Abstract
The gram-positive bacterium Mycobacterium sp. strain ESD is able to use the cyclodiene insecticide endosulfan as a source of sulfur for growth. This activity is dependent on the absence of sulfite or sulfate in the growth medium. A cosmid library of strain ESD DNA was constructed in a Mycobacterium-Escherichia coli shuttle vector and screened for endosulfan-degrading activity in Mycobacterium smegmatis, a species that does not degrade endosulfan. Using this method, we identified a single cosmid that conferred sulfur-dependent endosulfan-degrading activity on the host strain. An open reading frame (esd) was identified within this cosmid that, when expressed behind a constitutive promoter in a mycobacterial expression vector, conferred sulfite- and sulfate-independent β-endosulfan degradation activity on the recombinant strain. The translation product of this gene (Esd) had up to 50% sequence identity with an unusual family of monooxygenase enzymes that use reduced flavins, provided by a separate flavin reductase enzyme, as cosubstrates. An additional partial open reading frame was located upstream of the Esd gene that had sequence homology to the same monooxygenase family. A flavin reductase gene, identified in the M. smegmatis genome, was cloned, expressed, and used to provide reduced flavin mononucleotide for Esd in enzyme assays. Thin-layer chromatography and gas chromatography analyses of the enzyme assay mixtures revealed the disappearance of β-endosulfan and the appearance of the endosulfan metabolites, endosulfan monoaldehyde and endosulfan hydroxyether. This suggests that Esd catalyzes the oxygenation of β-endosulfan to endosulfan monoaldehyde and endosulfan hydroxyether. Esd did not degrade either α-endosulfan or the metabolite of endosulfan, endosulfan sulfate.
Organochlorine pesticides generally are mineralized very slowly or not at all in the environment. This recalcitrance has led to the banning of the use of most of these compounds throughout the world. An exception to this is the insecticide endosulfan, which has the same primary action and target site as other organochlorines (3). However, it has a relatively reactive cyclic sulfite diester group (27) and, as a consequence, has chemical and physical properties significantly different from those of other organochlorine insecticides that affect both its environmental fate and its biological fate. In particular, the environmental persistence of endosulfan is lower than that of other chemicals of the same class, although it is higher than that of many other insecticides. Since deregistration of most organochlorine insecticides in many countries, the continued availability of endosulfan has been important because of its use in integrated pest management and resistance management strategies. However, wastewater residue problems associated with the use of endosulfan have led to interest in postapplication detoxification of this insecticide.
Endosulfan can be detoxified in a single enzymatic reaction, making it an ideal candidate for bioremediation (24). Detoxification is thought to occur by removal of the sulfur moiety, and the resultant hexachloro product poses little environmental threat (6, 10). Two pathways for single-step detoxification of endosulfan by cultured soil microbial populations have been described. In the first pathway endosulfan is hydrolyzed to endosulfan diol. Because endosulfan is particularly susceptible to abiotic hydrolysis and forms endosulfan diol at pH values of 7.0 and above (18), it is difficult to estimate the contribution of enzymatic hydrolysis rather than chemical hydrolysis to this pathway of degradation. This is especially problematic because often the growth of microbes increases the alkalinity of the medium in which they are grown (2, 18, 20). This uncertainty needs to be resolved before isolation of the enzymes responsible for this reaction is attempted. The second pathway of metabolism involves the formation of endosulfan monoaldehyde (24) and has been found in the soil bacterium Mycobacterium sp. strain ESD and described (25). The formation of endosulfan monoaldehyde occurs only enzymatically, and therefore this pathway has been the focus of endosulfan bioremediation studies in our laboratory.
The commercial endosulfan insecticide is a mixture of two diastereoisomers, α-endosulfan and β-endosulfan, at a ratio of 7:3. Mycobacterium sp. strain ESD metabolizes the isomers to endosulfan sulfate and endosulfan monoaldehyde (24). However, the α isomer is predominantly oxidized to endosulfan sulfate, and the β isomer is predominantly converted to endosulfan monoaldehyde (24). Mycobacterium sp. strain ESD was isolated by providing endosulfan as the sole source of sulfur in successive enrichment cultures initially inoculated with endosulfan-contaminated soil. The formation of both endosulfan sulfate and endosulfan monoaldehyde by Mycobacterium sp. strain ESD occurs as a response to sulfur starvation and does not occur in the presence of relatively low levels of inorganic sulfate or sulfite. In this report we describe the cloning, molecular characterization, and expression of a monooxygenase capable of degrading β-endosulfan from Mycobacterium sp. strain ESD. Mycobacterium-Escherichia coli shuttle vectors allowed isolation of the enzyme by selection of an active enzyme in a related Mycobacterium species, Mycobacterium smegmatis, which is normally unable to metabolize endosulfan. Reduced flavin was provided to this enzyme in cell-free enzyme assay mixtures by expression of a flavin reductase gene isolated from the M. smegmatis genome. The enzyme appears to catalyze successive oxygenation of β-endosulfan to endosulfan monoaldehyde and then endosulfan hydroxyether.
MATERIALS AND METHODS
Media and reagents.
Sulfur-free medium (SFM) (pH 6.9) was prepared by the method of Sutherland et al. (24). Endosulfan or endosulfan sulfate (Sigma-Aldrich, Sydney, New South Wales, Australia) was added to bacterial cultures at a concentration of 50 μM when it was required. Modified Luria broth (LB) contained 10 g of tryptone, 5 g of yeast extract, and 0.5 g of NaCl in 1 liter of 100 mM potassium phosphate buffer (pH 6.9). LB (23) and LB containing 0.05% Tween 80 (LBT) were also used. Ampicillin (100 μg · ml−1), hygromycin (100 μg · ml−1), and kanamycin (25 μg · ml−1) were included when they were required. Flavin mononucleotide (FMN), flavin adenine dinucleotide (FAD), tetrahydrobiopterin, and NAD(P)H were obtained from Sigma Diagnostics (St. Louis, Mo.).
Bacterial strains and DNA vectors.
All the bacterial strains and DNA vectors used in this study are described in Table 1. Mycobacterium sp. strain ESD was maintained in SFM containing either endosulfan or 200 μM sulfate as a sulfur source at 28°C with shaking at 180 rpm. M. smegmatis was maintained in LBT at 37°C or in SFM containing 50 μM endosulfan at 28°C with shaking at 200 rpm. E. coli strains were maintained in LB at 37°C with shaking at 200 rpm.
