Abstract
Lactic acid bacteria (LAB) were isolated from Greek traditional wheat sourdoughs manufactured without the addition of baker's yeast. Application of sodium dodecyl sulfate-polyacrylamide gel electrophoresis of total cell protein, randomly amplified polymorphic DNA-PCR, DNA-DNA hybridization, and 16S ribosomal DNA sequence analysis, in combination with physiological traits such as fructose fermentation and mannitol production, allowed us to classify the isolated bacteria into the species Lactobacillus sanfranciscensis, Lactobacillus brevis, Lactobacillus paralimentarius, and Weissella cibaria. This consortium seems to be unique for the Greek traditional wheat sourdoughs studied. Strains of the species W. cibaria have not been isolated from sourdoughs previously. No Lactobacillus pontis or Lactobacillus panis strains were found. An L. brevis-like isolate (ACA-DC 3411 t1) could not be identified properly and might be a new sourdough LAB species. In addition, fermentation capabilities associated with the LAB detected have been studied. During laboratory fermentations, all heterofermentative sourdough LAB strains produced lactic acid, acetic acid, and ethanol. Mannitol was produced from fructose that served as an additional electron acceptor. In addition to glucose, almost all of the LAB isolates fermented maltose, while fructose as the sole carbohydrate source was fermented by all sourdough LAB tested except L. sanfranciscensis. Two of the L. paralimentarius isolates tested did not ferment maltose; all strains were homofermentative. In the presence of both maltose and fructose in the medium, induction of hexokinase activity occurred in all sourdough LAB species mentioned above, explaining why no glucose accumulation was found extracellularly. No maltose phosphorylase activity was found either. These data produced a variable fermentation coefficient and a unique sourdough metabolite composition.
Bread is a staple food in many European diets. Moreover, it contributes to cultural and geographical identity. Artisan bread production, which often employs sourdough processes or the use of pre-ferments, provides a wide regional variety of breads and specialty products. Sourdough is a mixture of wheat or rye flour and water that is fermented with lactic acid bacteria (LAB) and yeasts (16, 56). It is used as an inoculum for sourdough bread and sometimes also for bread making. Sourdough fermentations improve dough properties and bread texture and flavor, retard the staling process, and protect bread from mould and bacterial spoilage (8, 23, 40). Sourdough can be prepared in bakeries or obtained from commercial suppliers (3, 26). Due to their artisan and region-dependent handling, sourdoughs are an immense source of diverse LAB and yeast species and strains. Unfortunately, LAB from traditional sourdoughs cannot be properly identified by phenotypic approaches, such as fermentation patterns and cell wall analysis, due to the physiological, biochemical, and ecological specificity of these bacterial strains (56, 59). Molecular techniques, including 16S ribosomal DNA (rDNA) sequencing, randomly amplified polymorphic DNA (RAPD) analysis, and other PCR-based techniques, are necessary for the taxonomic investigation of complex ecosystems (44). These polyphasic approaches have allowed the description of new species, including Lactobacillus pontis (55), Lactobacillus panis (59), and Lactobacillus frumenti (30).
Based on the technology applied in their production, sourdoughs have been grouped into three types (2, 23, 54). Type I sourdoughs are traditional doughs sustained by continuous propagation at ambient temperatures of 20 to 30°C (2). Generally, traditional three-stage fermentation processes are used (23). Lactobacillus sanfranciscensis and L. pontis are the predominant LAB in these doughs (56). Also, Lactobacillus fructivorans, Lactobacillus fermentum, Lactobacillus brevis, and Lactobacillus farciminis were found in some of these doughs (42, 56). In contrast, leavening of type II sourdoughs is achieved by the addition of baker's yeast to the dough. This is essential, since type II doughs employ a less time-consuming one-stage fermentation process at temperatures exceeding 30°C. Type II doughs are mostly used in industrial processes. In most cases, the fermentation broth is applied as a dough-souring supplement and aroma carrier (48). The dominant LAB strains in these doughs are L. panis, L. pontis, Lactobacillus reuteri, Lactobacillus johnsonii, L. sanfranciscensis, L. fermentum, Lactobacillus delbrueckii, Lactobacillus acidophilus, L. brevis, Lactobacillus amylovorus, and L. frumenti (23, 30, 56). Type III doughs are dried preparations of dough (23). They are made by (traditional) sourdough fermentation with subsequent water evaporation (38). Greek traditional wheat sourdoughs belong to type I, but information about the LAB strains involved is lacking. To date, only data concerning the chemical characteristics and the yeast flora of these sourdoughs have been reported (33).
In sourdough, maltose is the most abundant fermentable carbohydrate, and hence maltose catabolism is a key process during fermentation. Among some sourdough LAB, in particular L. sanfranciscensis, a constitutive, intracellular maltose phosphorylase catalyzes the phosphorolytic cleavage of maltose, yielding glucose and glucose-1-phosphate (23, 56). In cells growing exponentially in maltose-containing media, hexokinase activity (which catalyzes the conversion of glucose to glucose-6-phosphate) is virtually absent and the nonphosphorylated glucose is excreted. In these heterofermentative LAB, glucose-1-phosphate is converted by phosphoglucomutase to glucose-6-phosphate, which is further metabolized via the phosphogluconate pathway to yield lactate, ethanol, and carbon dioxide. In dough, a number of metabolic conversions occur as a result of cometabolic interactions among the sourdough LAB. For example, the presence in the dough of fructose, citrate, malate, or oxygen allows the formation of additional metabolites, such as mannitol, succinate, and acetate. The acetate formed is of major importance for the development of flavor and the microbial stability of the bread. The molar ratio of lactate to acetate in bread is defined as the fermentation quotient (FQ), and it is considered optimum in the range 2.0 to 2.7 (23).
The aim of this paper was to examine the biodiversity and metabolic potential of the wild LAB microflora from Greek traditional wheat sourdoughs.
MATERIALS AND METHODS
Sourdough preparation.
Seven Greek traditional wheat sourdoughs (referred to as A, B, C, D, E, F, and G) were sampled in different geographical areas of Greece. Sourdoughs A to F were prepared according to the most common traditional procedure used locally. Leaves of basil (Ocimum basilicum) were suspended in tepid water and left at room temperature for 1 h. Then, the basil leaves were removed, wheat flour was added, and the dough was incubated for approximately 15 h at 30 to 40°C. The first sourdough formed was again mixed with flour, tepid water, and salt and incubated for another 15 h at the same temperature. The second sourdough is used as an inoculum for bread making, without the use of commercial yeast. For sourdough G, crushed chickpeas (Cicer arietinum), leaves from basil (O. basilicum) and pelargonium (Pelargonium grandifloris), and a small portion of ouzo were suspended in boiling water and incubated overnight at 30 to 40°C. Then, the supernatant foam was collected and mixed with flour, water, and salt, and the mixture was incubated overnight at 30 to 40°C. The sourdough is used as an inoculum for bread making, without the use of commercial yeast.
Sampling, isolation, and selection of lactic acid bacteria.
Sourdough samples were collected aseptically, the pH was recorded, and then the samples were stored at 4°C. Microbiological analysis was performed within 24 h. Samples (10 g) were diluted 1:10 with 90 ml of NaCl (0.9% [wt/vol]). LAB were enumerated by serial dilutions in sterile saline plated on MRS agar (Oxoid, Basingstoke, United Kingdom) supplemented with 2% (wt/vol) maltose. The plates were incubated at 30°C for 48 h. Purification of the strains was performed on the same medium by successive subculturing at 30°C. From each different colony type, one colony was selected for further characterization. The selected colonies were subcultured and stored at −80°C in 80% (vol/vol) glycerol.
Identification of LAB. (i) Strains and cultivation conditions.
The strains used throughout this study are listed in Table 1. They represent the isolates mentioned above and were deposited in the ACA-DC culture collection (Agricultural University of Athens, Athens, Greece). L. pontis LMG 14187T, L. pontis LMG 14188, L. sanfranciscensis LMG 11477, L. sanfranciscensis LMG 16002T, L. sanfranciscensis C57, and L. sanfranciscensis E13 were used as reference strains. The last two strains of L. sanfranciscensis are well characterized and originate from Italian wheat sourdoughs (22). The L. pontis and L. sanfranciscensis LMG culture collection strains (http://www.belspo.be/bccm/lgm.htm) were chosen for comparison. Lactobacillus alimentarius LMG 9187T, L. farciminis LMG 9200T, Lactobacillus kimchii LMG 19822T, and Lactobacillus paralimentarius LMG 19152T were used in the DNA-DNA hybridization and G+C content analyses. L. amylovorus DCE 471 was used as a control strain for the enzyme assays (13). The strains were cultivated for 24 to 48 h in modified MRS (mMRS) medium and incubated at 30°C under microaerophilic conditions unless otherwise indicated. mMRS medium contained (per liter) 20 g of peptone (Oxoid), 16 g of meat extract (Lab-Lemco; Oxoid), 8 g of yeast extract, 2 g of K2HPO4, 5 g of CH3COONa · 3H2O, 2 g of (NH4)3C6H5O7, 0.5 g of cysteine-HCl, 0.2 g of MgSO4 · 7H2O, 0.038 g of MnSO4 · H2O, 1 ml of Tween 80, and 10 g of maltose plus 5 g of fructose as carbohydrate sources and was adjusted to pH 5.4.
TABLE 1.
