Abstract
An innovative method was developed for rapid sensitive detection and efficient structural characterization of lipopeptide biosurfactants by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry by using whole microbial cells and crude culture filtrates as targets in combination with surface tension measurements. This was done for a bacterial strain that was isolated from petroleum sludge and efficiently produces biosurfactants. This organism was identified by using biochemical, physiological, and genetic parameters as a Bacillus subtilis strain, designated B. subtilis C-1. This assignment was supported by a mass spectrometric investigation of the secondary metabolite spectrum determined by whole-cell MALDI-TOF mass spectrometry, which revealed three lipopeptide complexes, the surfactins, the iturins, and the fengycins, which are well-known biosurfactants produced by B. subtilis strains. These compounds were structurally characterized by in situ structure analysis by using postsource decay MALDI-TOF mass spectrometry. The isoforms were separated by miniaturized high-resolution reversed-phase high-performance liquid chromatography for mass spectrometric characterization. Iturin compounds which contain unusual fatty acid components were detected.
Biosurfactants are a structurally diverse group of surface-active molecules synthesized by microorganisms (5, 7, 9, 10, 32, 36). They have unique amphipathic properties derived from their complex structures, which include a hydrophilic moiety and a hydrophobic portion. The most efficient biosurfactants reduce the surface tension of water from 72 dynes/cm to values in the range of 25 to 30 dynes/cm. Biosurfactant production can be determined by measuring the change in surface tension of cell-free culture broth.
Microbial surfactants have commonly been classified into the following categories: (i) glycolipids, (ii) lipopeptides, (iii) fatty acids, neutral lipids, and phospholipids, (iv) polymeric surfactants, and (v) particulate biosurfactants (5, 7, 9, 10, 32, 36). The lipopeptides are an interesting class of microbial surfactants (36) because of their manifold attractive properties. Members of this group often possess antibiotic activity as well.
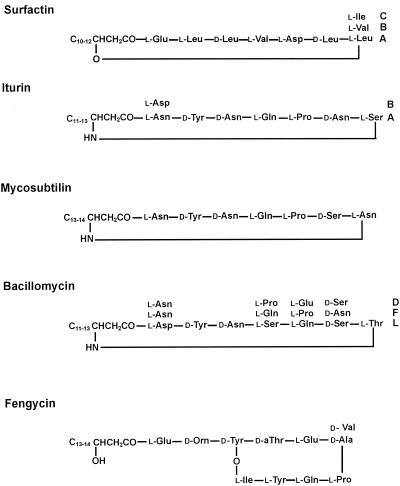
Bacillus subtilis strains produce a broad spectrum of bioactive peptides with great potential for biotechnological and biopharmaceutical applications. A well-known class of such compounds includes the lipopeptides surfactin (1, 13, 14, 17, 18), fengycin (35), and the iturin compounds (3) (iturins [26], mycosubtilins [25], and bacillomycins [27]), which are amphiphilic membrane-active biosurfactants and peptide antibiotics with potent antimicrobial activities. All these agents occur as families of closely related isoforms which differ in the length and branching of the fatty acid side chains and in the amino acid substitutions in the peptide rings (20, 36). The surfactin and iturin compounds are cyclic lipoheptapeptides which contain a β-hydroxy fatty acid and a β-amino fatty acid, respectively, as lipophilic components. Fengycin is a lipodecapeptide with a β-hydroxy fatty acid in its side chain. The structures of these biosurfactants are shown in Fig. 1.
FIG. 1.
Lipopeptide biosurfactants produced by B. subtilis strains.
Comparative studies of the interfacial and emulsifying properties of the lipopeptide biosurfactants produced by B. subtilis have been performed by Deleu et al. (6). All these compounds efficiently reduce the interfacial tension at the oil-water interface; the surfactins have the greatest effects, while the iturins and fengycins have lower surfactant strength. In particular, surfactin (30, 36) is one of the most thoroughly studied and best-characterized biosurfactants. Bacillus licheniformis (9, 11, 12, 39) and Bacillus pumilus (23) also produce lipopeptides similar to surfactin. These agents are natural compounds that have industrial importance and have attracted increasing biotechnological and pharmaceutical interest. They are distinguished by excellent surface- and membrane-active properties along with superior emulsifying and foaming properties (6, 31), which can be utilized in food biotechnology and in the agricultural sector. The lipopeptides belonging to the iturin family are potent antifungal agents which can be used as biopesticides for plant protection. In recent studies important biopharmaceutical applications of surfactins have been demonstrated. Surfactin shows potent antiviral and antimycoplasma activities (37, 38). Viruses which are covered by a lipoprotein membrane, such as herpes- and retroviruses, are efficiently inactivated by this biosurfactant. Surfactin is well qualified to maintain virus and mycoplasma safety in biotechnological products.
Matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry has been used as a novel, efficient method for identification and structural characterization of microbial secondary metabolites when whole cells are used as targets (8, 22); this method is well suited for rapid screening of unexplored microorganisms in different habitats in order to make novel bioactive compounds available for industrial exploitation. In this study we combined this technique with surface tension measurements to screen microorganisms for lipopeptide biosurfactants. This technique was used to examine a B. subtilis strain isolated by us from petroleum sludge.
MATERIALS AND METHODS
Microorganism.
The microbial culture selected for this study was isolated from petroleum sludge. Five different types of colonies were isolated on nutrient agar plates, and one strain was selected for further study; this strain showed maximal reduction of surface tension during growth in the culture supernatant. Cultures were stored frozen in 10% glycerol at −70°C. Working cultures were maintained on nutrient agar slants at 4°C.
Growth conditions.
For biosurfactant production, the isolated organism was grown aerobically on minimal salt medium containing (per liter) 2.0 g of KH2PO4, 5.0 g of K2HPO4, 3 g of (NH4)2SO4, 2 g of NaNO3, 0.1 g of NaCl, 0.2 g of MgSO4 · H2O, 0.01 g of FeSO4 · 7H2O, 0.01 g of CaCl2, and 1 ml of a trace element solution. The stock solution of trace elements contained (per liter) 2.32 g of ZnSO4 · 7H2O, 1.78 g of MnSO4 · 4H2O, 0.56 g of H3BO3, 1 g of CuSO4 · 5H2O, 0.39 g of Na2MoO4 · 7H2O, 0.42 g of CoCl2 · 6H2O, 1 g of EDTA, 0.004 g of NiCl2 · 6H2O, and 0.66 g of KI. The medium was supplemented with 0.05% yeast extract. Glucose was added as a carbon source at a concentration of 2% (wt/vol). The pH of the medium was 7.1 to 7.2. The organism was grown at 37°C for 48 h in 2-liter Erlenmeyer flasks containing 800 ml of medium with shaking at 200 rpm in a shaker incubator.
For MALDI-TOF mass spectrometry characterization, B. subtilis C-1 was cultivated both in surface and suspension cultures by using three different media, Landy medium (21), Difco sporulation medium, and sucrose-ammonium citrate medium, as reported by Leenders et al. (22). To prepare surface cultures, the strain was grown in petri dishes containing 1.5% (wt/vol) agar agar for 24 h and stored at room temperature prior to MALDI-TOF mass spectrometric analysis. Fermentation in liquid media was carried out in 500-ml Erlenmeyer flasks for various times at 30°C and 180 rpm in a New Brunswick shaker (New Brunswick Scientific Co., Edison, N.J.).
Surface tension measurement.
For detection of biosurfactants, B. subtilis C-1 was grown in minimal medium. The surface tension was measured with a Du-Nouy tensiometer (CSC, Fairfax, Va.). All measurements were made with culture supernatants obtained after centrifugation at 10,410 × g for 25 min at 4°C. For determination of the surface tension, the supernatants were diluted 10- and 100-fold. The surface tension of the minimal medium (71 dynes/cm) was used as the reference.