TABLE 1.
Bacterial strains and DNA vectors used in this study
| Strain or DNA vector | Description or genotypea | Reference or source |
|---|---|---|
| Mycobacterium sp. strain ESD | Endosulfan degrading | 25 |
| M. smegmatis mc2 | Not endosulfan degrading | H. Billman-Jacobe |
| E. coli strain TG1 | F′ traD36 lacIq Δ(lacZ)M15 proA+B+/supE Δ(hsdM-mcrB)5 (rk−mk− McrB−) thi Δ(lac-proAB), strain capable of growth in minimal media | New England Biolabs |
| E. coli strain EP1305 | F− Δ(mcrC-mrr) mcrA recA13 supE44 ara-14 galK2 lacY1 rpsL20 (Strr) xyl-5 mtl-1, strain used to amplify cosmid library | Epicenter Technologies |
| pGEM | Apr cloning vector | Promega |
| pYUB415 | Mycobacterium-E. coli shuttle cosmid vector, Apr Hyr, derivative of pYUB18 | William R. Jacobs, Jr., Albert Einstein College of Medicine, New York, N.Y |
| pMV216 | Mycobacterium-E. coli shuttle expression vector, Knr | |
| pET14b | E. coli expression vector, Apr | Novagen |
| pYUB-172 | Approximately 50-kb Mycobacterium sp. strain ESD DNA fragment in pYUB415 that confers endosulfan-degrading activity on M. smegmatis | This study |
| pYUB415-Apa3 | 3-kb ApaI fragment in pYUB415 that confers endosulfan- | This study |
| degrading activity on M. smegmatis from pYUB-172 | ||
| pMV216-esd | Esd mycobacterial expression construct in pMV216 | This study |
| pET14b-esd | Esd expression construct in pET14b | This study |
| pET14b-MsFR | MsFR expression construct in pET14b | This study |
Apr, contains ampicillin resistance gene; Hyr, contains hygromycin resistance gene; Knr, contains kanamycin resistance gene.
Isolation of genomic DNA.
Genomic DNA was extracted from Mycobacterium sp. strain ESD by using a method adapted from the method of Anderberg et al. (1). Cells were grown until the culture was turbid in LBT and then pelleted and resuspended in 1 ml of Tris-EDTA containing 200 μg of proteinase K per ml and 10 mg of lysozyme per ml. After 1 h of incubation at 37°C, the cells were pelleted and resuspended in 750 μl of a solution containing 4 M guanidine thiocyanate, 25 mM sodium citrate, and 0.5% Sarkosyl. Glass beads (diameter, 450 to 600 μm; Sigma) were added, and the cells and beads were mixed vigorously with a vortex mixer for 10 min. After the beads were settled by a brief centrifugal pulse, the supernatant was removed and extracted with phenol-chloroform several times. The DNA was precipitated with ethanol, resuspended in Tris-EDTA, and stored at −20°C.
Construction of cosmid library.
A Mycobacterium sp. strain ESD genomic library was constructed in the Mycobacterium-E. coli cosmid shuttle vector pYUB415 as follows. Genomic DNA of Mycobacterium sp. strain ESD was partially digested with Sau3AI restriction endonuclease and then separated by gel electrophoresis in a 1% low-melting-point agarose gel (23). An agarose gel slice containing 30- to 45-kb DNA fragments was melted, and the DNA was ligated in the gel to BamHI-digested, calf intestinal alkaline phosphatase-treated pYUB415 (23). After ligation, the GELase agarose gel-digesting enzyme preparation (Epicenter Technologies, Madison, Wis.) was added according to the manufacturer's instructions. The ligated DNA was packaged into MaxPlax packaging extracts (Epicenter Technologies) and used to infect E. coli strain EP1305 cells freshly prepared according to the manufacturer's instructions (Epicenter Technologies). Strains containing pYUB415 derivatives were grown in the presence of ampicillin (E. coli) or hygromycin (M. smegmatis).
Screening of cosmid clones and subsequent subclones.
Cosmid DNA was isolated from 2-ml cultures of individual library clones by using the alkaline lysis procedure of Sambrook et al. (23), purified by using the QIAquick system (Qiagen, Victoria, Australia), and then electroporated into freshly prepared M. smegmatis mc2 competent cells (prepared as described by Jacobs et al. [13]). The M. smegmatis used to screen the Mycobacterium sp. strain ESD cosmid library does not have detectable endosulfan-metabolizing activity (25) and can grow in SFM supplemented with either 50 μM magnesium sulfate or sodium sulfite. DNA (5 to 500 pg of in 1 μl of distilled H2O) was incubated with 50 μl of prepared cells for 1 min on ice and then transferred to a cuvette with a 0.2-cm gap (Bio-Rad, Sydney, New South Wales, Australia). This preparation was exposed to one electrical pulse consisting of 2,500 V and 25 μF with the resistance set at 1,000 Ω. Electroporated cells were incubated for 3 h in LBT and then plated onto LB agar containing hygromycin and incubated at 28°C for 4 days. Colonies were washed with SFM and used to inoculate SFM containing hygromycin and β-endosulfan. After 7 days of incubation at 28°C and 180 rpm, the culture was analyzed for endosulfan degradation by thin-layer chromatography (TLC) (24).
Construction of esd expression constructs.
To create a mycobacterial esd expression construct, the esd gene was amplified by PCR from the ApaI DNA fragment from Mycobacterium sp. strain ESD containing the Esd gene by using pYUB415-Apa3 as the template and CCTGCAGTGACCCGACAGCTACACCTC forward and CAAGCTTATTACGCGACCGCGTGCGCCA reverse oligonucleotide primers (PstI and HindIII sites, respectively, are underlined). PCR was performed by using Taq polymerase (Gibco BRL, Rockville, Md.) in the buffer provided by the manufacturer supplemented with 1.5 mM MgCl2. After incubation for 5 min at 95°C, 30 cycles of amplification (94°C for 15 s, 52.5°C for 30 s, and 72°C for 70 s) were performed, followed by a 5-min extension step at 72°C. The PCR product was cloned into the pGEM T Easy vector (Promega, Madison, Wis.) by following the manufacturer's instructions to create pGEM-esd. This plasmid was then digested with PstI and HindIII and cloned into similarly digested pMV261 (Table 1) to produce pMV261-esd.