Growth characteristics of various L. brevis, L. paralimentarius, L. pontis, L. sanfranciscensis, and W. cibaria strains after 16 h of fermentation at 30°Ca
| Strainb | pH | OD600 | Biomass concn
|
|
|---|---|---|---|---|
| Serial dilution count (log CFU ml−1) | Biomass (g [CDM] liter−1) | |||
| L. brevis | ||||
| ACA-DC 3365 | 4.47 (0.03) | 8.1 (0.5) | 9.1 (0.4) | 1.83 (0.07) |
| ACA-DC 3368 | 4.41 (0.06) | 7.6 (0.6) | 9.0 (0.3) | 2.02 (0.06) |
| ACA-DC 3401 | 4.43 (0.03) | 7.5 (0.5) | 9.1 (0.2) | 2.0 (0.4) |
| ACA-DC 3404 | 4.47 (0.04) | 7.8 (0.4) | 9.7 (0.4) | 2.0 (0.3) |
| ACA-DC 3406 | 4.46 (0.01) | 7.9 (0.4) | 9.7 (0.6) | 2.1 (0.3) |
| ACA-DC 3410 | 4.44 (0.01) | 9 (1) | 9.0 (0.2) | 2.0 (0.2) |
| ACA-DC 3416 | 4.53 (0.06) | 8.0 (0.9) | 9.0 (0.2) | 1.7 (0.3) |
| ACA-DC 3420 | 4.53 (0.01) | 8.1 (0.6) | 9.17 (0.08) | 1.78 (0.09) |
| ACA-DC 4003 | 4.61 (0.05) | 7.3 (0.1) | 9.07 (0.06) | 1.71 (0.02) |
| L. brevis-like | ||||
| ACA-DC 3411 t1 | 4.62 (0.03) | 4.57 (0.02) | 7.8 (0.4) | 1.01 (0.05) |
| L. paralimentarius | ||||
| ACA-DC 3363 | 4.53 (0.09) | 3.27 (0.01) | 8.6 (0.2) | 0.8 (0.1) |
| ACA-DC 3413 | 4.2 (0.2) | 5 (1) | 8.5 (0.2) | 1.3 (0.4) |
| ACA-DC 3415 | 4.5 (0.1) | 3.5 (0.9) | 8.35 (0.07) | 1.0 (0.3) |
| L. pontis | ||||
| LMG 14187T | 4.8 (0.3) | 4 (1) | 8.2 (0.2) | 1.3 (0.6) |
| LMG 14188 | 4.8 (0.2) | 4.5 (0.6) | 8.2 (0.4) | 1.6 (0.5) |
| L. sanfranciscensis | ||||
| ACA-DC 3370 | 4.30 (0.02) | 6.4 (0.3) | 9.0 (0.2) | 2.0 (0.1) |
| ACA-DC 3373 | 4.43 (0.04) | 5.2 (0.8) | 8.4 (0.4) | 1.6 (0.6) |
| ACA-DC 3378 | 4.45 (0.03) | 5.13 (0.08) | 9.1 (0.1) | 1.9 (0.2) |
| ACA-DC 3380 | 4.35 (0.03) | 7.0 (0.3) | 9.0 (0.4) | 2.2 (0.2) |
| ACA-DC 3382 | 4.35 (0.05) | 7.15 (0.06) | 8.4 (0.7) | 2.1 (0.4) |
| ACA-DC 3383 | 4.36 (0.04) | 5.5 (0.3) | 9.0 (0.3) | 1.6 (0.4) |
| ACA-DC 3408 | 4.40 (0.03) | 5.5 (0.1) | 8.33 (0.06) | 1.4 (0.3) |
| ACA-DC 3426 | 4.44 (0.02) | 3.4 (0.2) | 8.7 (0.1) | 1.5 (0.2) |
| LMG 11477 | 4.61 (0.04) | 4.6 (0.2) | 8.17 (0.06) | 1.2 (0.1) |
| LMG 16002T | 4.7 (0.1) | 3.4 (0.5) | 8.32 (0.07) | 1.0 (0.1) |
| C57 | 4.55 (0.04) | 5.6 (0.5) | 8.7 (0.4) | 1.7 (0.2) |
| E13 | 4.56 (0.05) | 5.3 (0.7) | 8.6 (0.4) | 1.3 (0.3) |
| W. cibaria | ||||
| ACA-DC 3385 | 4.37 (0.04) | 4.53 (0.05) | 8.86 (0.08) | 0.82 (0.05) |
| ACA-DC 3387 | 4.39 (0.06) | 5 (1) | 9.0 (0.1) | 0.86 (0.07) |
| ACA-DC 3392 | 4.52 (0.02) | 3.89 (0.09) | 8.79 (0.08) | 0.9 (0.2) |
| ACA-DC 3394 | 4.41 (0.03) | 4.6 (0.1) | 9.04 (0.05) | 1.0 (0.2) |
| ACA-DC 3396 | 4.53 (0.03) | 4.0 (0.3) | 8.92 (0.07) | 0.8 (0.2) |
| ACA-DC 3402 | 4.55 (0.01) | 4.1 (0.6) | 8.75 (0.07) | 0.8 (0.2) |
All fermentations were carried out in mMRS medium, adjusted to pH 5.4, containing (per liter) 20 g of peptone (Oxoid), 16 g of meat extract (Lab-Lemco; Oxoid), 8 g of yeast extract, 2 g of K2HPO4, 5 g of CH3COONa·3H2O, 2 g of (NH4)3C6H5O7, 0.5 g of cysteine-HCl, 0.2 g of MgSO4·7H2O, 0.038 g of MnSO4·H2O, 1 ml of Tween 80, and 10 g of maltose plus 5 g of fructose as carbohydrate sources. All determinations were performed in triplicate. The standard deviations are given in parentheses.
DCE, Department of Chemical Engineering (Brussels, Belgium); LMG, BCCM/LMG Bacteria Collection (Laboratory of Microbiology, University of Ghent, Ghent, Belgium).
(ii) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of whole-cell proteins.
Whole-cell protein extracts were prepared, and SDS-PAGE was performed as described by Pot et al. (35). Duplicate protein extracts were prepared to check the reproducibility of the growth conditions and the preparation of the extracts. Registration of the protein patterns, normalization of the densitometric traces, pattern storage, and grouping of the strains using the Pearson product moment correlation coefficient (r) and unweighted pair-group method using arithmetic averages (UPGMA) cluster analysis were performed as described by Pot et al., using Gel Compar version 4.2 software (Applied Maths, Sint-Martens-Latem, Belgium) (35).
(iii) RAPD analysis.
Five milliliters of an overnight culture at 30°C in MRS broth (Oxoid) supplemented with 2% (wt/vol) maltose were pelleted by centrifugation (5,000 × g; 15 min) and resuspended in 180 μl of ATL lysis buffer (QIAamp Tissue kit; Qiagen GmbH, Hilden, Germany). The cells were sonicated for 2 min (UP 50 H; Hielscher GmbH, Stahnsdorf, Germany). DNA isolation and purification were performed using the QIAamp tissue kit (Qiagen GmbH) according to the manufacturer's instructions.
RAPD-PCR was performed in a total volume of 50 μl. The arbitrary universal primer M13V (5′-GTT TCC CCA GTC ACG AC-3′) was used. The reaction mixture contained 20 pmol of primer (Interactiva, Ulm, Germany), 1.5 U of Taq polymerase, 5 μl of 10× reaction buffer, 200 nM each deoxynucleoside triphosphate, 1 μl of genomic DNA, and 5 mM MgCl2 (all components from Amersham Pharmacia Biotech, Freiburg, Germany). PCRs were carried out on a Hybaid Omni Gene (MWG-Biotech, Ebersberg, Germany) as follows: three cycles of 96°C for 3 min, 35°C for 5 min, and 75°C for 5 min and 32 cycles of 96°C for 1 min, 55°C for 2 min, and 75°C for 3 min.
PCR products were electrophoretically separated on a 1.5% (wt/vol) agarose gel (0.5× Tris-borate-EDTA buffer [45 mM Tris-borate, 1 mM EDTA]) in an MWG-Biotech chamber (20 by 25 cm) at a constant voltage of 170 V for 3 h. As a size marker encompassing the whole range of fragments, 1 μg of the BioSizer (AGS, Heidelberg, Germany) was loaded. After 15 min of staining with ethidium bromide, patterns were digitally saved by the EASY system (Herolab, Grieheim, Germany). Registration of the PCR patterns, normalization of the densitometric traces, pattern storage, grouping of the strains using the Pearson product moment correlation coefficient (r), and UPGMA cluster analysis were performed as described by Pot et al. using Gel Compar version 4.1 software (35).
(iv) 16S rDNA sequence analysis.
Genomic DNA was extracted by using the DNA isolation protocol of Niemann et al. (32). A fragment of the 16S rDNA gene (corresponding to positions 8 to 1541 in the Escherichia coli numbering system) was amplified by PCR using the conserved primers pA (5′ AGA GTT TGA TCC TGG CTC AG 3′) and pH (5′ AAG GAG GTG ATC CAG CCG CA 3′). The PCR product was purified using a Qiaquick PCR purification kit according to the manufacturer's instructions (Qiagen GmbH). Sequencing was performed with an Applied Biosystems 377 DNA sequencer and the protocols of the manufacturer using the ABI Prism Dye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems, Foster City, Calif.). Nearly complete sequences of strains ACA-DC 3363 (1,519 bp), ACA-DC 3411 t1 (1,518 bp), ACA-DC 3411 t2 (1,529 bp), ACA-DC 3413 (1,519 bp), ACA-DC 3415 (1,518 bp), C57 (1,460 bp), LMG 14187 (1,480 bp), and LMG 14188 (1,527 bp) were determined using the eight sequencing primers listed in Coenye et al. (6). Partial sequences of strains ACA-DC 3368 (696 bp), ACA-DC 3401 (985 bp), ACA-DC 3406 (708 bp), ACA-DC 3408 (804 bp), ACA-DC 3416 (708 bp), and ACA-DC 4003 (634 bp) were determined using three sequencing primers (gamma, *gamma, and pD). Sequence assembly was performed with the program AutoAssembler (Applied Biosystems). Phylogenetic analysis was performed using the software package Bionumerics (version 2.5; Applied Maths) and was based on the neighbor-joining method after including the consensus sequences in an alignment of small ribosomal subunit sequences collected from the international nucleotide sequence library EMBL. Unknown bases were excluded from the calculations.