E24.
Six milliliters of kerosene oil was added to 4 ml of the culture supernatant obtained after centrifugation of cells grown for 48 h in minimal medium and vortexed at high speed for 3 to 4 min. The emulsion stability was determined after 24 h. The emulsification index (E24) was calculated by measuring the height of the emulsion layer formed.
Surfactant isolation.
Bacterial cells were removed from the surfactant-containing medium by centrifugation (8,000 rpm for 25 min at 4°C). Lipopeptide surfactants were precipitated from the supernatant by adding 6 N HCl to obtain a final pH of 2.0. The acid precipitates were recovered by centrifugation (8,000 rpm for 15 min at 4°C) and were extracted with dichloromethane or methanol (lipopeptide fraction). When methanol was used as the solvent, the extract was neutralized immediately to avoid formation of methyl esters.
Purification of the lipopeptide biosurfactants of B. subtilis C-1 on an analytic scale.
A 10- to 50-μl portion of the lipopeptide fraction was loaded on a μRPC SC 2.1/10 column (Amersham Biosciences Europe GmbH, Freiburg, Germany) and separated by high-resolution reversed-phase high-performance liquid chromatography (HPLC) by using a Pharmacia Smart microseparation system. The products were eluted with a linear gradient of 20 to 100% acetonitrile-0.1% trifluoroacetic acid (TFA) in 60 min by using a flow rate of 100 μl/min. Solvent A was 20% acetonitrile in 0.1% TFA (vol/vol). Solvent B was acetonitrile containing 0.1% TFA (vol/vol). In the fractions obtained, the compounds were detected by MALDI-TOF mass spectrometry.
Separation of the lipopeptide biosurfactant products on a preparative scale.
After precipitation with HCl, the crude lipopeptide fraction dissolved in methanol or dichloromethane was evaporated in a rotary evaporator under a vacuum. The dried material was dissolved in a minimum volume of chloroform-methanol (1:1, vol/vol), applied to an LH-20 column (150 by 5 cm; Amersham Biosciences Europe GmbH), and fractionated by size exclusion chromatography by using the same solvent for elution. The products were monitored by determining the absorbance at 220 nm. Fractions were examined by thin-layer chromatography (TLC) on Silica Gel DC 60 plates obtained from Merck (Darmstadt, Germany). A chloroform-methanol-water mixture (65:25:4, vol/vol/vol) was used as the mobile phase. Spots were visualized by charring with H2SO4 and heating the plates at 200°C for 30 min. Fractions that eluted from the LH-20-column were pooled and concentrated with a rotary evaporator.
Isolated lipopeptides were purified further and fractionated into isoforms by reversed-phase HPLC on an octyldecyl silane (ODS) Hypersil C18 column (Knauer, Berlin, Germany) at room temperature. A linear 20 to 100% acetonitrile gradient was used for elution at a flow rate of 0.5 ml/min. Eluent A was 20% (vol/vol) acetonitrile in 100 mM ammonium acetate (pH 6.9), and eluent B was 100% acetonitrile. The eluted biosurfactants were detected by measuring the absorbance at 220 nm.
Amino acid analysis.
Surfactin isoforms obtained in pure form by LH-20 gel permeation chromatography in combination with reversed-phase HPLC were hydrolyzed with 6 N HCl for 20 h at 110°C. The water-soluble part of the hydrolysate was qualitatively analyzed to determine its amino acid composition by two-dimensional TLC on silica gel- and cellulose-coated TLC plates (Merck). Samples were spotted on TLC plates and separated by using the following two solvent systems: butanol-acetic acid-H2O (4:1:1, vol/vol/vol) and methanol-6 N HCl-H2O-pyridine (60:3:19.5:15, vol/vol/vol/vol). For detection of the amino acids, the plates were sprayed with a solution of 2% ninhydrin in acetone and kept at 110°C for 5 min. Quantitative amino acid analysis was performed with a Waters model 501amino acid analyzer.
Enantiospecific TLC of amino acids.
The separated amino acid spots of hydrolyzed surfactins were scraped off the TLC plate. One milliliter of a mixture containing methanol and water (1:1, vol/vol) was added to each sample. The silica gel was removed by centrifugation. The extracted amino acids were spotted on chiral TLC plates (Merck) along with standard l and d amino acids (Sigma Chemical Co., St. Louis, Mo.). The solvent system used was acetonitrile-methanol-water (4:1:1, vol/vol/vol). The spots were detected by spraying the plates with a solution of 2% ninhydrin in acetone.
MALDI-TOF mass spectrometry analysis.
MALDI-TOF mass spectra were recorded by using a Bruker Daltonik Reflex MALDI-TOF instrument containing a 337-nm nitrogen laser for desorption and ionization. For mass spectrometric analysis of isolated lipopeptide biosurfactants, 1- to 2-μl portions of fractions obtained after gel filtration on Sephadex LH-20 or after reversed-phase HPLC were each mixed with an equal volume of matrix medium (a saturated solution of α-cyano-4-hydroxycinnamic acid in 70% aqueous acetonitrile containing 0.1% [vol/vol] TFA). For product analysis with whole bacterial cells, cell material was picked from agar plates, spotted onto the sample target, covered with matrix medium, and air dried. Positive-ion detection and the reflector mode were used. The acceleration and reflector voltages were 20 and 23.4 kV in pulsed ion extraction mode. A molecular mass gate of 300 Da improved the measurement by filtering out most matrix ions. Postsource decay (PSD) mass spectra were obtained with the same samples.
RESULTS
Growth of and biosurfactant production by B. subtilis C-1 at different temperatures.
A bacterial strain that produced biosurfactants was isolated from petroleum sludge. This organism is a gram-positive, spore-forming, rod-like, oval bacterial strain which has been characterized by the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH Identification Service as a member of the B. subtilis group. This classification was based mainly on 16S ribosomal DNA data and the riboprint pattern. Partial sequencing of the 16S ribosomal DNA revealed 99.8% identity with B. subtilis. In addition, some biochemical and physiological parameters were determined which support this classification. This organism was designated B. subtilis C-1. It was cultivated in minimal medium at 25, 30, and 45°C for 48 h at 200 rpm in a shaker incubator. Growth curves at all temperatures were determined by monitoring the optical density at 600 nm at different times. Optimal growth was observed at 30°C. At 25 and 30°C the stationary phase was reached after 30 h. Biosurfactant compounds were detected by surface tension measurement. The surface tension values are shown in Table 1. The data obtained indicate that there was efficient biosurfactant production which was higher at 25 and 30°C than at 45°C.
TABLE 1.
Surface tensions of culture filtrates of B. subtilis C-1 grown at different temperaturesa
| Dilution | Surface tension (dynes/cm) of the cell-free broth at a growth temp of:
|
||
|---|---|---|---|
| 25°C | 30°C | 45°C | |
| None | 29.4 | 29.0 | 30.4 |
| 10-fold | 34.0 | 33.1 | 37.7 |
| 100-fold | 43.2 | 44.2 | 51.5 |
B. subtilis C-1 was grown in minimal medium for 48 h. The surface tension of this medium was 71 dynes/cm.
The dry biomass (0.33 g/100 ml) and the crude biosurfactant mass (54.7 mg/100 ml) obtained after growth of the organism at 30°C were significantly greater than the dry biomass and the crude biosurfactant mass obtained after the organism was grown at 25 and 45°C (Table 2).
TABLE 2.
Effects of temperatures on biosurfactant production after growth of B. subtilis C-1 for 48 h in minimal medium
| Growth temp (°C) | Surface tension (dynes/cm) | Dry biomass concn (g/100 ml) | Biosurfactant concn (mg/100 ml)a | E24 (%)b |
|---|---|---|---|---|
| 25 | 29.44 | 0.182 | 30.6 | 0 |
| 30 | 28.98 | 0.330 | 54.7 | 65 |
| 45 | 30.36 | 0.092 | 24.6 | 0 |
Precipitates obtained after acidification of 100-ml portions of the culture filtrates were redissolved in 20 ml of double-distilled water (pH 7.4) and lyophilized.
Four milliliters of the culture filtrate was mixed with 6 ml of kerosine oil and vortexed for 3 to 5 min. E24 was calculated as follows: [(height of the emulsion layer stable for 24 h)/(total height)] × 100.
The data shown in Table 2 also demonstrate that the culture supernatant was able to emulsify oil. The highest E24 value was observed at 30°C. On the basis of these data further studies to isolate and characterize the biosurfactant(s) produced were performed by growing the isolated B. subtilis strain at 30°C in different culture media.
Innovative method for detection and in situ structure analysis of lipopeptide biosurfactants by MALDI-TOF mass spectrometry of whole bacterial cells and of the crude fermentation broth.
To detect and identify the biosurfactants produced by B. subtilis C-1, we developed an innovative mass spectrometric method. The products were investigated by performing MALDI-TOF mass spectrometry of whole bacterial cells. To do this, B. subtilis C-1 was grown on agar plates at 30°C for 24 h by using Landy medium, Difco sporulation medium, or ammonium citrate-sucrose medium. Samples of cells were picked from the agar plates and embedded in an α-cyanocinnamic acid matrix solution directly on the target. After air drying, the secondary metabolite spectrum of the strain was determined with a Bruker Daltonix Reflex MALDI-TOF mass spectrometer.
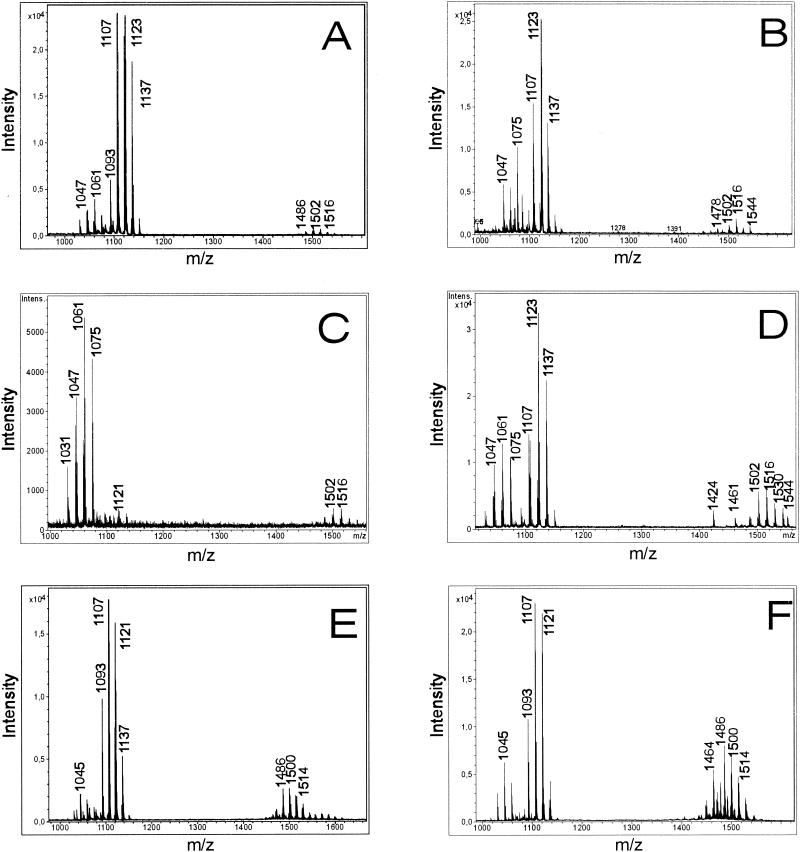
Figure 2A and B show the product patterns determined by MALDI-TOF mass spectrometry for whole B. subtilis C-1 cells grown as surface cultures on agar plates containing Landy medium and Difco sporulation medium, respectively. Mass spectra for whole cells obtained from suspension cultures were determined after different growth periods. Figure 2C and D show the mass spectra for cells harvested after 6 h in the exponential phase and after 30 h in the stationary phase, respectively. For comparison, Fig. 2E and F show the data for biosurfactant compounds released into the culture medium after growth for 30 h as detected by MALDI-TOF mass spectrometry of the crude fermentation broth (Fig. 2E) and of the methanolic extract of the lipopeptide fraction obtained by HCl precipitation (Fig. 2F). For the experiments whose results are shown in Fig. 2C to F, fermentation was performed in Landy medium. Cells were removed by centrifugation. All of the mass spectra in Fig. 2 show that there were three well-resolved groups of peaks at m/z values between 1000 and 1060, between 1070 and 1150, and between 1450 and 1550. The groups of peaks could be attributed to the isoform ensembles of surfactins, iturins, and fengycins, which represent the well-known biosurfactant families produced by B. subtilis strains (6, 22). In this way the classification of our isolate from petroleum sludge as a B. subtilis strain was corroborated by mass spectrometric characterization of its secondary metabolites.
FIG. 2.
MALDI-TOF mass spectrometric analysis of intact whole cells of B. subtilis C-1 grown on agar plates by using Landy (A) and Difco (B) sporulation media and of cells obtained from suspension cultures grown in Landy medium and harvested after 6 h (C) and 30 h (D). In addition, MALDI-TOF mass spectra were obtained for samples of the crude culture filtrate of B. subtilis C-1 grown in Landy medium (E) for 30 h and of the lipopeptide fraction obtained by precipitation with HCl (F). The fermentation temperature for all experiments was 30°C. Lipopeptide biosurfactants were detected in the range from m/z 900 to 1600.
Similar results were observed for all three culture media tested by us. From the MALDI-TOF mass spectra, it is apparent that biosurfactant production depends on the growth period. When Landy medium and a growth temperature of 30°C were used, the exponential phase extended over approximately 10 to 12 h. During this time predominantly surfactins were formed (Fig. 2C; growth time, 6 h). In this period the levels of iturin and fengycin production were rather low. These compounds appeared in the stationary phase, and the iturins were the main products during this phase (Fig. 2D to F; growth time, 30 h).
The quality of the mass spectra of the lipopeptide products of B. subtilis C-1 obtained for whole cells and for the crude, unfractionated culture supernatant is similar to the quality of the spectra obtained with solutions of the purified compounds. The results which are summarized in Fig. 2 demonstrate that by using MALDI-TOF mass spectrometry lipopeptide biosurfactants can be detected in minutes with high sensitivity, precision, and excellent resolution without a requirement for time-consuming isolation and chromatographic separation of the compounds.