To create an E. coli expression construct, the esd gene was amplified by PCR by using pYUB415-Apa3 as the template and CCATATGACCCGACAGCTACACCTC forward and CAGATCTATTACGCGACCGCGTGCGCCA reverse primers (NdeI and BglII sites, respectively, are underlined, and the initiation codon is indicated by boldface type). PCR was performed by using Taq polymerase (Life Technologies, Rockville, Md.) and buffer provided by the manufacturer supplemented with 1.5 mM MgCl2. After incubation for 5 min at 95°C, 30 cycles of amplification (95°C for 30 s, 65°C for 30 s, and 72°C for 30 s) were performed, followed by a 5-min extension step at 72°C. The PCR product was cloned into the pGEM T Easy vector (Promega) by following the manufacturer's instructions to create pGEM-esd2. This plasmid was digested with NdeI and BglII and cloned into NdeI-BamHI-digested pET14b (Novagen, Madison, Wis.), which contains an N-terminal His tag to aid recombinant protein purification, to produce pET14b-esd.
Construction of a flavin reductase expression plasmid.
The flavin reductase gene sequence of M. smegmatis was identified by using the BLAST program to identify sequences in the unannotated M. smegmatis genome sequence (obtained from The Institute for Genomic Research website [http://www.tigr.org]) homologous to the dszD gene sequence of Rhodococcus erythropolis strain D-1 (accession number AB051429 [19]). This sequence extends from position 137818 to position 138306 of the M. smegmatis gnl|TIGR-1772|msmeg-3271 genome sequence fragment (http://www.ncbi.nlm.nih.gov:80/cgi-bin/Entrez/framik?db=Genome&gi = 5073). An open reading frame (ORF) (MsFR) with 67% identity to the R. erythropolis dszD gene was amplified by PCR by using M. smegmatis total DNA as the template and the upstream GCCATATGGTCACCGCTGAGCAGTATCGCGCGGCG and downstream GCGGATCCTCAGGTGAGCGGACGGGTGCCCAGATA primers (NdeI and BamHI sites, respectively, are underlined, and the initiation codon is indicated by boldface type). PCR was performed by using Taq polymerase (Life Technologies) and buffer provided by the manufacturer supplemented with 1.5 mM MgCl2. After incubation for 5 min at 95°C, 30 cycles of amplification (95°C for 30 s, 65°C for 30 s, and 72°C for 30 s) were performed, followed by a 5-min extension step at 72°C. The PCR product was cloned into the pGEM T Easy vector (Promega) by following the manufacturer's instructions. The resultant construct was digested with NdeI and BamHI and cloned into pET14b (Novagen) similarly digested to produce pET14b-MsFR.
Expression and purification of recombinant proteins.
The flavin reductase and Esd proteins were expressed in E. coli in similar ways. Plasmid pET14b-MsFR or pET14b-esd was electroporated into E. coli BL21(DE3) cells and grown overnight on LB agar containing ampicillin. A single colony was then used to inoculate LB containing ampicillin and grown at 37°C until an optical density at 600 nm of 0.6 to 0.8 was reached. This culture was then diluted 50-fold in fresh LB containing ampicillin and incubated at 25°C overnight. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a concentration of 0.4 mM, and cells were grown for an additional 2 h. Cells were washed in 50 mM HEPES buffer (pH 6.9), resuspended in the same buffer to an optical density at 600 nm of 20, and disrupted by sonication. The sonicated cells were centrifuged at 5,000 × g for 20 min to remove cell debris, and then the membranes were removed by centrifugation at 100,000 × g in an ultracentrifuge. The soluble protein was bound and eluted from a His-bind column (Novagen) according to the manufacturer's instructions. After elution from the column, proteins were dialyzed against three changes of 50 mM HEPES buffer (pH 6.9), and the purified proteins were stored at −20°C. Protein expression and purification were confirmed by the presence of an approximately 21-kDa protein for MsFR and a 50-kDa protein for Esd by performing sodium dodecyl sulfate-polyacrylamide gel electrophoresis of the supernatant fraction and by analyzing enzyme activity (see below).
Enzyme assays.
Enzyme assays were performed in large glass test tubes (diameter, 2 cm) with shaking at 180 rpm and incubation at 28°C. The assay mixtures typically contained 20 μg of Esd, 12 μg of MsFR, 50 μM FMN, 4 mM NADH, and 500 μM β-endosulfan in 1 ml of 50 mM HEPES buffer (pH 6.9). Also included was either 7.5 mg of bovine serum albumin (BSA), 0.08% Triton X-100, or M. smegmatis cell extract containing 7.5 mg of protein (see below) and 10 mM MgCl2 to increase the apparent solubility of the insecticide. The presence of endosulfan-degrading activity was confirmed by the appearance of endosulfan monoaldehyde and endosulfan hydroxyether in a reaction mixture, as determined by the TLC method described by Sutherland et al. (24), after 4 h of incubation. A time course of the appearance of the metabolites determined by TLC revealed that endosulfan monoaldehyde appeared before endosulfan hydroxyether in the reaction mixture (data not shown). The activity was quantified by measuring the disappearance of endosulfan by the gas chromatography (GC) method described below after 5 min to 3 h of incubation.
The flavin reductase activity of MsFR was determined by a spectophotometric assay which measured the decrease in absorbance at 340 nm due to the disappearance of NAD(P)H. The assay mixture contained 7 μg of MsFR and electron acceptors in 1 ml (final volume) of 50 mM HEPES buffer. Electron acceptors were included at the following concentrations: 10 to 100 μM FMN; 10 to 100 μM riboflavin; 10 to 100 μM FAD; 50 μM methylene blue; and 50 μM tetrahydrobiopterin. The reaction was initiated by adding 0.5 mg of an electron donor (NADH or NADPH) and was performed for up to 5 min at 25°C. Enzyme activity was determined on the basis of the amount of NAD(P)H oxidized per minute at 25°C (E340 = 6,220 M−1 · cm−1).
Nonrecombinant M. smegmatis cell extracts were prepared by growing 400-ml cultures to the stationary phase, harvesting the cells by centrifugation, and then washing the cells in 50 ml of 50 mM HEPES buffer (pH 6.9) and resuspending the cells in the same buffer. Cells were lysed by sonication, and the cell debris was removed by centrifugation at 16,000 × g for 5 min. The supernatants were stored at −20°C until they were used.