(v) DNA base compositions and DNA-DNA hybridization experiments.
High-molecular-mass DNA was prepared from strains ACA-DC 3363, ACA-DC 3413, and ACA-DC 3415 and from the phylogenetically related reference strains L. alimentarius LMG 9187T, L. farciminis LMG 9200T, L. kimchii LMG 19822T, and L. paralimentarius LMG 19152T. The cells were cultivated in MRS broth supplemented with 2% (wt/vol) CaCO3 as a buffer component at 28°C under aerobic conditions for 24 h. DNA was extracted from 0.75 to 1.25 g (wet weight) of cells using the protocol described by Pitcher et al. (34) with the following modifications: the washed cell pellet was resuspended and lysed overnight in a buffer (10 mM Tris-HCl, 300 mM EDTA, pH 8.0) containing RNase (200 μg ml−1; Sigma-Aldrich, St. Louis, Mo.), mutanolysin (500 U ml−1; Sigma), and lysozyme (25 mg ml−1; Serva Electrophoresis GmbH, Heidelberg, Germany) at 37°C. Proteinase K (200 μg ml−1; Merck, Darmstadt, Germany) was added to the cell suspension for 15 min before the addition of guanidinium thiocyanate-EDTA-Sarkosyl reagent. The DNA was enzymatically degraded into nucleosides as described by Mesbah et al. (29). The obtained nucleoside mixture was then separated by high-performance liquid chromatography using a Symmetry Shield C8 column (Waters Corp., Milford, Mass.) with the thermostat at 37°C. The solvent was 0.02 M NH4H2PO4 (pH 4.0) with 1.5% acetonitrile. Nonmethylated lambda phage DNA (Sigma) was used as the calibration reference.
DNA-DNA hybridizations were performed with photobiotin-labeled probes in microplate wells as described by Ezaki et al., using a HTS7000 Bio Assay Reader (Perkin Elmer, Wellesley, Mass.) for the fluorescence measurements (14). Biotinylated DNA was hybridized with single-stranded unlabeled DNA noncovalently bound to microplate wells. Hybridizations were performed at 34°C in hybridization mixture (2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 5× Denhardt solution, 2.5% dextran sulfate, 50% formamide, 100 μg of denatured salmon sperm DNA per ml, and 1,250 ng of biotinylated DNA probe per ml).
Fermentation experiments. (i) Incubation conditions.
The fermentation experiments were performed in 100-ml glass bottles at an incubation temperature of 30°C in mMRS medium containing both maltose and fructose as carbohydrate sources. The experiments were also performed with fructose as the sole carbohydrate source to check fructose fermentation. Strains stored at −80°C were propagated twice in the appropriate medium before experimental use. The transfer inoculum was always 1% (vol/vol). After 16 h of incubation, samples were aseptically withdrawn for further analysis.
(ii) Growth assessment.
The pH of the culture medium was determined by using a pH meter (WTC pH 526 m; Euro-Scientific, Lint, Belgium). Optical density was measured at 600 nm (OD600), CFU were determined by serial dilution counts on mMRS agar, and biomass (as cell dry mass [CDM]) was determined gravimetrically after membrane filtration (0.45-μm-pore-size filters, type HA; Millipore Corp., Bedford, Mass.).
(iii) Determination of metabolites.
The residual sugar levels and metabolites formed were analyzed by high-performance liquid chromatography using a Waters chromatograph equipped with a Waters 410 differential refractometer, a Waters column oven, a Waters 717 plus autosampler, and Millenium software (version 2.10). Cells and solid particles were removed from 1.5-ml samples (appropriately diluted with ultrapure water) by microcentrifugation (13,000 rpm, 20 min; Biofuge, Meraeus, Hanau, Germany). Proteins were removed by the addition of an isovolume of 20% trichloroacetic acid, microcentrifugation (13,000 rpm; 20 min), and filtration through a nylon syringe filter (Euro-Scientific). A 30-μl portion was injected into a Polyspher OA KC column (Merck) held at 35°C. As the mobile phase, a 0.005 N H2SO4 solution was used at a fixed flow rate of 0.4 ml min−1. Ethanol was determined by an enzymatic method using an Ethanol Determination kit (STAG; R-Biopharm GmbH, Darmstadt, Germany). Mannitol and fructose were determined by high-performance anion-exchange chromatography with pulsed amperometric detection (Dionex, Sunnyvale, Calif.), using a Carbopac PA10 Column (Dionex). The elution was done isocratically with 50 mmol of NaOH liter−1.
Enzyme activities. (i) Preparation of cell extracts.
The strains L. brevis ACA-DC 3406, L. paralimentarius ACA-DC 3415, L. pontis LMG 14187, L. sanfranciscensis ACA-DC 3426, W. cibaria ACA-DC 3385, and L. amylovorus DCE 471 were propagated twice at 30°C in mMRS medium. The transfer inoculum was 1% (vol/vol). At the end of the exponential phase, the preculture was added to 1 liter of the same medium and incubated again at 30°C. The cell extract of 1 liter of culture was obtained as described by Stolz et al. and Degeest and De Vuyst with slight modifications (12, 51). When the culture reached the stationary growth phase (after 16 h), the cells were harvested by centrifugation at 20,000 × g and 5°C for 15 min. The harvested cells were washed twice with 100 mmol of ice-cold citrate-NaOH buffer liter−1 at pH 6.0 (buffer A) or 100 mmol of potassium phosphate buffer liter−1 at pH 6.0 (buffer B), depending on the subsequent enzyme assay. After being washed, the cells were resuspended in the same buffer to obtain 2 ml of total cell suspension per g (wet weight) of cells and further held on ice. The chilled cells were lysed using a Vibra-Cell sonicator (Sonic & Materials Inc., Danbury, Conn.) with a microtip setting (sonic power, 375 W; output control, 5) for 6 min and a 50% duty cycle consisting of 30 s of sonication pulses and 30 s of rest. Cell debris was removed by centrifugation (47,800 × g at 4°C for 15 min), and the supernatant fluid (the cell extract) was stored at −60°C until it was used for the enzyme and protein assays.
(ii) Enzyme assays.
Maltose phosphorylase activity in crude extracts was determined according to the method of Stolz et al. with a slight modification in that phosphoglucomutase was added (51). Two different buffers at pH 6.0, buffer A (100 mmol · liter−1 citrate-NaOH buffer) and buffer B (100 mmol · liter−1 potassium phosphate buffer), were used to check the presence of the enzyme maltose phosphorylase, which requires phosphate for activity. For the assay, the following components were mixed: 670 μl of buffer A or B, 50 μl of 20-mmol liter−1 MgSO4 · 7H2O, 20 μl of 1-mmol liter−1 glucose-1,6-bis-phosphate, 20 μl (10 U) of glucose-6-phosphate dehydrogenase, 20 μl (10 U) of phosphoglucomutase, 100 μl of 10-mmol liter−1 NADP+, and 20 μl of enzyme extract. After 5 min of preincubation at 30°C, 100 μl of either 0.5-mol liter−1 maltose or buffer A or B (controls) was added to start the reaction. NADPH formation at 30°C was determined at 340 nm. One unit of maltose phosphorylase activity catalyzes the reduction of 1 μmol of NADP+ per min.
The hexokinase activity was determined according to the method of Stolz et al. by mixing the following components: 610 μl of buffer B, 300 μl of solution 1 (NADP+-ATP solution; enzymatic glucose assay kit from Boehringer Mannheim, Mannheim, Germany), 20 μl (10 U) of glucose-6-phosphate dehydrogenase, and 20 μl of enzyme extract (51). After 5 min of preincubation at 30°C, 20 μl of either 0.5-mol liter−1 glucose or buffer B (control) was added. NADPH formation at 30°C was determined at 340 nm. One unit of hexokinase activity results in the reduction of 1 μmol of NADP+ per min.
The phosphoglucomutase activity was determined according to the method of Qian et al. and Degeest and De Vuyst with some modifications (12, 36). For the assay, the following components were mixed: 653 μl of 100-mmol liter−1 potassium phosphate buffer (pH 7.4), 67 μl of 10 mM NADP+, 20 μl of 1 mM glucose-1,6-bis-phosphate, 100 μl of 300 mM MgCl2, 100 μl of 430-mmol liter−1 l-cysteine, 20 μl (10 U) of glucose-6-phosphate dehydrogenase, and 20 μl of enzyme extract. After 5 min of preincubation at 30°C, 20 μl of either 0.5-mol liter−1 α-glucose-1-phosphate or 100-mmol liter−1 potassium phosphate buffer (pH 7.4) (control) was added. NADPH formation at 30°C was determined at 340 nm. One unit of phosphoglucomutase activity results in the reduction of 1 μmol of NADP+ per min.
All enzyme activities were measured within the linear range of initial velocity.
(iii) Protein assays.
Protein concentrations were determined using a Detergent Compatible Protein Assay kit (serial number 500-0116; Bio-Rad Laboratories, Nazareth-Eke, Belgium). Bovine serum albumin was used as a standard.
Nucleotide sequence accession numbers.
The sequences in this study were deposited in the EMBL database (EMBL-EBI; Hinxton, Cambridge, United Kingdom) (see Fig. 3 for accession numbers).
