Abstract
Sakacin G is a 37-amino-acid-residue-long class IIa bacteriocin produced by Lactobacillus sake 2512, which is encoded by the duplicated structural genes skgA1 and skgA2. Sakacin G appears to be unique and seems to be an intermediate between pediocin-like bacteriocins, according to its double-disulfide bridges required for antimicrobial activity, and mesentericin-like bacteriocins in terms of sequence homologies, inhibition spectrum, and specific activity.
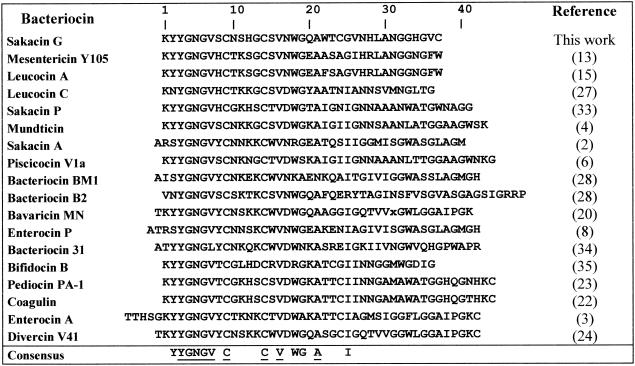
Many of the lactic acid bacteria are capable of inhibiting growth of some gram-positive bacteria, such as pathogenic species like Listeria monocytogenes, by secreting antimicrobial compounds, including peptides and proteins, also called bacteriocins. Among antibacterial peptides, lantibiotics, characterized in their primary structure by the presence of modified amino acid residues, have been widely studied and are used in many countries as preservatives in food products (29). Another group of peptides that do not bear any modified amino acid residue form a subclass called antilisterial bacteriocins, also known as class IIa bacteriocins (11) or cystibiotics (18). These peptides share strong structural homologies in their N-terminal domain (Fig. 1) with the presence of one disulfide bond. Their C-terminal domain is highly more variable. However, some of these bacteriocins, such as pediocin PA-1 (23), enterocin A (3), coagulin (22), and divercin V41 (24), are characterized by the presence of a second disulfide bond in the C-terminal region (Fig. 1). Moreover, in a previous work (14), we showed that these double-disulfide-bond bacteriocins exhibited higher specific activity than those with only two cysteine residues: i.e., mesentericin Y105 (13), sakacin A (2), and sakacin P (33).
FIG. 1.
Sequence alignment of class IIa bacteriocins. Leucocin A is identical to lecucocin B (12). Piscicocin V1a is identical to piscicolin 126 (19), sakacin A is identical to curvacin A (32), carnobacteriocin BM1 is identical to piscicocin V1b (6), and pediocin PA-1 is identical to pediocin AcH (26) and pediocin SJ-1 (30). Bavaracin A (21) is probably identical to sakacin P. Residues are numbered according to the sequence of sakacin G. The consensus sequence includes residues conserved by at least 75%. The residues conserved by more than 90% are underlined.
A new method for detection of antilisterial bacterial strains was recently developed and applied to select several strains from the Rhodia Food collection that produce bacteriocins (31). One strain, Lactobacillus sake 2512, appeared to produce an unknown bacteriocin. In the present paper, we report on the purification and biochemical characterization of a new and structurally original class IIa bacteriocin, sakacin G. Characterization of part of the genetic elements required for the production of this antimicrobial peptide is also described. Peculiar features in the proteic and nucleic structures found are discussed.
Bacteriocin characterization and purification.
Antilisterial activity of Lactobacillus sake 2512 was detected by a new luminometry-based method (31). Briefly, luminescent target strains (Listeria and Enterococcus) were obtained by cloning bacterial luciferase genes. When these strains were exposed to a culture supernatant containing antilisterial activity, their light emission was rapidly suppressed, allowing rapid detection of bacteriocins. The bacteriocin purification was conducted by a three-step procedure developed previously (14). The fraction of the major 23-min peak, collected during the reverse-phase high-performance liquid chromatography final purification step, was active against Listeria ivanovi BUG 496. The molecule, analyzed by electrospray ionization-mass spectrometry as previously described (14), showed a molecular mass of 3,834.32 ± 0.31 Da. Database screening revealed that the molecular mass of none of the previously described bacteriocins from lactic acid bacteria corresponds to this molecular mass. The amount of sakacin G purified from 100 ml of culture supernatant was estimated by protein titration with the bicinchoninic acid protein assay kit (Sigma) to be 120 μg and corresponds to a yield of 55% of recovered activity, measured with the critical dilution assay previously described (13).
Specific activity of sakacin G toward L. ivanovi BUG 496 was determined by the method used previously for other class IIa bacteriocins (14). The MIC of the new bacteriocin was 50.10−3 μg/ml, comparable to the MICs of mesentericin Y105, and sakacins A and P (33.10−3, 65.10−3, and 65.10−3 μg/ml, respectively) and significantly higher than those of the double-disulfide-bond-containing bacteriocins pediocin PA-1, enterocin A, and divercin V41 (4.10−3, 10.10−3, and 1.4.10−3 μg/ml, respectively).
Finally, the activity of the sakacin G toward 19 bacterial strains was tested by the well diffusion method, and the results are shown in Table 1. The inhibition spectrum of this bacteriocin appeared to be narrow and limited to the L. sake and Pediococcus cerevisiae strains tested, as far as lactic acid bacteria are concerned. This peptide, like the class IIa bacteriocins, appeared to be active toward all Listeria and Enterococcus faecalis strains tested. Compared to other class IIa bacteriocins tested against the same target strains (14), sakacin G is not active toward Lactobacillus casei DSM 20011 and Enterococcus faecium ENSAIA 631, as demonstrated for mesentericin-like peptides (mesentericin Y105, sakacin A, and sakacin P) and contrary to pediocin-like bacteriocins (pediocin PA-1, enterocin A, and divercin V41).
TABLE 1.
Inhibition spectrum of culture supernatant of L. sake 2512 containing sakacin G
| Indicator strain | Inhibition zone diam (mm)a |
|---|---|
| Lactococcus lactis ATCC 11454 | 0 |
| Leuconostoc paramesenteroides DSM 20288 | 0 |
| Leuconostoc mesenteroides DSM 20484 | 0 |
| Leuconostoc mesenteroides DSM 20240 | 0 |
| Lactobacillus delbrueckii DSM 20081 | 0 |
| Lactobacillus plantarum DSM 20174 | 0 |
| Lactobacillus brevis DSM 20054 | 0 |
| Lactobacillus casei DSM 20011 | 0 |
| Lactobacillus sake 2515 | 1 |
| Pediococcus acidilactici ENSAIA 583 | 0 |
| Pediococcus cerevisiae IP 5492 | 1 |
| Enterococcus faecium ENSAIA 631 | 0 |
| Enterococcus faecalis IP 5430 | 2 |
| Enterococcus faecalis ENSAIA 636 | 1 |
| Enterococcus durans ENSAIA 630 | 2 |
| Listeria inocua 8811 | 3 |
| Listeria monocytogenes EGDe | 2 |
| Listeria monocytogenes LO28 | 2 |
| Listeria ivanovi BUG 496 | 6 |
Fifty microliters of a 550-AU/ml active culture supernatant was tested.
Structural characterization.
The first 34 amino acid residues of the N-terminal sequence of sakacin G were determined by Edman degradation (“Centre Commun d'Analyses du CNRS,” IBCP, Lyon, France). The sequence KYYGNGVSXNSHGXSVNWGQAWTXGVNHLANGGH was obtained.
The class IIa consensus sequence YGNGV showed that sakacin G belongs, as expected according to its inhibition spectrum, to class IIa bacteriocins (Fig. 1). Moreover, the three undetermined residues at positions 9, 14, and 24 could be attributed to three cysteines by homology with other class IIa bacteriocin sequences (Fig. 1). However, the calculated molecular mass of this 34-residue peptide with three cysteines was lower than the measured molecular weight of 3,834.32, indicating that this sequence was incomplete and convincing us to clone and sequence the sakacin G structural gene.
Cloning and sequencing.
The sequence of the locus involved in bacteriocin expression was analyzed by a reverse genetic strategy. The bacterial strains, genetic constructs, and oligonucleotide sequences used in this study are described in Table 2. First, the structural bacteriocin gene was amplified by PCR with two degenerated primers, SakG01 and SakG02, based on the N-terminal amino acid sequence of sakacin G. The amplification led to a 100-bp DNA fragment, which was cloned into the pGEM-T cloning vector, leading to pJMBYC01. The 560-bp PvuII fragment from pJMBYC01, including the cloned fragment, was used as a hybridization probe in Southern blot experiments to locate the structural gene on the L. sake2512 DNA (data not shown). From the plasmid preparation of L. sake2512, the probe revealed that a bacteriocin gene was carried by a plasmid with an approximate size of 35 kbp, named pJMB35. After restriction, the probe hybridized to a 2-kbp HindIII fragment and to a 9-kbp EcoRI fragment. The HindIII fragment was purified and cloned into the pZERO2 plasmid, generating pJMBYC02.
TABLE 2.
Plasmids and oligonucleotides used in this study
| Plasmid or oligonucleotide | Relevant characteristicsa | Source or reference |
|---|---|---|
| Plasmids | ||
| pGEM-T | Apr, PCR product cloning vector, 3 kb | Promega |
| pZErO-2 | Kanr, cloning vector, 3.3 kb | Invitrogen |
| pJMBYC01 | pGEM-T::100-bp skgA1 PCR product, 3.1 kb | This work |
| pJMBYC02 | pZErO-2::2.1-kb HindIII fragment of a plasmid from L. sake 2512, 5.4 kb | This work |
| Oligonucleotides | ||
| SakG01 | 5′ AAR TAT TAT GGN AAY GGN GT 3′ | |
| SakG02 | 5′ ACA TGA TGN CCN CCR TTN GC 3′ |
Apr, ampicillin resistant; Kanr, kanamycin resistant.
The 2,047-bp cloned sequence was determined and analyzed. Analysis revealed two complete open reading frames (ORFs), skgA1 and skgA2, in addition to two truncated ORFs, skgI and skgD located on both sides of the cloned fragment (Fig. 2). The presumed skgA1, skgA2, and skgI genes were in the opposite orientation to skgD. Each ORF is preceded by a potential ribosome binding site (RBS).
FIG. 2.
Schematic representation of the sequenced 2.1-kbp HindIII fragment isolated from plasmid pJMB35. A size bar (0.5 kbp) is shown in the upper left corner.
Pre-sakacin G coding genes.
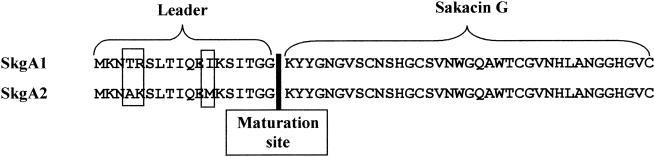
The skgA1 and skgA2 genes both code for a 55-amino-acid protein sharing an identical sequence (residues 19 to 55) (Fig. 3).
FIG. 3.
Sequence alignments and relevant features from the proteic sequences deduced from the skgA1 and skgA2 genes. Differences between the two sequences are framed.
Positions 1 to 18 in the sequences of the SkgA1 and SkgA2 proteins (Fig. 3) differed only by 3 residues and showed strong homologies with the sequences of class IIa bacteriocin peptide leaders, which are involved in peptide transport by specific ABC transporters (11). More particularly, the GG terminal unit (17, 18) is characteristic of these leader sequences and is the maturation site of these bacteriocins.
The sequence from positions 19 to 52 corresponds exactly to the amino acid sequence obtained by Edman degradation mentioned above. The hypothetical presence of three cysteines at positions 9, 14, and 24 was confirmed. Moreover, three additional residues of glycine, valine, and cysteine, respectively, appeared to complete the protein sequence at its C terminus. Thus, the difference of 4 Da between the calculated molecular mass of this peptide (3,838.20 Da) and the measured molecular mass (3,834.32 Da) first indicated the presence of two disulfide bonds within sakacin G, as was already described for other antilisterial bacteriocins and second confirmed the presence of the three additional amino acids that were not initially detected.
This sakacin G locus exhibits a specific feature, namely, the duplication of the sakacin G gene. This is the first example of a bacteriocin gene “duo” in the same operon. The mature sakacin G protein sequences deduced from the skgA1 and skgA2 genes are identical, whereas the leader sequences differ in three residues. Comparison of the nucleotide sequences of the skgA1 and skgA2 genes showed 95% homologies for mature bacteriocin sequences, whereas the leader sequence genes were only 78% identical. This indicates that conservation of the mature peptide sequence was more important than that of the leader and/or that the three amino acid positions modified in the leader are poorly involved in sakacin G maturation and secretion. Moreover, experiments (e.g., alternative skgA1 and skgA2 gene disruptions and/or fusion with two different reporter genes) must be conducted to determine whether both sakacin G structural genes are functional.
Role of disulfide bonds.
The role of disulfide bonds in antilisterial activity of class IIa bacteriocins was studied in various peptides (11). Particularly, in comparative studies (10, 14), it has been demonstrated that bacteriocins with two disulfide bonds are more efficient antimicrobial agents and have broader spectra of activity than those that possess a single disulfide bridge. Surprisingly, despite two disulfide bonds, sakacin G displays a low specific activity and a quite narrow spectrum of activity, close to that of mesentericin-like bacteriocins. However, it has been shown that the disulfide bond reduction dramatically reduced the activities of pediocin-like bacteriocins (5, 7, 10, 25), whereas reduction treatments of mesentericin-like bacteriocins (10, 15, 16, 28) resulted in only a moderate decrease in activity. According to the ambiguous status of sakacin G, reduction experiments with dithiothreitol (12.5 mg of dithiothreitol per ml added to a 0.5-mg/ml bacteriocin solution) were conducted with sakacin G, pediocin PA-1, and mesentericin Y105 (data not shown). The results showed, as expected, that disulfide bond reduction had no effect on mesentericin Y105 activity, even though the same treatment induced a dramatic loss (about 10 times) of antilisterial activity for pediocin PA-1 and, interestingly, for sakacin G.
Moreover, sequence comparison between sakacin G and other class IIa bacteriocins showed better identities with mesentericin-like bacteriocins—i.e., leucocin A (65%) and mesentericin Y105 (62%)—than with pediocin-like bacteriocins, such as pediocin PA-1 (52%).
Consequently, we propose that sakacin G belongs to a new subgroup of class IIa bacteriocins with two disulfide bonds, one of them (C terminal) required for antibacterial activity like that of pediocin-like peptides, but with primary sequences, sizes, homologies, and specific activities closer to those of mesentericin-like bacteriocins.
Sequence analysis of other genes.
The incomplete ORF skgI (Fig. 2) encodes a 54-residue protein. Comparison of this sequence with those in databases showed good homologies of the gene coding for SkgI with those coding for LccI (47%) and MesI (46%), which are the immunity proteins of leucocin A (15) and mesentericin Y105 (13), respectively. We can thus suggest that skgI encodes a sakacin G immunity protein.
The last incomplete gene, skgD (Fig. 2), encodes a 394-amino-acid protein. The position of the putative RBS points toward a TTG start codon, instead of a classical ATG. A search in protein databases revealed that SkgD shows great homologies with the following proteins of the ABC transporter family involved in bacteriocin secretion: pediocin PA-1, PedD and PapD (23, 26); sakacin P, SppT (17); sakacin A, SapT (1); and mesentericin Y105, MesD (13). The strongest homology was found with PlnG, potentially involved in expression of plantaricin A, a non-antilisterial class II bacteriocin (9). Thus, skgD may encode an ABC transporter specific for sakacin G.
New experiments to sequence the region of pJMB35 flanking the 2.1-kbp HindIII fragment would allow us to determine the complete sequences of skgI and skgD. This analysis could also show a downstream gene coding for SkgD, an accessory factor that is generally present near the ABC transporter-coding gene (11), with the exception of the gene coding for divercin V41, which contains a locus apparently devoid of the accessory factor gene (24). Subsequent cloning experiments with such operons in nonproducer strains would confirm that SkgI is the immunity protein to sakacin G and that SkgD is an ABC transporter dedicated to the secretion of sakacin G.
Nucleotide sequence accession number.
The 2,047-bp cloned sequence of sakacin G was determined and analyzed. This nucleotide sequence, corresponding to partial skg locus, has been deposited in the GenBank database under accession no. AF395533.
Acknowledgments
We thank Véronique Laffitte from Rhodia Food for providing bacterial strains, Anne Venisse for critical review of the manuscript, and George Hette for technical assistance.
REFERENCES
- 1.Axelsson, L., and A. Holck. 1995. The genes involved in production of and immunity to sakacin A, a bacteriocin from Lactobacillus sake Lb706. J. Bacteriol. 177:2125-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Axelsson, L., A. Holck, S.-E. Birkeland, T. Aukrust, and H. Blom. 1993. Cloning and nucleotide sequence of a gene from Lactobacillus sake Lb706 necessary for sakacin A production and immunity. Appl. Environ. Microbiol. 59:2868-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aymerich, T., H. Holo, L. S. Håvarstein, M. Hugas, M. Garriga, and I. F. Nes. 1996. Biochemical and genetic characterization of enterocin A from Enterococcus faecium, a new antilisterial bacteriocin in the pediocin family of bacteriocins. Appl. Environ. Microbiol. 62:1676-1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennik, M. H., B. Vanloo, R. Brasseur, L. G. Gorris, and E. J. Smid. 1998. A novel bacteriocin with a YGNGV motif from vegetable-associated Enterococcus mundtii: full characterization and interaction with target organisms. Biochim. Biophys. Acta 1373:47-58. [DOI] [PubMed] [Google Scholar]
- 5.Bhugaloo-Vial, P., J.-P. Douliez, D. Mollé, X. Dousset, P. Boyaval, and D. Marion. 1999. Delineation of key amino acid side chains and peptide domains for antimicrobial properties of divercin V41, a pediocin-like bacteriocin secreted by Carnobacterium divergens V41. Appl. Environ. Microbiol. 65:2895-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhugaloo-Vial, P., X. Dousset, A. Metivier, O. Sorokine, P. Anglade, P. Boyaval, and D. Marion. 1996. Purification and amino acid sequences of piscicocins V1a and V1b, two class IIa bacteriocins secreted by Carnobacterium piscicola V1 that display significantly different levels of specific inhibitory activity. Appl. Environ. Microbiol. 62:4410-4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chikindas, M. L., M. J. Garcia-Garcerá, A. J. M. Driessen, A. M. Ledeboer, J. Nissen-Meyer, I. F. Nes, T. Abee, W. N. Konings, and G. Venema. 1993. Pediocin PA-1, a bacteriocin from Pediococcus acidilactici PAC1.0, forms hydrophilic pores in the cytoplasmic membrane of target cells. Appl. Environ. Microbiol. 59:3577-3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cintas, L. M., P. Casaus, L. S. Hávarstein, P. E. Hernández, and I. F. Nes. 1997. Biochemical and genetic characterization of enterocin P, a novel sec-dependent bacteriocin from Enterococcus faecium P13 with a broad antimicrobial spectrum. Appl. Environ. Microbiol. 63:4321-4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diep, D. B., L. S. Hávarstein, and I. F. Nes. 1996. Characterization of the locus responsible for the bacteriocin production in Lactobacillus plantarum C11. J. Bacteriol. 178:4472-4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eijsink, V. G. H., M. Skeie, P. H. Middelhoven, M. B. Brurberg, and I. F. Nes. 1998. Comparative studies of class IIa bacteriocins of lactic acid bacteria. Appl. Environ. Microbiol. 64:3275-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ennahar, S., T. Sashihara, K. Sonomoto, and A. Ishizaki. 2000. Class IIa bacteriocins: biosynthesis, structure and activity. FEMS Microbiol. Rev. 24:85-106. [DOI] [PubMed] [Google Scholar]
- 12.Felix, J. V., M. A. Papathanasopoulos, A. A. Smith, A. von Holy, and J. W. Hastings. 1994. Characterization of leucocin B-Ta11a: a bacteriocin from Leuconostoc carnosum Ta11a isolated from meat. Curr. Microbiol. 29:207-212. [DOI] [PubMed] [Google Scholar]
- 13.Fremaux, C., Y. Hechard, and Y. Cenatiempo. 1995. Mesentericin Y105 gene clusters in Leuconostoc mesenteroides Y105. Microbiology 141:1637-1645. [DOI] [PubMed] [Google Scholar]
- 14.Guyonnet, D., C. Fremaux, Y. Cenatiempo, and J. M. Berjeaud. 2000. Method for rapid purification of class IIa bacteriocins and comparison of their activities. Appl. Environ. Microbiol. 66:1744-1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hastings, J. W., M. Sailer, K. Johnson, K. L. Roy, J. C. Vederas, and M. E. Stiles. 1991. Characterization of leucocin A-UAL 187 and cloning of the bacteriocin gene from Leuconostoc gelidum. J. Bacteriol. 173:7491-7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holck, A., L. Axelsson, S. E. Birkeland, T. Aukrust, and H. Blom. 1992. Purification and amino acid sequence of sakacin A, a bacteriocin from Lactobacillus sake Lb706. J. Gen. Microbiol. 138:2715-2720. [DOI] [PubMed] [Google Scholar]
- 17.Huhne, K., L. Axelsson, A. Holck, and L. Krockel. 1996. Analysis of the sakacin P gene cluster from Lactobacillus sake Lb674 and its expression in sakacin-negative Lb. sake strains. Microbiology 142:1437-1448. [DOI] [PubMed] [Google Scholar]
- 18.Jack, R. W., J. R. Tagg, and B. Ray. 1995. Bacteriocins of gram-positive bacteria. Microbiol. Rev. 59:171-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jack, R. W., J. Wan, J. Gordon, K. Harmark, B. E. Davidson, A. J. Hillier, R. E. H. Wettenhall, M. W. Hickey, and M. J. Coventry. 1996. Characterization of the chemical and antimicrobial properties of piscicolin 126, a bacteriocin produced by Carnobacterium piscicola JG126. Appl. Environ. Microbiol. 62:2897-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaiser, A. L., and T. J. Montville. 1996. Purification of the bacteriocin bavaricin MN and characterization of its mode of action against Listeria monocytogenes Scott A cells and lipid vesicles. Appl. Environ. Microbiol. 62:4529-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larsen, A. G., F. K. Vogensen, and J. Josephsen. 1993. Antimicrobial activity of lactic acid bacteria isolated from sour doughs: purification and characterization of bavaricin A, a bacteriocin produced by Lactobacillus bavaricus MI401. J. Appl. Bacteriol. 75:113-122. [DOI] [PubMed] [Google Scholar]
- 22.Le Marrec, C., B. Hyronimus, P. Bressollier, B. Verneuil, and M. C. Urdaci. 2000. Biochemical and genetic characterization of coagulin, a new antilisterial bacteriocin in the pediocin family of bacteriocins, produced by Bacillus coagulans I4. Appl. Environ. Microbiol. 66:5213-5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marugg, J. D., C. F. Gonzalez, B. S. Kunka, A. M. Ledeboer, M. J. Pucci, M. Y. Toonen, S. A. Walker, L. C. M. Zoetmulder, and P. A. Vandenbergh. 1992. Cloning, expression, and nucleotide sequence of genes involved in production of pediocin PA-1, and bacteriocin from Pediococcus acidilactici PAC1.0. Appl. Environ. Microbiol. 58:2360-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Metivier, A., M. F. Pilet, X. Dousset, O. Sorokine, P. Anglade, M. Zagorec, J. C. Piard, D. Marion, Y. Cenatiempo, and C. Fremaux. 1998. Divercin V41, a new bacteriocin with two disulphide bonds produced by Carnobacterium divergens V41: primary structure and genomic organization. Microbiology 144:2837-2844. [DOI] [PubMed] [Google Scholar]
- 25.Miller, K. W., R. Schamber, O. Osmanagaoglu, and B. Ray. 1998. Isolation and characterization of pediocin AcH chimeric protein mutants with altered bactericidal activity. Appl. Environ. Microbiol. 64:1997-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Motlagh, A., M. Bukhtiyarova, and B. Ray. 1994. Complete nucleotide sequence of pSMB 74, a plasmid encoding the production of pediocin AcH in Pediococcus acidilactici. Lett. Appl. Microbiol. 18:305-312. [DOI] [PubMed] [Google Scholar]
- 27.Papathanasopoulos, M. A., G. A. Dykes, A. M. Revol-Junelles, A. Delfour, A. von Holy, and J. W. Hastings. 1998. Sequence and structural relationships of leucocins A-, B- and C-TA33a from Leuconostoc mesenteroides TA33a. Microbiology 144:1343-1348. [DOI] [PubMed] [Google Scholar]
- 28.Quadri, L. E., M. Sailer, K. L. Roy, J. C. Vederas, and M. E. Stiles. 1994. Chemical and genetic characterization of bacteriocins produced by Carnobacterium piscicola LV17B. J. Biol. Chem. 269:12204-12211. [PubMed] [Google Scholar]
- 29.Schillinger, U., R. Geisen, and W. H. Holzapfel. 1996. Potential of antagonistic microorganisms and bacteriocins for the biological preservation of food. Trends Food Sci. Technol. 7:158-164. [Google Scholar]
- 30.Schved, F., A. Lalazar, P. Lindner, and B. J. Juven. 1994. Interaction of the bacteriocin produced by Pediococcus acidilactici SJ-1 with the cell envelope of Lactobacillus spp. Lett. Appl. Microbiol. 19:281-283. [Google Scholar]
- 31.Simon, L., C. Fremaux, Y. Cenatiempo, and J. M. Berjeaud. 2001. Luminescent method for the detection of antibacterial activities. Appl. Microbiol. Biotechnol. 57:757-763. [DOI] [PubMed] [Google Scholar]
- 32.Tichaczek, P. S., R. F. Vogel, and W. P. Hammes. 1993. Cloning and sequencing of curA encoding curvacin A, the bacteriocin produced by Lactobacillus curvatus LTH1174. Arch. Microbiol. 160:279-283. [DOI] [PubMed] [Google Scholar]
- 33.Tichaczek, P. S., R. F. Vogel, and W. P. Hammes. 1994. Cloning and sequencing of sakP encoding sakacin P, the bacteriocin produced by Lactobacillus sake LTH 673. Microbiology 140:361-367. [DOI] [PubMed] [Google Scholar]
- 34.Tomita, H., S. Fujimoto, K. Tanimoto, and Y. Ike. 1996. Cloning and genetic organization of the bacteriocin 31 determinant encoded on the Enterococcus faecalis pheromone-responsive conjugative plasmid pYI17. J. Bacteriol. 178:3585-3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yildirim, Z., D. K. Winters, and M. G. Johnson. 1999. Purification, amino acid sequence and mode of action of bifidocin B produced by Bifidobacterium bifidum NCFB 1454. J. Appl. Microbiol. 86:45-54. [DOI] [PubMed] [Google Scholar]