GC analysis.
Endosulfan sulfate (50 μg) was added to enzyme assay mixtures at the conclusion of the incubation time as an internal standard, and then each reaction mixture was extracted with 2 ml of ethyl acetate. The organic phase was passed through a 6-cm MgSO4 column in a Pasteur pipette stoppered with glass wool to remove any residual water, gently evaporated under a dry nitrogen stream, and then dissolved in 200 μl of dichloromethane prior to GC analysis.
GC was performed with a Varian model 3400 GC with a cool on-column injector, a Varian model 8200 autosampler, a flame ionization detector, and a computer with data acquisition and processing software. The capillary column (length, 30 m; inside diameter, 0.32 mm; film thickness, 0.25 μm) contained 5% phenyl methylsilicone (SE54; Alltech Econocap) preceded by a retention gap of deactivated silica (2 m) and was run with a helium flow rate of 2 ml min−1. The temperature program for analysis of β-endosulfan hydrolysis consisted of an initial period after injection of 2 min at 40°C; a temperature gradient that increased at a rate of 20°C min−1 to 200°C, followed by a 10-min holding period at 200°C; and then a temperature gradient that increased at a rate of 10°C min−1 to 300°C, followed by a 10-min holding period at 300°C.
Nucleotide sequence accession number.
The nucleotide sequence of esd and surrounding regions has been deposited in the GenBank database under accession number AF537302.
RESULTS
Isolation of a gene encoding an endosulfan-degrading protein.
A gene encoding a protein capable of degrading β-endosulfan was isolated after we screened 370 clones from the Mycobacterium sp. strain ESD cosmid library in M. smegmatis mc2, a strain that does not metabolize the insecticide. A single cosmid (pYUB-172) was identified that conferred β-endosulfan-degrading activity on M. smegmatis. When M. smegmatis containing pYUB-172 was grown in SFM with β-endosulfan as the sole source of sulfur, all the insecticide disappeared and the metabolites endosulfan monoaldehyde and endosulfan hydroxyether were produced. When α-endosulfan was provided as the sole source of sulfur, no degradation was apparent and no metabolites were observed. In Mycobacterium sp. strain ESD the endosulfan-degrading activity was absent when the bacterium was grown in the presence of a low concentration of sulfate (20 μM) (25). When sulfate was provided to M. smegmatis containing pYUB-172 as an additional source of sulfur in the medium, the recombinant did not degrade endosulfan, demonstrating that sulfur regulation is maintained on the cosmid. E. coli TG1 containing pYUB-172 did not degrade endosulfan irrespective of the presence of an alternative sulfur source.
DNA fragments generated by digestion of pYUB-172 with various restriction nucleases were cloned into pYUB415 and screened for endosulfan-degrading activity in M. smegmatis. Fragments with activity were further subcloned and screened in the same manner until the DNA fragment containing activity was reduced to a 3.0-kb ApaI DNA fragment (pYUB415-Apa3).
Analysis of a single ORF within the ApaI fragment and homology of its product to other proteins.
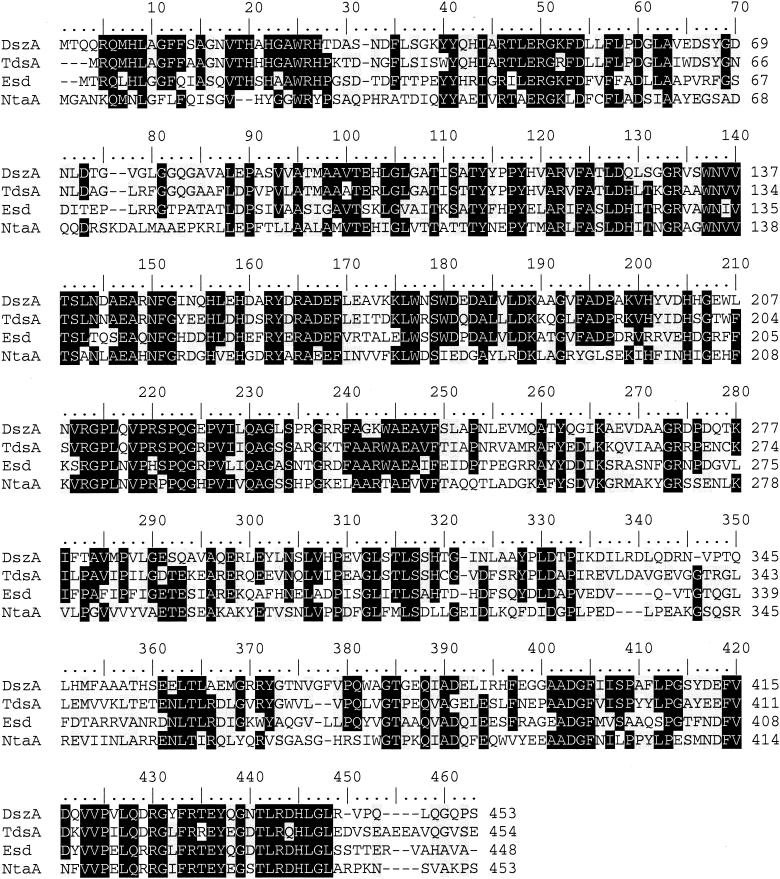
DNA sequence analysis of the 3.0-kb ApaI pYUB415-Apa3 fragment revealed a 1,347-bp ORF (esd), which encoded a 448-amino-acid protein (Esd, with a predicted molecular mass of 49.8 kDa) with a ribosome binding site (GGGAG) 9 bp upstream from the initiation codon (data not shown). As expected for sulfur-regulated proteins, Esd contained a low number of sulfur-containing amino acids compared to the number of sulfur-containing amino acids contained by non-sulfur-regulated proteins. The protein contained only a single methionine (frequency of sulfur-containing amino acids in the enzyme, 0.22%). The deduced amino acid sequence of the ORF was compared with the amino acid sequences of other proteins in the SwissProt and SpTrEMBL databases and was found to have significant similarity to the sequences of several other proteins (Fig. 1). The highest level of identity (50%) was to the tdsA product, a thermophilic flavomonoxygenase of Paenibacillus sp. strain A11-2 that catalyzes the conversion of dibenzothiophene 5,5-dioxide to 2-(2′-hydroxyphenyl)benzene sulfinate (12). The ORF product also had significant similarity (46% identity) to the product of dszA (formerly soxA), the tdsA homologue in Rhodococcus sp. strain IGTS8 (4), and 38% identity to component A of a nitrilotriacetate monooxygenase of Chelatobacter heintzii ATCC 29600 (16). The four proteins had similar lengths (448 to 453 amino acids), and similarities occurred throughout the proteins (Fig. 1). The regions conserved in the other proteins were also conserved in Esd, and no major insertion or deletion differences specific to Esd were identified.