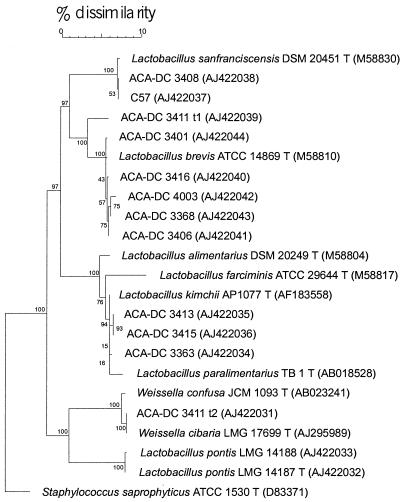
FIG. 3.
Neighbor-joining dendrogram showing the phylogenetic positions of Greek sourdough LAB isolates and related reference taxa based on 16S rDNA sequence comparisons. Staphylococcus saprophyticus was used as the outgroup, and bootstrap probability values are indicated at the branch points (100 trees were resampled). The EMBL accession numbers are given in parentheses after the strain designations.
RESULTS
Chemical and microbiological characterization of the sourdoughs.
Results concerning pHs and LAB populations of the prepared sourdoughs are given in Table 2.
TABLE 2.
Acidities and microbial counts of Greek wheat sourdoughs tested
| Sourdough | pH | No. of cocci (107 CFU) | No. of bacilli (107 CFU) |
|---|---|---|---|
| A | 3.75 | 7 | |
| B | 3.58 | 0.8 | 19.2 |
| C | 3.73 | 82 | |
| D | 4.09 | 0.06 | |
| E | 3.78 | 1 | 609 |
| F | 3.70 | 0.05 | |
| G | 4.75 | 0.5 | 5.3 |
A pH of 4.75 was determined for sample G. In the other samples, the pH values ranged from 4.09 to 3.58, which were similar to those reported in the literature (38). The rather high pH of sample G could be attributed to the dominance of low-acidifying species of LAB because of the different technology applied in sourdough preparation as well as the buffering capacity of the flour used.
Among the LAB, lactobacilli were the predominant microbes. Cocci were isolated only from sourdoughs B, E, and G, and in rather low numbers (Table 2).
PAGE of whole-cell proteins.
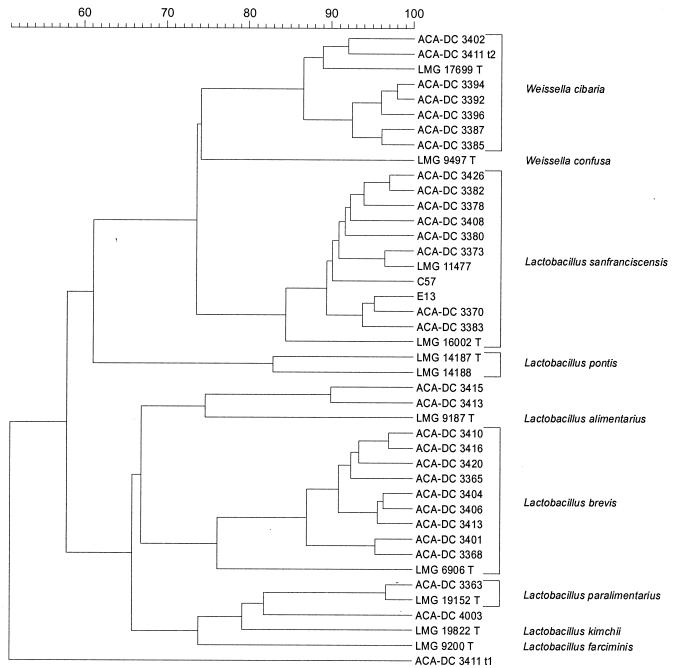
The correlation level for duplicate protein patterns was an r value of >94%. The whole-cell protein profiles of the newly isolated strains were initially compared with a database of reference patterns representing all validly described LAB species (data not shown). Based on these data, the majority of the isolates were identified at the species level. As visualized in a summarizing dendrogram (Fig. 1), three major clusters were delineated. Eight isolates (ACA-DC 3365, ACA-DC 3368, ACA-DC 3401, ACA-DC 3404, ACA-DC 3406, ACA-DC 3410, ACA-DC 3416, and ACA-DC 3420) were assigned to the species L. brevis, and another eight isolates (ACA-DC 3370, ACA-DC 3373, ACA-DC 3378, ACA-DC 3380, ACA-DC 3382, ACA-DC 3383, ACA-DC 3408, and ACA-DC 3426), as well as strains C57 and E13, were identified as L. sanfranciscensis. Seven isolates (ACA-DC 3385, ACA-DC 3387, ACA-DC 3392, ACA-DC 3394, ACA-DC 3396, ACA-DC 3402, and ACA-DC 3411 t2) belonged to the recently described species W. cibaria (1). One isolate, ACA-DC 3363, was identified as L. paralimentarius. The positions of the other four strains (ACA-DC 3411 t1, ACA-DC 3413, ACA-DC 3415, and ACA-DC 4003) were unclear.
FIG. 1.
Cluster analysis of digitized SDS-PAGE whole-cell protein profiles of Greek sourdough LAB isolates and representative reference strains. Similarities were expressed by the Pearson product moment correlation coefficient, r, and converted to percentage values, and clustering was performed by UPGMA analysis.
RAPD analysis.
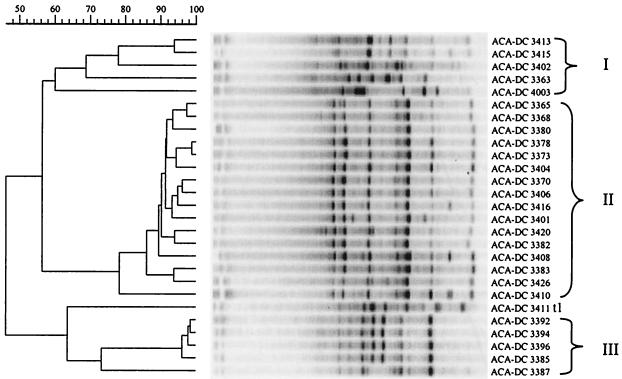
Cluster analysis of the RAPD-PCR patterns is shown in Fig. 2. One major (cluster II) and two minor (clusters I and III) clusters were formed. Sixteen out of the 27 strains, ACA-DC 3365, ACA-DC 3368, ACA-DC 3380, ACA-DC 3378, ACA-DC 3373, ACA-DC 3404, ACA-DC 3370, ACA-DC 3406, ACA-DC 3416, ACA-DC 3401, ACA-DC 3420, ACA-DC 3382, ACA-DC 3408, ACA-DC 3383, ACA-DC 3426, and ACA-DC 3410, were grouped in cluster II with a correlation of 79%. These strains were identified by SDS-PAGE analysis as either L. sanfranciscensis or L. brevis. In this context, the RAPD analysis with the M13V primer could not discriminate between the two species. Cluster III included six strains at a correlation of 62%; five of them, ACA-DC 3387, ACA-DC 3385, ACA-DC 3392, ACA-DC 3394, and ACA-DC 3396, were identified as W. cibaria by SDS-PAGE analysis, while the identification of strain ACA-DC 3411 t1 was unclear. Finally, strains ACA-DC 3413, ACA-DC 3415, ACA-DC 3402, ACA-DC 3363, and ACA-DC 4003 grouped with a correlation of 60% in cluster I. According to the SDS-PAGE analysis, strain ACA-DC 3402 was identified as W. cibaria and strain ACA-DC 3363 was identified as L. paralimentarius, while strains ACA-DC 3413, ACA-DC 3415, and ACA-DC 4003 could not be identified.
FIG. 2.
Cluster analysis of RAPD-PCR patterns of Greek sourdough LAB isolates. Similarities were expressed by the Pearson product moment correlation coefficient, r, and converted to percentage values, and clustering was performed by UPGMA analysis.
16S rDNA sequence analysis.
To establish the phylogenetic positions of isolates which could not be assigned to a particular species by SDS-PAGE of whole-cell proteins and to confirm the species status of the delineated and named protein electrophoretic clusters, the 16S rDNA genes of representative strains were amplified (completely or partially) and compared to deposited sequences available in the EMBL database. A phylogenetic tree is presented in Fig. 3. Strains ACA-DC 3368, ACA-DC 3401, ACA-DC 3406, ACA-DC 3416, and ACA-DC 4003 showed high levels of similarity (>99.0%) to reference strains of the species L. brevis, and strains ACA-DC 3408 and C57 showed high levels of similarity to L. sanfranciscensis. No similarities significant for possible relatedness at the species level (>97.0%) were obtained with any other validly described LAB species (47). Strain ACA-DC 3411 t1, which grouped separately after protein profiling, showed a 16S rDNA similarity of 97.1% to the type strain of L. brevis. This value does not exclude relatedness at the species level, and for this reason we will refer to the strain as L. brevis-like. Of the homogeneous W. cibaria cluster, strain ACA-DC 3411 t2 was selected for 16S rDNA sequencing and showed the highest similarity to the type strain of the species W. cibaria (99.9%). Although similarities significant for possible species relatedness were also obtained with the closely related species W. confusa (99.4%), the characteristic protein pattern of strains of W. cibaria confirms the species status as W. cibaria. Strain ACA-DC 3363, identified as L. paralimentarius by protein profiling, and the unidentified but analogous strains ACA-DC 3413 and ACA-DC 3415 yielded similar 16S rDNA sequences. High levels of similarity were obtained with L. kimchii (99.5 to 99.6%), L. paralimentarius (98.5 to 98.8%), L. alimentarius (97.5 to 97.8%), and L. farciminis (96.9 to 97.1%). No conclusions concerning the species status of strains ACA-DC 3413 and ACA-DC 3415 could be drawn from these data.
DNA base compositions and DNA-DNA hybridization experiments.
