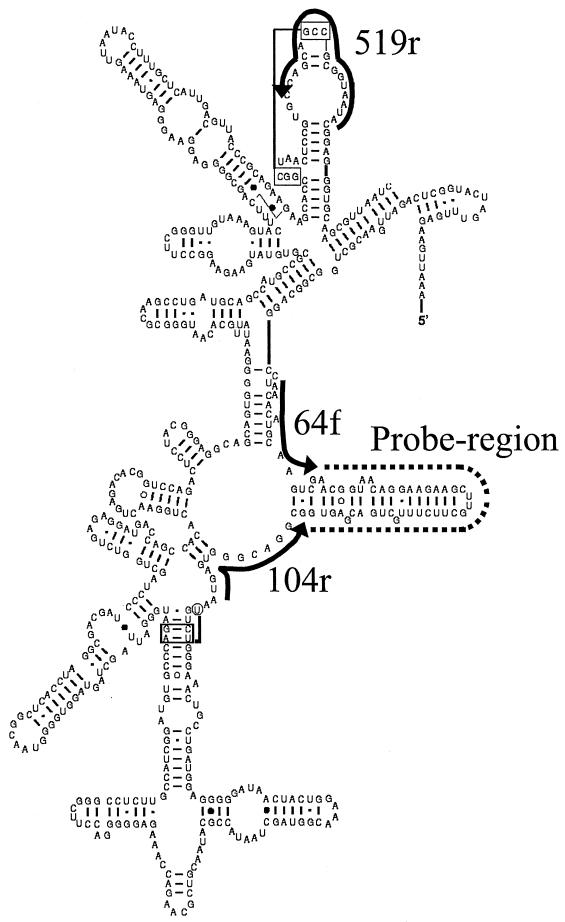
FIG. 2.
Partial secondary structure of 16S rRNA from E. coli (4), with target regions for the three Bacteria universal primers (64f, 104r, and 519r) used in the probe synthesis indicated by solid lines. The dashed line indicates the variable probing region.
TABLE 3.
Primer 519r similarity to 16S rRNA sequences in the RDP II database
| Nucleotide | Mismatchesa for the following nucleotide in the 519r primer target (sense, 3′→5′):
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C | A/U | U | A | A | U | G | G | C | G | C | C | G | C/A | C | G | A | C | |
| A | 0/0 | 16/0.8 | 0/0 | 1/0.0 | 0/0 | 0/0 | 0/0 | 2/0.1 | 0/0 | 1/0.0 | 0/0 | 1/0.0 | 1/0.0 | |||||
| T | 1/0.0 | 0/0 | 0/0 | 1/0.0 | 0/0 | 3/0.1 | 0/0 | 12/0.6 | 8/0.4 | 1/0.0 | 2/0.0 | 0/0 | 0/0 | 0/0 | 0/0 | |||
| C | 0/0 | 67/2.8 | 1/0.0 | 4/0.2 | 1/0.0 | 0/0 | 1/0.0 | 2/0.1 | 0/0 | 0/0 | 0/0 | |||||||
| G | 1/0.0 | 0/0 | 87/3.7 | 3/0.1 | 10/0.5 | 0/0 | 1/0.0 | 2/0.1 | 1/0.0 | 2/0.0 | 0/0 | 0/0 | 0/0 | |||||
| Primer 519, reverse (5′→3′) | G | A/T | A | T | T | A | C | C | G | C | G | C | G | G/T | G | C | T | G |
Absolute number/percentage of nucleotide-specific mismatches when the primer target consensus sequence was compared against 2,375 16S rRNA sequences retrieved from the RDP II database. (See footnote b, Table 1.)
Quantification of nucleic acids.
Genomic DNA and total nucleic acids were quantified by using UV absorbance at 260 nm, with an absorbance of 1 corresponding to 50 mg of DNA liter−1 and 40 mg of total nucleic acids liter−1 (16). Single-stranded DNA and oligonucleotides were quantified fluorometrically by using the Oligreen single-stranded-DNA quantitation reagent (Molecular Probes, Eugene, Oreg.). Assays were performed in 96-well microplates (Nunc) with synthetic oligonucleotides as standards (0 to 5 mg liter−1). Five-microliter subsamples of PCR-generated oligonucleotides or standards were added to individual wells and diluted in 150 μl of assay working solution (10 mM Tris, 1 mM EDTA, 1× Oligreen reagent, pH 8). Green fluorescence (485-nm excitation and 530-nm emission) was recorded after 5 min of incubation at room temperature by using a Fusion microplate reader (Packard Instruments, Meriden, Conn.), and concentrations of PCR-generated oligonucleotides were estimated from background-corrected standard curves generated by a 7- to 10-point linear regression with a coefficient of determination of >0.95.
