Abstract
Multiresistant Shiga toxin-producing Escherichia coli (STEC) O118:H16 and O118 nonmotile strains (designated O118:[H16]) were detected by examination of 171 STEC isolates for their antimicrobial sensitivity. Of 48 STEC O118:[H16] strains, 98% were resistant to sulfonamide, 96% were resistant to streptomycin, 79% were resistant to kanamycin, 75% were resistant to tetracycline, 67% were resistant to ampicillin, 60% were resistant to chloramphenicol, 48% were resistant to trimethoprim, and 10% each were resistant to gentamicin and nalidixic acid. Nalidixic acid resistance and reduced susceptibility to ciprofloxacin were associated with the mutation gyrALEU-83. The STEC O118:[H16] strains were found to belong to a single genetic clone as investigated by multilocus enzyme electrophoresis and by multilocus sequence analysis of E. coli housekeeping genes. The STEC O118:[H16] strains originated from humans and cattle and were isolated in seven different countries of Europe between 1986 and 1999. Strains showing multiresistance to up to eight different antimicrobials predominated among the more recent STEC O118:[H16] strains. The genes in parentheses were associated with resistance to kanamycin (aphA1-Ia), chloramphenicol (catA1), tetracycline [tet(A)], and ampicillin (blaTEM-1). Class 1 integrons containing sulI (sulfonamide resistance), aadA1a (streptomycin resistance), or dfrA1 (trimethoprim resistance)-aadA1a gene cassettes were detected in 28 strains. The blaTEM-1b gene was present in 18 of 21 strains that were examined by nucleotide sequencing. Class 1 integrons and blaTEM genes were localized on plasmids and/or on the chromosome in different STEC O118:[H16] strains. Hybridization of XbaI-digested chromosomal DNA separated by pulsed-field gel electrophoresis revealed that blaTEM genes were integrated at different positions in the chromosome of STEC O118:[H16] strains that could have occurred by Tn2 insertion. Our data suggest that strains belonging to the STEC O118:[H16] clonal group have a characteristic propensity for acquisition and maintenance of resistance determinants, thus contrasting to STEC belonging to other serotypes.
Escherichia coli O157 and certain other types of Shiga toxin-producing E. coli (STEC) are pathogenic for humans and can cause bloody diarrhea and hemolytic-uremic syndrome (HUS). Most of the human pathogenic STEC strains belong to a small number of O serogroups and carry the genes for production of Shiga toxins (stx1 and/or stx2), for intimin (eae), and for enterohemorrhagic E. coli (EHEC) hemolysin (19).
In Germany, STEC belonging to O groups 157, 26, 103, 111, 118, and 145 were most frequently isolated from children with severe diarrhea and HUS (6). Some of these strains are also known as pathogens in calves (35, 36). Both STEC O118:H16 and O118:nonmotile (NM) (designated as STEC O118:[H16]) were most prevalent among STEC strains, which were isolated from diarrheic calves in Germany between 1986 and 1996 (35). STEC O118:[H16] strains from cattle and humans were found to be identical for their virulence markers and other traits, and two cases of transmission of STEC O118 from cattle to humans are described elsewhere (4, 34). The first cases of human infections with STEC O118:[H16] reported from Germany occurred in 1996 (6). Between 1996 and 1998, 23 patients infected with STEC O118 were identified in our laboratory. All cases were sporadic, most of the patients were children, and most of them were living in a rural environment (4).
By routine examination, we found that almost all STEC O118:[H16] strains from Germany were similar in the pattern of resistance to a number of antimicrobial drugs. We have therefore investigated 52 different motile and nonmotile STEC O118 strains from cattle and humans that were originally isolated between 1986 and 1999 in seven different European countries. Our results show that the STEC O118:[H16] strains belong to a distinct clonal group and that the isolates from different European countries are characterized by an increase of antimicrobial resistance that was observed over the the period of sampling.
MATERIALS AND METHODS
Bacteria.
Forty-one STEC O118:H16 strains, seven STEC O118:NM strains, and four STEC O118:H12 strains were investigated, together with 119 STEC strains belonging to non-O118 serogroups. The sources and relevant properties of STEC isolates are described elsewhere (4, 6, 35). Of the 48 STEC O118:[H16] strains, 32 were from Germany, and the remaining 16 strains were obtained from Austria (Franz Allerberger, Universität Innsbruck), Belgium (Denis Piérard, Vrije Universiteit Brussel, and Jacques Mainil, Université de Liège), Denmark (Flemming Scheutz, Statens Seruminstitut, Copenhagen, Denmark), Italy (Alfredo Caprioli, Istituto Superiore di Sanità, Rome, Italy), Spain (Jorge Blanco, Universidad de Santiago de Compostela), and Switzerland (Louis Corboz, Universität Zürich).
Detection of STEC virulence markers.
All strains were investigated for production of Shiga toxins (Stx) and for stx-specific DNA sequences, as well as for their hemolytic phenotypes and for the EHEC hly gene as described previously (4). Determination of the presence of the eae gene and genetic subtyping of eae variants were performed by PCR and restriction endonuclease digestion of PCR products as described previously (22).
Antimicrobial susceptibility tests.
Antimicrobial susceptibility testing of bacteria was done by the disk diffusion method by using commercial disks (Oxoid, Wesel, Germany), according to the guidelines of the National Committee for Clinical Laboratory Standards (NCCLS) (20). Adjusted inocula of bacteria (ca. 5 × 104 CFU/ml) were inoculated on Mueller-Hinton agar (Oxoid) and incubated for 18 h at 35°C. Strains were considered resistant or sensitive by measuring the diameter of the growth inhibition zone, and interpretation of the results was done as recommended by the NCCLS (20). The antimicrobial agents tested, the loads of the disks, and sizes of growth inhibition of resistant strains, respectively, were as follows: sulfonamide (SUL; 300 μg, ≤12 mm), streptomycin (STR; 10 μg, ≤13 mm), kanamycin (KAN; 30 μg, ≤13 mm), tetracycline (TET; 30 μg, ≤14 mm), ampicillin (AMP; 10 μg, ≤13 mm), chloramphenicol (CHL; 30 μg, ≤12 mm), trimethoprim (TMP; 5 μg, ≤10 mm), gentamicin (GEN; 10 μg, ≤12 mm), and nalidixic acid (NAL; 30 μg, ≤13 mm). The STEC O118 strains were additionally investigated for the MICs of NAL and ciprofloxacin (CIP) by broth microdilution assay (21). The MIC was defined as the lowest concentration producing no visible growth. The breakpoints used to consider a strain as resistant were those recommended by the NCCLS (21) (NAL resistance, MIC ≥ 32 μg/ml; CIP resistance, MIC ≥ 4 μg/ml). In all tests, the susceptible strain E. coli ATCC 25922 was included for quality control.
Multilocus enzyme electrophoresis.