The G+C contents (in moles percent) of all isolates and reference strains of the L. alimentarius phylogenetic group were investigated, and the bacteria were included in a DNA-DNA hybridization study. The results are summarized in Table 3. The DNA base composition, with values in a narrow range of 35 to 37 mol% G+C, did not differentiate among species within this phylogenetic group. Low DNA-DNA binding values between L. alimentarius and L. farciminis and the other taxa confirm their separate species status. An intermediate level of DNA-DNA binding of 68% was observed between L. paralimentarius and L. kimchii. The hybridization results confirmed the assignment of strain ACA-DC 3363 to the species L. paralimentarius (binding value, 99%), although intermediate homology levels were also obtained with L. kimchii (66%). The taxonomic position of the strains ACA-DC 3413 and ACA-DC 3415 is less clear: intermediate binding values are obtained with L. paralimentarius (70 to 74%) and L. kimchii (58%). As 70% is generally accepted as the borderline for species delineation, we classified these strains in the species L. paralimentarius.
TABLE 3.
Percent DNA-DNA binding values of Greek sourdough isolates and related reference strains
| Strain | % G+C | DNA-DNA binding value
|
||||||
|---|---|---|---|---|---|---|---|---|
| LMG 9187T | LMG 9200T | LMG 19152T | LMG 19822T | ACA-DC 3363 | ACA-DC 3413 | ACA-DC 3415 | ||
| L. alimentarius LMG 9187T | 36.5 | 100 | ||||||
| L. farciminis LMG 9200T | 37.0 | 26 | 100 | |||||
| L. paralimentarius LMG 19152T | 36.2 | 36 | 27 | 100 | ||||
| L. kimchii LMG 19822T | 35.1 | 30 | 27 | 68 | 100 | |||
| ACA-DC 3363 | 35.7 | 34 | 29 | 99 | 66 | 100 | ||
| ACA-DC 3413 | 36.6 | 33 | 27 | 74 | 58 | 66 | 100 | |
| ACA-DC 3415 | 36.5 | 31 | 25 | 70 | 58 | 67 | 96 | 100 |
Growth and metabolite formation.
The growth characteristics of and metabolites formed by 11 L. brevis strains, 3 L. paralimentarius strains, 2 L. pontis strains, 12 L. sanfranciscensis strains, and 6 W. cibaria strains after 16 h of growth at 30°C in mMRS medium with maltose and fructose as sole carbohydrate sources are displayed in Tables 1 and 4.
TABLE 4.
Major metabolites formed by cells of various L. brevis, L. paralimentarius, L. pontis, L. sanfranciscensis, and W. cibaria strains in culture supernatant after 16 h of fermentation at 30°Ca
| Strain | Amt of metabolite (mmol liter−1)
|
Fructose fermentationb | FQ (lactic acid/ acetic acid) | |||||
|---|---|---|---|---|---|---|---|---|
| Maltose | Fructose | Lactic acid | Acetic acid | Ethanol | Mannitol | |||
| L. brevis | ||||||||
| ACA-DC 3365 | −31 (1) | −27.0 (0.3) | 58 (3) | 15 (1) | 32 (1) | 18.4 (0.8) | Positive | 3.9 (0.2) |
| ACA-DC 3368 | −30.5 (0.7) | −27 (2) | 61 (2) | 13 (3) | 34 (2) | 19.7 (0.8) | Positive | 4.7 (0.7) |
| ACA-DC 3401 | −30.5 (0.2) | −27.2 (0.1) | 60.7 (0.7) | 13 (3) | 35 (1) | 19 (2) | Positive | 4.6 (0.9) |
| ACA-DC 3404 | −30.3 (0.2) | −29.5 (0.1) | 60.6 (0.4) | 15 (3) | 31.9 (0.9) | 19 (1) | Positive | 4.2 (0.9) |
| ACA-DC 3406 | −30.4 (0.2) | −28.7 (0.6) | 62 (1) | 15 (3) | 35 (2) | 19 (1) | Positive | 4.3 (0.9) |
| ACA-DC 3410 | −30.3 (0.2) | −26.9 (0.2) | 59.1 (0.8) | 16 (2) | 34 (2) | 18 (2) | Positive | 3.6 (0.4) |
| ACA-DC 3416 | −28 (3) | −26 (2) | 57 (5) | 15 (5) | 29 (4) | 18 (1) | Positive | 4 (2) |
| ACA-DC 3420 | −30.8 (0.6) | −29.7 (0.3) | 62.0 (0.7) | 15 (3) | 37 (4) | 20 (2) | Positive | 4.3 (0.9) |
| ACA-DC 4003 | −29.5 (0.1) | −29 (2) | 61.7 (0.6) | 12.7 (0.5) | 38 (1) | 17.6 (0) | Positive | 4.9 (0.2) |
| L. brevis-like | ||||||||
| ACA-DC 3411 t1 | −20 (1) | −3 (3) | 49 (4) | 19 (9) | 21 (3) | 1.0 (0.1) | Positive | 3 (1) |
| L. paralimentarius | ||||||||
| ACA-DC 3363 | −2.0 (0.7) | −24 (6) | 48 (3) | 0 | 0.5 (0.3) | 3 (5) | Positive | NRc |
| ACA-DC 3413 | −15.8 (0.4) | −15 (3) | 103 (2) | 0 | 0.7 (0.2) | 0.2 (0.4) | Positive | NR |
| ACA-DC 3415 | −1.40 (0.09) | −22 (7) | 61.2 (0.3) | 0 | 0.10 (0.06) | 0.2 (0.4) | Positive | NR |
| L. pontis | ||||||||
| LMG 14187T | −18 (12) | −26 (3) | 28 (22) | 16 (3) | 11 (9) | 24 (2) | Positive | 2 (2) |
| LMG 14188 | −19 (6) | −26 (1) | 37 (12) | 16 (3) | 19 (12) | 23 (1) | Positive | 2.3 (0.6) |
| L. sanfranciscensis | ||||||||
| ACA-DC 3370 | −31 (1) | −29 (1) | 62.6 (0.7) | 28 (1) | 24 (2) | 25.1 (0.9) | Negative | 2.25 (0.06) |
| ACA-DC 3373 | −28 (2) | −29.3 (0.8) | 65 (2) | 30 (3) | 20 (2) | 24 (1) | Negative | 2.2 (0.2) |
| ACA-DC 3378 | −27 (2) | −28 (2) | 63 (1) | 26 (2) | 19 (2) | 25.5 (0.4) | Negative | 2.4 (0.2) |
| ACA-DC 3380 | −30.6 (0.9) | −29.6 (0.4) | 63 (1) | 28.5 (0.9) | 23 (1) | 25 (2) | Negative | 2.21 (0.03) |
| ACA-DC 3382 | −30.2 (0.2) | −29 (1) | 61.6 (0.3) | 29 (3) | 23 (2) | 26 (1) | Negative | 2.2 (0.2) |
| ACA-DC 3383 | −29 (1) | −29.5 (0.1) | 58 (1) | 30 (5) | 20 (5) | 26 (2) | Negative | 2.0 (0.4) |
| ACA-DC 3408 | −31 (2) | −29.9 (0.5) | 72 (4) | 28 (2) | 23 (3) | 23.2 (0.3) | Negative | 2.6 (0.2) |
| ACA-DC 3426 | −29.8 (0.2) | −33.8 (0.1) | 64.7 (0.9) | 27 (2) | 20.3 (0.5) | 23.4 (0.7) | Negative | 2.4 (0.2) |
| LMG 11477 | −21 (2) | −29.7 (0.3) | 45 (2) | 31 (2) | 12 (1) | 24 (3) | Negative | 1.4 (0.2) |
| LMG 16002T | −17 (4) | −29 (2) | 34 (4) | 17 (4) | 10 (2) | 24 (4) | Negative | 2.0 (0.3) |
| C57 | −28 (3) | −1 (2) | 69 (7) | 19 (9) | 30 (4) | 0.1 (0.1) | Negative | 4 (2) |
| E13 | −28 (3) | −2 (1) | 65 (4) | 13 (9) | 28 (2) | 0.1 (0.1) | Negative | 6 (3) |
| W. cibaria | ||||||||
| ACA-DC 3385 | −27 (2) | −11 (1) | 69.5 (0.6) | 3 (2) | 29 (3) | −2 (1) | Positive | 27 (12) |
| ACA-DC 3387 | −30 (2) | −13 (1) | 77 (2) | 3 (1) | 31 (2) | 0.0 (0.8) | Positive | 25 (7) |
| ACA-DC 3392 | −21.8 (0.7) | −9.6 (0.7) | 56 (2) | 3 (2) | 26 (1) | −2 (2) | Positive | 19 (9) |
| ACA-DC 3394 | −28 (1) | −13 (2) | 71 (2) | 4.2 (0.7) | 30.9 (0.8) | −1 (2) | Positive | 17 (2) |
| ACA-DC 3396 | −22 (1) | −8.7 (0.2) | 58 (3) | 5 (2) | 26 (4) | −1 (2) | Positive | 14 (5) |
| ACA-DC 3402 | −22 (2) | −14 (2) | 65 (6) | 5 (5) | 29 (3) | 0.2 (0.6) | Positive | 18 (10) |
All fermentations were carried out in mMRS medium (see note a to Table 1). All determinations were performed in triplicate. The standard deviation is given in parentheses. Negative values indicate consumption of substrates; positive values indicate products formed.
Fructose fermentation was determined by analyzing the fructose consumption in the fermentation medium lacking maltose.
NR, not relevant.
During growth, the pH of the medium decreased from 5.4 to between 4.3 and 4.7 and to between 4.1 and 4.5 for the heterofermentative and homofermentative strains, respectively. The OD value was between 3.3 and 9.0, the number of CFU varied between 7.8 and 9.7 log units, and the biomass averaged from 0.8 to 2.2 g (CDM) liter−1. All sourdough LAB strains tested consumed maltose, which is converted to, among others, lactic acid, because the amount of lactic acid that could be derived from fructose alone was exceeded. In contrast, two L. paralimentarius strains (ACA-DC 3363 and ACA-DC 3415) hardly fermented maltose in the presence of fructose. Except for the L. sanfranciscensis strains, all sourdough LAB strains were able to grow on fructose as the sole carbohydrate source. In addition, all sourdough LAB strains, except the L. paralimentarius and W. cibaria strains, produced mannitol by using fructose as an electron acceptor. Whereas the L. paralimentarius and W. cibaria strains converted fructose into lactic acid homofermentatively, all other fructose-positive sourdough LAB strains fermented maltose to a mixture of lactic acid, acetic acid, and ethanol, and they converted fructose mainly to mannitol. Interestingly, the L. sanfranciscensis C57 and E13 strains did not produce mannitol. All other L. sanfranciscensis strains formed mannitol in amounts nearly equimolar to the fructose consumption (average conversion, 86%). This was also observed for the L. pontis strains (conversion, 88%). However, the L. pontis strains produced less lactic acid and acetic acid (lower maltose conversion). All L. brevis strains produced less acetic acid, less mannitol (conversion, 68%), and more ethanol than the L. sanfranciscensis strains. Since a smaller amount of fructose was used as an electron acceptor by L. brevis than by L. sanfranciscensis or L. pontis, the FQ of the first was higher (4.2 compared to 2.2). Remarkably, the L. brevis-like strain ACA-DC3411 t1 showed a metabolite pattern different from those of the other L. brevis strains: it did not use fructose as an electron acceptor, and it produced less lactic acid and ethanol but more acetic acid from maltose. Finally, no maltose phosphorylase activity could be detected in any of the strains tested. In contrast, low hexokinase and phosphoglucomutase activities were found, except for the W. cibaria ACA-DC 3385 strain (Table 5). Indeed, glucose was not observed in the culture medium after 12 h of growth for any of the strains studied. This indicates that glucose was not released into the medium during growth, as was confirmed by an analysis of the entire fermentation course of L. sanfranciscensis ACA-DC 3426 (results not shown).
TABLE 5.
Hexokinase and phosphoglucomutase activities in crude extracts of L. brevis ACA-DC 3406, L. paralimentarius ACA-DC 3415, L. pontis LMG 14187T, L. sanfranciscensis ACA-DC 3426, W. cibaria ACA-DC 3385, and L. amylovorus DCE 471 after 16 h of fermentation at 30°C in mMRS medium
| Strain | Protein content (mg ml−1) | Hexokinase (U mg−1) | Phosphoglucomutase (U mg−1) |
|---|---|---|---|
| L. brevis ACA-DC 3406 | 12.5 | 2.04 | 0.042 |
| L. paralimentarius ACA-DC 3415 | 33.3 | 1.31 | 0.026 |
| L. pontis LMG 14187T | 28.0 | 3.17 | 0.042 |
| L. sanfranciscensis ACA-DC 3426 | 12.8 | 4.42 | 0.095 |
| W. cibaria ACA-DC 3385 | 9.6 | 11.68 | 0.112 |
| L. amylovorus DCE 471 | 7.8 | 4.02 | 0.037 |
DISCUSSION
Recent research on sourdoughs has been focused on the identification of the microflora and investigation of the carbohydrate and nitrogen metabolism of the yeasts and LAB involved and of flavor generation during fermentation (18). Studies dealing with the identification and characterization of LAB from traditional sourdoughs revealed the dominance of L. sanfranciscensis strains in type I sourdoughs, probably selected only by the environmental conditions induced by the sourdough fermentation technology; L. pontis and L. panis strains are typical for type II sourdoughs (2, 54, 55, 56, 59). However, many researchers still report the existence of unidentifiable and perhaps new sourdough LAB species and/or strains (41), such as the L. brevis-like strain ACA-DC 3411 t1 isolated during this study.
In general, the spectrum of LAB involved in sourdough fermentations includes many different species of lactobacilli, in particular L. sanfranciscensis and L. brevis, as is the case in the Greek wheat sourdoughs examined. However, although the RAPD-PCR technique has proven to be valuable for the routine identification and rapid differentiation of several species of lactobacilli (5, 53, 61), the primer M13V used in the present study did not allow us to distinguish L. sanfranciscensis and L. brevis. No fructose-positive isolates of L. sanfranciscensis were found either. On the other hand, it has been reported that L. sanfranciscensis strains strongly differ in, for instance, sugar fermentation patterns (15, 24, 26). In the present study, the application of SDS-PAGE of total cell protein, RAPD-PCR, DNA-DNA hybridization, and 16S rDNA sequence analysis, in combination with physiological traits such as fructose fermentation and mannitol production, allowed us to classify the Greek wheat sourdough LAB isolates into the species L. sanfranciscensis, L. brevis, L. paralimentarius, and W. cibaria. No L. pontis strains were found, although these LAB are frequently isolated from European sourdoughs (54, 56). From an ecological point of view, the presence of W. cibaria is interesting and, to our knowledge, new for sourdough fermentations. The only Weissella species detected in previous studies is W. confusa (9, 54). It may be assumed that isolates assigned to that species are misidentified and actually belong to the very recently described species W. cibaria, a species which is both genomically and phenotypically highly similar to W. confusa (1). A common characteristic of both species that makes it possible to distinguish them from the other Weissella species is the ability to grow at 45°C, which may be favorable for type II sourdough fermentation processes. Both taxa have been isolated from fermented foods (1). A misidentification may also have occurred for sourdough isolates previously assigned to the species L. alimentarius. Recently, two phylogenetically highly related species, L. paralimentarius and L. kimchii, were described (4, 60). In the description of the most recently described species, L. kimchii, the taxon L. alimentarius is included for comparison, but not L. paralimentarius. In the present study, it is shown that L. kimchii and L. paralimentarius are genomically related at a binding level close to the borderline of species delineation (68%). The observed instability of some phenotypic features of, for instance, L. alimentarius, included as a reference strain in both studies, may raise the question of whether there was enough evidence to create two distinct species for these taxa. The new isolates ACA-DC 3363, ACA-DC 3413, and ACA-DC 3415 from the present study were assigned to the species L. paralimentarius because of a DNA-DNA binding value higher than 70%.
The species diversity of the Greek sourdoughs studied is different from that of other European sourdoughs. L. sanfranciscensis is the predominant bacterial species in Italian wheat and rye sourdoughs, a strain that is associated either with L. plantarum or with L. alimentarius (11). The dominance of L. paracasei is rather uncommon, although this species has been found in Altamura bread, in Portuguese sourdoughs, and in pizza doughs (7, 37). L. sanfranciscensis and L. brevis are the species most frequently isolated from German rye sourdoughs (45). Strains of L. plantarum, L. brevis, and L. fermentum dominate in Russian rye sourdoughs (25), and in Finnish rye sourdoughs, L. acidophilus and L. plantarum are the most frequently isolated species (43). In Swedish rye sourdoughs, L. fermentum is the most frequently isolated species (46). However, it is often seen that sourdoughs contain a very complex association of several species, e.g., Italian Pugliese sourdoughs contain L. plantarum, L. brevis, L. fermentum, and L. fructivorans, and Foggia sourdoughs contain L. brevis, L. sanfranciscensis, Leuconostoc citreum, and W. confusa (11). Differences in wheat and rye flour, other ingredients, and processing technology are all factors that greatly influence the microbial composition of sourdoughs and, consequently, the characteristics of the baked end products. These data underline the uniqueness of the Greek wheat sourdoughs examined in this study.
Of the 33 sourdough LAB isolates studied in detail, the growth, final pH, and metabolite formation (e.g., the lactic acid/acetic acid ratio) differed significantly. All heterofermentative strains produced lactic acid, acetic acid, and ethanol. Lactic acid, ethanol, and carbon dioxide are the main end products of sourdough LAB cultivated under anaerobic conditions (38). Under microaerophilic conditions, both oxygen and fructose can be used as electron acceptors. This gives rise to the formation of additional metabolites, such as acetate and mannitol (23). Therefore, the amounts of acetate and ethanol formed, in particular, differed among the isolates. In addition to glucose, almost all of the LAB isolates fermented maltose, while fructose as the sole carbohydrate source was fermented by all sourdough LAB tested except L. sanfranciscensis. The capacity to ferment several wheat flour carbohydrates may reduce metabolic competition with yeasts and may influence the acidification rate, titratable acidity, and acetic acid content (11). Two of the L. paralimentarius isolates tested did not ferment maltose; all strains were homofermentative.
Microbial associations of maltose-positive and maltose-negative LAB strains are typical for sourdoughs dominated by L. sanfranciscensis (18). The species hydrolyzes maltose and accumulates glucose in the medium in a molar ratio of ∼1:1 (21, 50). However, we have shown that no glucose accumulation occurred in the fermentation broth and that no maltose phosphorylase activity could be detected in cell extracts prepared from cells grown in the presence of both maltose and fructose. In experiments performed with growing cells of L. sanfranciscensis, Neubauer et al. and Stolz et al. did not observe any significant accumulation of glucose in the medium, as reported for resting cells of L. sanfranciscensis, L. reuteri, and L. pontis (31, 49, 52). Also, it is believed that hexokinase activity is induced in the presence of glucose or fructose in the medium (51). Hence, the nonphosphorylated glucose formed upon the cleavage of maltose by maltose phosphorylase was possibly immediately converted by hexokinase activity, which in turn was induced by fructose in the medium. Thus, in the presence of both maltose and fructose in the medium, induction of hexokinase activity does occur, explaining why no glucose accumulation was found. Here, due to the lack of maltose phosphorylase activity, an alternative mechanism may be responsible for maltose uptake by L. sanfranciscensis as well. Furthermore, the capacity for a maltose-fructose cofermentation has been shown before (19, 20). All L. sanfranciscensis strains used in our study were unable to grow on fructose as the sole carbohydrate source; they used fructose as an electron acceptor. According to Röcken and Voysey, oxygen was proved to be the preferred hydrogen acceptor for the L. sanfranciscensis strains (38). When oxygen is depleted, fructose is used as an electron acceptor in the presence of maltose and fructose as carbohydrate sources and is thus converted into mannitol (20, 49). In the W. cibaria strains studied that do not use fructose as an electron acceptor, fructose fermentation went hand in hand with high hexokinase activity. All these data give rise to a variable FQ, which is optimal in the range 2.0 to 2.7 (23). The FQ of sourdough can be decreased by the addition of fructose to the dough when L. brevis is used as a starter culture, resulting in increased acetate titers (28, 39). Whereas the organic acids and ethanol contribute to the aroma of sourdoughs, it has been shown that the production of acetic and lactic acid by sourdough LAB, as well as specific inhibitors active against moulds or rope-forming bacilli, delays or prevents the growth of spoilage organisms during bread storage (8, 10, 17, 27). Furthermore, they strongly affect the rheological properties of wheat doughs (57, 58). Finally, biological acidification of the dough delays starch retrogradation effectively, and this effect is dependent on the selection of LAB used to initiate the fermentation (9). All of the characteristics mentioned above will contribute to a rational choice of new sourdough LAB strains to be used as industrial starter cultures.
In this paper, the biodiversity of Greek traditional wheat sourdough products has been unraveled, which will enable us to understand their functions in sourdoughs. The ultimate aim of our study is the development of novel starter cultures on the one hand and the improvement of traditional sourdough technology on the other. A rational choice of sourdough starter cultures will allow the optimization of sourdough processes and the deliberate adjustment of the lactate/acetate ratio and other metabolites, as well as technological characteristics, affecting the organoleptic properties of bread in artisan and industrial sourdough production.
Acknowledgments
The research presented in this paper was financially supported by the Institute for the Encouragement of Innovation through Science and Technology in Flanders (IWT), in particular the STWW project Functionality of Novel Starter Cultures in Traditional Fermentation Processes. We further acknowledge financial support from the Research Council of the Vrije Universiteit Brussel and the Fund for Scientific Research—Flanders.
M. Gobbetti from the Institute of Dairy Microbiology (Agricultural Faculty of Perugia, Perugia, Italy) is acknowledged for donating the strains L. sanfranciscensis C57 and L. sanfranciscensis E13. S.P. thanks M. Ehrmann and Rudi Vogel from the Technische Universität München (Weihenstephan, Germany) for performing RAPD-PCR analyses in their laboratory. We thank K. Lefebvre, K. Vandemeulebroecke, and C. Snauwaert for technical assistance.
REFERENCES
- 1.Björkroth, K. J., U. Schillinger, R. Geisen, N. Weiss, B. Hoste, W. H. Holzapfel, H. J. Korkeala, and P. Vandamme. 2002. Taxonomic study of Weissella confusa and description of Weissella cibaria, a novel species detected in food and clinical samples. Int. J. Syst. Evol. Microbiol. 52:141-148. [DOI] [PubMed] [Google Scholar]
- 2.Böcker, G., P. Stolz, and W. P. Hammes. 1995. Neue Erkenntnisse zum Ökosystem Sauerteig und zur Physiologie des sauerteig-typischen Stämme Lactobacillus sanfrancisco und Lactobacillus pontis. Getreide Mehl Brot 49:370-374. [Google Scholar]
- 3.Böcker, G., R. F. Vogel, and W. P. Hammes. 1990. Lactobacillus sanfrancisco als stabiles Element in einem Reinzucht-Sauerteig-Präparat. Getreide Mehl Brot 44:269-274. [Google Scholar]
- 4.Cai, Y., H. Okada, H. Mori, Y. Benno, and T. Nakase. 1999. Lactobacillus paralimentarius sp. nov., isolated from sourdough. Int. J. Syst. Bacteriol. 49:1451-1455. [DOI] [PubMed] [Google Scholar]
- 5.Cocconcelli, P. S., D. Porro, S. Gelandini, and L. Senini. 1995. Development of RAPD protocol for typing of strains of lactic acid bacteria and enterococci. Lett. Appl. Microbiol. 21:376-379. [DOI] [PubMed] [Google Scholar]
- 6.Coenye, T., E. Falsen, M. Vancanneyt, B. Hoste, J. R. W. Govan, K. Kersters, and P. Vandamme. 1999. Classification of Alcaligenes faecalis-like isolates from the environment and human clinical samples as Ralstonia gilardii sp. nov. Int. J. Syst. Bacteriol. 49:405-413. [DOI] [PubMed] [Google Scholar]
- 7.Coppola, S., O. Pepe, P. Masi, and M. Sepe. 1996. Characterization of leavened doughs for pizza in Naples. Adv. Food Prot. 18:160-162. [Google Scholar]
- 8.Corsetti, A., M. Gobbetti, F. Balestrieri, F. Paoletti, L. Russi, and J. Rossi. 1998. Sourdough lactic acid bacteria effects on bread firmness and staling. J. Food Sci. 63:347-351. [DOI] [PubMed] [Google Scholar]
- 9.Corsetti, A., M. Gobbetti, B. De Marco, F. Balestrieri, F. Paoletti, L. Russi, and J. Rossi. 2000. Combined effect of sourdough lactic acid bacteria and additives on bread firmness and staling. J. Agric. Food Sci. 48:3044-3051. [DOI] [PubMed] [Google Scholar]
- 10.Corsetti, A., M. Gobbetti, and E. Smacchi. 1996. Antimicrobial activity of sourdough lactic acid bacteria: isolation of a bacteriocin-like inhibitory substance from Lactobacillus sanfrancisco C57. Food Microbiol. 13:447-456. [Google Scholar]
- 11.Corsetti, A., P. Lavermicocca, M. Morea, F. Baruzzi, N. Tosti, and M. Gobbetti. 2001. Phenotypic and molecular identification and clustering of lactic acid bacteria and yeasts from wheat (species Triticum durum and Triticum aestivum) sourdoughs of Southern Italy. Int. J. Food Microbiol. 64:95-104. [DOI] [PubMed] [Google Scholar]
- 12.Degeest, B., and L. De Vuyst. 2000. Correlation of activities of the enzymes α-phosphoglucomutase, UDP-galactose 4-epimerase, and UDP-glucose pyrophosphorylase with exopolysaccharide biosynthesis by Streptococcus thermophilus LY03. Appl. Environ. Microbiol. 66:3519-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Vuyst, L., R. Callewaert, and B. Pot. 1996. Characterization of the antagonistic activity of Lactobacillus amylovorus DCE 471 and large scale isolation of its bacteriocin amylovorin L471. Syst. Appl. Microbiol. 19:9-20. [Google Scholar]
- 14.Ezaki, T., Y. Hashimoto, and E. Yabuuchi. 1989. Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int. J. Syst. Bacteriol. 39:224-229. [Google Scholar]
- 15.Foschino, R., C. Arrigoni, C. Picozzi, D. Mora, and A. Galli. 2001. Phenotypic and genotypic aspects of Lactobacillus sanfranciscensis strains isolated from sourdoughs in Italy. Food Microbiol. 18:277-285. [Google Scholar]
- 16.Gänzle, M. G., M. Ehrmann, and W. P. Hammes. 1998. Modeling of growth of Lactobacillus sanfranciscensis and Candida milleri in response to process parameters of sourdough fermentation. Appl. Environ. Microbiol. 64:2616-2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gänzle, M. G., A. Höltzel, J. Walter, G. Jung, and W. P. Hammes. 2000. Characterization of reutericyclin produced by Lactobacillus reuteri LTH 2584. Appl. Environ. Microbiol. 66:4325-4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gobbetti, M. 1998. The sourdough microflora: interactions of lactic acid bacteria and yeasts. Trends Food Sci. Technol. 9:267-274. [Google Scholar]
- 19.Gobbetti, M., and A. Corsetti. 1997. Lactobacillus sanfrancisco a key sourdough lactic acid bacterium: a review. Food Microbiol. 14:175-187. [Google Scholar]
- 20.Gobbetti, M., A. Corsetti, and J. Rossi. 1995. Maltose-fructose co-fermentation by Lactobacillus brevis subsp. lindneri CB1 fructose-negative strain. Appl. Microbiol. Biotechnol. 42:939-944. [Google Scholar]
- 21.Gobbetti, M., A. Corsetti, and J. Rossi. 1994. The sourdough microflora. Interactions between lactic acid bacteria and yeasts: metabolism of carbohydrates. Appl. Microbiol. Biotechnol. 41:456-460. [DOI] [PubMed] [Google Scholar]
- 22.Gobbetti, M., A. Corsetti, J. Rossi, F. De Rosa, and S. De Vincenzi. 1994. The sourdough microflora. Identification and clustering of lactic acid bacteria and yeasts from sourdoughs of Central Italy. Ital. J. Food. Sci. 1:85-94. [Google Scholar]
- 23.Hammes, W. P., and M. G. Gänzle. 1998. Sourdough breads and related products, p. 199-216. In B. J. B. Woods (ed.), Microbiology of fermented foods, vol. 1. Blackie Academic & Professional, London, United Kingdom.
- 24.Hammes, W. P., P. Stolz, and M. Gänzle. 1996. Metabolism of lactobacilli in traditional sourdoughs. Adv. Food Sci. 18:176-184. [Google Scholar]
- 25.Kazanskaya, L. N., O. V. Afanasyeva, and V. A. Patt. 1983. Microflora of rye sours and some specific features of its accumulation in bread baking plants of the USSR, p. 759-763. In J. Holas and F. Kratochvil (ed.), Developments in food science, vol. 5B. Progress in cereal chemistry and technology. Elsevier, London, United Kingdom.
- 26.Kline, L., and T. F. Sugihara. 1971. Microorganisms of the San Francisco sour dough bread process. II. Isolation and characterization of undescribed bacterial species responsible for the souring activity. Appl. Microbiol. 21:459-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lavermicocca, P., F. Valerio, A. Evidente, S. Lazzaroni, A. Corsetti, and M. Gobbetti. 2000. Purification and characterization of novel antifungal compounds from the sourdough Lactobacillus plantarum strain 21B. Appl. Environ. Microbiol. 66:4084-4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martinez-Anaya, M. A., M. L. Llin, M. L. Macias, and C. Collar. 1994. Regulation of acetic acid production by homo- and heterofermentative lactobacilli in whole-wheat sourdoughs. Z. Lebensm. Unters. Forsch. 199:186-190. [DOI] [PubMed] [Google Scholar]
- 29.Mesbah, M., U. Premachandran, and W. B. Whitman. 1989. Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int. J. Syst. Bacteriol. 38:159-167. [Google Scholar]
- 30.Müller, M. R. A., M. A. Ehrmann, and R. F. Vogel. 2000. Lactobacillus frumenti sp. nov., a new lactic acid bacterium isolated from rye-bran fermentations with a long fermentation period. Int. J. Evol. Microbiol. 50:2127-2133. [DOI] [PubMed] [Google Scholar]
- 31.Neubauer, H., E. Glaasker, W. P. Hammes, B. Poolman, and W. N. Konings. 1994. Mechanism of maltose uptake and glucose excretion in Lactobacillus sanfrancisco. J. Bacteriol. 176:3007-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niemann, S., A. Puehler, H.-V. Tichy, R. Simon, and W. Selbitschka. 1997. Evaluation of the resolving power of three different DNA fingerprinting methods to discriminate among isolates of a natural Rhizobium meliloti population. J. Appl. Microbiol. 82:477-484. [DOI] [PubMed] [Google Scholar]
- 33.Paramithiotis, S., M. R. A. Mueller, M. A. Ehrmann, E. Tsakalidou, H. Seiler, R. Vogel, and G. Kalantzopoulos. 2000. Polyphasic identification of wild yeast strains isolated from Greek sourdough. Syst. Appl. Microbiol. 23:156-164. [DOI] [PubMed] [Google Scholar]
- 34.Pitcher, D. G., N. A. Saunders, and R. J. Owen. 1989. Rapid extraction of bacterial genomic DNA with guanidinium thiocyanate. Lett. Appl. Microbiol. 8:151-156. [Google Scholar]
- 35.Pot, B., P. Vandamme, and K. Kersters. 1994. Analysis of electrophoretic whole-organism protein fingerprints, p. 493-521. In M. Goodfellow and A. G. O'Donnell (ed.), Chemical methods in prokaryotic systematics. J. Wiley and Sons, Chichester, United Kingdom.
- 36.Qian, N., G. A. Stanley, B. Hahn-Hägerdal, and P. Rådström. 1994. Purification and characterisation of two phosphoglucomutases from Lactococcus lactis subsp. lactis and their regulation in maltose- and glucose-utilizing cells. J. Bacteriol. 176:5304-5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rocha, J. M., and F. X. Malcata. 1999. On the microbiological profile of traditional Portuguese sourdough. J. Food Prot. 62:1416-1429. [DOI] [PubMed] [Google Scholar]
- 38.Röcken, W., and P. A. Voysey. 1995. Sourdough fermentation in bread making. J. Appl. Bacteriol. Symp. Suppl. 79:38S-48S. [Google Scholar]
- 39.Röcken, W., M. Rick, and M. Reinkemeier. 1992. Controlled production of acetic acid in wheat sour doughs. Z. Lebensm. Unters. Forsch. 195:259-263. [Google Scholar]
- 40.Rosenquist, H., and A. Hansen. 1998. The antimicrobial effect of organic acids, sour dough and nisin against Bacillus subtilis and B. licheniformis isolated from wheat bread. J. Appl. Microbiol. 85:621-631. [Google Scholar]
- 41.Rosenquist, H., and A. Hansen. 2000. The microbial stability of two bakery sourdoughs made from conventionally and organically grown rye. Food Microbiol. 17:241-250. [Google Scholar]
- 42.Salovaara, H. 1998. Lactic acid bacteria in cereal-based products, p. 115-137. In S. Salminen, and A. von Wright (ed.), Lactic acid bacteria—microbiology and functional aspects. Marcel Dekker, New York, N.Y.
- 43.Salovaara, H., and H. Katunpää. 1984. An approach to the classification of lactobacilli isolated from Finnish sour rye dough ferments. Acta Aliment. Pol. 10:231-239. [Google Scholar]
- 44.Schleifer, K. H., M. Ehrmann, C. Beimfohr, E. Brockmann, W. Ludwig, and R. Amann. 1995. Application of molecular methods for the classification and identification of lactic acid bacteria. Int. Dairy J. 5:1081-1094. [Google Scholar]
- 45.Spicher, G. 1987. The microflora of sourdough. XXII. Communication: the Lactobacillus species of wheat sourdough. Z. Lebensm. Unters. Forsch. 184:300-303. [Google Scholar]
- 46.Spicher, G., and C. Lönner. 1985. Die Mikroflora des Sauerteiges. XXII. Mitteilung: Die in Sauerteigen schwedischer Bäckereien vorkommenden Lactobacillen. Z. Lebensm. Unters. Forsch. 181:9-13. [Google Scholar]
- 47.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 48.Stolz, P., and G. Böcker. 1996. Technology, properties and applications of sourdough products. Adv. Food Sci. 18:234-236. [Google Scholar]
- 49.Stolz, P., G. Böcker, W. P. Hammes, and R. F. Vogel. 1995. Utilization of electron acceptors by lactobacilli isolated from sourdough. I. Lactobacillus sanfranciscensis. Z. Lebensm. Unters. Forsch. 201:91-96. [Google Scholar]
- 50.Stolz, P., G. Böcker, R. F. Vogel, and W. P. Hammes. 1993. Utilisation of maltose and glucose by lactobacilli isolated from sourdough. FEMS Microbiol. Lett. 109:237-242. [Google Scholar]
- 51.Stolz, P., W. P. Hammes, and R. F. Vogel. 1996. Maltose-phosphorylase and hexokinase activity in lactobacilli from traditionally prepared sourdoughs. Adv. Food Sci. 18:1-6. [Google Scholar]
- 52.Stolz, P., R. F. Vogel, and W. P. Hammes. 1995. Utilization of electron acceptors by lactobacilli isolated from sourdough. I. Lactobacillus pontis, L. reuteri, L. amylovorus, and L. fermentum. Z. Lebensm. Unters. Forsch. 201:402-410. [Google Scholar]
- 53.Tynkkynen, S., R. Satokari, M. Saarela, T. Mattila-Sandholm, and M. Saxelin. 1999. Comparison of ribotyping, randomly amplified polymorphic DNA analysis, and pulsed-field gel electrophoresis in typing of Lactobacillus rhamnosus and L. casei strains. Appl. Environ. Microbiol. 65:3908-3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vogel, R., M. Müller, P. Stolz, and M. Ehrmann. 1996. Ecology in sourdoughs produced by traditional and modern technologies. Adv. Food Sci. 18:152-159. [Google Scholar]
- 55.Vogel, R. F., G. Böcker, P. Stolz, M. Ehrmann, D. Fanta, W. Ludwig, B. Pot, K. Kersters, K. H. Schleifer, and W. P. Hammes. 1994. Identification of lactobacilli from sourdough and description of Lactobacillus pontis sp. nov. Int. J. Syst. Bacteriol. 44:223-229. [DOI] [PubMed] [Google Scholar]
- 56.Vogel, R. F., R. Knorr, M. R. A. Müller, U. Steudel, M. G. Gänzle, and M. Ehrmann. 1999. Non-dairy lactic fermentations: the cereal world. Antonie Leeuwenhoek 76:403-411. [PubMed] [Google Scholar]
- 57.Wehrle, K., and E. K. Arendt. 1998. Rheological changes in wheat sour dough during controlled and spontaneous fermentation. Cereal Chem. 75:882-886. [Google Scholar]
- 58.Wehrle, K., H. Grau, and E. K. Arendt. 1997. Effects of lactic acid, acetic acid, and table salt on fundamental rheological properties of wheat dough. Cereal Chem. 74:739-744. [Google Scholar]
- 59.Wiese, B. G., W. Strohmar, F. A. Rainey, and H. Diekmann. 1996. Lactobacillus panis sp. nov., from sourdough with a long fermentation period. Int. J. Syst. Microbiol. 46:449-453. [DOI] [PubMed] [Google Scholar]
- 60.Yoon, J.-H., S.-S. Kang, T.-I. Mheen, J.-S. Ahn, H.-J. Lee, T.-K. Kim, C.-S. Park, Y. H. Kho, K. H. Kang, and Y.-H. Park. 2000. Lactobacillus kimchii sp. nov., a new species from kimchi. Int. J. Syst. Evol. Microbiol. 50:1789-1795. [DOI] [PubMed] [Google Scholar]
- 61.Zapparoli, G., S. Torriani, and F. Dellaglio. 1998. Differentiation of Lactobacillus sanfranciscensis strains by randomly amplified polymorphic DNA and pulsed-field gel electrophoresis. FEMS Microbiol. Lett. 166:325-332. [Google Scholar]