Abstract
The alkane hydroxylase systems of two Rhodococcus strains (NRRL B-16531 and Q15, isolated from different geographical locations) were characterized. Both organisms contained at least four alkane monooxygenase gene homologs (alkB1, alkB2, alkB3, and alkB4). In both strains, the alkB1 and alkB2 homologs were part of alk gene clusters, each encoding two rubredoxins (rubA1 and rubA2; rubA3 and rubA4), a putative TetR transcriptional regulatory protein (alkU1; alkU2), and, in the alkB1 cluster, a rubredoxin reductase (rubB). The alkB3 and alkB4 homologs were found as separate genes which were not part of alk gene clusters. Functional heterologous expression of some of the rhodococcal alk genes (alkB2, rubA2, and rubA4 [NRRL B-16531]; alkB2 and rubB [Q15]) was achieved in Escherichia coli and Pseudomonas expression systems. Pseudomonas recombinants containing rhodococcal alkB2 were able to mineralize and grow on C12 to C16 n-alkanes. All rhodococcal alkane monooxygenases possessed the highly conserved eight-histidine motif, including two apparent alkane monooxygenase signature motifs (LQRH[S/A]DHH and NYXEHYG[L/M]), and the six hydrophobic membrane-spanning regions found in all alkane monooxygenases related to the Pseudomonas putida GPo1 alkane monooxygenase. The presence of multiple alkane hydroxylases in the two rhodococcal strains is reminiscent of other multiple-degradative-enzyme systems reported in Rhodococcus.
Although many microorganisms are capable of degrading aliphatic hydrocarbons and are readily isolated from contaminated and noncontaminated sites, relatively little is known about the genetic characteristics of their alkane-degradative systems. Indeed, until recently, only the alkane-degradative genes of a small number of gram-negative bacteria, namely, Pseudomonas and Acinetobacter, have been described in detail. Of these, the alk system found in Pseudomonas putida GPo1, which degrades C5 to C12 n-alkanes, remains the most extensively characterized alkane hydroxylase system (44, 47). The initial terminal oxidation of the alkane substrate to a 1-alkanol is catalyzed by a three-component alkane hydroxylase complex consisting of a particulate nonheme integral-membrane alkane monooxygenase (AlkB) and two soluble proteins, rubredoxin (AlkG) and rubredoxin reductase (AlkT) (47). The P. putida alk genes are located in two different loci (alkBFGHJKL and alkST) on the OCT plasmid, separated by 10 kb of DNA (44). Five chromosomal genes (alkM, rubA, rubB, alkR, and xcpR) in at least three different loci are required for degradation of C12 to C18 alkanes in Acinetobacter sp. strain ADP1 (12, 30, 31). Similar to P. putida GPo1, the initial terminal alkane oxidation is also catalyzed by a three-component alkane hydroxylase system, which comprises an alkane monooxygenase (AlkM), rubredoxin (RubA), and rubredoxin reductase (RubB). More recently, Acinetobacter sp. strain M-1 was shown to possess two alkane monooxygenase genes (alkMa and alkMb), as well as single copies of rubA and rubB, located in three different loci (40).
Much less is known about the alkane-degradative systems of gram-positive bacteria. A putative alkane monooxygenase gene has been identified in the finished genome sequence of Mycobacterium tuberculosis H37Rv (6), while other alkB homologs were amplified from Rhodococcus erythropolis NRRL B-16531 and Prauserella rugosa NRRL B-2295 DNAs using highly degenerate primers (37). Using the same primers, a C6 to C8 alkane-inducible alkB-homolog was cloned from Nocardioides sp. strain CF8 (13). The M. tuberculosis and P. rugosa alkB homologs could be functionally expressed in an alkB knockout derivative of Pseudomonas fluorescens CHA0 and in P. putida GPo12(pGEc47ΔB) and were shown to oxidize alkanes ranging from C10 to C16 (36).
Rhodococcus and other closely related high-G+C, mycolic acid-containing actinomycetes, such as Mycobacterium, Corynebacterium, Gordona, and Nocardia, are increasingly recognized as ideal candidates for the biodegradation of hydrocarbons because of their ability to degrade a wide range of organic compounds (4), their hydrophobic cell surfaces, their production of biosurfactants, and their ubiquity and robustness in the environment (23, 48). Considerable interest is being devoted to using bacterial alkane oxidation systems as biocatalysts for the production of fine chemicals and pharmaceuticals (15, 18, 20, 23-25, 29).
In the present study, we describe the isolation and characterization of multiple alkane monooxygenase genes found in two rhodococci from different geographical locations, Rhodococcus sp. strain Q15 (51, 53), isolated from Lake Ontario sediment, and R. erythropolis strain NRRL B-16531 (ATCC 15960; formerly Corynebacterium hydrocarboclastus p-9) (17), isolated from petroleum-contaminated soil in Japan. Rhodococcus sp. strain Q15 degrades a broad range of aliphatics (C8 to C32 n-alkanes, branched alkanes, and a substituted cyclohexane) at temperatures ranging from 0 to 30°C and oxidizes alkanes by both the terminal and the subterminal oxidation pathways (51). R. erythropolis NRRL B-16531 degrades C6 to C36 n-alkanes (46) and was one of eight strains able to stereospecifically oxidize the alkyl side chain of cumene in a collection of 1,229 bacteria, yeasts, and fungi (15).
In both bacteria, four alkane hydroxylase gene homologs were found, two of which are parts of gene clusters containing rubredoxin and rubredoxin reductase genes. Functional heterologous expression of some of these genes was achieved. The alkB gene clusters of NRRL B-16531 and Q15 were initially cloned and characterized independently by a Swiss and a Canadian laboratory, respectively. Subsequent communication between the two groups revealed the similarity of their results, and consequently, the groups continued this research as a collaborative effort.
MATERIALS AND METHODS
Bacterial strains, plasmids, and general methods.
The bacterial strains and plasmids used or constructed in this study are listed in Table 1. Rhodococcus strains Q15 and Q15 NP (plasmid cured) were grown on trypticase soy agar at 28°C and maintained at 4°C. R. erythropolis NRRL B-16531 was grown on Luria broth (LB) medium at 30°C and maintained at 4°C. Plasmid pGEc47 carries all of the genes necessary to convert n-alkanes into the corresponding fatty acids (10). Escherichia coli and Pseudomonas strains containing deletion derivatives of pGEc47 (pGEc47ΔB [45], pGEc47ΔG [43], and pGEc47ΔT [42]) cannot grow on n-octane unless the deleted gene, or an equivalent gene from another organism, is supplied in trans on additional plasmids. P. fluorescens KOB2Δ1 is an alkB1 deletion derivative of P. fluorescens CHA0 which no longer grows on C12 to C16 n-alkanes. KOB2Δ1 can be complemented for growth on these alkanes by pCom8 derivatives containing alkB genes from other bacteria (36). Plasmid pCom8 is a broad-host-range vector based on pUCP25 and the P. putida GPo1 alkBp promoter (38). Plasmid pKKPalk is an E. coli expression vector with the same promoter (38).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics (genotype) | Reference or source |
|---|---|---|
| Strains | ||
| P. fluorescens KOB2Δ1 | alkB1 deletion; C12-C16 Alk−; C18-C28 Alk+ | 36 |
| R. erythropolis NRRL B-16531 | Wild type; C6-C36 Alk+ | 15, 17, 46 |
| Rhodococcus sp. strain Q15 | Wild type; C8-C32 Alk+ | 51, 53 |
| Rhodococcus sp. strain Q15 NP | Plasmid cured; C8-C32 Alk+ | 51 |
| P. putida GPo12(pGEc47ΔB) | Alk− (alkB BamHI deletion); Tetr | 36 |
| E. coli GEc137(pGEc47) | C6-C12 Alk+ (P. putida alkBFGHJKL alkST); Tetr | 10 |
| E. coli GEc137(pGEc47ΔB) | Alk− (alkB BamHI deletion); Tetr | 45 |
| E. coli GEc137(pGEc47ΔT) | Alk− (alkT HpaI-AccI deletion); Tetr | 47 |
| E. coli GEc137(pGEc47ΔG) | Alk− (alkG HindIII deletion); Tetr | 43 |
| Cloning and expression vectors | ||
| pBluescript II KS(+/−) | Apr | Stratagene |
| pGEM7-Zf(+) | Cloning vector; Apr | Promega |
| pZErO-2.1 | Cloning vector; Kmr; ccdB | Invitrogen |
| pKKPalk | Expression vector; alkB promoter; Apr | 38 |
| pCom8 | Expression vector; alkB promoter; gentamicinr; broad host range | 38 |
| Plasmids containing DNA from Rhodococcus sp. strain Q15 | ||
| pKS1 | 6,389-kb EcoRI-EcoRI Q15 NP alkB1+ genomic DNA fragment cloned in pBluescript II KS(+/−) EcoRI-EcoRI | This study |
| pKS2 | 4.1-kb BglII-BglII Q15 NP alkB2+ genomic DNA fragment cloned in pBluescript II KS(+/−) BamHI-BamHI | This study |
| pCom8Q15alkB1 | 1,179-bp EcoRI-SmaI fragment from pTQ15alkB1 cloned in pCom8/EcoRI/SmaI | This study |
| pCom8Q15alkB2 | 1,269-bp EcoRI-HindIII PCR fragment cloned in pCom8/EcoRI/HindIII | This study |
| pKKPalkQ15rubA1 | 203-bp EcoRI-BamHI PCR fragment cloned in pKKPalk/EcoRI/BamHI | This study |
| pKKPalkQ15rubA2 | 235-bp EcoRI-BamHI PCR fragment cloned in pKKPalk/EcoRI/BamHI | This study |
| pKKPalkQ15rubB | 1,287-bp EcoRI-HindIII PCR fragment cloned in pKKPalk/EcoRI/HindIII | This study |
| Plasmids containing DNA from R. erythropolis NRRL B-16531 | ||
| p16531 | pGEM7-Zf(+) with 550-bp NRRL B-16531 alkB1 fragment | 37 |
| p23-D1 | pGEM7-Zf(+) with 550-bp NRRL B-16531 alkB2 fragment | 46 |
| p62-O | pGEM7-Zf(+) with 550-bp NRRL B-16531 alkB3 fragment | 46 |
| p23-D2 | pGEM7-Zf(+) with 550-bp NRRL B-16531 alkB4 fragment | 46 |
| pAlkB1 | 3,040-bp BamHI fragment cloned in pZErO2.1 | This study |
| pRubB | 4,560-bp PstI fragment cloned in pZErO2.1 | This study |
| pAlkB2 | 3,392-bp EcoRI fragment cloned in pZErO2.1 | This study |
| pAlkB3 | 1,967-bp BamHI fragment cloned in pZErO2.1 | This study |
| palkB3 (Sau3) | 1.2-kb Sau3A partial fragment cloned in pZErO2.1 | This study |
| pAlkB4 (Sau1-8) | 8 different Sau3A partial fragments in pZErO2.1 | This study |
| pCom8-alkB1 (Rer) | pCom8 with R. erythropolis NRRL B-16531 alkB1 | This study |
| pCom8-alkB2 (Rer) | pCom8 with R. erythropolis NRRL B-16531 alkB2 | This study |
| pCom8-alkB3 (Rer) | pCom8 with R. erythropolis NRRL B-16531 alkB3 | This study |
| pCom8-alkB4 (Rer) | pCom8 with R. erythropolis NRRL B-16531 alkB4 | This study |
| pKKPalk-rubA1 (Rer) | pKKPalk with R. erythropolis NRRL B-16531 rubA1 | 43 |
| pKKPalk-rubA2 (Rer) | pKKPalk with R. erythropolis NRRL B-16531 rubA2 | 43 |
| pKKPalk-rubA3 (Rer) | pKKPalk with R. erythropolis NRRL B-16531 rubA3 | 43 |
| pKKPalk-rubA4 (Rer) | pKKPalk with R. erythropolis NRRL B-16531 rubA4 | 43 |
| pKKPalk-rubB (Rer) | pKKPalk with R. erythropolis NRRL B-16531 rubB | This study |
For the NRRL B-16531 experiments, E. coli JM101 [endA hsdR supR thi-1 Δ(lac-proAB) F′(traD36 proAB lacIq lacZM15)] (55) and DH10B (Gibco BRL) were used for cloning and the production of plasmid DNA for sequencing. LB (32) and E2 medium (22) supplemented with carbon sources and/or antibiotics were used throughout. To culture NRRL B-16531 on n-octane, petri dishes with E2 medium were incubated in a sealed container together with an open Erlenmeyer flask containing n-octane. n-dodecane, n-tetradecane, and n-hexadecane were supplied by placing a Whatman 3MM filter disk with 200 μl of n-alkane in the lid of the petri dish. All cultures were grown aerobically at 30 or 37°C.
For the Q15 experiments, E. coli DH10B or JM110 (JM101dam, dcm) was used as a host for recombinant plasmids. The E. coli strains were routinely cultured in LB at 37°C. When necessary, the LB medium was supplemented with ampicillin (50 μg/ml). Plasmid and chromosomal DNA purifications, enzymatic digests, ligations, and E. coli transformations were performed using standard molecular techniques (3). PCRs were performed with Taq DNA polymerase (Amersham Pharmacia Biotech, Piscataway, N.J.) or Pfu DNA polymerase (Stratagene, La Jolla, Calif.) when PCR products were cloned. Nonradioactive DNA probe labeling and Southern and colony hybridizations (DIG System; Roche Molecular Biochemicals, Rotkreuz, Switzerland) were performed according to the manufacturer's instructions.
Cloning and sequence analysis of NRRL B-16531 and Q15 alk genes.
Chromosomal DNA was isolated from NRRL B-16531 according to the method of Desomer et al. (8). To clone the four full-length NRRL B-16531 alkane hydroxylase genes, suitable restriction fragments in the range of 2 to 5 kb were selected by Southern blotting using probes obtained from PCR fragments cloned into plasmids p16531 (alkB1) (37), p62-O (alkB2), p23-D1 (alkB3), and p23-D2 (alkB4) (46). Chromosomal restriction fragments around the desired size were cut out from a preparative agarose gel, isolated by electroelution, ligated between the appropriate sites of pGEM7-Zf(+) (Promega, Madison, Wis.) or pZErO-2.1 (Invitrogen, Basel, Switzerland), and transformed into E. coli DH10B (Invitrogen) by electroporation (9). E. coli transformants were selected with ampicillin (200 μg/ml) or kanamycin (50 μg/ml). The transformants containing the desired genes were identified by colony blotting using the probes described above. The 16531-alkB1 probe was used to clone a 2.75-kb BamHI fragment. As this fragment included the start of a rubredoxin reductase gene, in addition to two rubredoxin genes and the complete alkB1 gene, we also cloned an overlapping 3.9-kb PstI fragment containing the complete rubredoxin reductase gene. In the same way, the 23-D1-alkB2 and 62-O-alkB3 probes were used to clone a 3.4-kb EcoRI and a 1.6-kb BamHI fragment, respectively. The EcoRI fragment contained an alkB homolog and two rubredoxin genes, while the BamHI fragment contained an incomplete alkB homolog, the N-terminal 50 amino acids of which were missing. An overlapping 1.2-kb Sau3A fragment yielded the missing part of alkB3. The 23-D2-alkB4 probe was used to select several overlapping Sau3A clones. The resulting sequence contained the complete alkB homolog (alkB4). Plasmid DNA was isolated using the High Pure plasmid isolation kit (Roche Diagnostics). Both strands of the inserts were completely sequenced on a Li-Cor 4000L sequencer using IRD800-labeled −40 forward (AGGGTTTTCCCAGTCACGACGTT) and −40 reverse (GAGCGGATAACAATTTCACACAGG) primers and the Amersham Thermosequenase cycle-sequencing kit (Amersham Pharmacia Biotech Europe GmbH, Freiburg, Germany).
For the Q15 experiments, the PCR primer Mt-alkB-F1 (AACACCGCCCACGAAATGGGGC) and the reverse primer Mt-alkB-R1 (GGCGTGGTGATCGCTGTGTCGCTG), derived from the corresponding DNA sequences from the first and third highly conserved histidine motif boxes in M. tuberculosis H37Rv alkB (6), yielded a PCR fragment of the expected size (548 bp) from Q15. The fragment, designated alkB1, was cloned and sequenced as previously described (49). In order to clone the complete Q15 alkB1 gene, Southern analysis was done on total DNA from the Q15 NP strain restricted with different enzymes and probed with the 548-bp digoxigenin-dUTP-labeled PCR fragment (Q15 alkB1 probe). Appropriately sized Q15 alkB1 probe-positive EcoRI fragments were gel purified and used to construct an enriched DNA library in pBluescript II KS(+/−) (Stratagene). E. coli DH10B clones were screened by colony hybridization using the Q15 alkB1 probe. The recombinant plasmid, designated pKS1, was purified from an alkB1+ clone, and the complete nucleotide sequence (6,389 bp) of the EcoRI insert was determined on both strands by primer walking with a T7 DNA-sequencing kit (Applied Biosystems, Foster City, Calif.). The primers Q15alkB1-2L (CAGCTGGAACAGTGATCGCATCTG; position 884 on alkB1) and Q15alkB1-5L (GACCTTCTCGCGGACGCCGCAGTC; position 1315 on alkB1) resulted in the amplification of an unexpected ∼430-bp PCR fragment that was subsequently gel purified and sequenced; it was homologous, but not identical, to Q15 alkB1. The 430-bp PCR fragment, designated alkB2, was used as a probe to clone a 4,145-bp BglII fragment from Q15 NP genomic DNA as described above for Q15 alkB1. Genes homologous to NRRL B-16531 alkB3 and alkB4 in Q15 were detected by PCR analyses using primers from within the NRRL B-16531 alkB3 sequence (Q15 alkB3-F2, GGTGTCGACGCTCCTGCATGGC, and Q15 alkB3-R2, CGCCTTGGTGTGAATGAGCTCG) and from the NRRL B-16531 alkB4 sequence (alkB4FWE, CGGAATTCACATGACGACCTTCGCGG, and alkB4RVH, GGTCGTACTAAAGCTTAGTCCGGC). A 1,282-nucleotide (nt) PCR amplification product for Q15 alkB3 and a 1,217-nt Q15 alkB4 amplification product were purified, cloned, and sequenced. DNA sequencing was performed with the 373 automated fluorescence sequencer (Applied Biosystems).
Nucleotide and amino acid sequences were compared with the EMBL, SwissProt, and GenBank databases using BLASTN and BLASTX at the National Center for Biotechnology Information (1). DNA and protein sequences were further analyzed using GeneWorks II software (Intelligenetics, Mountain View, Calif.) and LASERGENE Navigator from DNASTAR (Madison, Wis.).
Functional expression of rhodococcal alkB, rubA, and rubB genes in P. fluorescens and E. coli.
R. erythropolis NRRL B-16531 alkB2, alkB3, and alkB4 were amplified using primers alkB2FWE (GGAGGAATTCCATGTCGACGCACG), alkB2RVH (GGCGCGAAGCTTCTTTCTGCGGC), alkB3FWE (GCTCGAGAATTCTCGATGACAG), alkB3RVH (GGTGAAGCTTGCATGAGTCGGG), alkB4FWE (CGGAATTCACATGACGACCTTCGCGG), and alkB4RVH (GGTCGTACTAAAGCTTAGTCCGGC), respectively. As an EcoRI site is located immediately upstream of the ATG start codon of the alkB1 gene, this gene was cloned as an EcoRI-BamHI fragment from the BamHI genomic clone. All genes were inserted into pCom8 (38), using the EcoRI and HindIII sites introduced by the primers (underlined in the sequences) in the case of alkB2-4. For Q15, alkB1 and alkB2 were cloned into pCom8 like the corresponding NRRL B-16531 genes and transformed into E. coli. The pCom8 derivatives (Table 1) were isolated and then transformed into P. fluorescens KOB2Δ1 according to the method of Højberg et al. (14). E. coli and P. fluorescens KOB2Δ1 recombinants harboring pCom8 derivatives were selected with 10 and 100 μg of gentamicin/ml, respectively. PCR amplification and cloning of the NRRL B-16531 rubA1, rubA2, rubA3, and rubA4 genes in pKKPalk have been described elsewhere (43). Q15 rubA1, rubA2, and rubB and NRRL B-16531 rubB were also cloned in pKKPalk, and the recombinant plasmids were transformed into E. coli GEc137 containing pGEc47ΔG for rubA plasmids or pGEc47ΔT for rubB plasmids (42). E. coli transformants were selected for on LB supplemented with tetracycline (12.5 μg/ml) and carbenicillin (50 μg/ml) or ampicillin (200 μg/ml).
To measure in vivo alkane hydroxylase activity, the alkB, rubA, and rubB recombinants were assayed for the ability to mineralize 14C-radiolabeled alkanes (C12, C16, or C28) (51) in minimal salts medium (MSM) supplemented with 100 mg of unlabeled alkane/liter, 50 mg of yeast extract/liter, the indicated 14C-labeled alkane substrate, and 0.01% rhamnolipid surfactant for the Pseudomonas strains or 0.1% Triton X-100 surfactant for the E. coli strains. The recombinants were also monitored for growth on various alkanes at 30°C. For E. coli strains, growth on M9 agar plates supplemented with 0.001% thiamine was monitored; n-alkanes (C8, C10, and C12) were provided as vapor in a closed system as the sole C and energy source. The growth of P. fluorescens KOB2Δ1 recombinants on alkanes was monitored at 30°C and 200 rpm in 250-ml baffled Erlenmeyer flasks containing 50 ml of MSM (22) supplemented with 1% (vol/vol) n-alkanes (C8, C10, C12, C14, and C16). For optical density measurements, culture liquid (1 ml) was spun down in an Eppendorf 5415 C microcentrifuge (15,000 rpm), and 0.5 ml of supernatant containing the alkane droplets was removed. After the addition of 0.5 ml of water, the cell pellet was resuspended and the optical density was measured at 450 nm.
Nucleotide sequence accession numbers.
The sequences of the four R. erythropolis NRRL B-16531 alkane hydroxylase genes and flanking DNA have been submitted to GenBank and received the following accession numbers: alkB1, AJ009586; alkB2, AJ297269; alkB3, AJ301876; and alkB4, AJ301877. For Rhodococcus sp. strain Q15, the accession numbers are as follows: alkB1, AF388181; alkB2, AF388182; alkB3, AF388179; and alkB4, AF388180.
RESULTS
Identification of Rhodococcus strains NRRL B-16531 and Q15.
Comparison of the 16S ribosomal DNA sequences from Q15 (EMBL accession no. AF046885) and NRRL B-16531 (EMBL accession no. AJ009591) revealed 99% DNA sequence identity on an 809-nt overlap, indicating that the two rhodococci are closely related but not identical. The NRRL B-16531 16S sequence was identical to that of the R. erythropolis type strain, ATCC 4277. Strain NRRL B-16531 possesses a plasmid similar in size to the larger of the two large plasmids (∼90 and ∼115 kb) found in Q15 (51) and also possesses a smaller (∼3.5-kb) plasmid not found in Q15 (data not shown).
Cloning and sequence analyses of the NRRL B-16531 and Q15 alk genes.
Comparison of the regions cloned from Q15 and NRRL B-16531 revealed that their alk genes, including the spacer regions, are almost identical, with 94.7 to 100% DNA sequence identity. The derived amino acid sequences have 97 to 100% amino acid sequence identity and generally the same length (Table 2). Therefore, sequence comparisons for the alk genes and encoded proteins in the two rhodococcal strains are described together.
TABLE 2.
Comparison of Q15 and NRRL B-16531 alk systems
| Gene | No. of amino acid residues
|
% DNA identitya | % amino acid identityb | Best database match (% amino acid identity/no. of amino acids)c | |
|---|---|---|---|---|---|
| Q15 | NRRL B-16531 | ||||
| alkB1 cluster | |||||
| alkB1 | 391 | 391 | 97.4 | 99.7 | Mt AlkB (62/396-aa overlap) |
| rubA1 | 56 | 56 | 94.7 | 98.2 | Mt RubA (74/50) |
| rubA2 | 63 | 63 | 98.0 | 98.0 | Mt RubB (83/56) |
| rubB | 418 | 418 | 96.2 | 96.7 | Nocardia ferredoxin reductase (36/410) |
| alkU1 | 205 | 208 | 96.0 | 98.5 | Mt reg.? (38/192) |
| alkB2 cluster | |||||
| alkB2 | 408 | 408 | 99.3 | 99.3 | Mt AlkB (65/415) |
| rubA3 | 61 | 61 | 98.9 | 100 | Mt RubA (76/51) |
| rubA4 | 60 | 60 | 100 | 100 | Mt RubB (84/59) |
| alkU2 | 218 | 162d | 99.4 | 98.8 | Mt reg.? (63/191) |
| alkB3 | 383 | 383 | 98 | 97 | Mt AlkB (47/380) |
| alkB4 | 386 | 386 | 99.7 | 99.7 | Mt AlkB (49/386) |
DNA sequence identity between Q15 and NRRL B-16531 alk genes.
Amino acid sequence identity between Q15 and NRRL B-16531 Alk proteins.
reg.?, hypothetical protein Rv3249c, a TetR family protein; Mt, M. tuberculosis H37Rv.
Incomplete ORF.
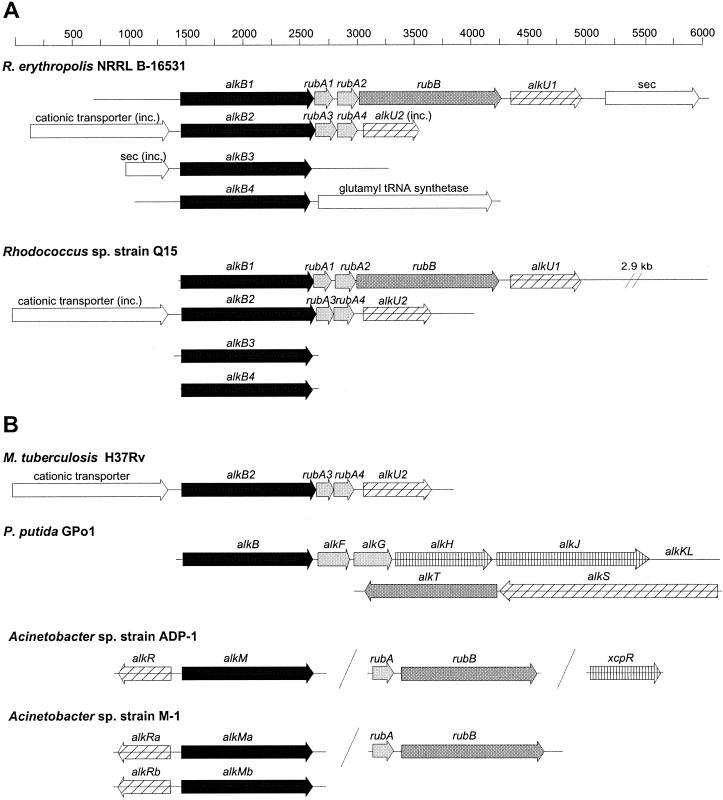
Sequence analysis of the DNA and deduced amino acid sequences of both the NRRL B-16531 and Q15 alkB1 gene regions revealed five consecutive open reading frames (ORFs) (Fig. 1) whose products possessed the greatest amino acid sequence identities with the complete sequences of alkane hydroxylase components and a putative regulatory protein. We have designated the five genes alkB1 (alkane monooxygenase), rubA1 (rubredoxin), rubA2 (rubredoxin), rubB (rubredoxin reductase), and alkU1 (putative TetR-related regulatory protein). The alkB2 gene regions in both strains contained four ORFs (Fig. 1) designated alkB2 (alkane monooxygenase), rubA3 (rubredoxin), rubA4 (rubredoxin), and alkU2 (putative TetR-related regulatory protein). The NRRL B-16531 alkB3 and alkB4 gene regions both contained an ORF encoding an alkane monooxygenase homolog. These genes were not flanked by rubredoxin or rubredoxin reductase genes. The flanking regions of Q15 alkB3 and alkB4 were not investigated.
FIG. 1.
(A) Cloned alk gene fragments from Rhodococcus strains NRRL B-16531 and Q15. (B) For comparison, the gene organizations of other bacterial alkane-degradative systems are shown (taken from references 36 and 40). Similar shading patterns of the arrows represent similar functions; solid, alkane monoxygenases; light shading, rubredoxins; dark shading, rubredoxin reductases; hatched, transcriptional regulatory proteins; vertically striped, other alk genes. The open arrows indicate genes that are not part of alk gene clusters. The directions of the arrows indicate the directions of transcription. inc., incomplete ORF. The 2.9-kb sequence downstream of alkU1 in Q15 does not encode proteins with detectable sequence identity to any sequence in GenBank.
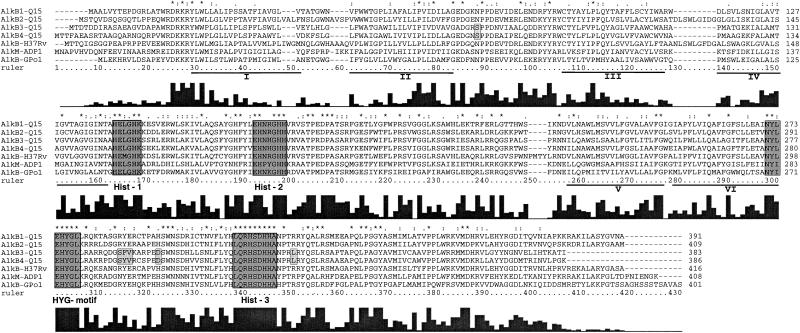
The four rhodococcal alkB genes encoded proteins similar in size and with the greatest amino acid sequence identities (47 to 65%) to the alkane monooxygenases of M. tuberculosis and P. rugosa and, to a lesser degree (41 to 50%), to other alkane monooxygenases. In addition to the high full-length homology, the four rhodococcal AlkB proteins possessed eight histidines which are highly conserved in nonheme iron integral membrane alkane hydroxylases and desaturases and which are believed to be required for catalytic activity by these enzymes (33, 34, 37). Sequences corresponding to the three histidine boxes (Hist1, HE[L/M]XHK; Hist2, EHXXGHH; and Hist3, LQRH[S/A]DHHA) are highly conserved in all bacterial alkane monooxygenases. The Hist3 box is the longest almost perfectly conserved stretch in all alkane hydroxylases but is not well conserved in other closely related hydrocarbon monooxygenases. An additional well-conserved histidine box (NYXEHYG[L/M], designated the HYG motif [Fig. 2]) is located about 30 amino acids [aa] upstream of the Hist3 box (37). This HYG motif is also quite well, but not perfectly, conserved in related hydrocarbon monooxygenases, such as three xylene monooxygenases (XylM), a nitrotoluene monooxygenase (NtnMa), and two cymene monooxygenases (CymAa) (NYXQHYG[L/Q]). Therefore, the Hist3 motif and the HYG motif can be used as apparent signature motifs specific for bacterial alkane monooxygenases. The positions and lengths of the six transmembrane helices initially reported in P. putida GPo1 AlkB (45) were also conserved in all rhodococcal AlkBs and all other alkane hydroxylases. However, there was relatively little amino acid homology within these hydrophobic stretches (Fig. 2, bar graph).
FIG. 2.
Manual alignment of predicted amino acid sequences corresponding to alkane monooxygenase genes from various bacteria. The three conserved histidine boxes of the eight-histidine motif (34) (Hist-1, Hist-2, and Hist-3) and the additional histidine motif NYXEHYGL (HYG-motif) are boxed and darkly shaded. The locations of the six putative transmembrane helices in the P. putida GPo1 AlkB sequence are underlined and marked by roman numerals. Amino acid residues conserved in all alkane monooxygenases are marked by asterisks above the alignment. The degree of conservation of each position is indicated by the bar graph below the alignment, created by Clustal X. Six positions in AlkB3-Q15 and AlkB4-Q15 that deviate from all other alkane monooxygenase are boxed and lightly shaded. AlkB-H37Rv, alkane hydroxylase from M. tuberculosis H37Rv (accession number CAB08323.1); Alkm-ADP-1, Acinetobacter sp. strain ADP1 AlkM (AJ002316); AlkB-GPo1, P. putida GPo1 AlkB (AJ245436).
The four rubredoxin genes found immediately downstream of the alkB1 and alkB2 genes encoded proteins with the greatest amino acid sequence identities to RubA and RubB of the alkane hydroxylase system in M. tuberculosis and to other bacterial rubredoxins involved in alkane oxidation (43). All four rubredoxins contained the PS00202 rubredoxin signature (PROSITE database) and were single-domain proteins, like the Acinetobacter sp. strain ADP1 rubredoxin (11). In a phylogenetic analysis, RubA2 and RubA4 of NRRL B-16531 and Q15 were more closely related to the ADP1 rubredoxin and the C-terminal domain of GPo1 AlkG than to rhodococcal RubA1 and RubA3. The last two proteins are more closely related to the N-terminal domains of P. putida GPo1 AlkG and AlkF (AlkG1 and AlkF1), which are not able to transfer electrons from the GPo1 rubredoxin reductase to the GPo1 alkane hydroxylase (21, 43). In both the NRRL B-16531 and Q15 strains, the alkB1 and rubA1 genes and the rubA2 and rubB genes had 3′-end-5′-end ATGA and TGATG sequence overlaps, respectively. The rubA1 and rubA2 genes did not overlap. For the alkB2 gene cluster, the NRRL B-16531 and Q15 alkB2 and rubA3 genes contained 3′-end-5′-end ATGA and TGATG sequence overlaps, respectively. Both the NRRL B-16531 and Q15 rubA3 and rubA4 genes contained 3′-end-5′-end ATGA sequence overlaps.
The next large ORF in the alkB1 cluster, rubB, encoded a large protein exhibiting significant full-length sequence identity to a variety of bacterial reductase subunits of hydroxylase systems, including the P. putida and Acinetobacter alkane hydroxylase systems (Table 2), dioxygenase systems, and cytochrome P-450 systems involved in hydrocarbon degradation. The greatest amino acid homologies were found with a Nocardia sp. ferredoxin reductase (39% amino acid identity; 408-aa overlap) (EMBL accession no. AB017795; phenanthrene degradation), R. erythropolis ThcD (38% amino acid identity; 395-aa overlap) (EMBL accession no. P43494; thiocarbamate degradation), and several hypothetical ferredoxin reductases found in M. tuberculosis.
The ORFs designated alkU1 and alkU2, which immediately follow the alkB1 and alkB2 gene clusters, respectively, encode proteins which have the greatest amino acid sequence identities to a hypothetical transcription regulatory protein (accession number CAB08350.1) (38 and 63% amino acid identity, respectively) that is located immediately downstream of the alkB-rubA-rubB gene cluster in M. tuberculosis H37Rv. Related peptides are also encoded immediately adjacent to the P. rugosa alkane hydroxylase gene (36) and the Nocardioides sp. strain CF8 alkB homolog (13) (up- and downstream, respectively). AlkU1 and AlkU2 possessed helix-turn-helix DNA binding motifs near the N terminus that are also present in known transcriptional regulatory proteins of the TetR/ArcR family (PFAM 00440; TetR; bacterial regulatory proteins).
The remaining parts of the Q15 and NRRL B-16531 alkB1, -2, -3, and -4 fragments did not encode proteins with similarity to known alkane alcohol dehydrogenase or aldehyde dehydrogenase genes. The ORFs downstream of NRRL B-16531 alkB1 and upstream of NRRL B-16531 alkB3 show weak sequence identity to (putative) exported proteins (Fig. 1). Interestingly, the corresponding DNA regions almost immediately downstream from the Q15 and NRRL B-16531 alkB1 gene clusters show relatively little DNA homology (data not shown) and have markedly different G+C contents: 60.2% in NRRL B-16531 versus 53.3% in Q15. The Q15 and NRRL B-16531 alkB2 and M. tuberculosis alkB gene clusters have the same gene organization and close to 70% DNA sequence identity over their entire lengths, including the putative transporter and alkU2 genes, indicating that these clusters have a common origin. The level of sequence identity is similar to the level of DNA sequence identity between alkB1 and alkB2 and between alkB3 and alkB4 but clearly higher than the DNA sequence identity between alkB1 or -2 and alkB3 or -4 (63%).
Heterologous expression of the rhodococcal alk genes in E. coli and P. fluorescens.
Efficient expression vectors and recombinant E. coli and Pseudomonas hosts that allowed the functional expression of the M. tuberculosis and P. rugosa AlkB homologs (36) and rubredoxins and rubredoxin reductases from several microorganisms were used to heterologously express the rhodococcal alk genes (Tables 3 and 4). The four NRRL B-16531 alkB genes and Q15 alkB1 and alkB2 were cloned in pCom8 (38). Only NRRL B-16531 and Q15 alkB2 allowed an alkB knockout mutant of P. fluorescens CHA0, KOB2Δ1, to grow on C12 to C16 n-alkanes (Table 3) and mineralize 14C-radiolabeled n-dodecane and n-hexadecane (Table 4). Mineralization of 14C-radiolabeled n-octacosane was observed in both the alkB2 clones and the alkB knockout mutant of P. fluorescens CHA0, due to an additional long-chain alkane hydroxylase system in this strain (36). None of the NRRL B-16531 alkB genes allowed E. coli GEc137 or P. putida GPo12 containing an alkB deletion derivative of pGEc47 to grow on C6 to C12 alkanes. Q15 alkB1 also could not be functionally expressed in the E. coli host.
TABLE 3.
Heterologous expression of the rhodococcal alk genes in E. coli and P. fluorescens as measured by growth on n-alkanes
| Straina | Growth on alkaneb
|
|||||
|---|---|---|---|---|---|---|
| C8 | C10 | C12 | C14 | C16 | C18 | |
| P. fluorescens KOB2Δ1 | ||||||
| pCom8 | − | − | − | − | +/− | ++++ |
| pCom8 + NRRL B-16531 alkB1 | − | − | − | − | +/− | ++++ |
| pCom8 + NRRL B-16531 alkB2 | − | + | ++ | +++ | +++ | ++++ |
| pCom8 + NRRL B-16531 alkB3 | − | − | − | − | +/− | ++++ |
| pCom8 + NRRL B-16531 alkB4 | − | − | − | − | +/− | ++++ |
| pCom8 + Q15 alkB1 | − | − | − | − | +/− | ++++ |
| pCom8 + Q15 alkB2 | − | + | ++ | +++ | +++ | ++++ |
| E. coli GEc137 | ||||||
| pGEc47: alk+ | +++ | +++ | +/− | |||
| ΔT | − | − | − | |||
| ΔT; pKKPalk Q15 rubB | ++ | +++ | +/− | |||
| ΔT; pKKPalk NRRL B-16531 rubB | − | − | − | |||
| ΔG | − | − | − | |||
| ΔG; pKKPalk NRRL B-16531 rubA1 | − | − | − | |||
| ΔG; pKKPalk NRRL B-16531 rubA2 | ++ | ++ | − | |||
| ΔG; pKKPalk NRRL B-16531 rubA3 | − | − | − | |||
| ΔG; pKKPalk NRRL B-16531 rubA4 | ++ | ++ | − | |||
| ΔG; pKKPalk Q15 rubA1 | − | − | − | |||
| ΔG; pKKPalk Q15 rubA2 | − | +/− | − | |||
| MSM control | − | − | − | |||
ΔG, pGEc47ΔG; ΔT, pGEc47ΔT.
−, (no growth); +/− (very slight growth) to ++++ (heavy growth) as determined by an increase in turbidity in liquid medium or growth on solid medium relative to the corresponding controls (not inoculated or not containing an alkane).
TABLE 4.
Heterologous expression of the rhodococcal alk genes in E. coli and P. fluorescens as measured by mineralization of various 14C-labeled n-alkanes
| Straina | Mineralization of alkaneb
|
||
|---|---|---|---|
| C12 | C16 | C28 | |
| P. fluorescens KOB2Δ1 | |||
| pCom8 | 1.2 | 1.9 | 18.5 |
| pCom8 + Q15 alkB1 | 2 | 2 | 13.8 |
| pCom8 + Q15 alkB2 | 16 | 32 | 16.2 |
| pCom8 + NRRL B-16531 alkB2 | 17 | 25 | 12 |
| E. coli GEc137 | |||
| pGEc47: Alk+ | 39 | ||
| ΔT | <1 | ||
| ΔT + pKKPalk Q15 rubB | 27 | ||
| ΔG | <1 | ||
| ΔG + pKKPalk NRRL B-16531 rubA2 | 13 | ||
| MSM control (not inoculated) | <1 | <1 | <1 |
ΔG, pGEc47ΔG; ΔT, pGEc47ΔT.
Values represent average percentages of 14C recovered as CO2 from duplicate samples after 2 weeks of incubation at 28°C. The values for the control represent the background radioactivity.
We tested whether the rhodococcal rubredoxin and rubredoxin reductase genes complement deletions of the corresponding GPo1 proteins (AlkG and AlkT, respectively) in E. coli, using pKKPalk (38). Here, we found that NRRL B-16531 rubA2 and rubA4, but not the rubA1 and rubA3 genes, complemented an E. coli recombinant containing pGEc47ΔG for growth on n-octane vapor (43). We were unable to obtain functional expression of the Q15 rubredoxin genes. The Q15 rubredoxin reductase gene (rubB), but not the NRRL B-16531 rubB, complemented an alkT deletion, as determined by growth on n-octane, n-decane, and n-dodecane (Table 3) and mineralization of n-dodecane (Table 4). Much greater levels of mineralization were observed when the media were supplemented with the appropriate surfactant (rhamnolipid or Triton X-100 for the Pseudomonas system; Triton X-100 for the E. coli system). Mineralization in the appropriate controls was minimal.
DISCUSSION
Multiple alkane monooxygenases and rubredoxins and a rubredoxin reductase in two similar rhodococcal strains isolated in Japan and Canada were cloned and characterized. This is the first report of a detailed genetic characterization of alkane hydroxylase systems in Rhodococcus, a genus considered to be an important component of hydrocarbon-containing microbial communities present in contaminated soils and sediments (39, 48). The presence of multiple alkane hydroxylases in Rhodococcus strains NRRL B-16531 and Q15 may be a common feature of Rhodococcus strains, as three to five alkB homologs were also found in eight other Rhodococcus strains (46). It is also reminiscent of the two separate alkM genes found in Acinetobacter sp. strain M-1 (40) and of other multiple degradative enzyme systems reported in Rhodococcus responsible for polychlorobiphenyl degradation (2) and indene bioconversion (41). In yeasts, multiple cytochrome P450 alkane hydroxylases with overlapping substrate ranges have been reported as well (16, 54).
Heterologous expression of some of the rhodococcal alk genes (NRRL B-16531 alkB2, rubA2, and rubA4 and Q15 alkB2 and rubB) was achieved in E. coli and Pseudomonas expression systems based on the P. putida GPo1 alkB promoter as shown by mineralization and growth assays, confirming their respective functions in n-alkane degradation. The mineralization and growth assays of Pseudomonas clones containing the alkB2 gene indicate that AlkB2 is at least partly responsible for the initial oxidation of C12 to C16 n-alkanes by the two rhodococcal strains, while RubA2, RubA4, and RubB are able to function as electron transfer components.
We were unable to show functional heterologous expression of rhodococcal AlkB1, AlkB3, and AlkB4 in the Pseudomonas or E. coli expression system. Several explanations can be put forward. (i) Functional expression in E. coli or Pseudomonas requires proper synthesis, correct folding, and proper assembly, which is not always ensured for rhodococcal and other heterologous proteins (5, 7, 19, 26-28). For example, AlkM, the Acinetobacter sp. strain ADP-1 alkane monooxygenase (30), could also not be functionally expressed with the same Pseudomonas or E. coli expression system (36). (ii) The three AlkB proteins may have substrate ranges that lie outside the range that can be tested in our recombinant host strains (between C6 and C12 for E. coli and P. putida or between C12 and C16 for P. fluorescens). Here, it should be noted that AlkB3 and AlkB4 homologs occur more frequently in gram-positive strains able to grow on very long chain alkanes (over C20) (46). (iii) AlkB1, AlkB3, and AlkB4 may not accept electrons from the rubredoxins in the host strains. However, this is unlikely, at least in the case of AlkB1, as this protein is encoded in an operon-like arrangement with a rubredoxin (RubA2) that could replace its P. putida GPo1 counterpart (43). (iv) The AlkB proteins could produce secondary alcohols by subterminal alkane oxidation. However, P. fluorescens KOB2Δ1 is able to grow well on secondary alcohols and ketones (data not shown). Therefore, complementation should be possible if AlkB1, AlkB3, and AlkB4 produce secondary alcohols from C12 to C16 alkanes. (v) It is possible that the three AlkB homologs are not alkane monooxygenases. However, all three have >50% full-length protein sequence identity to functional alkane monooxygenases from gram-positive bacteria even if several residues which are well conserved in all other alkane monooxygenase sequences are not conserved in AlkB3 and AlkB4 (Fig. 2). The three homologs also have >45% sequence identity to 13 functional alkane monooxygenases from gram-negative bacteria; the alkane monooxygenase sequences from gram-positive bacteria constitute just one branch of a more deeply rooted alkane monooxygenase tree (46). The closest relatives of the AlkBs, with a different but still closely related function, are the xylene and cymene monooxygenases, with only 25 to 30% sequence identity to the alkane monooxygenases (or homologs) described in this paper, followed by the desaturases with <20% sequence identity.
In conclusion, the first two explanations, that the three AlkB homologs are alkane monooxygenases that cannot be expressed in E. coli and Pseudomonas or have substrate ranges that lie outside the range that can be tested with these hosts, are the most likely and should be explored in more detail, e.g., by developing other expression hosts for rhodococcal proteins, such as Streptomyces lividans (35) or, ideally, alkane-negative Rhodococcus strains or mutants.
The genetic organization of the alk genes of various bacterial alkane hydroxylase systems is summarized in Fig. 1. The head-to-tail organization of the rhodococcal alkB1 and alkB2 gene clusters suggests that they may be transcribed as an operon. Moreover, several ORFs in these gene clusters have overlapping stop and start codons. This phenomenon is indicative of translational coupling and is thought to ensure the production of stoichiometric amounts of the involved proteins. Translational coupling has been observed in several rhodococcal operon-like structures from aromatic degradation pathways (23). The rhodococcal alkB1 gene clusters are the only bacterial alkane hydroxylase gene clusters identified to date that encode all three components of an alkane hydroxylase system in a single operon-like structure. The alkB1 and alkB2 gene organization is also reminiscent of the P. putida GPo1 alkBFGHJKL operon (47). This operon also encodes two rubredoxins, AlkF and AlkG (21). As in GPo1, only the second rubredoxins of NRRL B-16531 (RubA2 and RubA4) but not the first rubredoxins (RubA1 and RubA3) in each gene cluster are functional electron transfer components. In addition, RubA2 and RubA4 possess relatively greater amino acid sequence identity to rubredoxins known to be required for alkane utilization than RubA1 and RubA3. The functions of RubA1 and RubA3, and of other closely related rubredoxins in gram-negative and gram-positive alkane-degrading strains, remain unknown (43).
The alkB3 and alkB4 genes are not accompanied by rubredoxin or rubredoxin reductase genes. Probably, the rubredoxins and rubredoxin reductase encoded in the alkB1 and alkB2 gene clusters also serve as electron transfer components for AlkB3 and AlkB4. In this respect, the rhodococcal alk gene organization would be similar to that reported for Acinetobacter sp. strain M-1, where a single constitutively expressed rubredoxin and a rubredoxin reductase serve as electron transfer proteins for two differentially regulated alkane hydroxylases (40). For the rhodococcal alk genes, this implies that the alkB1 gene cluster may have to be expressed constitutively. The two putative TetR-type transcription regulation genes, found in the cloned alkB1 and alkB2 gene regions, and similar genes found adjacent to alk genes in other similar actinomycetes do not resemble previously identified alk gene regulatory proteins and thus may constitute a new class of regulatory proteins involved in alkane degradation by these bacteria. Neither of the alkB1 or alkB2 gene clusters contains alcohol dehydrogenase or aldehyde dehydrogenase genes, unlike the GPo1 alk system but similar to the situation in most other alkane-degrading bacteria.
Due to the low DNA sequence identity of the four rhodococcal alkB genes to the alkane monooxygenase genes of most of the known gram-negative bacteria, DNA probes based on the rhodococcal genes may be used for detecting and monitoring similar alkane-degradative rhodococci and other closely related high-G+C, mycolic acid-containing actinomycetes in hydrocarbon-contaminated sites. Screening by PCR and colony hybridization has already provided evidence that DNAs with high sequence identity to rhodococcal alkB1 and alkB2 exist in a variety of hydrocarbon-contaminated soils (52), as well as in previously isolated psychrotrophic alkane-degradative actinomycete strains (50) (data not shown). This indicates that these genotypes are widespread in nature and may be important components in hydrocarbon degradation at contaminated sites. However, the high-G+C, mycolic acid-containing actinomycetes contain several additional, highly divergent alkB genes that cannot be detected using probes based on the NRRL B-16531 and Q15 alkB genes (46). Therefore, a combination of hybridization experiments and PCR with highly degenerate primers based on the third histidine box and the highly conserved HYG box is likely to give a more comprehensive overview of the occurrence of alkB homologs in nature.