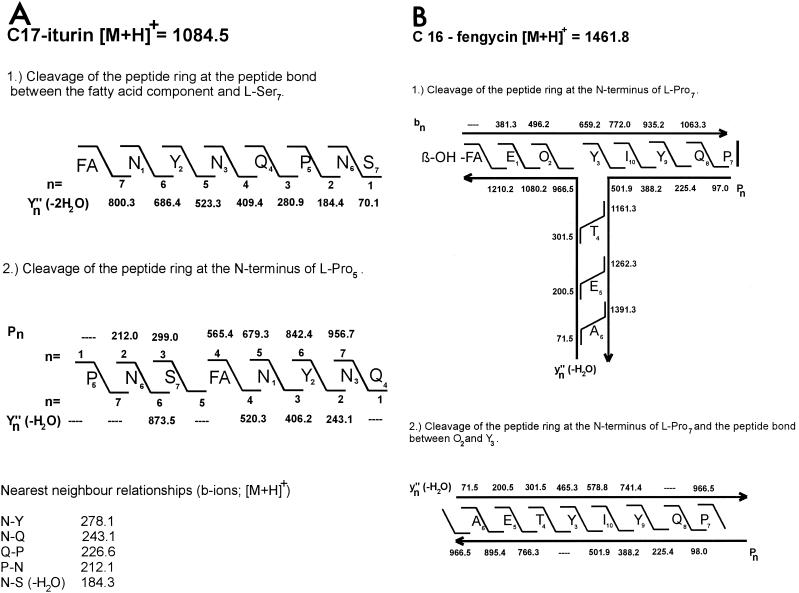
The mass numbers for the iturin and fengycin lipopeptide families observed in the MALDI-TOF mass spectra shown in Fig. 2 are summarized in Tables 3 and 4. Isoforms of these lipopeptides were identified and characterized by in situ structure analysis by using PSD MALDI-TOF mass spectrometry. For example, in Fig. 3 this is demonstrated in detail for an iturin species and a fengycin species with the mass numbers m/z 1084.5 and 1461.8, respectively, when whole cells were used as targets. The structures of these compounds were derived from a series of bn, Yn" (-H2O)- and proline-directed Pn fragment ions derived from the PSD MALDI-TOF mass spectra. The data obtained show that the species with a mass number of m/z 1084.5 can be attributed to a protonated C17 isoform of iturin A (Fig. 3A).
TABLE 3.
Assignments of all iturin mass peaks obtained by MALDI-TOF mass spectrometry of whole cells of B. subtilis C-1, the culture filtrate, and the lipopeptide fractiona
| Mass peak (m/z) | Assignment |
|---|---|
| 1070.6 | C16 iturin, [M+H]+ |
| 1084.6 | C17 iturin, [M+H]+ |
| 1098.6 | C18 iturin, [M+H]+ |
| 1112.6 | C19 iturin, [M+H]+ |
| 1092.6 | C16 iturin, [M+Na]+ |
| 1106.6b | C17 iturin, [M+Na]+ |
| 1120.6b | C18 iturin, [M+Na]+ |
| 1134.6 | C19 iturin, [M+Na]+ |
| 1108.6 | C16 iturin, [M+K]+ |
| 1122.6b | C17 iturin, [M+K]+ |
| 1136.6b | C18 iturin, [M+K]+ |
| 1150.6 | C19 iturin, [M+K]+ |
The mass data represent monoisotopic mass numbers.
Mass numbers of the main isoforms.
TABLE 4.
Assignments of all fengycin mass peaks obtained by MALDI-TOF mass spectrometry of whole cells of B. subtilis C-1, the culture filtrate, and the lipopeptide fractiona
| Mass peak (m/z) | Assignment | nb | Amino acid at position 6 |
|---|---|---|---|
| 1447.7 | C15 fengycin, [M+H]+ | 1 | Ala |
| 1449.8 | C15 fengycin, [M+H]+ | 0 | Ala |
| 1461.8 | C16 fengycin, [M+H]+ | 1 | Ala |
| 1463.9 | C16 fengycin, [M+H]+ | 0 | Ala |
| 1469.8 | C15 fengycin, [M+Na]+ | 1 | Ala |
| 1471.8 | C15 fengycin, [M+Na]+ | 0 | Ala |
| 1475.8 | C17 fengycin, [M+H]+ | 1 | Ala |
| 1477.8 | C17 fengycin, [M+H]+ | 0 | Ala |
| 1483.8 | C16 fengycin, [M+Na]+ | 1 | Ala |
| 1485.8c | C16 fengycin, [M+Na]+ | 0 | Ala |
| 1489.9 | C16 fengycin, [M+H]+ | 1 | Val |
| 1491.7 | C16 fengycin, [M+H]+ | 0 | Val |
| 1497.8 | C17 fengycin, [M+Na]+ | 1 | Ala |
| 1499.9 | C17 fengycin, [M+Na]+ | 0 | Ala |
| 1501.8c | C16 fengycin, [M+K]+ | 0 | Ala |
| 1503.8 | C17 fengycin, [M+H]+ | 1 | Val |
| 1505.8 | C17 fengycin, [M+H]+ | 0 | Val |
| 1513.9 | C16 fengycin, [M+Na]+ | 0 | Val |
| 1515.9c | C17 fengycin, [M+K]+ | 0 | Ala |
| 1527.9 | C17 fengycin, [M+Na]+ | 0 | Val |
| 1529.9c | C16 fengycin, [M+K]+ | 0 | Val |
The mass data represent monoisotopic mass numbers.
n is the number of double bonds in the fengycin species.
Mass numbers of the main isoforms.
FIG. 3.
In situ structure analysis of two lipopeptide biosurfactant products of B. subtilis C-1 having mass numbers of m/z 1084.5 (A) and m/z 1461.8 (B) by PSD MALDI-TOF mass spectrometry of whole cells of B. subtilis C-1. The structures were derived from a series of N- and C-terminal fragments [bn and Yn" (-H2O) ions, as well as proline-directed Pn fragments]. FA, fatty acid; β-OH-FA, β-hydroxy fatty acid.
Obviously, the peptide ring of this product was cleaved both at the peptide bond between the fatty acid residue and the serine at position 7 and at the N terminus of proline-5. In the first case a series of Yn" (-2H2O) ions was generated which originated from the Yn" (-H2O) ions by elimination of water at serine-7. The most intense ions in the PSD MALDI-TOF mass spectrum were the proline-directed Pn ions and the corresponding Yn" (-H2O) ion fragments. Figure 3A clearly shows that all three Asx residues in this iturin compound are asparagines. Aspartic acid was not found. In this way the amino acid sequence of iturin A could definitely be established. Our conclusion was corroborated by detection of dipeptide fragment ions (bn ions) for neighboring amino acids in the peptide ring of iturin A, which are included in Fig. 3A. Similar results were obtained for the corresponding C16 species having a mass number of m/z 1070.6. Remarkably, the fatty acid components of these iturin compounds have unusual features. For all isoforms listed in Tables 3 and 5, the mass numbers of the fatty acid constituents are 1 mass unit lower than those of the corresponding saturated compounds that have been described so far (3, 15, 25, 26, 28). For example, the C16 and C17 iturin species with mass numbers of m/z 1070.6 and 1084.5 contain fatty acid components with m/z 270.4 and 284.4, respectively. Their structures will be clarified in another paper.
TABLE 5.
Separation of the lipopeptide biosurfactants produced by B. subtilis C-1 by high-resolution reversed-phase HPLC and analysis by MALDI-TOF mass spectrometrya
| Fraction(s) | Main mass peak(s) (m/z) | Assignment |
|---|---|---|
| 23, 24 | 1070.6, 1092.6 | C16 iturin, [M+H and Na]+ |
| 25 | 1106.6 | C17 iturin, [M+Na]+ |
| 26, 27 | 1120.6 | C18 iturin, [M+Na]+ |
| 28 | 1435.8 | C14 fengycin, [M+H]+ |
| 29 | 1449.8 | C15 fengycin, [M+H]+ |
| 30 | 1463.9, 1485.8 | C16 fengycin, [M+H and Na]+ |
| 31 | 1491.8, 1499.8, 1529.7 | C16 and C17 fengycins |
| 32 | 1477.8 | C17 fengycin, [M+H]+ |
| 1491.9, 1505.9 | C16 and C17 fengycins, [M+H]+ | |
| 33 | 1447.9, 1475.9, 1505.9 | C15 and C17 fengycins, [M+H]+ |
| 34 | 1461.8, 1475.9 | C16 and C17 fengycins, [M+H]+ |
| 35 | 1489.9 | C16 fengycin, [M+H]+ |
| 36 | 1475.8, 1497.8 | C17 fengycin, [M+H and Na]+ |
| 37 | 1046.6 | C13 surfactin, [M+K]+ |
| 38 | 1030.6 | C13 surfactin, [M+Na]+ |
| 39 | 1044.6, 1060.5 | C14 surfactin, [M+Na and K]+ |
| 40 | 1058.6 | C15 surfactin, [M+Na]+ |
| 41, 42 | 1058.7, 1074.6 | C15 surfactin, [M+Na and K]+ |
The mass data represent monoisotopic mass numbers. Twenty microliters of the crude lipopeptide fraction was separated on a μRPC SC 2.1/10 column by using a Pharmacia Smart microseparation system, as described in Materials and Methods.
The mass peak at m/z 1461.8 (Fig. 3B) could be attributed to a fengycin isoform containing a β-hydroxy fatty acid with a chain length of 16 carbon atoms containing one double bond. An alanine was detected at position 6 of the peptide ring. Also, ring cleavage was observed at the Ala-Pro bond, resulting in a T-like structure, as revealed by specific series of bn, Yn" (-H2O) and Pn ions derived from the PSD MALDI-TOF mass spectra. In a similar way the structures of some other fengycin species were determined. For example, the compounds with mass numbers of m/z 1485.8 and 1505.8 were identified as fengycins with β-hydroxy fatty acid components having chain lengths of 16 and 17 carbon atoms. The first species (m/z 1485.8) is a sodium adduct of a C16 isoform with an alanine at position 6. The other compound (m/z 1505.8) is a protonated form of a C17 isoform with a valine instead of an alanine at position 6.
All these results demonstrate the high potential of MALDI-TOF mass spectrometry for detection and analysis of the structure of secondary metabolites, such as lipopeptide surfactants, in situ when whole cells and crude fermentation broth are used as targets. This method is well suited for rapid primary screening of new microbial isolates for novel natural compounds. In combination with our mass spectrometric strategy, below we describe efficient procedures for fractionation of such lipopeptide complexes on an analytic scale by using a high-resolution separation method in order to investigate pure isoforms in detail and for the provision of lipopeptide biosurfactants on a preparative scale for biological and biotechnological testing.
Fractionation of the lipopeptide complexes of B. subtilis C-1 on an analytic scale by high-resolution reversed-phase HPLC.
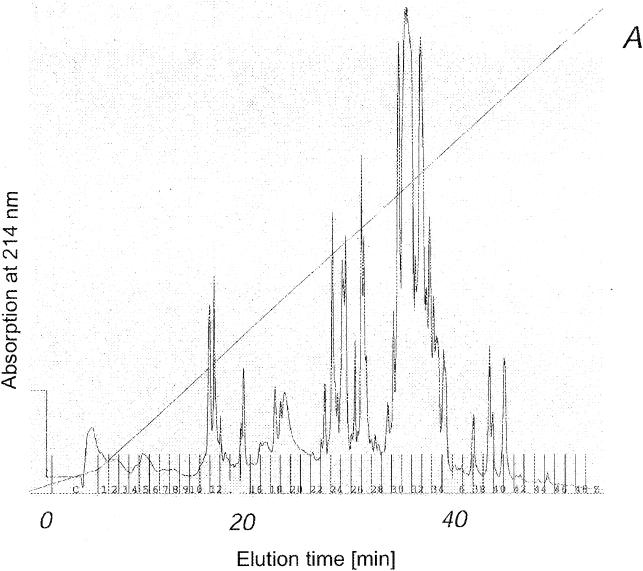
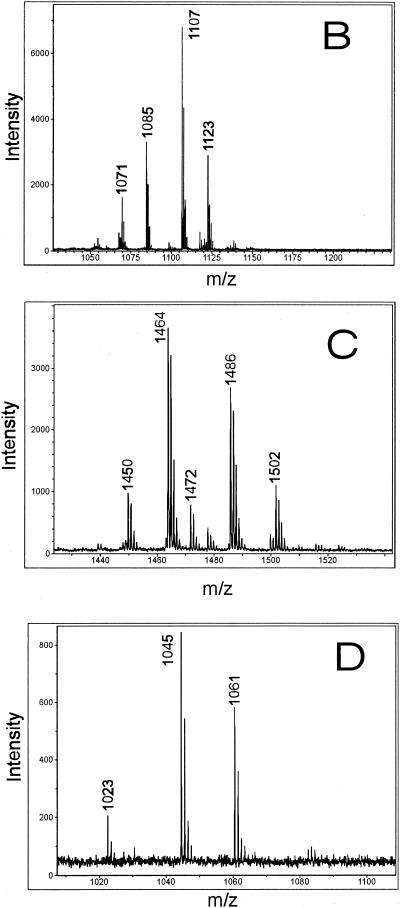
As shown in Fig. 4A, an efficient miniaturized procedure was used to separate the isoforms of the lipopeptide complexes from B. subtilis C-1 by reversed-phase HPLC on a μRPC column by using a Pharmacia Smart microseparation system, as described in Materials and Methods. The lipopeptide products were found in fractions 23 to 42. Iturins were eluted first, in fractions 23 to 27, followed by the fengycins in fractions 28 to 36 and the surfactins in fractions 37 to 42. The isoforms of these biosurfactant families were well resolved according to their hydrophobicities, which are determined mainly by the lengths of the fatty acid moieties. The identities of the lipopeptide isoforms eluted from the μRPC column are summarized in Table 5. The excellent resolution of this microseparation technique was demonstrated by MALDI-TOF mass spectrometric analysis of fractions 25, 30, and 39 (Fig. 4B to D), which contained purified forms of a C17 iturin, a C16 fengycin, and a C14 surfactin, respectively. The mass spectra of these lipopeptides have peaks which can be attributed to the protonated forms, as well as to the sodium and potassium adducts. Apparently, the alkali forms are the most abundant species. By using this rapid, efficient procedure purified isoforms could be obtained for extensive structural characterization.
FIG. 4.
Separation of the biosurfactant products of B. subtilis C-1 on an analytical scale by high-resolution reversed-phase HPLC of a 20-μl aliquot of the methanolic extract of the lipopeptide fraction obtained by precipitation from the culture filtrate with concentrated HCl on a μRPC SC 2.1/10 column by using a Pharmacia Smart microseparation system. (A) HPLC chromatogram. Lipopeptides were eluted with a gradient of 20 to 100% acetonitrile in the presence of 0.1% TFA. (B to D) MALDI mass spectra of aliquots of fractions 25, 30, and 39, respectively.
Purification on a preparative scale and structural characterization of the surfactin biosurfactant products of B. subtilis C-1.
For provision of lipopeptide biosurfactants on a preparative scale, B. subtilis C-1 was grown at 30°C for 48 h. On average, approximately 100 mg of crude material per liter of culture supernatant could be recovered after extraction of acid precipitates with dichloromethane. This product was fractionated and purified by gel permeation chromatography on a Sephadex LH-20 column. For example, purification of the surfactin compounds which showed the highest biosurfactant activities among the lipopeptides produced by B. subtilis C-1 is described below. The collected fractions were examined by TLC as described in Materials and Methods. Surfactin appeared as a single elongated spot on the TLC plates at Rf values of 0.62 (solvent A) and 0.52 (chloroform-methanol-water, 65:25:4 [vol/vol/vol]; solvent B). Several of the fractions obtained contained essentially pure surfactin, and the yield was approximately 25 to 30 mg of the entire surfactin complex per liter of culture supernatant; the surfactin was characterized by amino acid and mass spectrometric analyses.
Hydrolyzed samples of purified surfactin were subjected to one-dimensional and two-dimensional TLC. TLC analysis with two different solvent systems resulted in four spots which matched the spots for the standard amino acids leucine and/or isoleucine, valine, glutamic acid, and aspartic acid which were chromatographed on the same plate (data not shown). Leucine and isoleucine could not be separated on TLC plates. The hydrolysate of surfactin was also applied to a Waters model 501 amino acid analyzer. The biosurfactant product contained Glu, Asp, Val, Ile, and Leu at a ratio of 1:1:1:1:3. Enantiospecific TLC on chiral TLC plates revealed the presence of both the l and d forms of Leu, whereas for Glu, Asp, Val, and Ile, only the l forms were detected.
The surfactin biosurfactant complex was investigated by MALDI-TOF mass spectrometry. Pure fractions of the compound obtained by LH-20 gel permeation chromatography produced mass peaks at m/z 1016.8, 1030.8, 1044.8, and 1058.8 at a ratio of 1:5.5:8.3:5.2, indicating that the surfactin was a mixture of structural analogs with mass differences of 14 Da. The components of the surfactin complex were separated by reversed-phase HPLC on an ODS Hypersil C18 column. The HPLC chromatogram (data not shown) had three prominent, nearly baseline, separated large peaks and some minor peaks. Most of the peaks could be attributed to essentially pure surfactin isoforms by MALDI-TOF mass spectrometry. The distribution of the molecular ions found in these fractions and assignment of the ions to the different surfactin species are summarized in Table 6. From the mass spectrometric data it is apparent that the surfactin isoforms eluted according to their hydrophobicities. For the small peak A that appeared in fractions 24 to 26 in front of the three main peaks (peaks B to D), a main parent ion at m/z 1016.7 was found that was attributed to a small amount of the sodium adduct of a valine-7 surfactin (20). The low-mass peak at m/z 1032.7 represented the potassium adduct of this species. Obviously, in a minor part of the surfactin complex the Leu/Ile residue at position 7 of the peptide ring is replaced by valine, as previously reported by other authors (2, 20, 29). Large peaks B and C comprising fractions 27 to 29 and 30 to 32 contained essentially pure C13 and C14 surfactin species, respectively. In fractions 37 to 39 (peak D) a mixture of C14 and C15 surfactins was found. The structure of the components of the surfactin complex was investigated by PSD MALDI-TOF mass spectrometry by using surfactin isoforms isolated from B. subtilis OKB 105 as reference compounds (22). In this way the species with mass numbers of m/z 1030.7, 1044.7, and 1058.7 could be identified by their bn ions as the [M+Na]+ ions of surfactins with C13, C14, and C15 β-hydroxy fatty acid side chains.
TABLE 6.
MALDI-TOF mass spectrometric characterization of the surfactin biosurfactants produced by B. subtilis C-1 after separation by reversed-phase HPLC on ODS Hypersil C18a
| Fractions | Mass peak (m/z) | Amplitude (cm) | Assignment (surfactin species) |
|---|---|---|---|
| 24-26 | 994.2 | 4.5 | Val-7 C13, [M+H]+ |
| 1016.7 | 14.8 | Val-7 C13, [M+Na]+ | |
| 1032.7 | 3.6 | Val-7 C13, [M+K]+ | |
| 27-29 | 1030.7 | 14.8 | Leu/Ile-7 C13, [M+Na]+ |
| 1046.7 | 5.0 | Leu/Ile-7 C13, [M+K]+ | |
| 30-32 | 1022.7 | 1.3 | Leu/Ile-7 C14, [M+H]+ |
| 1044.7 | 14.9 | Leu/Ile-7 C14, [M+Na]+ | |
| 1060.7 | 8.0 | Leu/Ile-7 C14, [M+K]+ | |
| 37-39 | 1036.7 | 1.1 | Leu/Ile-7 C15, [M+H]+ |
| 1044.7 | 1.2 | Leu/Ile-7 C14, [M+Na]+ | |
| 1058.7 | 12.4 | Leu/Ile-7 C15, [M+Na]+ |
Surfactins were fractionated by using a 20 to 100% acetonitrile linear gradient for 60 min. Eluent A was 20% (vol/vol) acetonitrile in 100 mM ammonium acetate (pH 6.9). Eluent B was 100% acetonitrile. The UV absorbance was monitored at 220 nm. The flow rate was 0.5 ml/min. In the collected fractions surfactin isoforms were identified by MALDI-TOF mass spectrometry. The mass data represent monoisotopic mass numbers.
DISCUSSION
Recently, we reported that MALDI-TOF mass spectrometry is an innovative, highly efficient technique for rapid typing of microorganisms by analysis of their secondary metabolite spectra (22), which is also well suited for studying the production of natural compounds in both surface and suspension cultures. The detection limit is usually in the upper femtomolar to picomolar range. The molecular masses can be determined with an accuracy of 0.01 to 0.02%.
In this study we used this method to screen for microbial biosurfactants and to characterize their molecular structures. Production of these compounds can be monitored efficiently by measuring the surface tension of culture supernatants. However, structural characterization requires techniques that involve high-resolution structure analysis. In particular, an advanced mass spectrometric method is well qualified for this task. In this study we used MALDI-TOF mass spectrometry to detect and characterize biosurfactants directly in microbial cells, as well as in cellular extracts and crude culture filtrates. As previously reported (22), by using whole-cell MALDI-TOF mass spectrometry cellular products can be detected which either are attached to the cell surface or are integrated into the outer cell membrane. In this way information on the secondary metabolites produced by a microorganism can be obtained in minutes with high precision and sensitivity with no need to fractionate and purify the detected compounds. Depending on the structural complexity of the molecules investigated, in some cases rapid in situ structure analysis is possible by interpretation of fragment spectra obtained from PSD MALDI-TOF mass spectrometry. Often this technique can be used successfully for structural characterization of natural compounds with a regular polymeric structure, such as bioactive peptides, which have linear or cyclic sequences of amino acids. Elucidation of the structure can be started by using immonium ions, which provide information on the amino acid composition of a peptide, followed by a search for dipeptide fragment ions which indicate the nearest neighbors in the peptide moiety. On the basis of the data obtained the entire peptide sequence can be estimated. In essence, MALDI-TOF mass spectrometry is a powerful tool for investigating large biological structures and solutions with complex compositions.
This novel method was used for characterization of a bacterial strain isolated by us from petroleum sludge, which was identified by its genetic, physiological, and morphological features as a B. subtilis strain designated B. subtilis C-1. As Tables 1 and 2 show, this organism was found to be an efficient biosurfactant producer based on surface tension measurements for culture supernatants obtained from fermentations performed at various temperatures. The surface tension measured for the pure culture medium (71 dynes/cm) was lowered in the culture filtrates by the production of surface-active compounds to approximately 29 dynes/cm, a value that has been reported for the most efficient biosurfactants which have been isolated and studied so far (4, 5, 7, 16). Such compounds have great potential for industrial exploitation (5, 7, 9, 10, 32). In particular, during the last decade interesting, promising biopharmaceutical applications have been developed for surfactin (37, 38), which is the most efficient lipopeptide biosurfactant produced by B. subtilis. Therefore, the availability of innovative, efficient procedures for detection, structure analysis, functional characterization, and biological testing of such compounds has great biotechnological relevance.
Figure 2 shows the MALDI mass spectra of whole cells of B. subtilis C-1 grown in surface and suspension cultures, as well as the MALDI mass spectra of aliquots of the culture supernatants and the crude lipopeptide fraction obtained by precipitation with HCl and extraction of the pellet with methanol. Prominent groups of mass peaks were observed in the mass range between m/z 1000 and 1500. By comparing the mass data summarized in Tables 3 to 6 with the mass numbers reported for the lipopeptide complexes from other B. subtilis strains (19, 20, 22, 25-27, 33, 34) and by analyzing the fragment patterns derived from PSD MALDI-TOF mass spectra (Fig. 3), the lipopeptide products of B. subtilis C-1 could be identified as surfactins, iturins, and fengycins. All these biosurfactants appeared as families of closely related isoforms which differed in the lengths of their fatty acid side chains, as well as in the amino acid substitutions in their peptide rings (Fig. 1). In this way the identification of our isolate as a B. subtilis strain was corroborated by mass spectrometric analysis of its secondary metabolite spectrum.
The lipopeptide families produced by B. subtilis strains are prominent examples that demonstrate the capacity of MALDI-TOF mass spectrometry for detection and in situ structure analysis. This novel mass spectrometric method was combined with an efficient miniaturized separation procedure involving the Pharmacia Smart microseparation system, which is well suited for rapid, high-resolution fractionation of most of the lipopeptide isoforms on a small scale for analytical purposes, as described for mass spectrometric analysis. By using this procedure small aliquots (10 to 50 μl) of the culture filtrate or crude cellular extracts could be fractionated with excellent resolution and could be analyzed with high sensitivity and precision (Fig. 4). The mass spectrometric investigation of B. subtilis C-1 revealed the following lipopeptide pattern.
The iturin A compounds formed by this organism as the main products in the stationary phase of growth contain unusual fatty acid components with chain lengths of 16 to 19 carbon atoms. C16 and C17 species are the predominant isoforms. These results are different from the data obtained by Hourdou et al. (15) and Peypoux et al. (26) in their extensive studies of iturin compounds. The B. subtilis strains used by these authors produce iturins with saturated β-amino C14 to C16 fatty acid side chains (15, 26). The fatty acid components of our isolate resemble those found in mycosubtilins (15, 25, 28). All of them showed mass numbers which are 1 mass unit lower than the value expected in the case of saturated β-amino fatty acid side chains. This result is compatible with the assumption of iturin B molecules which contain one aspartic acid residue instead of an asparagine residue, as well as an unsaturated β-amino fatty acid moiety with one double bond. However, this is certainly not the case, because an aspartic acid was not found in the fragment patterns derived from the PSD MALDI-TOF mass spectra of the new iturin compounds. Another possibility to explain our data is the hypothesis that β-hydroxy rather than β-amino fatty acid constituents contain one double bond, but this hypothesis has to be clarified by a detailed structure analysis, which is being done.
The fengycin species of our isolate comprise C15 to C17 variants which have a characteristic Ala-Val dimorphy at position 6 of the peptide ring, as previously reported for other fengycin producers (24, 33-35). As Table 4 shows, two kinds of variants were observed, which differ by 2 mass units. This feature is due to two series of compounds, one with saturated lipid moieties and the other with fatty acid side chains containing one double bond whose position remains to be elucidated by further studies.
Testing of the biological activities and development of industrial applications for these interesting compounds require that they be isolated on a preparative scale. To do this, efficient techniques, such as solid-phase extraction and dispersion and gel permeation and adsorption chromatography in combination with preparative reversed-phase HPLC, are available. In this paper we report on a two-step procedure for the fractionation of the surfactin lipopeptide complex of B. subtilis C-1, which is the predominant biosurfactant product in the exponential phase of growth. Surfactin isoforms were isolated in pure form by a combination of gel permeation chromatography on Sephadex LH-20 and reversed-phase HPLC on Hypersil C18. Surfactin isoforms were identified by MALDI-TOF mass spectrometry and by correlation with previous results (20). The data obtained are summarized in Table 6. The surfactin complex of B. subtilis C-1 comprised C13 to C15 isoforms which occurred at a ratio similar to the ratios observed for other surfactin producers (19, 20, 22), and C14 and C15 surfactins were the main species.
The innovative method presented in this paper is very important for making available new microbial biosurfactant compounds for industrial exploitation, and in particular, for developing specific biopharmaceutical applications. This has recently been demonstrated for the surfactins showing potent antivirus and antimycoplasma properties (37, 38), which can be efficiently utilized for inactivation of virus and mycoplasma contaminants in biotechnological and biopharmaceutical products.
Acknowledgments
We are grateful for the facilities provided by the Director, IMTECH.
Financial support was received from the Council for Scientific and Industrial Research (CSIR), the Department of Biotechnology (DBT), the Government of India, and the Deutsche Forschungsgemeinschaft (grant Va 63/4-4).
REFERENCES
- 1.Arima, K., A. Kakinuma, and G. Tamura. 1968. Surfactin, a crystalline peptidelipid surfactant produced by Bacillus subtilis: isolation, characterization and its inhibition of fibrin clot formation. Biochem. Biophys. Res. Commun. 31:488-494. [DOI] [PubMed] [Google Scholar]
- 2.Baumgart, F., B. Kluge, C. Ullrich, J. Vater, and D. Ziessow. 1991. Identification of amino acid substitutions in the lipopeptide surfactin using 2D NMR spectroscopy. Biochem. Biophys. Res. Commun. 177:998-1005. [DOI] [PubMed] [Google Scholar]
- 3.Besson, F., F. Peypoux, G. Michel, and L. Delcambe. 1978. Identification of antibiotics of iturin group in various strains of Bacillus subtilis. J. Antibiot. (Tokyo) 31:284-288. [DOI] [PubMed] [Google Scholar]
- 4.Cooper, D. G., C. R. Macdonald, J. B. Duff, and N. Kosaric. 1981. Enhanced production of surfactin from Bacillus subtilis by continuous product removal and metal cation additions. Appl. Environ. Microbiol. 42:408-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper, D. G., and J. E. Zajic. 1980. Surface-active compounds from microorganisms. Adv. Appl. Microbiol. 26:229-256. [Google Scholar]
- 6.Deleu, M., H. Razafindralambo, Y. Popineau, P. Jacques, P. Thonart, and M. Paquot. 1999. Interfacial and emulsifying properties of lipopeptides from Bacillus subtilis. Colloids Surf. A Physicochem. Eng. Aspects 152:3-10. [Google Scholar]
- 7.Desai, J. D., and I. M. Banat. 1997. Microbial production of surfactants and their commercial potential. Microbiol. Mol. Biol. Rev. 61:47-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erhard, M., H. von Döhren, and P. Jungblut. 1997. Rapid typing and elucidation of new secondary metabolites of intact cyanobacteria using MALDI-TOF mass spectrometry. Nat. Biotechnol. 15:906-909. [DOI] [PubMed] [Google Scholar]
- 9.Fiechter, A. 1992. Biosurfactants: moving towards industrial application. TIBTECH 10:208-217. [DOI] [PubMed] [Google Scholar]
- 10.Haferburg, D., R. Hommel, R. Claus, and H.-P. Kleber. 1986. Extracellular microbial lipids as biosurfactants. Adv. Biochem. Eng. Biotechnol. 33:53-93. [Google Scholar]
- 11.Horowitz, S., J. N. Gilbert, and W. M. Griffin. 1990. Isolation and characterization of a surfactant produced by Bacillus licheniformis 86. J. Ind. Microbiol. 6:243-248. [Google Scholar]
- 12.Horowitz, S., and W. M. Griffin. 1991. Structural analysis of Bacillus licheniformis 86 surfactant. J. Ind. Microbiol. 7:45-52. [DOI] [PubMed] [Google Scholar]
- 13.Hosono, K., and H. Suzuki. 1983. Acylpeptides, the inhibitors of cyclic adenosine 3′,5′-monophosphate phosphodiesterase. I. Purification, physicochemical properties and structures of fatty acid residues. J. Antibiot. (Tokyo) 36:667-673. [DOI] [PubMed] [Google Scholar]
- 14.Hosono, K., and H. Suzuki. 1983. Acylpeptides, the inhibitors of cyclic adenosine 3′,5′-monophosphate phosphodiesterase. II. Amino acid sequence and location of lactone linkage. J. Antibiot. (Tokyo) 36:674-678. [DOI] [PubMed] [Google Scholar]
- 15.Hourdou, M.-L., F. Besson, I. Tenoux, and G. Michel. 1989. Fatty acids and β-amino acid syntheses in strains of Bacillus subtilis producing iturinic antibiotics. Lipids 24:940-944. [DOI] [PubMed] [Google Scholar]
- 16.Jenny, K., O. Käppeli, and A. Fiechter. 1991. Biosurfactants from Bacillus licheniformis: structural analysis and characterization. Appl. Microbiol. Biotechnol. 36:5-13. [DOI] [PubMed] [Google Scholar]
- 17.Kakinuma, A., M. Hori, M. Isono, G. Tamura, and K. Arima. 1969. Determination of amino acid sequence in surfactin, a crystalline peptide lipid surfactant produced by Bacillus subtilis. Agric. Biol. Chem. 33:971-972. [Google Scholar]
- 18.Kakinuma, A., H. Sugino, M. Isono, G. Tamura, and K. Arima. 1969. Determination of fatty acid in surfactin and elucidation of the total structure of surfactin. Agric. Biol. Chem. 33:973-976. [Google Scholar]
- 19.Kakinuma, A., A. Ouchida, T. Shima, H. Sugino, M. Isono, G. Tamura, and K. Arima. 1969. Confirmation of the structure of surfactin by mass spectrometry. Agric. Biol. Chem. 33:1669-1671. [Google Scholar]
- 20.Kowall, M., J. Vater, B. Kluge, T. Stein, P. Franke, and D. Ziessow. 1998. Separation and characterization of surfactin isoforms produced by Bacillus subtilis OKB 105. J. Colloid Interface Sci. 203:1-8. [DOI] [PubMed] [Google Scholar]
- 21.Landy, M., G. H. Warren, S. B. Rosenman, and L. G. Colio. 1948. Bacillomycin, an antibiotic from Bacillus subtilis active against pathogenic fungi. Proc. Soc. Exp. Biol. Med. 67:539-541. [DOI] [PubMed] [Google Scholar]
- 22.Leenders, F., T. H. Stein, B. Kablitz, P. Franke, and J. Vater. 1999. Rapid typing of Bacillus subtilis strains by their secondary metabolites using matrix-assisted laser desorption/ionization mass spectrometry of intact cells. Rapid Commun. Mass Spectrom. 13:943-949. [Google Scholar]
- 23.Naruse, N., O. Tenmyo, S. Kobaru, H. Kamei, T. Miyaki, M. Konishi, and T. Oki. 1990. Pumilacidin, a complex of new antiviral antibiotics—production, isolation, chemical properties, structure and biological activity. J. Antibiot. (Tokyo) 43:267-280. [DOI] [PubMed] [Google Scholar]
- 24.Nishikori, T., H. Naganawa, Y. Muraoka, T. Aoyagi, and H. Umezawa. 1986. Plipastatins: new inhibitors of phopholipase A2, produced by Bacillus cereus BMG302-fF67. II. Structure of fatty acid residue and amino acid sequence. J. Antibiot. (Tokyo) 39:745-754. [DOI] [PubMed] [Google Scholar]
- 25.Peypoux, F., G. Michel, and L. Delcambe. 1976. Structure de la mycosubtiline, antibiotique isolé de Bacillus subtilis. Eur. J. Biochem. 63:391-398. [DOI] [PubMed] [Google Scholar]
- 26.Peypoux, F., M. Guinand, G. Michel, L. Delcambe, B. C. Das, and E. Lederer. 1978. Structure of iturin A, a peptidolipid antibiotic from Bacillus subtilis. Biochemistry 17:3992-3996. [DOI] [PubMed] [Google Scholar]
- 27.Peypoux, F., M.-T. Pommier, B. C. Das, F. Besson, L. Delcambe, and G. Michel. 1984. Structures of bacillomycin D and bacillomycin L peptidolipid antibiotics from Bacillus subtilis. J. Antibiot. (Tokyo) 77:1600-1604. [DOI] [PubMed] [Google Scholar]
- 28.Peypoux, F., M. T. Pommier, D. Marion, M. Ptak, B. C. Das, and G. Michel. 1986. Revised structure of mycosubtilin, a peptidolipid antibiotic from Bacillus subtilis. J. Antibiot. (Tokyo) 39:636-641. [DOI] [PubMed] [Google Scholar]
- 29.Peypoux, F., J.-M. Bonmatin, H. Labbé, B. C. Das, M. Ptak, and G. Michel. 1991. Isolation and characterization of a new variant of surfactin, the [Val-7] surfactin. Eur. J. Biochem. 202:101-106. [DOI] [PubMed] [Google Scholar]
- 30.Peypoux, F., J. M. Bonmatin, and J. Wallach. 1999. Recent trends in the biochemistry of surfactin. Appl. Microbiol. Biotechnol. 51:553-563. [DOI] [PubMed] [Google Scholar]
- 31.Razafindralambo, H., Y. Popineau, M. Deleu, C. Hbid, P. Jacques, P. Thonart, and M. Paquot. 1998. Foaming properties of lipopeptides produced by Bacillus subtilis: effects of lipid and peptide structural attributes. J. Agric. Food Chem. 46:911-916. [Google Scholar]
- 32.Rosenberg, E. 1986. Microbial surfactants. Crit. Rev. Biotechnol. 3:109-132. [Google Scholar]
- 33.Schneider, J. 1999. Doctoral thesis. University of Cologne, Cologne, Germany.
- 34.Schneider, J., K. Taraz, H. Budzikiewicz, P. Jacques, and P. Thonart. 1999. The structure of two fengycins from Bacillus subtilis S499. Z. Naturforsch. 54:859-865. [DOI] [PubMed] [Google Scholar]
- 35.Vanittanakom, N., W. Loeffler, U. Koch, and G. Jung. 1986. Fengycin-a novel antifungal lipopeptide antibiotic produced by Bacillus subtilis F-29-3. J. Antibiot. (Tokyo) 39:888-901. [DOI] [PubMed] [Google Scholar]
- 36.Vater, J. 1986. Lipopeptides, an attractive class of microbial surfactants. Prog. Colloid Polymer Sci. 72:12-18. [Google Scholar]
- 37.Vollenbroich, D., G. Pauli, M. Özel, and J. Vater. 1997. Antimycoplasma properties and application in cell culture of surfactin, a lipopeptide antibiotic from Bacillus subtilis. Appl. Environ. Microbiol. 63:44-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vollenbroich, D., M. Özel, J. Vater, R. M. Kamp, and G. Pauli. 1997. Mechanism of inactivation of enveloped viruses by the biosurfactant surfactin from Bacillus subtilis. Biologicals 25:289-297. [DOI] [PubMed] [Google Scholar]
- 39.Yakimov, M. M., K. N. Timmis, V. Wray, and H. L. Fredrickson. 1995. Characterization of a new lipopeptide surfactant produced by thermotolerant and halotolerant subsurface Bacillus licheniformis BAS 50. Appl. Environ. Microbiol. 61:1706-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]