PCR generation of oligonucleotide probes.
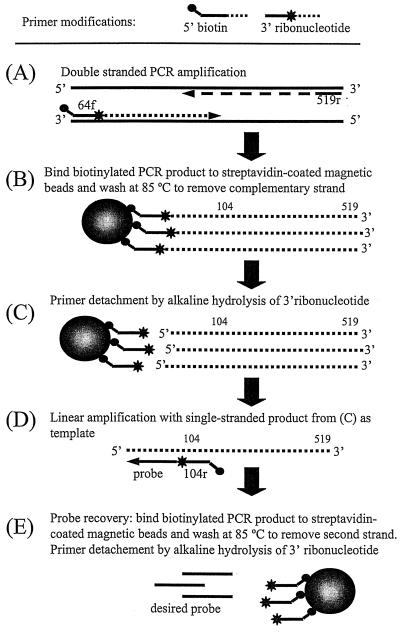
The oligonucleotide probes were produced by using the following five-step procedure outlined in Fig. 3, including two PCR amplifications with detachable primers.
FIG. 3.
Outline of PCR-based method to generate population-specific oligonucleotide probes without prior sequence information.
(i) Double-stranded PCR amplification (step A).
Approximately 100 ng of genomic DNA was used as a template in a PCR with the primer pair 64f-519r, where the 64f primer contained a 5′ biotin modification and a 3′ ribonucleotide. PCR was performed in 50-μl aliquots by using a Stratagene Robocycler with 1 cycle of 3 min at 94°C; 35 cycles of 1 min at 94°C, 1 min at 50°C, and 2 min at 72°C; and finally 1 cycle of 10 min at 72°C. Each tube contained 1× PCR buffer (10 mM Tris-HCl [pH 9], 50 mM KCl, 0.1% Triton X-100, 2 mM MgCl2), a 200 μM concentration of each deoxynucleoside triphosphate, a 100 nM concentration of each primer (Integrated DNA Technologies, Coralville, Iowa), and 2.5 U of Taq DNA polymerase (Promega, Madison, Wis.).
(ii) Binding of PCR product (step B).
Biotinylated products were bound to 0.1 mg of paramagnetic and streptavidin-coated Dynabeads M-280 (Dynal, Skøyen, Norway) according to the instructions supplied by the manufacturer after excess primer and nucleotides were removed by using the QIAquick PCR purification kit (Qiagen, Valencia, Calif.). To remove the nonbiotinylated complementary strand, beads were washed twice at 85°C for 1 min in autoclaved Q-grade water (Millipore), using an MPC magnet (Dynal) to separate beads from liquid.
(iii) Primer detachment by alkaline hydrolysis of ribonucleotide (step C).
The beads with the bound PCR product were washed once at room temperature in sterile Q-grade water before the ribonucleotide was hydrolyzed by incubation of the beads for 12 h in 100 μl of 0.2 M NaOH at 37°C with gentle mixing. After hydrolysis, primers bound to beads were removed from solution with the MPC magnet. The aqueous solution containing the single-stranded PCR product was diluted to 400 μl in 10 mM Tris (pH 8) and precipitated with sodium acetate and ethanol (15). The precipitated DNA was dissolved in 50 μl of Tris (10 mM, pH 8) and stored at −20°C.
(iv) Linear amplification (step D).
The single-stranded fragment from step C served as the template in a linear amplification of the population-specific probing region with primer 104r modified with a 5′ biotin and 3′ ribonucleotide. Approximately 0.5 pmol of single-stranded template was added to each tube. Amplification conditions and reagents were as described above.
(v) Recovery of oligonucleotide probe (step E).
The reaction product from the linear amplification was bound to 0.2 mg of magnetic beads, which were washed twice at 85°C in Q-grade water to remove the complementary strand. The amplified product was separated from biotinylated primers by alkaline hydrolysis and removal of the magnetic beads as described above. The final product, which is complementary to the probing region of the 16S rRNA (Fig. 2), was precipitated, dried, and dissolved in 10 mM Tris (pH 8) for long-term storage at −20°C.
Hydrolysis efficiency.
To test the efficiency of the primer detachment by alkaline hydrolysis, two permutations of the 104r primer, one with and one without the 3′ ribonucleotide (Synthetic Genetics, San Diego, Calif.), were 5′ end labeled with 32P by using T4 polynucleotide kinase (New England Biolabs, Beverly, Mass.) (15). Primers were separated from unreacted nucleotides in Sephadex G-50 spin columns (Amersham Pharmacia Biotech, Piscataway, N.J.), and both were used in separate PCRs with nonmodified primer 64f.
PCRs were performed in 40-μl volumes containing 100 ng of genomic DNA from E. coli K-12 by using primer pair 64f-104r or 64f-104r-3′ ribonucleotide and the same conditions and reagents as described above. PCR products were agarose gel purified from unreacted nucleotides and primers by using QIAquick gel extraction kits (Qiagen). Half of the volume of each purified product (64f-104r and 64f-104r-3′ ribonucleotide) was subjected to alkaline hydrolysis for 12 h (0.2 M NaOH, 37°C), whereas the second half was kept frozen in 10 mM Tris (pH 7.5) as a reference. Both hydrolyzed and reference samples were precipitated with ammonium acetate and ethanol (16), resuspended in 20 μl of Q-grade water, and separated in a 10% denaturing acrylamide gel on a Mini-Protean 3 system (Bio-Rad, Hercules, Calif.) at 9 V cm−1. Gels were vacuum dried on blot paper, and bands were identified by 32P-phosphorimaging with multipurpose plates and a Cyclone phosphorimager (Packard Instruments).
Linear amplification optimization.
The yield of the linear amplification for the probe synthesis (Fig. 3D) was determined with PCR mixtures containing 32P-labeled primer 104r. Single-stranded PCR product was generated from 40 ng of Shewanella baltica genomic DNA as outlined in Fig. 3A to C and used as a template. Individual reaction tubes were removed from the thermocycler after 3 to 40 amplification cycles. Products were separated on a 10% denaturing acrylamide gel and detected as described above. The intensity of the radioactive signal was estimated by image densitometry with background correction by using the Optiquant 3.1 software (Packard Instruments).
Determination of probe purity and Td.
The size and purity of a PCR-generated oligonucleotide from E. coli was compared to those of purchased synthetic probes that had been either desalted or gel purified. Purified oligonucleotides were labeled with 32P as described above and separated by size on 10% denaturing acrylamide gels, followed by phosphorimaging detection.
For determination of dissociation temperatures (Td), total nucleic acids extracted from E. coli were blotted on Zetaprobe nylon membranes (Bio-Rad) with a Minifold II slot blot apparatus (Schleicher & Schuell, Keene, N.H.) after a 30-s alkaline denaturation (10 mM NaOH, 1 mM Tris) at 85°C. Each slot contained 150 ng of total nucleic acids (approximately 50 ng of 16S rRNA). After each slot was washed with 200 μl of diethyl pyrocarbonate-treated water (16), nucleic acids were UV-cross-linked (120 mJ) in a Stratalinker 2400 (Stratagene) and rinsed in diethyl pyrocarbonate-treated water before drying.
The Td of the PCR-generated and the purchased synthetic probes were determined as described by Polz and Cavanaugh (15). Membrane strips containing triplicate slots of E. coli nucleic acids were prewetted in 2 ml of hybridization buffer (5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 1% SDS) at 37°C for 30 min, followed by addition of 10 μl of buffer containing 1 pmol of probe per slot. Membranes were hybridized overnight at 37°C, and unbound probe was removed by two 15-min washes at the same temperature in washing buffer (1× SSC, 0.1% SDS). Subsequently, individual slots were cut out and washed sequentially at gradually increasing temperatures (37 to 87°C) by transferring them to 7-ml scintillation vials containing 2 ml of washing buffer for 5 min at a time. The radioactivity remaining in the wash solutions and activity remaining on the filter after the final wash were determined by liquid scintillation after adding 3 ml of Scintisafe Plus 50% (Packard) to each vial. The total activity in the wash solutions and the activity remaining on the membrane were summed, and the Td was estimated as the temperature at which 50% of the probe was dissociated from the membranes.
Hybridization specificity.