FIG. 1.
Multiple sequence alignment of the Esd monooxygenase with other monooxygenase enzymes with significant amino acid sequence similarity. The numbers above the sequence indicate the total numbers of residues; the numbers at the ends of lines indicate the positions of residues in the sequences of the individual proteins. Black shading indicates identical residues, and grey shading indicates similarly charged residues. DszA, dibenzothiophene 5,5′-dioxide monooxygenase from R. erythropolis (accession number AF048979); TdsA, dibenzothiophene 5,5′-dioxide monooxygenase from thermophilic Paenibacillus sp. strain A11-2 (accession number AB033997); Esd, endosulfan-degrading monooxygenase described in this paper; NtaA, nitrilotriacetate monooxygenase from C. heintzii (accession number U39411).
The ORF was amplified by PCR and cloned behind the constitutive promoter of the mycobacterial expression vector pMV261. The resulting construct (designated pMV261::esd) transferred the β-endosulfan-degrading phenotype to M. smegmatis without the sulfate regulation observed with pYUB415-Apa3, demonstrating that sulfate did not have a direct effect on enzymatic activity. When α-endosulfan or endosulfan sulfate was included in the medium rather than β-endosulfan, no disappearance of substrate was observed and no endosulfan metabolites were detected, suggesting that the enzyme was specific for the β isomer of endosulfan.
Sequence analysis of the 3.0-kb ApaI fragment of pYUB415-Apa3 did not reveal any ORF in the 400-bp region downstream from esd whose product had significant sequence homology to any protein in the SwissProt or SpTrEMBL database. Upstream of esd was a region for which it was difficult to generate sequence data, presumably due to an extensive secondary structure. This region contained a 53-bp palindromic sequence that contained a pair of mismatched base pairs midway through the sequence. An additional ORF (870 bp) was located upstream of esd and extended into the end of the 3.0-kb fragment. The amino acid sequence of the product of this ORF had 24% identity to the amino acid sequence of the dszC product of R. erythropolis extending from the amino acid at position 64 to the end. The dszC gene is located in an operon with dszA and encodes an enzyme that oxidizes dibenzothiophene to dibenzothiophene 5,5′-dioxide (4). The dszC homologue of Mycobacterium sp. strain ESD extended into the region that we were unable to sequence immediately upstream of esd.
Effect of flavin reductase gene expression on endosulfan degradation in enzyme assays.
Initially, we did not detect β-endosulfan-degrading activity in cell-free enzyme assay mixtures containing E. coli-expressed Esd (data not shown). It has been shown that the activity of DszA of Rhodococcus was enhanced by the presence of either endogenous or heterologous flavin reductase (9, 11, 12, 17, 19, 28). As sequence analysis suggested that Esd was homologous to DszA, we isolated and overexpressed a flavin reductase gene from M. smegmatis to supply flavin reductase in Esd enzyme assays.
An ORF (MsFR) in the M. smegmatis genome sequence contained a 487-bp region whose translated product had 61% identity to the protein sequence encoded by dszD, a flavin reductase gene from R. erythropolis. In contrast to the low frequency of sulfur-containing amino acids in Esd, MsFR contained three cysteine and three methionine residues (frequency of sulfur-containing amino acids, 3.8%). DszD is a flavin reductase enzyme that is part of a two-component flavin-diffusible monooxygenase family of enzymes. Galán et al. (9) compared the amino acid sequences of the flavin reductase components of this family and identified several conserved residues. MsFR contains the first of these residues, a serine residue located before a pair of proline residues in the N terminus of the protein. However, the highly conserved GDH motif located in the C termini of members of this family is a GDS motif in MsFR. This motif was speculated to be involved in NAD(P)H binding (9). The relevance of this is not known.
We amplified the MsFR gene using PCR, inserted the amplicon into pET14b (pET14b-MsFR), and overexpressed the protein (MsFR) in E. coli. Purification with a nickel column resulted in a yield of approximately 0.5 mg of MsFR or 0.9 mg of Esd from 100-ml overexpressing cultures with approximately 95 and 90% purity, respectively. The purified protein expressed utilized FMN and NADH as an electron acceptor and an electron donor, respectively (Table 2). FMN was a better electron acceptor for MsFR than riboflavin and FAD were, but the latter two compounds could support some enzyme activity. Methylene blue and tetrahydrobiopterin were not utilized as electron acceptors, and NADPH could not replace NADH as the electron donor.
TABLE 2.
Substrate specificity of the flavin reductase protein, MsFRa
| Flavin substrates | Activity (μmol · min−1 · μg of MsFR−1) | Coupled reaction with Esdb | |
|---|---|---|---|
| Donor | Acceptor | ||
| NADH | FMN | 463.2 ± 39.9 | Yes |
| Riboflavin | 92.6 ± 23.1 | NRc | |
| FAD | 58.0 ± 14.7 | Yes | |
| Methylene blue | 0 | NDd | |
| Tetrahydrobiopterin | 0 | ND | |
| NADPH | FMN | 0.63 ± 0.21 | NR |
| NADH | FMN, no MsFR | 0.84 ± 0.12 | NR |
Electron acceptors were included at the following concentrations: 10 to 100 μM FMN; 10 to 100 μM riboflavin; 10 to 100 μM FAD; 50 μM methylene blue; and 50 μM tetrahydrobiopterin. The reaction was initiated by adding 0.5 mg of electron donor (NADH or NADPH) as described in Materials and Methods.
Electron donors (5 mg52ml−1) and acceptors (50 μM) were included in standard Esd enzyme assay mixtures, and endosulfan degradation was analyzed by TLC as described in Materials and Methods.
NR, no reaction detected.
ND, not done.