To analyze enzyme electrophoretic variation, bacterial cell lysates were prepared and subjected to multilocus enzyme electrophoresis (28). Seventeen enzymes were examined for allelic variation as described previously (33). For each enzyme, electromorphs were determined through comparisions to standard mobility variants and assigned numbers by their rate of anodal migration. Isolates that lacked detectable enzyme activity were designated as a null allele at the locus in question. To estimate the genetic relationship among isolates, electromorphs were equated with alleles at the corresponding enzyme locus, so that each bacterial strain was characterized by its multilocus genotype (allele combination) for the enzyme-encoding loci assayed. Distinctive multilocus genotypes were designated electrophoretic types (ETs) and were numbered by their inferred relationships from phylogenetic analysis (33).
Multilocus sequencing methods.
Oligonucleotide primers designed to amplify internal fragments for 13 housekeeping genes were used in multilocus sequence analysis (C. L. Tarr, T. M. Large, A. C. Bumbaugh, D. W. Lacher, and T. S. Whittam, unpublished data). Six of these genes were shown to be useful for identifying clonal frames in a previous study of pathogenic E. coli (24). The housekeeping genes include arcA, aroE, aspC, clpX, cyaA, dnaG, fadD, grpE, icd, lysP, mdh, mtlD, and rpoS (23). Amplification reactions consisted of 5 μl of 10× PCR buffer, 8 μl of deoxynucleoside triphosphate mix, 0.5 μl of each primer, 0.5 μl of Boehringer Mannheim Taq polymerase, and 34.5 μl of dH2O (23). PCR was performed for 35 cycles under the following conditions: 1 min of denaturation at 92°C, 1 min of primer annealing at 57°C, and 15 s of extension at 72°C with an initial denaturing step of 94°C for 10 min. Amplicons were purified by using the Qiagen QiaQuick PCR purification kit. Cycle sequencing reactions (6 μl of CEQ mix, 1 μl of 20 μM primer and 3 μl of PCR product) were performed for 30 cycles under the following conditions: 96°C for 20 s, 50°C for 20 s, and 60°C for 4 min, with an initial denaturing step of 94°C for 1 min. Products were purified by using Sephadex columns and dried under vacuum centrifugation at room temperature. The samples were then rehydrated in 40 μl of formamide and sequenced by using a Beckman CEQ L automated sequencer. Data were analyzed by using the CEQ software and exported for analysis by using the SeqMan module of the DNAStar Lasergene software. Consensus sequences were aligned with CLUSTALX and the output files were modified for use in MEGA2 (14).
PCR for detection of antimicrobial resistance determinants.
The detection of drug resistance genes (Table 1) was performed by PCR amplication and sequencing as described above. PCRs were performed with 5 μl of boiled bacterial cells (ca. 5 × 106 bacteria), or bacterial DNA (1 ng) in a 50-μl reaction mixture (1 U of Amplitaq Gold Polymerase; Perkin-Elmer Applied Biosystems, Weiterstadt, Germany; 5 μl of 10× buffer; 200 μM deoxynucleoside triphosphate; 1.25 mM MgCl2; 5% dimethyl sulfoxide; 0.5 μM concentrations of each primer). DNA amplification was carried out in a GenAmp PCR system 9700 (PE Applied Biosystems, Foster City, Calif.). The PCR conditions were as follows: an initial hot start cycle at 94°C for 10 min, followed by 35 cycles of denaturation of 94°C for 1 min, primer annealing for 30 s at 55 to 65°C, and primer extension for 1 min at 72°C. A final incubation at 72°C for 10 min was performed after the last cycle. The PCR primers and the corresponding annealing temperatures are listed in Table 1. Detection of class 1 integron and gene cassettes located therein was performed according to conditions described elsewhere (11, 15). The specificity of the PCR products was determined by comparing the size of DNA fragments obtained by digestion with appropriate restriction endonucleases with the expected size deduced from the nucleotide sequence deposited in the GenBank (Table 1) and as described by Guerra et al. (11).
TABLE 1.
Primers used for the detection of integrons and resistance determinants
| Target genea (resistance) | Primer | Nucleotide sequence (5′-3′) | Reference | Ta (°C)b | Accession no. | Product size (bp) |
|---|---|---|---|---|---|---|
| Integronc | 5′CS | GGC ATC CAA GCA GCA AG | 15 | 55 | AY007807 | Variable |
| 3′CS | AAG CAG ACT TGA CCT GA | |||||
| Integrase | intI1-F | CCT CCC GCA CGA TGA TC | 4 | 58.5 | AF071413 | 280 |
| intI1-R | TCC ACG CAT CGT CAG GC | |||||
| blaTEM (AMP) | TEM-9F | CAT TTC CGT GTC GCC CTT ATT CC | This study | 59 | J01749 | 828 |
| TEM-9R | GGC ACC TAT CTC AGC GAT CTG TCT A | |||||
| catA1 (CHL) | cat F1-F | AAG TTG GCA GCA TTC ACC CG | This study | 61 | J01841 | 573 |
| cat B2-R | TCG TGG TAT TCA CTC CAG AGC G | |||||
| tet(A) (TET) | tet(A)-F | GGT CTT GCT CGT CTC GCT GG | This study | 62 | X61367 | 690 |
| tet(A)-R | AAC GCC ATC CAT CCC CGT G | |||||
| aadA1 (STR) | aadA1a-F | AAC GAC CTT TTG GAA ACT TCG G | This study | 60 | X12870 | 352 |
| aadA1a-R | TTC GCT CAT CGC CAG CCC AG | |||||
| strA (STR) | strA-F | CCA ATC GCA GAT AGA AGG CAA G | This study | 65 | AJ249779 | 580 |
| strA-R | ATC AAC TGG CAG GAG GAA CAG G | |||||
| aphA1-Ia (KAN) | aph(3′)I-F | AAC GTC TTG CTC GAG GCC GCG | 25 | 65 | J01839 | 670 |
| aph(3′)I-R | GGC AAG ATC CTG GTA TCG GTC TGC | |||||
| aadB (GEN) | aadB-Fd | GGG CGC GTC ATG GAG GAG TT | 25 | 65 | X04555 | 329 |
| aadB-Rd | TAT CGC GAC CTG AAA GCG GC | |||||
| sulI (SUL) | sul1-F | TGG TGA CGG TGT TCG GCA TTC | 17 | 62 | U49101 | 790 |
| sul1-R | GCG AAG GTT TCC GAG AAG GTG | |||||
| QRDR gyrA | gyrA-P1 | TGT CCG AGA TGG CCT GAA GC | -e | 58 | U28377 | 374 |
| gyrA-P3 | TGC CGT CAT AGT TAT CAA CGA | |||||
| QRDR parC | parC-3 | CCG TGC GTT GCC GTT TAT TG | -e | 58 | U28377 | 368 |
| parC-4 | AAGTGCCGTCGAAGTTTGGCA |
That is, it confers resistance to AMP, CHL, TET, STR, KAN, GEN, SUL, or it is the QRDR (quinolone resistance-determining region).
Ta, annealing temperature.
The dfrA1 gene was identified by nucleotide sequencing and restriction enzyme digestion of PCR products generated with primers 5′CS and 3′CS.
ant(2")I-F and ant(2")I-R (25).
Unpublished data (B. Malorny, Federal Institute for Health Production of Consumer and Veterinary Medicine, Berlin, Germany).
Nucleotide sequencing of the PCR products.
The generated PCR products of blaTEM-1, gyrA, and parC genes and class 1 integrons were purified with the QIAquick kit (Qiagen, Hilden, Germany). Sequencing reactions were carried out by using the Big Dye terminator cycle sequencing ready reaction kit (PE Applied Biosystems) according to the instructions given by the manufacturer. Automated nucleotide sequencing was performed with an ABI 377 Prism DNA Sequencer (PE Applied Biosystems) and documentation of sequence results was obtained by using the program AutoAssembler (PE Applied Biosystems). Sequences obtained were compared to those registered in the GenBank.
DNA hybridization analysis.
Plasmid DNA was prepared as described previously (12). Conjugational transfer of resistance plasmids was performed as described by using the plasmid-free E. coli K-12 JC3272 as recipient (3). Plasmid DNA was separated on 0.7% agarose gels for Southern blotting. Preparation of samples for pulsed-field gel electrophoresis (PFGE) and Southern hybridizations of blotted PFGE gels were performed as described previously (5). The 828-bp PCR fragment generated by primers TEM-9F1/R1 (Table 1) from plasmid pBR322 was used as blaTEM-specific DNA probe and the 1.6-kb fragment generated by primers 5′CS/3′CS (Table 1) from STEC O118:[H16] strain CB6525 was taken as the integron-specific DNA probe (15, 29; the present work). PCR probes were labeled with digoxigenin-11-dUTP (Roche Applied Science, Mannheim, Germany) as described and Southern hybridizations were conducted under conditions of high stringency (6).
RESULTS
A multiresistant clone of STEC O118:[H16] strains occurs in both humans and cattle.
The investigation of STEC strains which were isolated from patients in Germany between 1996 and 1999 revealed that 19 of 26 (73.1%) of STEC O118:[H16] isolates were resistant to AMP, CHL, KAN, STR, SUL, and TET. In contrast, only a few of 119 examined non-O118 STEC strains that were isolated at the same time period showed resistance to one or more of these antibiotics, and the resistant strains were not associated with a particular serotype (Table 2). Human infections with multiresistant STEC O118:[H16] strains were observed over a period of several years. Because the cases were not epidemiologically linked (4), we became interested in the origin of the multiresistant STEC O118 strains. We have therefore investigated 41 STEC O118:H16, seven O118:NM, and four O118:H12 strains that were isolated in Austria (n = 1), Belgium (n = 11), Denmark (n = 1), Germany (n = 35), Italy (n = 1), Spain (n = 2), and Switzerland (n = 1) between 1986 and 1999.
TABLE 2.
Antimicrobial drug resistance in 48 STEC O118:[H16] ET-A and in 119 non-O118 STEC strainsa
| Antimicrobial drug | No. of resistant strains (%)
|
|
|---|---|---|
| STEC O118:[H16] ET-A (n = 48) | STEC non-O118a (n = 119) | |
| SUL | 47 (97.9) | 18 (15.1) |
| STR | 46 (95.8) | 18 (15.1) |
| KAN | 38 (79.2) | 3 (2.5) |
| TET | 36 (75.0) | 13 (10.9) |
| AMP | 32 (66.7) | 4 (3.4) |
| CHL | 29 (60.4) | 2 (1.7) |
| TMP | 23 (47.9) | 6 (5.0) |
| GEN | 5 (10.4) | 0 (<0.8) |
| NAL | 5 (10.4) | 0 (<0.8) |
Belonging to O serogroups O5 (n = 1), O18 (n = 1), O26 (n = 23), O76 (n = 3), O78 (n = 1), O84 (n = 2), O91 (n = 5), O103 (n = 6), O111 (n = 3), O113 (n = 3), O128 (n = 5), O157 (n = 33), O145 (n = 6), O146 (n = 6), O163 (n = 1), Orough (n = 18), and OX178 (n = 2).
By multilocus enzyme electrophoresis, the 52 STEC O118 strains were divided into two ETs (ET-A and ET-B), showing mobility differences in 7 of 17 employed enzymes. A total of 48 O118:[H16] strains from humans (n = 28) and cattle (n = 20) belonged to the ET-A clone. All ET-A strains were positive for stx1 and for the intimin gene eaeβ1 (22). Three (6.25%) of the 48 ET-A strains carried an stx2 gene and 44 (91.7%) were positive for plasmid-encoded EHEC hemolysin (EHEC-hlyA). Four strains belonging to the ET-B clone (O118:H12) were isolates from humans in Germany and were positive for the stx2d-OUNT variant gene but negative for eae and EHEC-hlyA (4; data not shown).
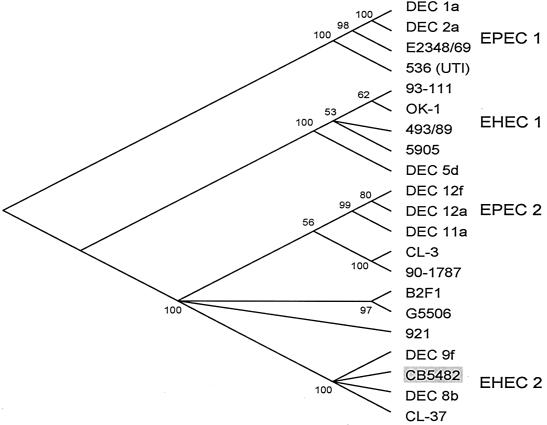
To determine the genetic relatedness of the ET-A O118:[H16] clone to other pathogenic groups E. coli associated with diarrheal disease, we determined the nucleotide sequences for coding regions of 13 chromosomal loci encoding proteins with housekeeping functions. Comparison of the multilocus sequence data to the reference strain 93-111 (serotype O157:H7, clonal group EHEC 1), DEC 8b (O111:H8, EHEC 2), and B2F1 (O91:H21, STEC) indicated that the ET-A O118:[H16] strain CB5482 was most closely related to the EHEC 2 clonal group. The percentage of nucleotide difference in the total of 6,720 bp was 2.04 ± 0.18 (strain 93-111), 0.03 ± 0.02 (DEC 8b), and 0.40 ± 0.08 (B2F1). Comparison of the differences per 100 synonymous sites gave 6.65 ± 0.62 (strain 93-111, EHEC 1), 0.11 ± 0.08 (DEC 8b), and 1.35 ± 0.28 (B2F1). Thus, CB5482 has the lowest pairwise genetic distance to DEC 8b of the EHEC 2 group and is most distant from EHEC O157:H7. A phylogenetic analysis with these sequences shows that CB5482 belongs to the EHEC 2 group (Fig. 1), which also contains STEC strains with serotypes O26:H11 and O111:H8. Bootstrap confidence limits indicate that 100% of 1,000 randomly generated phylogenies place CB5482 in the EHEC 2 complex.
FIG. 1.
Clonal phylogeny inferred from distances at synonymous sites (dS) in gene sequences in 13 concatenated loci and inferred by the neighbor-joining algorithm. This is a consensus tree with the numbers at each node representing the percentage of boostrapped trees in which the node was observed. In addition to the O118:[H16] strain (CB5482), the pathogenic strains included in the figure are described by Reid et al. (23).
Among STEC O118 strains, resistance to the tested antimicrobials was only found in O118:[H16] ET-A strains (Table 2). Only one of the 48 STEC O118:[H16] ET-A strains and all four STEC O118:H12 ET-B strains were susceptible to all antimicrobials. STEC O118:[H16] ET-A strains showing resistance to six or more of the drugs predominated among the isolates collected after 1994. The oldest isolates showing antibiotic resistance were from 1986 (STR and SUL), 1987 (KAN and TMP), 1988 (AMP), 1989 (CHL, GEN, and TET), and 1992 (NAL). Isolates from humans and cattle were not different from each other with regard to their resistance patterns and genes (Table 3).
TABLE 3.
Antibiotic resistance phenotypes and genes in multidrug-resistant STEC O118:[H16] ET-A strains from humans and cattle in Europe
| Resistance patterna (no. of strains) | Resistance gene(s) (no. of strains carrying a given gene) | No. of isolates (country, yr of isolation)b
|
|
|---|---|---|---|
| Human | Cattle | ||
| STR-SUL (5) | strA | 1 (DK, 1999) | 4 (B, 1986-1987) |
| STR-SUL-TET (1) | aadA1a, sulI, tetA | 1 (A, 1997) | |
| STR-SUL-TMP-KAN (5) | strA, dfrA1-aadA1a, sulI, aphA1-1a | 5 (B, 1987) | |
| STR-SUL-KAN-TET-NAL (1) | strA, aadA1a, sulI, aphA1-Ia, tetA, gyrALEU-83 | 1 (I, 1992) | |
| SUL-TMP-TET-GEN (1) | dfrA1-aadA1a, sulI | 1 (D, 1997) | |
| STR-SUL-TMP-KAN-TET-CHL (1) | dfrA1-aadA1a, sulI, aphA1-Ia, tetA, catA1 | 1 (D, 1997) | |
| STR-SUL-TET-CHL-GEN (1) | strA, aadA1a, sulI, catA1, aadB | 1 (D, 1989) | |
| STR-SUL-KAN-TET-AMP (4) | strA, aphA1-Ia, tetA, blaTEM-1 | 3 (D, 1999) | 1 (D, 1997) |
| STR-SUL-TMP-KAN-TET-AMP (1) | strA, dfrA1-aadA1a, sulI, aphA1-IA, tetA, blaTEM-1 | 1 (CH, 1988) | |
| STR-SUL-TMP-KAN-TET-AMP-CHL-GEN (3) | strA, dfrA1-aadA1a, sulI, aphA1-Ia, tetA, blaTEM-1, catA1, aadB (1) | 3 (D, 1994 and 1998) | |
| STR-SUL-KAN-TET-AMP-CHL-GEN (1) | strA, sulI, aphAI-Ia, tetA, blaTEM-1, catA1 | 1 (D, 1997) | |
| STR-SUL-TMP-AMP-CHL-NAL (1) | strA, dfrA1-aadAla, sulI, blaTEM-1, gyrALEU-83 | 1 (D, 1996) | |
| STR-SUL-KAN-TET-AMP-CHL (11)c | strA, aadA1a (4), sulI (4), aphA1-Ia, tetA, blaTEM-1, catA1, gyrALEU-83 (3) | 8 (D, 1996-1999; B, 1991; SP, 1998) | 3 (D, 1994-1998) |
| STR-SUL-TMP-KAN-TET-AMP-CHL (11) | strA, dfrA1-aadA1a (9), sulI (9), aphA1-Ia, tetA, blaTEM-1, catAI | 9 (D, 1996-1998; SP, 1996) | 2 (D, 1994 and 1996) |
The relation gene and/or antimicrobial resistance is given in Table 1, except for dfrA1, which is implicated in TMP resistance.
The country and year of isolation are indicated in parentheses: B, Belgium; D, Germany; I, Italy; CH, Switzerland; A, Austria; SP, Spain; DK, Denmark.
Three strains were resistant to NAL and carried a gyrALEU-83 gene.
Genes associated with antibiotic resistance in STEC O118:[H16] ET-A strains.
The following genes were identified in STEC O118:[H16] ET-A strains resistant to the following: KAN (aphA1-Ia, 38 of 38 strains), CHL (catA1, 28 of 29 strains), TET [tet(A), 34 of 36 strains], and AMP (blaTEM, 32 of 32 strains). A strA gene was found in 43 of the 46 STR-resistant strains and an aadB gene in two of the five strains that were resistant to GEN. A sulI gene was found in 29 of the 47 strains resistant to SUL. The three STR-resistant strains which were negative for the strA gene carried aadA1a genes, which are described as associated with integron structures (7).
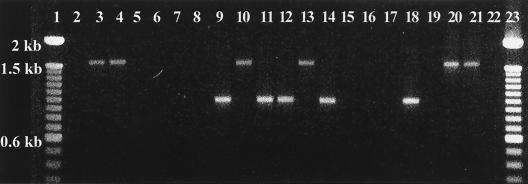
We have therefore investigated the STEC O118:[H16] ET-A strains for the presence of class 1 integrons with the specific primers 5′CS/3′CS (15), and 28 of the 48 ET-A strains were found positive. Twenty-one strains generated amplification products of 1.6 kb that contained dfrA1 and aadA1a gene cassettes (dfrA1-aadA1a), and the other seven strains generated PCR products of ca. 1.0 kb, which contained only the aadA1a gene cassette (Fig. 2). The 28 strains carrying the class 1 integron also carried the sulI gene. The other strain positive for sulI (CB8153) was positive for the integrase-specific PCR primers (intI1-F/R) but carried no aadA1a or dfrA1 gene (Fig. 2).
FIG. 2.
Amplification of class 1 integrons from STEC O118:[H16] ET-A strains with primers 5′CS/3′CS (Table 1). Nineteen representative integron-positive and -negative strains from humans and cattle are shown. Lanes 1 and 23, 100-bp ladder; lane 2, CB7503 (negative); lane 3, CB7512 (dfrA1-aadA1a, 1.6 kb); lane 4, CB7882 (dfrA1-aadA1a, 1.6 kb); lane 5, CB8153 (negative with primers 5′CS/3′CS, positive for integrase and sulI); lane 6, CB8220 (negative); lane 7, CB8255 (negative); lane 8, CB8271 (negative); lane 9, E-D143 (aadA1a, 1.0 kb); lane 10, VTH28 (dfrA1-aadA1a, 1.6 kb); lane 11, VTH62 (aadA1a, 1.0 kb); lane 12, T17968 (aadA1a, 1.0 kb); lane 13, EC970130 (dfrA1-aadA1a, 1.6 kb); lane 14, 666/89 (aadA1a, 1.0 kb); lane 15, CB6069 (O118: H12, ET-B, negative); lane 16, 1874-99 (negative); lane 17, EH78 (negative); lane 18, RW2017 (aadA1a, 1.0 kb); lane 19, RW2030 (negative); lane 20, RW2173 (dfrA1-aadA1a, 1.6 kb); lane 21, RW2266 (dfrA1-aadA1a, 1.6 kb); lane 22, negative PCR control.
Five strains showed resistance to NAL (MIC of >128 μg/ml) and reduced susceptibility to CIP (MICs of 0.5 and 0.25 μg/ml; one and four strains, respectively). Nucleotide sequence analysis of the quinolone resistance-determining region of the gyrA and parC genes of these strains showed a single mutation (Ser→Leu) in the codon 83 of the gyrA gene (gyrALEU-83).
STEC O118:[H16] ET-A strains are similar in their blaTEM gene sequences.
blaTEM genes are found in a number of different genetic variants in E. coli and other Enterobacteriaceae (10). We used this polymorphism to investigate the relationships between AMP-resistant STEC O118:[H16] strains by nucleotide sequencing of an 828-bp stretch of the amplified blaTEM gene in 21 STEC O118:[H16] ET-A strains that were isolated in Germany (n = 17), Belgium (n = 1), Switzerland (n = 1), and Spain (n = 2) (Table 4). Of the 21 STEC O118:[H16] strains, 18 were identical in that their blaTEM genes all showed mutations at positions 436 (C→T) and 604 (G→T). According to the updated blaTEM gene nomenclature the mutated genes are designated blaTEM-1b (10). One strain (CB6525) showed only a base substitution at position 436 (C→T) corresponding to the blaTEM-1c derivative and two other strains (CB7099 and VTH62) carried the original blaTEM-1a sequence (10) (Table 4).
TABLE 4.
blaTEM genotypes of 21 STEC O118:[H16] ET-A strains and location of blaTEM genesa
| Strain | Source (origin, isolation date [yr]) | blaTEM gene | blaTEM gene location (size [kb]) | Position in Fig. 3 |
|---|---|---|---|---|
| EC970130 | c (CH, 1988) | blaTEM-1b | pl | |
| EH78 | h (B, 1991) | blaTEM-1b | pl | |
| VTH28 | h (SP, 1996) | blaTEM-1b | chr (ND) | |
| CB5482 | h (D, 1996) | blaTEM-1b | chr (459) | Lane 7, panels A and B |
| CB6175 | h (D, 1996) | blaTEM-1b | chr (459) | Lane 8, panels A and B; lane 9, panels C and D |
| CB7109 | c (D, 1996) | blaTEM-1b | chr (476) | Lane 8, panels C and D |
| CB6236 | h (D, 1996) | blaTEM-1b | chr (459) | Lane 6, panels A and B |
| CB6365 | h (D, 1996) | blaTEM-1b | pl and chr (331) | Lane 3, panels A and B; lane 1, panels C and D |
| CB6525 | h (D, 1996) | blaTEM-1c | pl and chr (105) | Lane 4, panels C and D |
| CB6585 | h (D, 1996) | blaTEM-1b | chr (459) | Lane 5, panels A and B; lane 5, panels C and D |
| CB6586 | h (D, 1996) | blaTEM-1b | chr (502) | Lane 4, panels A and B; lane 7, panels C and D |
| CB6981 | c (D, 1996) | blaTEM-1b | chr (502) | Lane 6, panels C and D |
| CB6980 | c (D, 1997) | blaTEM-1b | chr (ND) | |
| CB6888 | h (D, 1997) | blaTEM-1b | pl | |
| CB7014 | h (D, 1997) | blaTEM-1b | chr (476) | Lane 3, panels C and D |
| CB7035 | h (D, 1997) | blaTEM-1b | chr (47) | Lane 2, panels C and D |
| CB7099 | h (D, 1997) | blaTEM-1a | chr (ND) | |
| VTH62 | h (SP, 1998) | blaTEM-1a | chr (ND) | |
| CB7451 | h (D, 1998) | blaTEM-1b | chr (ND) | |
| CB7727 | h (D, 1998) | blaTEM-1b | chr (ND) | |
| CB7834 | h (D, 1998) | blaTEM-1b | chr (ND) |
pl, plasmid > 93 kb; chr, chromosome; ND, not done; c, cattle; h, human. In parentheses are shown the sizes of the XbaI DNA fragments hybridizing with the blaTEM gene probe.
Apart from the STEC O118:[H16] ET-A strains, only four STEC O26:H11 strains from our STEC collection were positive for blaTEM genes (Table 2). We examined the blaTEM genotypes of these strains by nucleotide sequence analysis. In contrast to the STEC O118 strains, all of these strains were found to be positive for the blaTEM-1a gene (data not shown), which suggests that blaTEM-1a has been acquired at least twice in the divergence of the EHEC 2 group.
Genetic location of class 1 integrons and blaTEM genes in STEC O118:[H16] strains.
The genetic location of class 1 integrons was investigated in all 28 positive STEC O118:[H16] strains by Southern blot hybridization with a gene probe covering the dfrA1-aadA1a genes. Plasmid-encoded class 1 integrons were detected in 14 of the 28 strains. Further search for transferrable antimicrobial resistance revealed conjugative plasmids carrying blaTEM genes that encoded AMP resistance in the recipient strains. blaTEM genes harbored on large plasmids (>93 kb) were detected in 15 of the 32 AMP-resistant, blaTEM-positive STEC O118:[H16] strains by DNA hybridization. Plasmids carrying both blaTEM genes and class 1 integrons were found in four strains (data not shown).
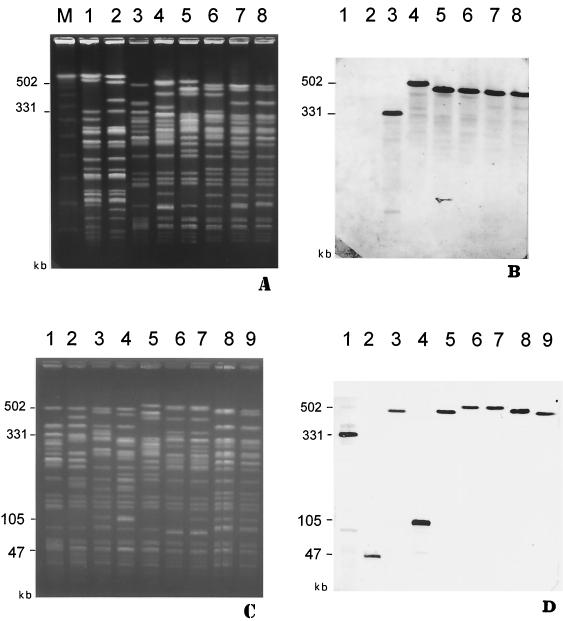
Of 32 blaTEM-positive strains, 17 hybridized with the blaTEM probe only with chromosomal DNA. The similarity between strains with chromosomally inherited blaTEM genes was further investigated by determining the size of the hybridizing XbaI fragments from chromosomal DNA. XbaI fragments obtained from 11 strains were separated by PFGE, and the gels were blotted onto nylon membranes. With all 11 strains, only one XbaI fragment hybridized with the blaTEM gene probe. The probe hybridizing fragments were of different sizes: 502 kb (two strains), 476 kb (two strains), 459 kb (four strains), 331 kb (one strain), 105 kb (one strain), and 47 kb (one strain) (Fig. 3A and C and Table 4).
FIG. 3.
(A) PFGE analysis of XbaI-digested total DNA of STEC O118:[H16] ET-A strains. Lanes: M, molecular weight standard (lambda concatemers; Bio-Rad Laboratories); 1, CB6591 (O118:H12, blaTEM-negative); 2, CB6069 (O118:H12, blaTEM negative); 3, CB6365 (O118:H16, blaTEM-1b); 4, CB6586 (O118:NM, blaTEM-1b); 5, CB6585 (O118:H16, blaTEM-1b); 6, CB6236 (O118:H16, blaTEM-1b); 7, CB5482 (O118:H16, blaTEM-1b); 8, CB6175 (O118:H16, blaTEM-1b). The positions of some XbaI fragments are indicated in kilobases at the left side of the gel. (B) Hybridization of DNA from strains shown in panel A with the blaTEM-specific gene probe. (C) PFGE analysis of XbaI-digested total DNA of STEC O118:[H16] ET-A strains. Lanes: 1, CB6365 (O118:H16, blaTEM-1b); 2, CB7035 (O118:H16, blaTEM-1b); 3, CB7014 (O118:H16, blaTEM-1b); 4, CB6525 (O118:H16, blaTEM-1c); 5, CB6585 (O118:H16, blaTEM-1b); 6, CB6981 (O118:NM, blaTEM-1b); 7, CB6586 (O118:H16, blaTEM-1b); 8, CB7109 (O118:H16, blaTEM-1b); 9, CB6175 (O118:H16, blaTEM-1b). The positions of some XbaI fragments are indicated in kilobases at the left side of the gel. (D) Hybridization of DNA from strains shown in panel A with the blaTEM-specific gene probe.
One pair of human (CB6586) and cattle (CB6981) strains that were isolated from a case of transmission between animals and humans (4) shared blaTEM probe hybridizing fragments of the same size (502 kb) (Table 4 and Fig. 3C and D). Another pair of STEC O118:[H16] strains, which were isolated on a farm from a calf and from a human, had identical XbaI profiles except for the blaTEM probe hybridizing fragment, which was 459 kb in the human (CB6175) and 476 kb in the calf isolate (CB7109) (Table 4 and Fig. 3C and D). The remaining seven STEC O118:[H16] strains were from single human cases and showed heterogeneous XbaI patterns that were to some extent associated with size differences in the blaTEM probe hybridizing fragments (Table 4 and Fig. 3).
DISCUSSION
STEC O118:[H16] strains have been identified as important agents of diarrhea in calves in Belgium and Germany (16, 35) and have been increasingly isolated from humans with diarrhea and HUS in Germany beginning in 1996 (4, 6). Human infections with STEC O118 are frequently associated with rural environments and multiple cases of transmission from cattle to humans have been described (4). These findings suggest that cattle are the major source of human infections with this organism.
By analysis of serotypes, virulence markers, and macrorestriction patterns of genomic DNA, STEC O118 were divided into three groups which were represented by three serotypes, O118:[H16], O118:H12, and O118:H30 (34). Multilocus enzyme electrophoresis revealed that STEC O118:[H16] and STEC O118:H12 strains represent two genetically different clonal types, ET-A and ET-B, which differ at 6 of 17 enzyme loci assayed.
The STEC O118:[H16] ET-A clone is a member of the EHEC 2 clone complex based on multilocus sequence analysis. Thus, the clone marked by ET-A is most closely related to STEC clones with serotypes O26:H11 and O111:H8 (33). The fact that all STEC O118:[H16] ET-A strains were positive for intimin β is consistent with other members (O26:H11 and RDEC) of the EHEC 2 group.
Resistance to two or more classes of antimicrobials was found as a characteristic trait in 47 (97.9%) of 48 STEC O118:[H16] ET-A strains that were collected from humans and cattle in Germany and in some other European countries between 1996 and 1999. In contrast, antimicrobial resistance was less frequently (15.1%) expressed by STEC belonging to other serotypes that were isolated at the same time period, and the resistant strains were not associated with a particular serotype. Similar findings were made in studies performed in the United States (13.3% of STEC were resistant) and Japan (8.7% of STEC were resistant) (13, 27).
The origin of the strains and the differences found in their XbaI PFGE patterns indicate that most of STEC O118:[H16] ET-A strains from our study were epidemiologically unrelated to each other; however, similar antimicrobial resistance patterns were found in strains of different origin and source (4; this work). Analysis of genes encoding resistance to KAN, CHL, TET, and AMP revealed a remarkable homology between STEC O118:[H16] strains showing resistance to these compounds. This finding could indicate that the resistant strains are clonally related, having originated once from a common ancestor with these resistance determinants. An alternative hypothesis is that resistance has evolved multiple times in the STEC O118:[H16] ET-A clone because members of the clone have a particular propensity for acquiring and incorporating resistance (R) genes. R genes could have been aquired by uptake of R plasmids, which were found to be present in many of the STEC O118:[H16] ET-A strains. In a further step, the R genes could have been incorporated into the chromosome of the bacteria as was found to be the case with many strains for class 1 integron-associated R genes (dfrA1, aadA1a, and sulI) and for the blaTEM genes. A chromosomal location of blaTEM genes was only rarely described in strains of E. coli (31) and is therefore a remarkable feature of STEC O118:[H16] ET-A strains. blaTEM genes were reported to be associated with different transposons, including Tn2 (10). By using a PCR with one primer specific for tnpR of Tn2 in combination with a blaTEM-1-specific primer we have found that transposon-specific sequences were closely linked to chromosomally and plasmid-encoded blaTEM-1 genes in 12 of the STEC O118:[H16] ET-A strains (data not shown). It was reported that Tn2 can insert at multiple sites in the bacterial chromosome (30) and the heterogeneous hybridization patterns that were obtained with the blaTEM gene probe and XbaI-digested genomic DNA of STEC O118:[H16] ET-A strains indicate that the blaTEM genes are integrated at multiple sites in the chromosome of these strains.
The finding that strains showing resistance to eight different antimicrobial agents predominated among the most recently isolated STEC O118:[H16] strains supports the hypothesis that STEC O118:[H16] ET-A strains have a tendency to acquire and accumulate R determinants in nature. The presence of resistance to NAL and reduced susceptibility to CIP in 9.4% of the strains is a cause of concern. Additional amino acid changes within the quinolone resistance-determining region of the gyrase (gyrA and gyrB) and the topoisomerase IV (parC) genes could give rise to fluoroquinolone resistance (18, 32). In E. coli this resistance is generally associated with mutations affecting the amino codon Ser83 or Asp87 of gyrA in a first step and with Ser80 of parC in a secondary step (18, 32). The five STEC O118:[H16] ET-A strains of our series presented a single point mutation in gyrA, which conferred the reduced susceptibility to ciprofloxacin, and thus we could consider these strains as progenitors for the evolution of full resistance.
Resistance properties can provide a selective advantage if the bacteria are frequently exposed to antimicrobial substances, which is often the case in livestock on farms. Indeed, multiresistant STEC O118:[H16] strains were isolated from cattle housed at different farms in Belgium, Germany, and Switzerland. For more than four decades, it has been a common practice on farms to use antimicrobial agents for disease prevention and growth promotion of animals. The widespread use of antimicrobial agents would select for resistance and may have promoted the increasing frequency of STEC O118:[H16] ET-A strains in the bovines (16, 35). As a consequence, humans became more likely to be exposed to these organisms via the food chain and by direct and indirect transmission from cattle, and this could explain why infections with this pathogen have increased particularly in the rural population (4). It has been shown that STEC can survive for long time periods in fecally contaminated soil and water (1, 8, 9). Further dissemination of STEC O118 to farmland and to noninfected cattle could have occurred by irrigation of soils with contaminated surface water and by direct and indirect transmission of bacteria to noninfected animals.
Our findings show that certain types of STEC, such as STEC O118:[H16] ET-A strains, are well adapted to survive under antibiotic pressure. A study performed at the University Hospital in Dusseldorf, Dusseldorf, Germany, has revealed that class 1 integron-carrying E. coli and other Enterobacteriaceae have increased from 4.7 to 17.4% between 1993 and 1999 (26). Furthermore, our own investigations show that 12.5% of a series of 300 German E. coli strains, belonging to different serotypes, isolated between 1999 and 2001 from animal sources (cattle, avian, and swine), carried class 1 integrons. At least five different gene cassette arrays were found within these integrons. Among them, dfrA1-aadA1a and aadA1a were the most frequently encountered (unpublished data). Integron-associated antimicrobial resistance was also found in other STEC belonging to different serotypes, including O157:H7, which were isolated from humans, animals, and food in the United States (37). Although antimicrobial therapy is generally not recommended for treatment of STEC infections in humans, the indirect selection for multiresistant strains will contribute to the increase of emerging, antimicrobial-resistant pathogens, such as STEC O118:[H16], and facilitate the spread of these mobile resistance elements to other bacteria. An increased surveillance and the development of adequate prevention strategies are needed for public health reasons.
Acknowledgments
We acknowledge the technical assistance provided by E. Junker (Federal Institute for Health Production of Consumers and Veterinary Medicine, NRL-Salm, Berlin, Germany). We are grateful to Franz Allerberger (Universität Innsbruck, Innsbruck, Austria), Jorge Blanco (Universidad de Santiago de Compostela, Santiago de Compostela, Spain), Alfredo Caprioli (Istituto Superiore di Sanità, Rome, Italy), Louis Corboz (Universität Zürich, Zurich, Switzerland), Jacques Mainil (Université de Liège, Liège, Belgium), Denis Piérard (Vrije Universiteit Brussel, Brussels, Belgium), Flemming Scheutz (Statens Seruminstitut, Copenhagen, Denmark), and Lothar Wieler (Freie Universität Berlin, Berlin, Germany) for supplying E. coli O118 strains used in the present study.
This work was supported by the German Federal Ministry of Health, by funds from the European Commission project “Attaching and Effacing Escherichia coli Infections” (QLK2-2000-00600), and by the Federal Institute for Health Production of Consumers and Veterinary Medicine, Berlin, Germany (BgVV F501-28). Work in the Whittam Laboratory was supported by the Enteric Pathogens Research Unit (N01-AI-65299) at the University of Maryland Medical School.
REFERENCES
- 1.Barker, J., T. J. Humphrey, and M. W. Brown. 1999. Survival of Escherichia coli O157 in a soil protozoan: implications for disease. FEMS Microbiol. Lett. 173:291-295. [DOI] [PubMed] [Google Scholar]
- 2.Bass, L., C. A. Liebert, M. D. Lee, A. O. Summers, D. G. White, S. G. Thayer, and J. J. Maurer. 1999. Incidence and characterization of integrons, genetic elements mediating multiple-drug resistance, in avian Escherichia coli. Antimicrob. Agents Chemother. 43:2925-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beutin, L., and M. Achtman. 1979. Two Escherichia coli chromosomal cistrons, sfrA and sfrB, which are needed for expression of F factor tra functions. J. Bacteriol. 139:730-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beutin, L., M. Bulte, A. Weber, S. Zimmermann, and K. Gleier. 2000. Investigation of human infections with verocytotoxin-producing strains of Escherichia coli (VTEC) belonging to serogroup O118 with evidence for zoonotic transmission. Epidemiol. Infect. 125:47-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beutin, L., D. Geier, S. Zimmermann, S. Aleksic, H. A. Gillespie, and T. S. Whittam. 1997. Epidemiological relatedness and clonal types of natural populations of Escherichia coli strains producing Shiga toxins in separate populations of cattle and sheep. Appl. Environ. Microbiol. 63:2175-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beutin, L., S. Zimmermann, and K. Gleier. 1998. Human infections with Shiga toxin-producing Escherichia coli other than serogroup O157 in Germany. Emerg. Infect. Dis. 4:635-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fluit, A. C., and F. J. Schmitz. 1999. Class 1 integrons, gene cassettes, mobility, and epidemiology. Eur. J. Clin. Microbiol. Infect. Dis. 18:761-770. [DOI] [PubMed] [Google Scholar]
- 8.Fukushima, H., K. Hoshina, and M. Gomyoda. 1999. Long-term survival of Shiga toxin-producing Escherichia coli O26, O111, and O157 in bovine feces. Appl. Environ. Microbiol. 65:5177-5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gagliardi, J. V., and J. S. Karns. 2000. Leaching of Escherichia coli O157:H7 in diverse soils under various agricultural management practices. Appl. Environ. Microbiol. 66:877-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goussard, S., and P. Courvalin. 1999. Updated sequence information for TEM β-lactamase genes. Antimicrob. Agents Chemother. 43:367-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guerra, B., S. Soto, S. Cal, and M. C. Mendoza. 2000. Antimicrobial resistance and spread of class 1 integrons among Salmonella serotypes. Antimicrob. Agents Chemother. 44:2166-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kado, C. I., and S. T. Liu. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobayashi, H., J. Shimada, M. Nakazawa, T. Morozumi, T. Pohjanvirta, S. Pelkonen, and K. Yamamoto. 2001. Prevalence and characteristics of Shiga toxin-producing Escherichia coli from healthy cattle in Japan. Appl. Environ. Microbiol. 67:484-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2000. MEGA 2: molecular evolutionary genetics analysis software. Arizona State University, Tempe, Ariz.
- 15.Levesque, C., L. Piche, C. Larose, and P. H. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mainil, J. 1999. Shiga/verocytotoxins and Shiga/verotoxigenic Escherichia coli in animals. Vet. Res. 30:235-257. [PubMed] [Google Scholar]
- 17.Mazel, D., B. Dychinco, V. A. Webb, and J. Davies. 2000. Antibiotic resistance in the ECOR collection: integrons and identification of a novel aad gene. Antimicrob. Agents Chemother. 44:1568-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDonald, L. C., F. J. Chen, H. J. Lo, H. C. Yin, P. L. Lu, C. H. Huang, P. Chen, T. L. Lauderdale, and M. Ho. 2001. Emergence of reduced susceptibility and resistance to fluoroquinolones in Escherichia coli in Taiwan and contributions of distinct selective pressures. Antimicrob. Agents Chemother. 45:3084-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.National Committee for Clinical Laboratory Standards. 2000. Performance standards for antimicrobial disk susceptibility tests. Approved standard, 7th ed. NCCLS M2-A7. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 21.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard, 5th ed. NCCLS M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 22.Oswald, E., H. Schmidt, S. Morabito, H. Karch, O. Marches, and A. Caprioli. 2000. Typing of intimin genes in human and animal enterohemorrhagic and enteropathogenic Escherichia coli: characterization of a new intimin variant. Infect. Immun. 68:64-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reid, S. D., C. J. Herbelin, A. C. Bumbaugh, R. K. Selander, and T. S. Whittam. 1999. MULTILOCUS sequence typing (MLST) of pathogenic Escherichia coli. [Online.] http://www.shigatox.net/stec/mlst.
- 24.Reid, S. D., C. J. Herbelin, A. C. Bumbaugh, R. K. Selander, and T. S. Whittam. 2000. Parallel evolution of virulence in pathogenic Escherichia coli. Nature 406:64-67. [DOI] [PubMed] [Google Scholar]
- 25.Sandvang, D., and F. M. Aarestrup. 2000. Characterization of aminoglycoside resistance genes and class 1 integrons in porcine and bovine gentamicin-resistant Escherichia coli. Microb. Drug Resist. 6:19-27. [DOI] [PubMed] [Google Scholar]
- 26.Schmitz, F. J., D. Hafner, R. Geisel, P. Follmann, C. Kirschke, J. Verhoef, K. Kohrer, and A. C. Fluit. 2001. Increased prevalence of class I integrons in Escherichia coli, Klebsiella species, and Enterobacter species isolates over a 7-year period in a German university hospital. J. Clin. Microbiol. 39:3724-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schroeder, C. M., C. Zhao, C. DebRoy, J. Torcolini, S. Zhao, D. G. White, D. D. Wagner, P. F. McDermott, R. D. Walker, and J. Meng. 2002. Antimicrobial resistance of Escherichia coli O157 isolated from humans, cattle, swine, and food. Appl. Environ. Microbiol. 68:576-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Selander, R. K., D. A. Caugant, H. Ochman, J. M. Musser, M. N. Gilmour, and T. S. Whittam. 1986. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl. Environ. Microbiol. 51:873-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sutcliffe, J. G. 1978. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc. Natl. Acad. Sci. USA 75:3737-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, A., X. Dai, and D. Lu. 1980. The transposition properties of Tn2 in Escherichia coli. Cell 21:251-255. [DOI] [PubMed] [Google Scholar]
- 31.Weber, D. A., C. C. Sanders, J. S. Bakken, and J. P. Quinn. 1990. A novel chromosomal TEM derivative and alterations in outer membrane proteins together mediate selective ceftazidime resistance in Escherichia coli. J. Infect. Dis. 162:460-465. [DOI] [PubMed] [Google Scholar]
- 32.White, D. G., L. J. Piddock, J. J. Maurer, S. Zhao, V. Ricci, and S. G. Thayer. 2000. Characterization of fluoroquinolone resistance among veterinary isolates of avian Escherichia coli. Antimicrob. Agents Chemother. 44:2897-2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whittam, T. S., M. L. Wolfe, I. K. Wachsmuth, F. Orskov, I. Orskov, and R. A. Wilson. 1993. Clonal relationships among Escherichia coli strains that cause hemorrhagic colitis and infantile diarrhea. Infect. Immun. 61:1619-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wieler, L. H., B. Busse, H. Steinruck, L. Beutin, A. Weber, H. Karch, and G. Baljer. 2000. Enterohemorrhagic Escherichia coli (EHEC) strains of serogroup O118 display three distinctive clonal groups of EHEC pathogens. J. Clin. Microbiol. 38:2162-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wieler, L. H., A. Schwanitz, E. Vieler, B. Busse, H. Steinruck, J. B. Kaper, and G. Baljer. 1998. Virulence properties of Shiga toxin-producing Escherichia coli (STEC) strains of serogroup O118, a major group of STEC pathogens in calves. J. Clin. Microbiol. 36:1604-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wieler, L. H., E. Vieler, C. Erpenstein, T. Schlapp, H. Steinruck, R. Bauerfeind, A. Byomi, and G. Baljer. 1996. Shiga toxin-producing Escherichia coli strains from bovines: association of adhesion with carriage of eae and other genes. J. Clin. Microbiol. 34:2980-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao, S., D. G. White, B. Ge, S. Ayers, S. Friedman, L. English, D. Wagner, S. Gaines, and J. Meng. 2001. Identification and characterization of integron-mediated antibiotic resistance among Shiga toxin-producing Escherichia coli isolates. Appl. Environ. Microbiol. 67:1558-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]