In summary, four alkane monooxygenase homologs (two part of alkane gene clusters and two occurring as separate genes) were identified in two closely related Rhodococcus spp. based on (i) the significant full-length amino acid sequence identity of their components with other genetically characterized alkane hydroxylase systems; (ii) the conservation in the Rhodococcus alkane monooxygenases of the eight-histidine motif, including the apparent alkane monooxygenase signature motifs, and hydrophobic membrane-spanning regions found in all known alkane monooxygenases; and (iii) functional heterologous expression of some of these genes in E. coli and Pseudomonas alk expression systems. The most likely explanation for the presence of four alkane monooxygenases in one strain (assuming that all four oxidize alkanes) is that each alkane monooxygenase is specific for a certain range of alkanes. For example, P. putida GPo1 AlkB does not act on alkanes longer than C12, while the M. tuberculosis and P. rugosa AlkBs do not efficiently oxidize alkanes shorter than C12 (36). As the Rhodococcus strains studied here oxidize alkanes up to C32 to C36, AlkB1, AlkB3, and AlkB4 may each cover a part of the C18 to C36 range. Unfortunately, it is not yet possible to link sequence features, for example, specific amino acid residues or sequences within the hydrophobic transmembrane regions, with an alkane oxidation range. To make this possible, further studies will focus on the specific role of each of the rhodococcal alk genes in the degradation of alkane or other compounds. The observation that other rhodococci also possess multiple, but not always the same, alkane monooxygenase homologs (46) may help to answer this and other questions related to horizontal gene transfer and the evolution of alkane monooxygenase genes in actinomycetes.
Acknowledgments
This research was supported in part by the Swiss National Science Foundation through the Swiss Priority Program in Biotechnology, grant no. 5002-037023.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Asturias, J. A., and K. N. Timmis. 1993. Three different 2,3-dihydroxybiphenyl-1,2-dioxygenase genes in the gram-positive polychlorobiphenyl-degrading bacterium Rhodococcus globerulus P6. J. Bacteriol. 175:4631-4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1990. Current protocols in molecular biology, vol. 2. Wiley Interscience, New York, N.Y.
- 4.Beard, T. M., and M. I. Page. 1998. Enantioselective biotransformations using rhodococci. Antonie Leeuwenhoek 74:99-106. [DOI] [PubMed] [Google Scholar]
- 5.Cherbrou, H., Y. Hurtubise, D. Barriault, and M. Sylvestre. 1999. Heterologous expression of the purified oxygenase component of Rhodococcus globerulus P6 biphenyl dioxygenase and of chimeras derived from it. J. Bacteriol. 181:4805-4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLeah, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Soeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrett. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 7.De Schrijver, A., I. Nagy, G. Schoofs, P. Proost, J. Vanderleyden, K. H. van Pee, and R. De Mot. 1997. Thiocarbamate herbicide-inducible non-heme haloperoxidase of Rhodococcus erythropolis NI86/21. Appl. Environ. Microbiol. 63:1911-1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desomer, J., M. Crespi, and M. Van Montagu. 1991. Illegitimate integration of non-replicative vectors in the genome of Rhodococcus fascians upon electrotransformation as an insertional mutagenesis system. Mol. Microbiol. 5:2115-2124. [DOI] [PubMed] [Google Scholar]
- 9.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eggink, G., R. G. Lageveen, B. Altenburg, and B. Witholt. 1987. Controlled and functional expression of Pseudomonas oleovorans alkane utilizing system in Pseudomonas putida and Escherichia coli. J. Biol. Chem. 262:17712-17718. [PubMed] [Google Scholar]
- 11.Geiβdörfer, W., S. C. Frosch, G. Haspel, S. Ehrt, and W. Hillen. 1995. Two genes encoding proteins with similarities to rubredoxin and rubredoxin reductase are required for conversion of dodecane to lauric acid in Acinetobacter calcoaceticus ADP1. Microbiology 141:1425-1432. [DOI] [PubMed] [Google Scholar]
- 12.Geiβdörfer, W., R. B. Kok, A. Ratajczak, K. J. Hellingwerf, and W. Hillen. 1999. The genes rubA and rubB for alkane degradation in Acinetobacter sp. strain ADP1 are in an operon with estB, encoding an esterase, and oxyR. J. Bacteriol. 181:4292-4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamamura, N., C. M. Yeager, and D. J. Arp. 2001. Two distinct monooxygenases for alkane oxidation in Nocardioides sp. strain CF8. Appl. Environ. Microbiol. 67:4992-4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Højberg, O., U. Schnider, H. V. Winteler, J. Sørensen, and D. Haas. 1999. Oxygen-sensing reporter strain of Pseudomonas fluorescens for monitoring the distribution of low-oxygen habitats in soil. Appl. Environ. Microbiol. 65:4085-4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hou, C. T., M. A. Jackson, M. O. Bagby, and L. A. Becker. 1994. Microbial oxidation of cumene by octane-grown cells. Appl. Microbiol. Biotechnol. 41:178-182. [Google Scholar]
- 16.Iida, T., T. Sumita, A. Ohta, and M. Takagi. 2000. The cytochrome P450ALK multigene family of an n-alkane-assimilating yeast. Yarrowia lipolytica: cloning and characterization of genes coding for new CYP 52 family members. Yeast 16:1077-1087. [DOI] [PubMed] [Google Scholar]
- 17.Iizuka, H., and K. Komagata. 1964. Microbiological studies on petroleum and natural gas. I. Determination of hydrocarbon-utilizing bacteria. J. Gen. Appl. Microbiol. 10:207-221. [Google Scholar]
- 18.Johnstone, S. L., G. T. Phillips, B. W. Robertson, P. D. Watts, M. A. Bertola, H. S. Koger, and A. F. Marx. 1986. Stereoselective synthesis of S-(−)-β-blockers via microbially produced epoxide intermediates, p. 387-392. In C. Laane, J. Tramper, and M. D. Lilly (ed.), Biocatalysis in organic media. Elsevier, Amsterdam, The Netherlands.
- 19.Kessler, M., E. R. Dabbs, B. Averhoff, and G. Gottschalk. 1996. Studies on the isopropylbenzene 2,3-dioxygenase and 3-isopropylcatechol 2,3-dioxygenase genes encoded by the linear plasmid of Rhodococcus erythropolis BD2. Microbiology 142:3241-3251. [DOI] [PubMed] [Google Scholar]
- 20.Kiener, A., and T. Zimmermann. September 1992. Microbiological process for terminal hydroxylation of ethyl-groups on aromatic 5- or 6-membered heterocycles. European patent EP502524.
- 21.Kok, M., R. Oldenhuis, M. P. G. van der Linden, C. H. C. Meulenberg, J. Kingma, and B. Witholt. 1989. The Pseudomonas oleovorans alkBAC operon encodes two structurally related rubredoxins and an aldehyde dehydrogenase. J. Biol. Chem. 264:5442-5451. [PubMed] [Google Scholar]
- 22.Lageveen, R. G., G. W. Huisman, H. Preusting, P. E. F. Ketelaar, G. Eggink, and B. Witholt. 1988. Formation of polyester by Pseudomonas oleovorans: the effect of substrate on the formation and composition of poly-(R)-3-hydroxyalkanoates and poly-(R)-3-hydroxyalkenoates. Appl. Environ. Microbiol. 54:2924-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larkin, M. J., R. De Mot, L. A. Kulakov, and I. Nagy. 1998. Applied aspects of Rhodococcus genetics. Antonie Leeuwenhoek 74:133-153. [DOI] [PubMed] [Google Scholar]
- 24.Mathys, R. G., A. Schmid, and B. Witholt. 1999. Integrated two-liquid phase bioconversion and product-recovery processes for the oxidation of alkanes: process design and economic evaluation. Biotechnol. Bioeng. 64:459-477. [DOI] [PubMed] [Google Scholar]
- 25.Matsui, T., and K. Furuhashi. 1995. Asymmetric oxidation of isopropyl moieties of aliphatic and aromatic hydrocarbons by Rhodococcus sp. 11B. Biosci. Biotech. Biochem. 59:1342-1344. [Google Scholar]
- 26.McKay, D. B., M. Seeger, M. Zielinski, B. Hofer, and K. N. Timmis. 1997. Heterologous expression of biphenyl dioxygenase-encoding genes from a gram-positive broad-spectrum polychlorinated biphenyl degrader and characterization of chlorobiphenyl oxidation by the gene products. J. Bacteriol. 179:1924-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagy, I., G. Schoofs, F. Compernolle, P. Proost, J. Vanderleyden, and R. De Mot. 1995. Degradation of the thiocarbamate herbicide ETTC (S-ethyl dipropylcarbamothioate) and biosafening by Rhodococcus sp. strain NI86/21 involve an inducible cytochrome P-450 system and aldehyde dehydrogenase. J. Bacteriol. 177:676-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagy, I., S. Verheigen, A. De Schrijver, J. van Damme, P. Proost, G. Schoofs, J. Vanderleyden, and R. De Mot. 1995. Characterization of the Rhodococcus sp. NI86/21 gene encoding N,N′-dimethyl-4-nitrosoaniline oxidoreductase inducible by atrazine and thiocarbamate herbicides. Arch. Microbiol. 163:439-446. [DOI] [PubMed] [Google Scholar]
- 29.Panke, S., A. Meyer, C. M. Huber, B. Witholt, and M. G. Wubbolts. 1999. An alkane-responsive expression system for the production of fine chemicals. Appl. Environ. Microbiol. 65:2324-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ratajczak, A., W. Geiβdörfer, and W. Hillen. 1998. Alkane hydroxylase from Acinetobacter sp. strain ADP-1 is encoded by alkM and belongs to a new family of bacterial integral-membrane hydrocarbon hydroxylases. Appl. Environ. Microbiol. 64:1175-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ratajczak, A., W. Geiβdörfer, and W. Hillen. 1998. Expression of alkane hydroxylase from Acinetobacter sp. strain ADP1 is induced by a broad range of n-alkanes and requires the transcriptional activator AlkR. J. Bacteriol. 180:5822-5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Shanklin, J., C. Achim, H. Schmidt, B. G. Fox, and E. Münck. 1997. Mössbauer studies of alkane ω-hydroxylase: evidence for a diiron cluster in an integral-membrane protein. Proc. Natl. Acad. Sci. USA 94:2981-2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shanklin, J., E. Whittle, and B. G. Fox. 1994. Eight histidine residues are catalytically essential in a membrane-associated iron enzyme, stearoyl-CoA desaturase, and are conserved in alkane hydroxylase and xylene monooxygenase. Biochemistry 33:12787-12794. [DOI] [PubMed] [Google Scholar]
- 35.Smith, T. J., J. S. Lloyd, S. C. Gallagher, W. L. Fosdike, J. C. Murrell, and H. Dalton. 1999. Heterologous expression of alkene monooxygenase from Rhodococcus rhodochrous B-276. Eur. J. Biochem. 260:446-452. [DOI] [PubMed] [Google Scholar]
- 36.Smits, T. H. M., S. B. Balada, B. Witholt, and J. B. van Beilen. 2002. Functional analysis of alkane hydroxylases from gram-negative and gram-positive bacteria. J. Bacteriol. 184:1733-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smits, T. H. M., M. Röthlisberger, B. Witholt, and J. B. van Beilen. 1999. Molecular screening for alkane hydroxylase genes in Gram-negative and Gram-positive strains. Environ. Microbiol. 1:307-318. [DOI] [PubMed] [Google Scholar]
- 38.Smits, T. H. M., M. A. Seeger, B. Witholt, and J. B. van Beilen. 2001. New alkane-responsive expression vectors for E. coli and Pseudomonas. Plasmid 46:16-24. [DOI] [PubMed] [Google Scholar]
- 39.Stephens, G. M., and H. Dalton. 1987. Is toxin production by coryneform bacteria linked to their ability to utilize hydrocarbons? Trends Biotechnol. 5:5-7. [Google Scholar]
- 40.Tani, A., T. Ishige, Y. Sakai, and N. Kato. 2001. Gene structures and regulation of the alkane hydroxylase complex in Acinetobacter sp. strain M-1. J. Bacteriol. 183:1819-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Treadway, S. L., K. S. Yanagimachi, E. Lankenau, P. A. Lessard, G. Stephanopoulus, and A. J. Sinskey. 1999. Isolation and characterization of indene bioconversion genes from Rhodococcus strain 124. Appl. Microbiol. Biotechnol. 51:786-793. [DOI] [PubMed] [Google Scholar]
- 42.van Beilen, J. B. 1994. Alkane oxidation by Pseudomonas oleovorans: genes and proteins. Ph.D. thesis. University of Groningen, Groningen, The Netherlands.
- 43.van Beilen, J. B., M. Neuenschwander, T. H. M. Smits, C. Roth, S. B. Balada, and B. Witholt. 2002. Rubredoxins involved in alkane oxidation. J. Bacteriol. 184:1722-1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Beilen, J. B., S. Panke, S. Lucchini, A. G. Franchini, M. Röthlisberger, and B. Witholt. 2001. Analysis of Pseudomonas putida alkane degradation gene clusters and flanking insertion sequences: evolution and regulation of the alk-genes. Microbiology 147:1621-1630. [DOI] [PubMed] [Google Scholar]
- 45.van Beilen, J. B., D. Penninga, and B. Witholt. 1992. Topology of the membrane-bound alkane hydroxylase of Pseudomonas oleovorans. J. Biol. Chem. 267:9194-9201. [PubMed] [Google Scholar]
- 46.van Beilen, J. B., T. H. M. Smits, L. G. Whyte, S. Schorcht, M. Röthlisberger, T. Plaggemeier, K.-H. Engesser, and B. Witholt. Alkane hydroxylases in Gram-positive strains. Environ. Microbiol., in press. [DOI] [PubMed]
- 47.van Beilen, J. B., M. G. Wubbolts, and B. Witholt. 1994. Genetics of alkane oxidation by Pseudomonas oleovorans. Biodegradation 5:161-174. [DOI] [PubMed] [Google Scholar]
- 48.Warhurst, A. W., and C. A. Fewson. 1994. Biotransformations catalyzed by the genus Rhodococcus. Crit. Rev. Biotechnol. 14:29-73. [DOI] [PubMed] [Google Scholar]
- 49.Whyte, L. G., L. Bourbonniere, and C. W. Greer. 1997. Biodegradation of petroleum hydrocarbons by psychrotrophic Pseudomonas strains possessing both alkane (alk) and naphthalene (nah) catabolic pathways. Appl. Environ. Microbiol. 63:3719-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whyte, L. G., C. W. Greer, and W. E. Inniss. 1996. Assessment of the biodegradation potential of psychrotrophic microorganisms. Can. J. Microbiol. 42:99-106. [DOI] [PubMed] [Google Scholar]
- 51.Whyte, L. G., J. Hawari, E. Zhou, L. Bourbonnière, W. E. Inniss, and C. W. Greer. 1998. Biodegradation of variable-chain-length alkanes at low temperatures by a psychrotrophic Rhodococcus sp. Appl. Environ. Microbiol. 64:2578-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whyte, L. G., A. Schultz, J. B. van Beilen, A. P. Luz, D. Pellizari, D. Labbé, and C. W. Greer. 2002. Prevalence of alkane monooxygenase genes in arctic and Antarctic hydrocarbon-contaminated and pristine soils. FEMS Microbiol. Ecol. 41:141-150. [DOI] [PubMed] [Google Scholar]
- 53.Whyte, L. G., S. J. Slagman, F. Pietrantonio, L. Bourbonnière, S. F. Koval, J. R. Lawrence, W. E. Inniss, and C. W. Greer. 1999. Physiological adaptations involved in alkane assimilation at low temperatures by Rhodococcus sp. strain Q15. Appl. Environ. Microbiol. 65:2961-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yadav, J. S., and J. C. Loper. 1999. Multiple P450alk (cytochrome P450 alkane hydroxylase) genes from the halotolerant yeast Debaryomyces hansenii. Gene 226:139-146. [DOI] [PubMed] [Google Scholar]
- 55.Yanish-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]