When MsFR was included in assay mixtures containing Esd overexpressed from pET14b-esd in E. coli along with NADH, FMN, or FAD and a solubilizing agent (Table 3), β-endosulfan disappeared and the metabolites, endosulfan monoaldehyde and endosulfan hydroxyether, were detected. Endosulfan is very hydrophobic, and initial assays demonstrated that in the absence of added detergent, BSA, or M. smegmatis cell extract the insecticide partitioned to the glass surface of the reaction vessel and was not available to the enzyme (data not shown). Activity was greatly enhanced in the presence of M. smegmatis cell extract compared to the activity in the presence of BSA or Triton X-100 (Table 3). Furthermore, the enhancing activity of the M. smegmatis cell extract depended on both the age of the culture from which it was prepared (an extract from a stationary-phase culture had a significantly greater enhancing effect than an extract prepared from log-phase cells [Table 3]) and the presence of 10 mM MgCl2. TLC analysis indicated that the relative proportions of the two metabolites, endosulfan monoaldehyde and endosulfan hydroxyether, were similar in the presence of added detergent, BSA, and M. smegmatis cell extract. When either α-endosulfan or endosulfan sulfate was included in the assay mixture rather than β-endosulfan, no metabolites were observed.
TABLE 3.
Activity of the endosulfan-degrading gene product, Esd, expressed in E. coli
| Reaction mixture | Solubilizing factor added | Amt of β-endosulfan metabolized (μmol · min−1) |
|---|---|---|
| Completea | BSAb | 0.5 ± 0.1 |
| 0.08% Triton X-100 | 4.1 ± 1.4 | |
| M. smegmatis stationary-phase cell extractb | 18.4 ± 6.9 | |
| M. smegmatis log-phase cell extractb | 0.8 ± 0.3 | |
| None | NDc | |
| No Esd protein | M. smegmatis stationary-phase cell extractb | ND |
| No MsFR protein | M. smegmatis stationary-phase cell extractb | 2.6 ± 0.8 |
| No NADH | M. smegmatis stationary-phase cell extractb | 2.2 ± 0.3 |
The complete reaction mixture contained 0.4 nmol of Esd (20 μg), 0.5 nmol of MsFR (12 μg), 50 μM FMN, 4 mM NADH, and 500 μM β-endosulfan in 1 ml of 50 mM HEPES buffer (pH 6.9).
The amount added was 7.5 mg of protein per ml.
ND, none detected.
Pathway analysis.
The metabolites endosulfan monoalde-hyde (recovered by TLC on alumina) and endosulfan hydroxyether (a gift from I. Kennedy) were included in enzyme assay mixtures as substrates (under the conditions described in Materials and Methods and in the presence of 0.08% Triton X-100). No degradation of either compound was observed by TLC analysis after 16 h of incubation.
DISCUSSION
We exploited the reactive nature of the cyclic sulfite diester group of endosulfan to isolate a strain of Mycobacterium (strain ESD) that degrades the insecticide in order to access the sulfur as a nutrient source (24, 25). From this strain we isolated esd, a gene that encodes an enzyme capable of degrading β-endosulfan when it is provided with reduced flavin. A sequence comparison revealed that esd was most similar to the tdsA gene of Paenibacillus sp. strain A11-2 (12) and its homologue, dszA of Rhodococcus sp. strain IGTS8 (4). The products of these genes are members of the two-component flavin-diffusible monooxygenase family (TC-FDM) of enzymes that require reduced FMN supplied by an NAD(P)H-dependent flavin reductase. This family differs from other monooxygenase families as the enzymes do not contain bound flavin but rather use the compound as a cosubstrate. The flavin reductase enzymes that provide reduced FMN are interchangeable, and physical contact with the monooxygenase is not required for activity (9, 11, 28).
Our early attempts to isolate a gene-enzyme system with endosulfan-degrading activity from Mycobacterium sp. strain ESD by screening a cosmid library in E. coli were unsuccessful (data not shown). We now suspect that this was because, at least in part, degradation involves both monooxygenase and flavin reductase components and that the E. coli host cells did not have appropriate flavin reductase activity to support the activity of recombinant cells containing the monooxygenase enzyme. Isolation of the monooxygenase component was successful after a Mycobacterium sp. strain ESD genomic DNA cosmid library was screened in M. smegmatis, an organism that did not have endosulfan-degrading activity but did possess appropriate flavin reductase activity. When it was found that the esd product had sequence similarity to other monooxygenases that require a separate flavin reductase, we searched the M. smegmatis genome for sequences with homology to the sequences of genes encoding enzymes that are potentially capable of providing reduced flavin to this type of enzyme. A flavin reductase gene was identified by analysis of the M. smegmatis genome sequence, and this sequence was used as a template for active flavin reductase (MsFR). Recombinant MsFR could reduce FMN, FAD, and riboflavin with NADH as the electron donor, but only reduced FMN and reduced FAD could be utilized by the monooxygenase as electron donors in the degradation of endosulfan (Table 2).
The specific activity of MsFR is similar to the specific activities of other flavin reductase enzymes belonging to the TC-FDM family that reduce flavin as a substrate rather than as a prosthetic group (7, 8, 9, 14, 19, 21, 26). The substrate specificities of these enzymes vary; some of them act on FAD and riboflavin better than on FMN (8, 21), some of them have similar activities with the three flavins (9, 26), and some of them, like MsFR, have higher activity with FMN (19). Similarly, some of the flavin reductases utilize NADPH in preference to NADH (7), while others are inert with NADPH and exhibit activity with NADH (8, 9, 14, 19, 26).
An additional problem with the initial screening in an E. coli host may have been at the level of regulation of gene expression. Expression of the monooxygenase component is strongly regulated by sulfur levels in the parent strain, and based on similar studies (4, 22), this regulatory mechanism is unlikely to be recognized in the E. coli host strain. Genes encoding other monooxygenases of the TC-FDM family are also controlled by sulfur-regulated promoters not recognized in E. coli (4, 22). Like esd, these related genes are efficiently expressed under the control of alternate promoters to produce active protein. The proteins encoded by these genes are involved in the desulfurization of methanesulfonates and other alkanesulfonates that are part of the response of the bacterium to sulfur limitation (for a review see reference 15). This involves expression of a set of 7 to 14 proteins, which allows the bacterium to utilize organosulfur-containing compounds in the absence of more bioavailable sulfur sources, such as sulfate and sulfite. Sulfur regulation is retained by the 3-kb ApaI fragment that confers endosulfan-degrading activity on M. smegmatis but was absent when esd was expressed behind a constitutive mycobacterial promoter, suggesting that the regulation is cis to esd, most likely in the 53-bp palindromic region upstream of the gene that we have had difficulty sequencing. Expression of the dszA gene from R. erythropolis IGTS8 is also strongly repressed by sulfate, as well as by sulfur-containing amino acids. It has been proposed that two possible regions at bases −263 to −244 and −93 to −38 immediately upstream from the initiator codon (positions 1 to 3) are responsible for this regulation (17). A potential hairpin structure was also located between bases −75 and −57.
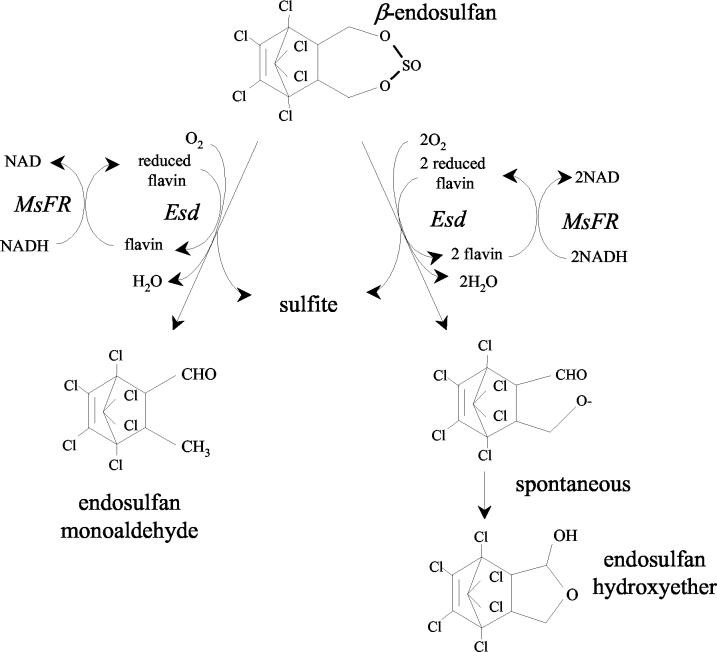
We were surprised to find that Esd was a member of a monooxygenase family and that catalysis required reduced flavin, as it has previously been proposed that the mechanism for endosulfan metabolism by Mycobacterium sp. strain ESD is most likely to be base-catalyzed hydrolysis, like the hydrolysis that occurs under alkaline conditions to produce endosulfan diol (24). Esd appears to perform oxygenation on β-endosulfan to produce endosulfan monoaldehyde and endosulfan hydroxyether. As neither metabolite acts as a substrate for the other, it appears that the reactions occur in tandem. We propose that the steric requirements of the enzyme allow access of both methylene groups of β-endosulfan to possible oxidation. If only one of these methylene groups reacts, then the subsequent proton rearrangement releases endosulfan monoaldehyde from the active site. However, if both methylene groups are oxidized, then the organic moiety cyclizes to a hemiacetal (that is, endosulfan hydroxyether) as shown in Fig. 2. The esd gene does not confer the ability to degrade the α isomer of endosulfan on M. smegmatis, and presumably there is another gene in the Mycobacterium sp. strain ESD genome that is responsible for the observed α-endosulfan-degrading activity of this strain (24). Although α-endosulfan and β-endosulfan are diastereoisomers with distinct physical and chemical properties, they are both degraded to endosulfan monoaldehyde in Mycobacterium sp. strain ESD (24), and we were surprised to find that the esd product was apparently inert with the α isomer.
FIG. 2.
Proposed mechanism for the reduced flavin-dependent metabolism of β-endosulfan catalyzed by Esd and MsFR. MsFR reduces FMN or FAD by using NADH as an electron donor. The reduced flavin is a cofactor in the metabolism of endosulfan that releases the sulfur moiety and endosulfan monoaldehyde or endosulfan monoaldehyde.
The activity of the purified protein expressed by E. coli was significantly enhanced when M. smegmatis cell extracts were included in the cell-free assay mixture, although the products of the reaction remained the same. Endosulfan is very hydrophobic (Kow log p, 4.8; solubility in water, 0.32 mg · liter−1) and partitioned rapidly out of the aqueous phase to the glass surfaces of our reaction vessels when it was added to enzyme assay mixtures at concentrations that were required for detection (which were significantly higher than the solubility limit of the insecticide). Detergent, BSA, or mycobacterial cell extract was an essential component of the reaction mixture and increased the apparent solubility of the insecticide and allowed degradative activity to be detected. In the absence of these supplements the endosulfan partitioned to the glass surface of the reaction vessel and degradative activity was not detected. Many soil bacteria produce biosurfactants to enhance degradation of hydrophobic compounds (5). The M. smegmatis extracts increase the apparent solubility and therefore the bioavailability of endosulfan to the soluble esd-encoded enzyme in our system. The enhanced degradation in the presence of a natural biosurfactant suggests that the Esd activity in our enzyme assays is limited by endosulfan solubility rather than the activity of the enzyme. The bioavailability of the enzyme and the ability of the enzyme to degrade endosulfan at concentrations present in aqueous solutions are currently being investigated.
The genes encoding DszA in R. erythropolis and TdsA in Paenibacillus sp. strain A11-2, two of the three proteins with significant sequence similarity to Esd, are located in operon-like structures within the genomes of the two organisms (4, 12). These operon-like structures each encode two other enzymes, DszB and DszC and TdsB and TdsC, which form pathways that convert dibenzothiophene to 2-hydroxybiphenyl and sulfite. The organization of these genes suggests that they are coordinately regulated by a single promoter. Regulation of this region of the dsz gene cluster by sulfur has been mentioned previously. Interestingly, the flavin reductase enzyme that is presumed to provide electrons to the DszA and DszC enzymes of this cluster is located elsewhere in the genome (11). This is also the case with other TC-FDM systems (see reference 15 for a review). The other enzyme with significant sequence similarity to Esd, the nitrilotriacetate monooxygenase of C. heintzii, is found in association with a flavin reductase, but the genes encoding these enzymes are not coordinately regulated. These genes are located close together in the genome but are orientated divergently, with a 307-bp intergenic region separating them (16). We did not find any evidence that there is an operon-like structure surrounding the esd gene of Mycobacterium sp. strain ESD; the 3.0-kb fragment from Mycobacterium sp. strain ESD that had endosulfan-degrading activity contained only the single complete gene, and this gene was expressed under sulfur-limited conditions, indicating that the fragment contained a promoter. No ORF was identified in the 400-bp region downstream from esd. We also did not find evidence that there is a proximal flavin reductase, although our search was limited to the 3-kb ApaI fragment containing esd. We did, however, identify an incomplete ORF upstream of esd, orientated in the same direction as esd, with sequence homology to dszC of R. erythropolis. This ORF is interesting in light of the fact that Mycobacterium sp. strain ESD could metabolize both α- and β-endosulfans to endosulfan monoaldehyde but esd is specific for the β isomer of endosulfan. Possibly more significant is the finding that the product of the ORF was most similar to DszC, an enzyme that oxidizes dibenzothiophene to dibenzothiophene 5,5′-dioxide. This reaction is very similar to the oxidation of endosulfan to endosulfan sulfate observed in Mycobacterium sp. strain ESD. It is thought that this oxidation is catalyzed by a non-cytochrome P450 monooxygenase as it is not inhibited by the cytochrome P450 inhibitor piperonyl butoxide (24). We are currently investigating whether this gene is involved in either α-endosulfan metabolism or oxidation of either isomer to endosulfan sulfate.
Acknowledgments
We are grateful for the financial support provided by the Cotton Research and Development Corporation (grant CSE 77C), the Horticultural Research and Development Corporation (grant HG97340), and Orica Australia Pty Ltd. Sequencing of M. smegmatis was accomplished with support from the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
We thank Chris Coppin, Kahli Weir, and Sue Dorrian for technical assistance.
REFERENCES
- 1.Anderberg, R. J., J. A. Strachan, and G. A. Cangelosi. 1995. Purification of DNA from Mycobacterium species without sonication or phenol. BioTechniques 18:217-219. [PubMed] [Google Scholar]
- 2.Awasthi, N., N. Manickam, and A. Kumar. 1997. Biodegradation of endosulfan by a bacterial coculture. Bull. Environ. Contam. Toxicol. 59:928-934. [DOI] [PubMed] [Google Scholar]
- 3.Casida, J. E. 1993. Insecticide action at the GABA-gated chloride channel: recognition, progress, and prospects. Arch. Insect Biochem. Physiol. 22:13-16. [DOI] [PubMed] [Google Scholar]
- 4.Denome, S. A., C. Oldfield, L. J. Nash, and K. D. Young. 1994. Characterization of the desulfurization genes from Rhodococcus sp. strain IGTSG. J. Bacteriol. 176:6707-6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desai, J. D., and I. M. Banat. 1997. Microbial production of surfactants and their commercial potential. Microbiol. Mol. Biol. Rev. 61:47-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dorough, H. W., K. Huhtanen, T. C. Marshall, and H. E. Bryant. 1978. Fate of endosulfan in rats and toxicological considerations of apolar metabolites. Pestic. Biochem. Physiol. 8:241-252. [Google Scholar]
- 7.Eichhorn, E., J. van der Ploeg, and T. Leisinger. 1999. Characterization of a two-component alkanesulfonate monooxygenase from Escherichia coli. J. Biol. Chem. 274:6707-6716. [DOI] [PubMed] [Google Scholar]
- 8.Fontecave, M., R. Eliasson, and P. Reichard. 1987. NAD(P)H:flavin oxidoreductase of Escherichia coli. A ferric iron reductase participating in the generation of the free radical of ribonucleotide reductase. J. Biol. Chem. 262:12325-12331. [PubMed] [Google Scholar]
- 9.Galán, B., E. Diaz, and G. L. Garcia. 2000. Enhanced desulphurization by engineering a flavin reductase-encoding gene cassette in recombinant biocatalysts. Environ. Microbiol. 2:687-694. [DOI] [PubMed] [Google Scholar]
- 10.Goebel, H., S. Gorbach, W. Knauf, R. H. Rimpau, and H. Huttenbach. 1982. Properties, effects, residues and analytics of the insecticide endosulfan. Residue Rev. 83:40-41. [DOI] [PubMed] [Google Scholar]
- 11.Grey, K. A., O. S. Prgrebinsky, G. T. Mrachko, L. Xi, D. J. Monticello, and C. H. Squires. 1996. Molecular mechanisms of biocatalytic desulfurization of fossil fuels. Nat. Biotechnol. 14:1705-1709. [DOI] [PubMed] [Google Scholar]
- 12.Ishii, Y., T. Ohshiro, Y. Aoi, M. Suzuki, and Y. Izumi. 2000. Identification of the gene encoding a NAD(P)H-flavin oxidoreductase coupling with dibenzothiophene (DBT)-desulfurizing enzymes from the DBT-nondesulfurizing bacterium Paenibacillus polymyxa A-1. J. Biosci. Bioeng. 90:220-222. [PubMed] [Google Scholar]
- 13.Jacobs, W. R., G. V. Kalpana, J. D. Cirillo, L. Pascopella, S. B. Snapper, R. A. Udani, W. Jones, R. G. Barletta, and B. R. Bloom. 1991. Genetic systems for mycobacteria. Methods Enzymol. 204:537-555. [DOI] [PubMed] [Google Scholar]
- 14.Kendrew, S. G., S. E. Harding, D. A. Hopwood, and E. N. G. Marsh. 1995. Identification of a flavin:NADH oxidoreductase involved in the biosynthesis of actinorhodin. Purification and characterisation of the recombinant enzyme. J. Biol. Chem. 270:17339-17343. [DOI] [PubMed] [Google Scholar]
- 15.Kertesz, M. A. 1999. Riding the sulfur-cycle—metabolism of sulfonates and sulfate esters in Gram-negative bacteria. FEMS Microbiol. Rev. 24:135-175. [DOI] [PubMed] [Google Scholar]
- 16.Knobel, H.-R., T. Egli, and J. R. van der Meer. 1996. Cloning and characterization of the genes encoding nitrilotriacetate monooxygenase of Chelatobacter heintzii ATCC 29600. J. Bacteriol. 178:6123-6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lei, B., and S.-C. Tu. 1996. Gene overexpression, purification, and identification of a desulfurization enzyme from Rhodococcus sp. strain IGTS8 as a sulfide/sulfoxide monooxygenase. J. Bacteriol. 178:5699-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martens, R. 1976. Degradation of [8,9-14C]endosulfan by soil microorganisms. Appl. Environ. Microbiol. 6:853-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsubara, T., T. Ohshiro, Y. