The increasing release of organic pollutants by industries cause many health-related problems. However, increased awareness of the harmful effects of environmental pollution has led to a dramatic increase in research on various strategies that may be employed to clean up the environment. It is now realized that microbial metabolism provides a safer, more efficient, and less expensive alternative to physicochemical methods for pollution abatement (27). In the past few decades, a vast range of xenobiotic compounds have been found to be susceptible to microbial mineralization. In most instances where mineralization has been demonstrated, the catabolic pathway and its regulation have also been determined.
Bacterial chemotaxis, movement under the influence of a chemical gradient, either toward (positive chemotaxis) or away (negative chemotaxis) from the gradient helps bacteria to find optimum conditions for their growth and survival. However, this aspect has received little attention, even though some microorganisms with the chemotactic ability toward different xenobiotic compounds have been isolated and characterized (8, 19, 23, 41, 53). In many cases, the chemoattractant is a compound that serves as carbon and energy source, whereas a chemorepellent is toxic for the bacteria.
We describe here recent discoveries in bacterial chemotaxis toward pollutants and how they may be explored and exploited for bioremediation applications.
BACTERIAL BEHAVIORAL RESPONSES
Responding to changes in the environment is a fundamental property of a living cell. It is especially important for unicellular organisms, which directly interact with the changing microenvironment. Through evolution, microorganisms have developed effective mechanisms that help them to regulate their cellular function in response to changes in its environment (29). Of these, chemotaxis, i.e., the migration of microorganisms under the influence of a chemical gradient, is the best-studied bacterial behavioral response that navigates the bacteria to niches that are optimum for their growth and survival. Bacteria swim toward or away from a chemical stimulant in a guided, nonrandom manner. The chemotactic swimming is a result of rotation of flagella at speeds of ca. 18,000 rpm, and it is powered by the proton motive force (14). Flagellar motors are reversible in nature that help to change bacterial tumbling into a directional swimming by reversing the flagellar rotation from clockwise to counterclockwise direction (44). An environmental stimulus, e.g., light, oxygen, chemical, etc., is sensed by a receptor and signal(s) in the form of two-component regulatory systems is transmitted to the flagellar motors, which then move in the required direction (10, 44).
In order to learn bacterial chemotaxis in response to the pollutants, it is important to understand the molecular mechanisms of well-characterized chemotaxis toward natural compounds such as amino acids, sugars, aromatic acids etc. Chemotaxis is broadly divided into two categories on the basis of signal transduction strategies. One kind of chemotaxis is independent of the metabolism of the chemoeffector molecule, whereas metabolism of the signaling molecule is a prerequisite for the other kind of chemotaxis (2). The two signal transduction strategies of chemotaxis that are relevant to this minireview are discussed briefly in the next two sections.
METABOLISM-INDEPENDENT CHEMOTAXIS
Although Bunning (11) and Pfeffer (46) were the first to identify bacterial chemotaxis, Adler (1) provided the first extensive biochemical and genetic evidence that chemotaxis in Escherichia coli is independent of uptake or metabolism of the chemical stimulus. In metabolism-independent chemotaxis: (i) essentially nonmetabolizable analogues of metabolizable attractants are also attractants, (ii) mutations in the metabolism of a chemical attractant do not affect chemotaxis, and (iii) a chemoattractant attracts bacteria even in the presence of metabolizable compounds. Since Adler's time research has revealed the molecular details of this kind of chemotaxis in Escherichia coli and Salmonella enterica serovar Typhimurium (4, 10). Metabolism-independent chemosensing occurs through transmembrane chemoreceptors (chemotaxis transducers) that transmit information to flagella via two-component regulatory systems that direct the cells to move in preferential directions (10, 44). Transmembrane signaling in E. coli is a paradigm for metabolism-independent chemotaxis (1, 16). Metabolism-independent chemical sensing is found in several bacterial species, including E. coli, Salmonella sp., Bacillus subtilis, and Pseudomonas sp. toward a number of chemicals (4, 7, 10, 16). Detailed genetic and biochemical investigations of E. coli have proven that four chemoreceptors out of five are transmembrane proteins involved in the signaling process: Tar for aspartate, Tsr for serine, Trg for ribose and galactose, and Tap for peptides (16). These four homologous methyl-accepting chemotaxis proteins (MCPs) have a periplasmic sensing domain and a cytoplasmic signaling domain. Binding of an effector molecule to the periplasmic sensing domain of these MCPs initiates a signal that is transduced to the flagellar motor in the form of a phosphorelay that is similar to the other two-component signal transduction systems of bacteria (10, 44). A naphthalene chemorecepter, NahY (discussed later), has been reported to have significant homology to the above-mentioned MCPs (20). Different signal transducer proteins involved in the chemotaxis of E. coli and Pseudomonas sp. and their mechanism of interaction have been identified (10, 33, 44).
METABOLISM-DEPENDENT CHEMOTAXIS
In contrast to the metabolism-independent chemotaxis discussed above, some chemotactic responses in bacteria require metabolism of the chemoeffector molecule. E. coli exhibits metabolism-dependent chemotaxis toward proline (12), glycerol (72), and succinate (9). Metabolism-dependent chemotaxis is best studied in an α-proteobacterium, Azospirillum brasilense, where (i) nonmetabolizable analogues of metabolizable attractants are not attractants, (ii) inhibition of the metabolism of a chemical attractant completely abolishes chemotaxis to that particular attractant, and (iii) presence of another metabolizable chemical prevents chemotaxis to all chemoattractants studied (3). Rhodobacter sphaeroides shows chemotaxis toward a wide range of amino acids, organic acids, and sugars (5). Although all attractants are metabolizable, direct correlation has only been shown toward chemotaxis to sugars and its metabolism (32). Similarly, Campylobacter jejuni (28), Sinorhizobium meliloti (5), and Rhodobacter sphaeroides (47) show metabolism-dependent chemotactic responses toward pyruvate, organic acids, carbohydrates, flavones, and ammonia. Metabolism-dependent chemotaxis shares signaling pathways with other bacterial behavioral responses collectively known as energy taxis, defined as a behavioral response to stimuli affecting cellular energy levels (61-63). Aerotaxis (61) and phototaxis (63) are its well-studied examples. The signal for this type of tactic movement originates within the electron transport chain, where a change in the rate of electron transport (or a related parameter defining cellular energy levels) is detected by a signal transduction system (2, 61, 63). In this phenomenon metabolizable substrates can stimulate a behavioral response as long as a receptor sensing the change in cellular energy level is present. Aerotaxis helps a bacterium to move to the niches that are optimum in terms of oxygen concentration (58, 61); this phenomenon could play an important role in oxidative biodegradation of different xenobiotics since different oxygenases that are important enzymes of the degradation pathways require molecular oxygen for the activity. It may also be helpful for anaerobic/microaerophilic microorganisms to move deeper into the soils and sediments (anaerobic/microaerophilic conditions) and degrade toxic contaminants.
BACTERIAL CHEMOTAXIS TOWARD POLLUTANTS
The enhancement of chemotaxis in Spirochaeta aurantia grown under conditions of nutrient limitation has been shown (64). In conditions of limited carbon and energy sources, it is possible that chemotaxis might have been selected as an advantageous behavior in bacteria along with xenobiotic degradation capabilities after exposure to such compounds. Although bacterial degradation capabilities in many cases have been proved to be efficient for remediation of contaminated sites, bacterial chemotaxis toward pollutants has received less attention. The first step in bioremediation, however, is the bioavailability of a compound to the bacterial cells which may be facilitated by chemotaxis.
Bioavailability of organic contaminants has been identified as a major limitation to efficient bioremediation of contaminated sites (26). Contaminated soils contain a separate nonaqueous-phase liquid (NAPL) that may be present as droplets or films on soil surfaces. Biodegradation takes place more readily when the target contaminants are dissolved in an aqueous medium (60). Many pollutants, especially those that are hydrophobic, are virtually insoluble in water and remain adsorbed in the NAPL (60). In order for biodegradation to occur, bacteria must have access to the target compounds either by dissolution of the target compounds in the aqueous phase or by adhesion of the bacteria directly to the NAPL water interface. In order to gain access to such adsorbed pollutants, degradative bacteria need to find and attach to surfaces possibly through biofilm formation. Chemotaxis has been shown to play an important role in biofilm formation in several microorganisms (40, 48, 49, 67) that may guide a bacterium to swim toward nutrients (hydrophobic pollutants) adsorbed to a surface, followed by attachment using its flagella. It has been shown that flagella are required for attachment to abiotic surfaces, thus facilitating the initiation of biofilm formation (48, 60). In addition, chemotaxis and/or motility might be required for the bacteria within a developing biofilm to move along the surface, thereby facilitating growth and spread of the biofilm (60).
CHEMOTAXIS TOWARD SIMPLE AROMATIC COMPOUNDS
Aromatic compounds are abundant in the biosphere due to natural and anthropogenic activities and some of them are pollutants (18, 35). Bacterial degradation of structurally simple, readily biodegradable aromatic compounds has been studied with the expectation that this will facilitate work on more recalcitrant members of the group. Consequently significant work has been done on the biodegradation of these compounds (13, 35); however, little attention has been given to the chemotaxis aspect.
Common soil bacteria such as Rhizobium sp. (15, 43), Bradyrhizobium sp. (43), Pseudomonas sp. (24), and Azospirillum sp. (36) have been shown to be chemotactically attracted toward different aromatic hydrocarbons. Many of these compounds are present in soils, sediments, and rhizosphere, and they serve as sources of carbon and energy for the microorganisms. Aromatic acids such as benzoate, p-hydroxybenzoate (PHB), methylbenzoates, the m-, p-, and o-toluates, salicylate, dl-mandelate, β-phenylpyruvate, and benzoylformate have been reported to be attractants for Pseudomonas putida PRS2000 (24). It has been argued that at least two sets of chemoreceptors are synthesized in P. putida PRS2000. The chemoreceptor of the benzoate chemotaxis system (responsible for chemotaxis to benzoate, toluates, PHB, and salicylate) is induced by growth on substrates that involves β-ketoadipate as an intermediate (i.e., with benzoate or PHB). Another chemorecepeptor is associated with chemotaxis toward mandelate, benzoylformate, and β-phenylpyruvate (24). A gene cluster, pcaRKF, was found to be involved in chemotaxis, biodegradation, and transport of PHB in this strain (22). The pcaK encodes a nonessential transporter for PHB but has been shown to be required for chemotaxis toward PHB and benzoate. Chemotaxis to PHB in this strain can be eliminated by disrupting pcaK without any accompanying effect on metabolism, suggesting that this chemotactic response is receptor-mediated (22). There are yet no conclusive results to show whether PcaK itself is a chemoreceptor for PHB chemotaxis or it plays an indirect role in chemotaxis; however, expression of the tactic response to aromatic acids in strain PRS2000 does not require the expression of enzymes for the degradation of these compounds.
Parales et al. (42) reported toluene as a chemoattractant for three toluene-degrading bacteria (P. putida F1, Ralstonia pickettii POK01, and Burkholderia cepacia G4). P. putida F1 was also reported to be chemotactic toward seven other organic pollutants, some of which served as sources of carbon and energy, but almost every compound was found to be a substrate for toluene dioxygenase (Table 1) (42). Bacterial chemotaxis toward toluene, benzene, ethylbenzene, isopropylbenzene, naphthalene, trifluorotoluene, perchloroethylene, dichloroethylene, and trichloroethylene was found to be induced during the growth of P. putida F1 on toluene (Table 1). This inducible nature of chemotactic response toward toluene and toluene dioxygenase substrates suggests a correlation between chemotaxis and their transformation and/or degradation. Although the P. putida F1 mutants defective in toluene dioxygenase, which cannot grow on toluene, are chemotactic toward toluene, an indirect correlation between chemotaxis and biodegradation was still shown by the mutation studies of todST genes that are positioned adjacent to the tod structural genes (todRXFC1C2BADEGIH that code for toluene degradation) (42). The todS and todT genes encode a two-component sensory transduction system required for induction of tod structural genes in the presence of toluene. It has been observed that inactivation of todS blocks growth on toluene and a mutation in todT results in a very slow growth on toluene. Interestingly, both of these mutants did not exhibit chemotaxis to toluene, suggesting that the genes required for the chemotactic response to toluene are coordinately regulated with those for toluene degradation by todS and todT.
TABLE 1.
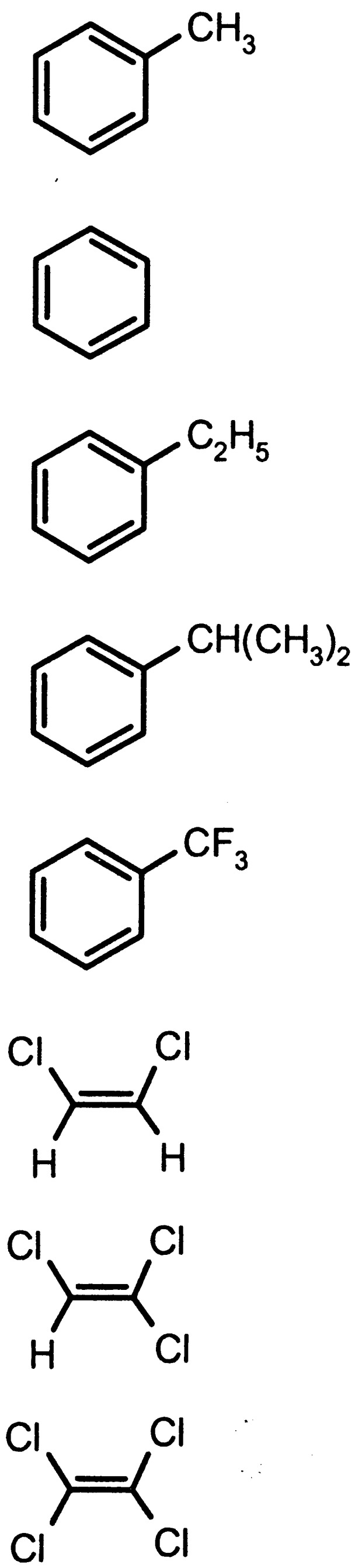
Adapted from reference 42 with permission of the publisher.
All chemicals are substrates for toluene dixoygenase except for PCE.
S, strong response; W, weak response.
Yes, growth substrate; No, not growth substrate; ND, not determined.
Abbreviations: TFT, trifluorotoluene; PCE, perchloroethylene; DCE, dichloroethylene; TCE, trichloroethylene.
CHEMOTAXIS TOWARD NAPHTHALENE
Naphthalene, listed as a priority pollutant by the U.S. Environmental Protection Agency (66), is commonly found in industrial effluents and is a constituent of coal tar (17). It is easily degraded by bacteria and is thus often used as a model compound for in situ biodegradation studies of polycyclic aromatic hydrocarbons (13). The high solubility of naphthalene compared to other polycyclic aromatic hydrocarbons and the fact that the naphthalene degradative genes are plasmid encoded has facilitated research on naphthalene degradation (71). As a result, a number of naphthalene-degrading microorganisms have been isolated and studied for mineralization (55, 71).
Grimm and Harwood (19) reported for the first time that naphthalene and its degradation pathway intermediate, salicylate, influence behavioral responses in two naphthalene-degrading motile bacteria, P. putida G7 and Pseudomonas sp. strain NCIB 9816-4. These responses are encoded by the catabolic plasmids NAH7 and DTG1 in strains G7 and NCIB 9816-4, respectively, and chemotaxis of these strains is induced during their growth on naphthalene. The naphthalene chemoreceptor, NahY, which is an MCP, has been characterized in P. putida G7 (20). NahY is encoded downstream of the naphthalene catabolic genes on the NAH7 plasmid and is cotranscribed with the degradation genes, as shown by reverse transcription-PCR analysis (20). The NahY chemoreceptor, along with degradation genes, was required to restore chemotaxis in P. putida G7:C1, an NAH7-cured derivative of strain G7 (20). Although there is some information about the role of catabolic genes and the associated receptor, the precise molecular mechanisms underlying the chemotactic response are yet to elucidated.
Another naphthalene-degrading plasmid, pRKJ1, has also been reported (54). The transfer of pRKJ1 into plasmid-free P. putida KT2442 resulted in the acquisition of chemotaxis and degradation properties (54), which were not present earlier. The recombinant plasmid pRKJ3 (containing 25-kb EcoRI fragment in vector pLAFR3) was transferred into plasmid-free strain of RKJ1, RKJ5, and was shown to be chemotactic toward naphthalene and salicylate. Neither strain KT2442 nor strain RKJ5 containing only the vector pLAFR3 showed chemotaxis. This established the role of the 25-kb EcoRI fragment in chemotaxis associated with complete degradation of these compounds. These results imply that chemotaxis toward naphthalene and/or salicylate might be due to a change in cellular energy levels that results from complete metabolism (metabolism-dependent chemotaxis) and/or because of intracellular receptors that recognize such contaminants or their degradation intermediates.
Marx and Aitken (39) evaluated the role of chemotaxis in naphthalene degradation by P. putida G7 in a heterogeneous aqueous system. Naphthalene degradation by the wild-type strain was compared to that of a nonmotile strain and a mutant strain deficient in naphthalene chemotaxis. These three strains degraded naphthalene at similar rates in a homogeneously mixed system. Studies conducted in an aqueous system with naphthalene concentration gradients have shown that the wild-type P. putida G7 is able to degrade naphthalene at a much faster rate compared to either of the two mutants, indicating that chemotaxis may enhance biodegradation. Different mathematical models have also been developed to quantify chemotaxis to naphthalene by this organism and its influence on naphthalene degradation (38, 45). It has been shown that the cell concentration for a nonchemotactic strain would have to be several orders of magnitude higher than for a chemotactic strain to achieve similar rates of naphthalene degradation (45).
CHEMOTAXIS TOWARD CHLOROAROMATIC COMPOUNDS
Toxic chlorinated compounds are prevalent in the biosphere due to ever-growing industrial activities (51). Some of these are used worldwide as herbicides, pesticides, and explosives (21, 51). Many chlorinated compounds are biodegradable by a wide range of bacterial strains, some of which have been isolated and characterized for potential bioremediation applications (50, 51). P. putida PRS2000 was also found to be attracted toward 3- and 4-chlorobenzoate, although these compounds are not metabolized by this organism (23). Evidence (discussed above) indicated that these two chemicals are recognized by the benzoate chemotaxis system.
2,4-Dichlorophenoxyacetate (2,4-D) is a widely used herbicide, and there are reports of bacterial strains that utilize 2,4-D (37). Hawkins and Harwood (25) recently reported the chemotaxis of R. eutropha JMP123(pJP4) toward this compound. Chemotaxis was induced by growth on 2,4-D and depended on the presence of the catabolic plasmid pJP4 that harbors tfd genes for 2,4-D degradation. The tfd cluster also encodes a nonessential permease TfdK for growth on 2,4-D. A tfdK mutant, which can grow at wild-type rates on 2,4-D, was found to be nonchemotactic, suggesting the decisive role of TfdK in 2,4-D chemotaxis. The chemoattraction of R. eutropha JMP123(pJP4) cells to 2,4-D and its concomitant degradation suggest that chemotaxis may be an essential feature in the biodegradation of 2,4-D by this organism.
CHEMOTAXIS TOWARD NITROAROMATIC COMPOUNDS
Nitroaromatic compounds (NACs) are used worldwide for the production of plastics, dyes, pesticides, insecticides, and explosives (59). Once released into the environment NACs undergo complex physical, chemical, and biological changes and sometimes are transformed into more harmful and toxic compounds. Due to the toxic, mutagenic, and carcinogenic nature of NACs and their incomplete degradation products, the U.S. Environmental Protection Agency has listed some of the NACs as priority pollutants (34). Some of these compounds are susceptible to microbial degradation (57, 59).
Chemotaxis of Ralstonia sp. strain SJ98 toward different NACs has been reported recently (8, 53). It was observed that strain SJ98 was chemotactic toward p-nitrophenol, 4-nitrocatechol, o-nitrobenzoate, p-nitrobenzoate, and 3-methyl-4-nitrophenol with subsequent degradation (8, 53). The ability of NACs to elicit a behavioral response of the strain SJ98 was also quantified (8, 53). In contrast to the compounds mentioned above, strain SJ98 utilized neither p-nitroaniline, 2,3-dinitrotoluene, naphthalene, phenanthrene, nor salicylic acid nor showed chemotaxis toward these compounds (53). These results therefore indicate a correlation between chemotaxis and biodegradation. Identification of the molecular mechanism operating behind chemotaxis in this strain is in progress in our laboratory.
POLLUTANTS AS CHEMOTACTIC REPELLENT
Negative chemotaxis, the movement of organisms away from chemicals, was discovered in bacteria, along with positive chemotaxis by Pfeffer (46). Thereafter, negative taxis of different bacterial species from acids, bases, salts, alcohols, hydrocarbons, heavy metals, oxygen, and phenol has been reported (68). Tso and Adler (65) demonstrated negative chemotaxis of E. coli against a wide range of chemicals such as fatty acids, amino acids, indoles and their analogues, aliphatic alcohols, aromatic compounds, acids, bases, sulfides, and inorganic ions. The molecular mechanisms operating behind such chemotactic responses has not been completely explored. They argue that the signal transduction pathway for positive and negative chemotaxis in E. coli is not totally separate; rather, they finally converge in a common pathway.
Phenol, an environmental pollutant, acts as a chemoattractant for E. coli, whereas it is a chemorepellent for serovar Typhimurium (31) and Vibrio albinolyticus (30). It is interesting that E. coli and serovar Typhimurium contain different chemorecepters that mediate both positive and negative chemotaxis in response to phenol. In E. coli the attractant response to phenol is mediated by Tar, and this response dominates the Trg- and Tap-mediated repellent response to phenol (69, 70). In S. enterica serovar Typhimurium, however, Tar-mediated attraction to phenol (31) is overcome by Tcp that is a specific chemoreceptor for taxis toward citrate and away from phenol (69). Tar, Trg, Tap, and Tcp are different MCPs, as discussed above. The need for the presence of receptors responsible for positive and negative chemotaxis for any chemoeffector molecule, along with their signal transduction mechanisms, remains to be determined.
The capacity of a compound to elicit a positive or negative chemotactic response is sometimes related to its nutritional properties. The same compound can act as an attractant to a microorganism that is able to utilize it as a growth substrate and also as a repellent to another microorganism for which it is toxic. This can be exemplified by the fact that E. coli and P. putida, which differ markedly in their nutritional properties, respond differently toward the presence of benzoate and salicylate in their microenvironment. These two pollutants are chemorepellents for E. coli (65). These membrane-permeable weak acids disturb the homeostasis of E. coli by lowering the intracellular pH, and this change in intracellular pH can impair numerous metabolic processes; as a result, the cells move away from these compounds as an advantageous behavioral response (52). Unlike the enteric bacterium E. coli, P. putida is able to utilize salicylate and benzoate amid a wide range of aromatic compounds as carbon and energy sources, thereby showing positive chemotaxis toward these compounds (24). A survival value of negative chemotaxis for E. coli that is a prey for neutrophils has also been argued in a separate study (6). Neutrophils actively search for bacteria by moving up gradients of N-formylated peptides that are released by the bacterial cells (56). Benov and Fridovich (6) demonstrated that E. coli flee by moving down gradients of the products of the neutrophil respiratory burst, i.e., H2O2, OCl−, and N-chlorotaurine. In the absence of any direct evidence, such a survival value of negative chemotaxis can be expected to influence biodegradation by (i) keeping biodegradation-competent cells away from high (toxic) concentrations and allowing access only to low concentrations of pollutants and (ii) keeping nondegrading cells away form the toxic pollutants and thereby allowing access only to degrading microorganisms so that competition for other nutrients is eliminated. Thus, negative chemotaxis may have potential in bioremediation applications.
CONCLUSION
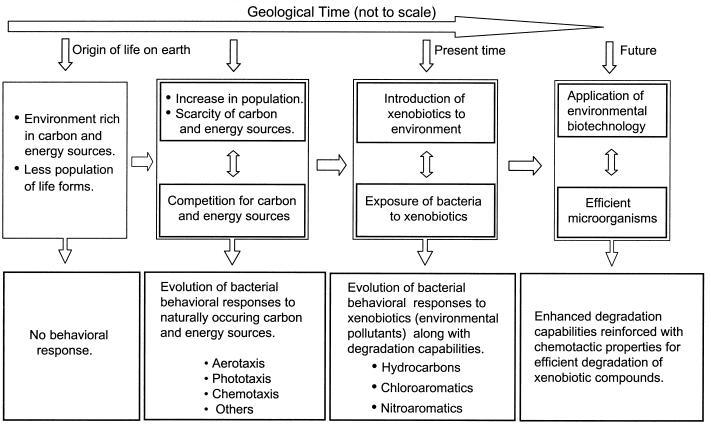
It is clear that chemotaxis is a selective advantage to the degradative bacteria for guiding them to sense and locate pollutants that are present in the environment (Fig. 1). Coordinated regulation of bacterial chemotaxis toward almost all toxic compounds and their respective mineralization and/or transformation indicate that this phenomenon might be an integral feature of degradation (Fig. 1). Studies related to chemotaxis with respect to degradation of environmental pollutants such as naphthalene, toluene, and NACs suggest that chemotaxis is related to the metabolism of the chemoattractant, although the phenomenon may not abide strictly with either of the two types of chemotactic responses discussed above, i.e., metabolism-independent chemotaxis and metabolism-dependent chemotaxis. Since the chemotaxis of bacteria helps them to approach and degrade toxic compounds in the environment, the molecular basis of this phenomenon is a fertile and useful area for future research.
FIG. 1.
Bacterial chemotaxis might have evolved as a selective advantage to bacteria for searching for the chemicals that could act as a source of carbon and energy to the cells. Knowledge of the bacterial behavior toward naturally occurring chemicals and molecular mechanisms underlying bacterial chemotaxis toward xenobiotics can be exploited for designing efficient systems for bioremediation of contaminated sites.
Acknowledgments
We are grateful to Amit Ghosh, Director, for encouragement. We are also thankful to Debarati Paul for reading the manuscript.
Support for this work was, in part, provided by the Indo-Swiss Collaboration in Biotechnology.
Footnotes
This is IMTECH communication no. 019/2002.
REFERENCES
- 1.Adler, J. 1969. Chemoreceptors in bacteria. Science 166:1588-1597. [DOI] [PubMed] [Google Scholar]
- 2.Alexander, G., and I. B. Zhulin. 2001. More than one way to sense chemicals. J. Bacteriol. 183:4681-4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexander, G., S. E. Greer, and I. B. Zhulin. 2000. Energy taxis is a dominant behavior in Azospirillum brasilense. J. Bacteriol. 182:6042-6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armitage, J. P. 1999. Bacterial tactic responses. Adv. Microb. Physiol. 41:229-289. [DOI] [PubMed] [Google Scholar]
- 5.Armitage, J. P., and R. Schmitt. 1997. Bacterial chemotaxis: Rhodobacter sphaeroides and Sinorhizobium meliloti: variations on a theme? Microbiology 143:3671-3682. [DOI] [PubMed] [Google Scholar]
- 6.Benov, L., and I. Fridovich. 1996. Escherichia coli exhibits negative chemotaxis in gradients of hydrogen peroxide, hypochlorite, and N-chlorotaurine: products of the respiratory bursts of phagocytic cells. Proc. Natl. Acad. Sci. USA 93:4999-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berg, H. C. 1975. Chemotaxis in bacteria. Annu. Rev. Biophys. Bioeng. 4:119-136. [DOI] [PubMed] [Google Scholar]
- 8.Bhushan, B., S. K. Samanta, A. Chauhan, A. K. Chakraborti, and R. K. Jain. 2000. Chemotaxis and degradation of 3-methyl-4-nitrophenol by Ralstonia sp. SJ98. Biochem. Biophys. Res. Commun. 275:129-133. [DOI] [PubMed] [Google Scholar]
- 9.Bibikov, S. I., R. Biran, K. E. Rudd, and J. S. Parkinson. 1997. A signal transducer for aerotaxis in Escherichia coli. J. Bacteriol. 179:4075-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bren, A., and M. Eisenbach. 2000. How signals are heard during bacterial chemotaxis: protein-protein interactions in sensory signal propagation. J. Bacteriol. 182:6865-6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bünning, E. 1989. Ahead of his time: Wilhelm Pfeffer, p. 49-53. In Early advances in plant biology. Carlton University Press, Ottawa, Ontario, Canada.
- 12.Clancy, M., K. A. Madill, and J. M. Wood. 1981. Genetic and biochemical requirements for chemotaxis to l-proline in Escherichia coli. J. Bacteriol. 146:902-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dables, S. 1986. Biochemistry of aromatic hydrocarbon degradation in pseudomonads, p. 527-556. In J. S. Sokatch (ed.), The bacteria, vol. 10. Academic Press, Inc., Orlando, Fla.
- 14.DeRosier, D. J. 1998. The turn of the screw: the bacterial flagellar motor. Cell 93:17-20. [DOI] [PubMed] [Google Scholar]
- 15.Dharmatilake, A. J., and W. D. Bauer. 1992. Chemotaxis of Rhizobium meliloti towards nodulation gene-inducible compounds from alfalfa roots. Appl. Environ. Microbiol. 58:1153-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falke, J. J., and G. L. Hazelbauer. 2001. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem. Sci. 26:257-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finalayson-Pitts, B. J., and J. N. Pitts, Jr. 1997. Tropospheric air pollution: ozone, airborne toxics, polycyclic aromatic hydrocarbons and particles. Science 276:1045-1052. [DOI] [PubMed] [Google Scholar]
- 18.Ghisalba, O. 1993. Microbial degradation of chemical waste, an alternate to physical method of waste disposal. Experientia 39:1247-1257. [Google Scholar]
- 19.Grimm, A. C., and C. S. Harwood. 1997. Chemotaxis of Pseudomonas sp. to the polycyclic aromatic hydrocarbon, naphthalene. Appl. Environ. Microbiol. 63:4111-4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grimm, A. C., and C. S. Harwood. 1999. NahY, a catabolic plasmid-encoded receptor is required for chemotaxis of Pseudomonas putida to the aromatic hydrocarbon naphthalene. J. Bacteriol. 181:3310-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harayama, S., M. Kok, and E. L. Neidle. 1992. Functional and evolutionary relationships among diverse oxygenases. Annu. Rev. Microbiol. 46:565-602. [DOI] [PubMed] [Google Scholar]
- 22.Harwood, C. S., N. N. Nichols, M.-K. Kim, J. L. Ditty, and R. E. Parales. 1994. Identification of a pcaRKF gene cluster from Pseudomonas putida: involvement in chemotaxis, biodegradation, and transport of 4-hydroxybenzoate. J. Bacteriol. 176:6479-6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harwood, C. S., R. E. Parales, and M. Dispensa. 1990. Chemotaxis of Pseudomonas putida toward chlorinated benzoates. Appl. Environ. Microbiol. 42:263-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harwood, C. S., M. Rivelli, and L. N. Ornston. 1984. Aromatic acids are chemoattractants for Pseudomonas putida. J. Bacteriol. 160:622-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hawkins, A. C., and C. S. Harwood. 2002. Chemotaxis of Ralstonia eutropha JMP123(pJP4) to the herbicide 2,4-dichlorophenoxyacetate. Appl. Environ. Microbiol. 68:968-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Head, I. M. 1998. Bioremediation: towards a credible technology. Microbiology 144:599-608. [DOI] [PubMed] [Google Scholar]
- 27.Hebes, S. E., and I. R. Schwall. 1987. Microbial degradation of polycyclic aromatic hydrocarbons in pristine and petroleum contaminated sediments. Appl. Environ. Microbiol. 35:306-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hendrixson, D. R., B. J. Akerley, and V. J. DiRita. 2001. Transposon mutagenesis of Campylobacter jejuni identifies a bipartite energy taxis system required for motility. Mol. Microbiol. 40:214-224. [DOI] [PubMed] [Google Scholar]
- 29.Hoch, J. A. 2000. Two-component and phosphorelay signal transduction. Curr. Opin. Microbiol. 3:165-170. [DOI] [PubMed] [Google Scholar]
- 30.Homma, M., H. Oota, S. Kojima, I. Kawagishi, and Y. Imae. 1996. Chemotactic response to an attractant and a repellent by the polar and lateral flagellar systems of Vibrio alginolyticus. Microbiology 142:2777-2783. [DOI] [PubMed] [Google Scholar]
- 31.Imae, Y., K. Oosawa, T. Mizuno, M. Kihara, and R. M. Macnab. 1987. Phenol: a complex chemoeffector in bacterial chemotaxis. J. Bacteriol. 169:371-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeziore-Sassoon, Y., P. A. Hamblin, C. A. Bootle-Wilbraham, P. S. Poole, and J. P. Armitage. 1998. Metabolism is required for chemotaxis to sugars in Rhodobacter sphaeroides. Microbiology 144:229-239. [DOI] [PubMed] [Google Scholar]
- 33.Kato, J., T. Nakamura, A. Kurodo, and H. Ohtake. 1999. Cloning and characterization of chemotaxis genes in Pseudomonas aeruginosa. Biosci. Biotechnol. Biochem. 63:155-161. [DOI] [PubMed] [Google Scholar]
- 34.Keith, L. H., and W. A. Telliard. 1976. Priority pollutants. I. A prospective view. Environ. Sci. Technol. 13:416-423. [Google Scholar]
- 35.Kirk, T. K., and R. L. Farrel. 1987. Enzymatic “combustion”: the microbial degradation of lignin. Annu. Rev. Microbiol. 41:465-505. [DOI] [PubMed] [Google Scholar]
- 36.Lopez-de-Victoria, G., and C. R. Lowell. 1993. Chemotaxis of Azospirillum species to aromatic compounds. Appl. Environ. Microbiol. 59:2951-2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.MacGowan, C., R. Fulthorpe, A. Wright, and J. M. Tiedje. 1998. Evidence for interspecies gene transfer in the evolution of 2,4-dichlorophenoxyacetic acid degraders. Appl. Environ. Microbiol. 64:4089-4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marx, R. B., and M. D. Aitken. 1999. Quantification of chemotaxis to naphthalene by Pseudomaonas putida G7. Appl. Enviorn. Microbiol. 65:2847-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marx, R. B., and M. D. Aitken. 2000. Bacterial chemotaxis enhances naphthalene degradation in a heterogeneous aqueous system. Environ. Sci. Technol. 34:3379-3383. [Google Scholar]
- 40.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 41.Parales, R. E., and C. S. Harwood. 2002. Bacterial chemotaxis to pollutants and plant-derived aromatic molecules. Curr. Opin. Microbiol. 5:266-273. [DOI] [PubMed] [Google Scholar]
- 42.Parales, R. E., J. L. Ditty, and C. S. Harwood. 2000. Toluene-degrading bacteria are chemotactic towards the environmental pollutants benzene, toluene, and trichloroethylene. Appl. Environ. Microbiol. 66:4098-4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parke, D., M. L. Rivelli, and L. N. Ornston. 1985. Chemotaxis to aromatic and hydroaromatic acids: comparison of Bradyrhizobium japonicum and Rhizobium trifolii. J. Bacteriol. 163:417-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parkinson, J. S. 1993. Signal transduction schemes of bacteria. Cell 73:857-871. [DOI] [PubMed] [Google Scholar]
- 45.Pedit, J. A., R. B. Marx, C. T. Miller, and M. D. Aitken. 2002. Quantitative analysis of experiments on bacterial chemotaxis to naphthalene. Biotechnol. Bioeng. 78:626-634. [DOI] [PubMed] [Google Scholar]
- 46.Pfeffer, W. 1883. Locomotorische richtungsbewegungen durch chemische rieze. Ber. Deutsch. Bot. Ges. 1:524-533. [Google Scholar]
- 47.Poole, P. S., and J. P. Armitage. 1989. Role of metabolism in the chemotactic response of Rhodobacter sphaeroides to ammonia. J. Bacteriol. 171:2900-2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 49.Prigent-Combaret, C., O. Vidal, C. Doral, and P. Lejeune. 1999. Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J. Bacteriol. 181:5993-6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reinke, W. 1984. Microbial degradation of halogenated aromatic compounds, p. 319-360. In D. T. Gibson (ed.), Microbial degradation of aromatic compounds. Marcel Dekker, Inc., New York, N.Y.
- 51.Reinke, W., and H.-J. Knackmuss. 1988. Microbial degradation of haloaromatics. Annu. Rev. Microbiol. 42:263-288. [DOI] [PubMed] [Google Scholar]
- 52.Repaske, D. R., and J. Adler. 1981. Changes in intracellular pH of Escherichia coli mediates the chemotactic response to certain attractants and repellents. J. Bacteriol. 145:1196-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Samanta, S. K., B. Bhushan, A. Chauhan, and R. K. Jain. 2000. Chemotaxis of a Ralstonia sp. SJ98 toward different nitroaromatic compounds and their degradation. Biochem. Biophy. Res. Commun. 269:117-123. [DOI] [PubMed] [Google Scholar]
- 54.Samanta, S. K., and R. K. Jain. 2000. Evidence for plasmid-mediated chemotaxis of Pseudomonas putida towards naphthalene and salicylate. Can. J. Microbiol. 28:683-691. [DOI] [PubMed] [Google Scholar]
- 55.Samanta, S. K., O. V. Singh, and R. K. Jain. 2002. Polycyclic aromatic hydrocarbons: environmental pollution and bioremediation. Trends Biotechnol. 20:243-248. [DOI] [PubMed] [Google Scholar]
- 56.Schiffmann, E., B. A. Corcoran, and S. M. Wahl. 1975. N-Formylmethionine peptides as chemoattractants for leucocytes. Proc. Natl. Acad. Sci. USA 72:1059-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shirley, F. N., Spain, J. C., and He. Zhongqi. 2000. Strategies for aerobic degradation of nitroaromatic compounds by bacteria: process discovery to field applications, p. 7-62. In J. C. Spain, J. B. Hughes, and H.-J. Knackmuss (ed.), Biodegradation of nitroaromatic compounds and explosives. Lewis Publishers, Washington, D.C.
- 58.Shoi, J., C. V. Dang, and B. L. Taylor. 1998. Oxygen as attractant and repellent in bacterial chemotaxis. J. Bacteriol. 169:3118-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Spain, J. C. 1995. Biodegradation of nitroaromatic compounds. Annu. Rev. Microbiol. 49:523-555. [DOI] [PubMed] [Google Scholar]
- 60.Stelmack, P. L., M. R. Gray, and M. A. Pickard. 1999. Bacterial adhesion to soil contaminants in the presence of surfactants. Appl. Environ. Microbiol. 65:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taylor, B. L. 1983. How do bacteria find the optimum concentration of oxygen? Trends Biochem. Sci. 8:438-441. [Google Scholar]
- 62.Taylor, B. L. 1993. Aerotaxis and other energy sensing behavior in bacteria. Annu. Rev. Microbiol. 53:103-128. [DOI] [PubMed] [Google Scholar]
- 63.Taylor, B. L., and I. B. Zhulin. 1998. In search of higher energy: metabolism-dependent behavior in bacteria. Mol. Microbiol. 28:683-690. [DOI] [PubMed] [Google Scholar]
- 64.Terracciano, J. S., and E. Canale-Parola. 1984. Enhancement of chemotaxis in Spirochaeta aurantia grown under conditions of nutrient limitation. J. Bacteriol. 159:173-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tso, W.-W., and J. Adler. 1974. Negative chemotaxis in Escherichia coli. J. Bacteriol. 118:560-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.U.S. Environmental Protection Agency. 1987. Summary review of health effects associated with naphthalene. Document EPA/600/8-87/055/7. Office of Health and Environmental Assessment, U.S. Environmental Protection Agency, Washington, D.C.
- 67.Watnick, P. I., and R. Kolter. 1999. Steps in the development of a Vibrio cholerae EI Tor biofilm. Mol. Microbiol. 34:586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weibull, C. 1960. Movement, p. 153-205. In I. C. Gunsalus and R. Y. Stainer (ed.), The bacteria, vol. 1. Academic Press, Inc., New York, N.Y.
- 69.Yamamoto, K., and Y. Imae. 1993. Cloning and characterization of the Salmonella typhimurium-specific chemoreceptor Tcp for chemotaxis to citrate and from phenol. Proc. Natl. Acad. Sci. USA 90:217-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yamamoto, K., R. M. Macnab, and Y. Imae. 1990. Repellent response functions of the Trg and Tap chemoreceptors of Escherichia coli. J. Bacteriol. 172:383-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yen, K.-M., and C. M. Serdar. 1998. Genetics of naphthalene catabolism by pseudomonads. Crit. Rev. Microbiol. 15:247-268. [DOI] [PubMed] [Google Scholar]
- 72.Zhulin, I. B., E. H. Rowsell, M. S. Johnson, and B. L. Taylor. 1997. Glycerol elicits energy taxis of Escherichia coli and Salmonella typhimurium. J. Bacteriol. 179:3196-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]