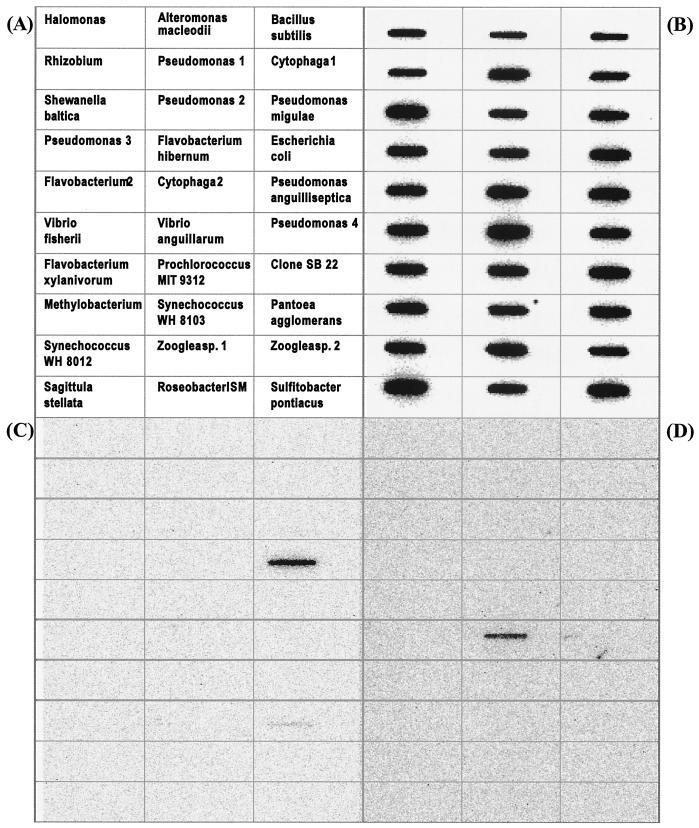
To test the hybridization specificity of the PCR-generated probes, nucleic acids from 30 different closely and distantly related bacterial strains (Fig. 1; see Fig. 7A) were blotted on Zetaprobe nylon membranes as described above. A eubacterial probe, EUB338 (20), and PCR-generated population-specific probes from E. coli and Vibrio anguillarum were 32P labeled at the 5′ termini with bacteriophage T4 polynucleotide kinase (16). Each membrane was incubated overnight at 37°C with 4 pmol of the respective probe. Subsequently, membranes were washed twice for 20 min at 37°C in washing buffer (1× SSC, 0.1% SDS) before a final stringent wash for 30 min at a higher temperature (EUB338 at 50°C and the longer PCR-generated probes at 70°C). Hybridization was detected by phosphorimaging as described above.
FIG. 7.
Membrane hybridization of total nucleic acids from 30 bacterial strains with 32P-labeled oligonucleotide probes. (A) Identity and location of immobilized nucleic acids on the membrane; (B) hybridizations with universal bacterial probe EUB338; (C) PCR-generated probe specific for E. coli; (D) PCR-generated probe specific for V. anguillarum.
RESULTS
Primers 64f, 104r, and 519r are a perfect match to almost all bacterial 16S rRNA sequences present in the RDP database of July 1996 (Tables 1 to 3). Tables 1 to 3 illustrate the design process. In the cases of 64f and 104r, the first permutation of the primers was based on the consensus sequence (sense) obtained from an alignment of a subset of phylogenetically representative sequences published by the RDP. This resulted in a 16-mer (64f) oligonucleotide and a 17-mer (104r) oligonucleotide, each with a single fourfold-degenerate position. In the subsequent inspection against the full alignment of the RDP, additional degeneracies were introduced in the primers if >1% of the RDP sequences contained mismatches. The only exceptions were the two positions at the 3′ end of 64f, where changes were avoided since less then 2% of the sequences were mismatched and degeneracies could have destabilized the primer-target hybrid. Overall, this process resulted in the introduction of two additional twofold degenerate positions in both primers (Tables 1 and 2). The same process confirmed the high conservation of the target site for the previously designed 519r primer (8) (Table 3). A comparison with a more recent version of the prokaryotic 16S rRNA RDP II database (version 8.1) showed that the three primers were perfectly matched to between 83 and 86% of the sequences in the database. The residual 14 to 17% were almost exclusively single mismatches against a few predominantly pathogenic groups which are overrepresented in the RDP II (e.g., Chlamydia, spirochetes, and Mycoplasma). The only systematic exception was multiple mismatches of primer 104r against representatives of the genus Planctomyces. All primers successfully amplified DNAs from the 44 environmental isolates of Bacteria listed in Fig. 1. This collection also contains several strains with a single mismatch against both the 64f and 104r primers.
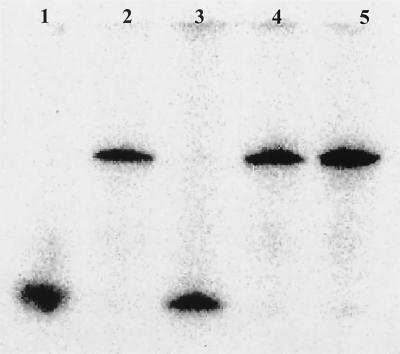
Alkaline hydrolysis of 3′ ribonucleotides is a robust and efficient way to detach primers from amplification products. This is evident from the complete hydrolysis of primer 104r following incubation in 0.2 M NaOH with no apparent damage to the DNA (Fig. 4) (17). The mechanism of hydrolysis ensures that the ribonucleotide is removed with the primer. An OH−-induced attack and subsequent breaking of the 3′ phosphodiester bond produces an OH group at the 5′ terminus of the synthesized probe (13). Experiments with primer 64f showed a similar efficiency of detachment (data not shown). In contrast to its sensitivity to alkali, the 3′ ribonucleotide is stable in the presence of RNase, since addition of the enzyme did not affect amplification and alkaline detachment of the primer (data not shown).
FIG. 4.
(A) Phosphorimaging scan showing detachment of the 5′-32P-labeled primer from the PCR product after alkaline hydrolysis of the ribonucleotide at the 3′ end of primer 104r. PCR products with primer 104r lacking the 3′ ribonucleotides are included for comparison. Lanes: 1, primer 104r; 2, PCR product with 3′ ribonucleotide-modified primer; 3, PCR product with 3′ ribonucleotide-modified primer after alkaline hydrolysis; 4, PCR product with nonmodified primer; 5, PCR product with nonmodified primer after alkaline hydrolysis.
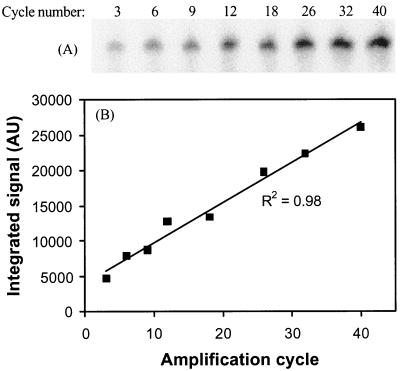
Quantification of product yield in the last amplification step with primer 104r showed that product accumulated linearly with cycle number to the assay maximum of 40 amplification cycles at an initial template concentration of about 0.25 pmol per 50-μl reaction mixture (Fig. 5). To ensure high product recovery, it is important to add sufficient beads to the binding reaction, since unincorporated primers are not removed and will compete with the product for binding sites on the beads. We observed that the product recovery at bead concentrations of lower than 25 μg per pmol of total biotinylated nucleic acid was positively related to the concentration of beads, with 74 and 85% lower probe recovery at bead additions of 10 and 5 μg per pmol of biotinylated nucleic acid. Using 20 μg of streptavidin-coated beads per pmol of biotinylated nucleic acid, the recovery of oligonucleotide probes in our regular experiments ranged from 3 to 6 pmol for a 50-μl reaction mixture, and we recommend this as a minimum bead addition. Alternatively, the reaction can be set up so that only little unincorporated primer remains to compete with the product binding, e.g., by increasing either the template concentration or the number of amplification cycles.
FIG. 5.
Linear amplification with 5′-32P-labeled primer 104r. (A) Gel electrophoresis and phosphorimaging detection of amplified fragments after 3 to 40 cycles. (B) Quantification by image analysis using local background subtraction shows a linear accumulation of amplification product with cycle number. AU, absorbance units.
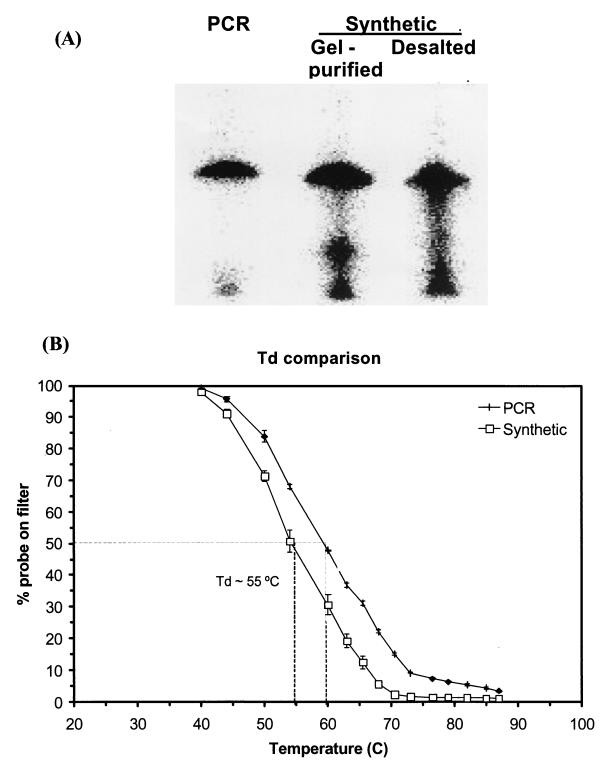
The purity of the PCR-generated probes compared favorably to that of purchased synthetic oligonucleotides with identical sequence that had either been gel purified or desalted (Synthetic Genetics) (Fig. 6A). The size of the PCR generated probe was similar to the synthetic probes; however, the purity of the PCR-generated probe appeared superior as less smearing was consistently observed (Fig. 6A). The comparison of melting behavior of the same set of probes against total nucleic acids of E. coli revealed that the Td was 5°C higher for the PCR-generated probe than for the gel-purified synthetic probe (Fig. 6B). A similar difference was observed when the desalted synthetic oligonucleotide was compared to the PCR generated probe (data not shown).
FIG. 6.
(A) Gel electrophoresis of a 5′-32P-labeled PCR-produced oligonucleotide probe targeting 16S rRNA of E. coli. Two synthetic oligonucleotides of identical sequence (desalted or purified by acrylamide gel electrophoresis) are included as a reference. (B) Dissociation curves for a commercially produced, gel-purified oligonucleotide probe and a PCR-generated oligonucleotide probe with identical sequence (5′ACTCGTCAGCAAAGAGCAAGCTTCTTCCTGTTACCGTT3′). Each symbol represents the mean dissociation of probe hybridized to triplicate slot blots with total nucleic acids from E. coli as a target (∼50 ng of 16S rRNA). Error bars represent ±1 standard deviation, and dashed lines indicate the Td for the two probes.
Sequence comparison of the region located between the two primers 64f and 104r shows substantial variation in both composition and length among most of the 44 phylogenetically representative bacterial isolates tested (Fig. 1 and 7). The length typically varied between 20 and 35 nucleotides; however, a few strains had probing regions of up to 49 nucleotides (Fig. 1). The potential use of this region to differentiate even closely related bacterial strains is illustrated by the Roseobacter-affiliated strains used in this study (Fig. 1). Even though the overall 16S rRNA sequences for these strains vary by only a few percent, most strains can be differentiated in silico by using the ∼20-nucleotide probing region. The only exceptions in this group are the two Sulfitobacter strains, S. pontiacus and Sulfitobacter sp. strain EE-36 which have an identical probing region and show only 0.4% variation in their 16S rRNA sequences. Other strains that were not well resolved at the isolate level include three of the Synechococcus and Prochlorococcus isolates (1 to 3.9% sequence variation in 16S rRNA), the two Zooglea isolates (1.5% sequence variation in 16S rRNA), and five Pseudomonas isolates (1.1 to 4.5% sequence variation in16S rRNA). This overall picture of probe specificity was confirmed by detailed comparison of deeply branching and distantly related bacterial groups, such as the thermophilic oxygen reducers, Thermotogales, green nonsulfur bacteria, deinococci, fusobacteria, and actinomycetes (data not shown).
The PCR-generated probes also showed good ability to differentiate 30 closely and distantly related bacterial strains in slot blot hybridization of total nucleic acids (Fig. 7). Each slot contained 150 ng of total nucleic acids, and the relative abundances of 16S rRNA molecules in each slot, assessed by hybridization with the 32P-labeled Bacteria-specific probe EUB338, appeared to be similar in magnitude (Fig. 7B). Hybridizations of PCR-generated probes targeting E. coli and V. anguillarum showed high specificity for the respective target under the tested conditions (Fig. 7C and D), with the only exception of slight cross-hybridization of the E. coli probe with Pantoea agglomerans nucleic acids. This was surprising and remains unexplained at this point, since the target sequences are much more dissimilar than other sequences that were perfectly differentiated in the hybridization.
DISCUSSION
The method presented here is a cost-effective and rapid complement to sequencing-dependent design and production of synthetic oligonucleotide probes that target variable regions in the 16S rRNA of bacteria. The general approach is based on a PCR-based amplification of a variable sequence motif in the 16S rDNA (Fig. 2) carried out in two steps that ultimately result in a single-stranded fragment complementary to the variable region (Fig. 3). To achieve sufficient hybridization specificity, the flanking universal primers need to be removed after each amplification step. This is accomplished by using modified primers carrying a biotin linked to the 5′ terminus and a 3′ ribonucleotide residue. The PCR products can then be bound to streptavidin-coated beads before the primer is detached from the product by alkaline hydrolysis of the ribonucleotide (Fig. 3 and 4).
The generation of population-specific probes by PCR has previously been attempted. Ludwig et al. (9) used a 148- to 358-nucleotide-long variable region within bacterial 23S rRNA flanked by conserved primers as hybridization probes, with good success. However, variable regions within the 16S rRNA are significantly shorter, making primer removal necessary. This was recently achieved by enzymatic removal of primers carrying a 3′ phosphorothioate followed by digestion of the remaining complementary overhang by mung bean nuclease (6). The resultant probe was a double-stranded polynucleotide that was used to identify 16S rDNA in temperature gradient gel electrophoresis gels for microbial community analysis. Unfortunately, this procedure did not appear to generate probes of uniform size, and the hybridization efficiency is likely impaired by the presence of a complementary strand that competes with the target. Thus, we propose the protocols presented here as a generally applicable improvement to generation of hybridization probes via PCR.
The probing region identified in the present study represents one of the most variable stem structures in the 16S rRNA (http://rrna.uia.ac.be/) (22) and is thus a suitable target for identification of bacterial populations. Nonetheless, there is no single region within the 16S rRNA that will differentiate all closely related sequences (18). This is evident in our analysis of several isolates of Pseudomonas, Roseobacter, Prochlorococcus, and Synechococcus, which display on the order of 0.4 to 4.2% sequence variation along the entire 16S rRNA but are not distinguishable in the probing region. Thus, if population-level identification of a specific microorganism were required, it would be prudent, as in any other hybridization approach, to use more than one probe. This can be accomplished by using additional PCR-generated probes, since we have recently identified at least two suitable regions within the 23S rRNA (unpublished).
Several steps within the probe generation protocol were optimized to ensure reproducible production of high-quality, uniform oligonucleotides. First, it is important to generate a single-stranded product by removing the complementary strand from the PCR product after the first amplification prior to hydrolysis of the ribonucleotide (Fig. 2B). This is necessary to avoid regeneration of the priming region in subsequent amplification when the strand shortened by the detachment of the primer can act as a primer for the complementary strand. The removal is efficiently achieved by washing the bead-bound PCR product for 1 min at 85°C. Washes at higher temperature or at 37°C in presence of 50 to 100% formamide as recommended by the manufacturer resulted in the loss of most bound product, probably due to dissociation of the biotin-streptavidin bond. Second, to maximize the final yield of probe after the second linear amplification, it is desirable to have a nearly complete incorporation of biotinylated primer into products, since free primers may compete with PCR product for available streptavidin binding sites on the beads. It is possible to compensate for this by adding a larger amount of beads, but this will increase costs for production of probes substantially.
The PCR-generated probes displayed melting behavior typical of oligonucleotides (Fig. 5B). However, the Td of PCR-generated probes were 5°C higher than those of gel-purified synthetic probes of identical sequence (Fig. 5A). This may be due to the lower purity of the synthetic probes (Fig. 5B), but the difference could also be due to some unknown structural differences between synthetic and PCR-generated nucleic acids. Polz and Cavanaugh (15) and McMahon et al. (11) observed a similar difference in Td between native and in vitro-transcribed 16S rRNAs, but the observations also remained unexplained. The substantial length heterogeneity of the probing region (Fig. 1) may result in Td variability between different probes. This may complicate the use of the PCR-generated probes in modern chip technologies, and further development of the method may be required. One possible solution to this problem is to use other variable regions with less length heterogeneity for PCR production of probes.
In summary, the PCR-based probe synthesis outlined in this study provides the means to rapidly generate oligonucleotide probes for identification of bacteria at a low cost. The region identified in this study is not perfect in differentiation of all closely related bacteria, but the specificity of the analysis can readily be improved by combined use of similarly produced probes targeting other variable regions. The procedure can easily be adapted to a 96-well microplate format with only minor modifications, facilitating high-throughput production of probes. Both genomic DNAs from pure cultures and cloned 16S rDNA fragments can be used as templates for probe synthesis, and hence the method is suitable for rapid hybridization-based screening of samples. Other possible applications are the high-intensity labeling of fluorescent in situ hybridization probes by incorporation of fluorescent nucleotides during the PCR or the production of oligonucleotide probes for microarray-based identification of bacterial populations (3).
Acknowledgments
We thank Mary Ann Moran (University of Georgia), Veljo Kisand (University of Umeå), and William Burkholder (Massachusetts Institute of Technology) for providing bacterial strains.
This work was supported by the K&A Wallenberg Foundation (postdoctoral fellowship to S.B.) and by grants from the NSF to C.M.C. and M.F.P.
REFERENCES
- 1.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashelford, K. E., A. J. Weightman, and J. C. Fry. 2002.. PRIMROSE: a computer program for generating, and estimating the phylogenetic range of 16S rRNA oligonucleotide probes and primers in conjunction with the RDP-II database. Nucleic Acids Res. 30:3481-3489. [DOI] [PMC free article] [PubMed]
- 3.Guschin, D. Y., B. K. Mobarry, D. Proudnikov, D. A. Stahl, B. E. Rittman, and A. D. Mirzabekov. 1997. Oligonucleotide microchips as genosensors for determinative and environmental studies in microbiology. Appl. Environ. Microbiol. 63:2397-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gutell, R. R., J. J. Cannone, Z. Shang, Y. Du, and M. J. Serra. 2000. A story: unpaired adenosine bases in ribosomal RNA. J. Mol. Biol. 304:335-354. [DOI] [PubMed] [Google Scholar]
- 5.Head, I. M., J. R. Saunders, and R. W. Pickup. 1998. Microbial evolution, diversity and ecology; a decade of ribosomal RNA analysis of uncultivated microorganisms. Microb. Ecol. 35:1-21. [DOI] [PubMed] [Google Scholar]
- 6.Heuer, H., K. Hartung, G. Wieland, I. Kramer, and K. Smalla. 1999. Polynucelotide probes that target a hypervariable region of 16S rRNA genes to identify bacterial isolates corresponding to bands of community fingerprints. Appl. Environ. Microbiol. 65:1045-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kisand, V., R. Cuadros, and J. Wikner. 2002. Phylogeny of culturable estuarine bacteria catabolizing riverine organic matter in the northern Baltic Sea. Appl. Environ. Microbiol. 68:379-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lane, D. L., B. Pace, G. J. Olsen, D. A. Stahl, M. L. Sogin, and N. R. Pace. 1985. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. USA 82:6955-6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ludwig, W., S. Dorn, N. Springer, G. Kirchhof, and K.-H. Schleifer. 1994. PCR-based preparation of 23S rRNA-targeted group-specific polynucleotide probes. Appl. Environ. Microbiol. 60:3236-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McMahon, K. D., D. A. Stahl, and L. Raskin. 1998. A comparison of the use of in vitro-transcribed and native rRNA for the quantification of microorganisms in the environment. Microb. Ecol. 36:362-371. [DOI] [PubMed] [Google Scholar]
- 12.Moore, L. R., A. F. Post, G. Rocap, and S. W. Chisholm. 2002.. Utilization of different nitrogen sources by the marine cyanobacteria, Prochlorococcus and Synechococcus. Limnol. Oceanogr. 44:628-638.
- 13.Nelson D. L., and M. M. Cox. 2000. Lehninger principles of biochemistry, 3rd ed. Worth Publishers, New York, N.Y.
- 14.Pace, N. R., D. A. Stahl, D. J. Lane, and G. J. Olsen. 1986. The analysis of natural microbial populations by ribosomal RNA sequences. Adv. Microb. Ecol. 9:1-55. [Google Scholar]
- 15.Polz, M. F., and C. M. Cavanaugh. 1997. A simple method for quantification of uncultured microorganisms in the environment based on in vitro transcription of 16S rRNA. Appl. Environ. Microbiol. 63:1028-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 17.Silveira, M. H., and L. E. Orgel. 1995. PCR with detachable primers. Nucleic Acids Res. 23:1083-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stackebrandt, E., and B. M. Goebel. 1994. A place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 19.Stahl, D. A. 1995. Application of phylogenetically based hybridization probes to microbial ecology. Mol. Ecol. 4:535-542. [Google Scholar]
- 20.Stahl, D. A., and R. Amann. 1991. Development and application of nucleic acid probes, p. 205-248. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, New York, N.Y.
- 21.Suzuki, M. T., and E. F. Delong. 2002. Marine prokaryotic diversity, p. 209-234. In J. T. Staley and A.-L. Reysenbach (ed.), Biodiversity of microbial life. Wiley Liss, New York, N.Y.
- 22.Van de Peer, Y., S. Chapelle, and R. Wachter. 1996. A quantitative map of nucleotide substitution rates in bacterial rRNA. Nucleic Acids Res. 24:3381-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]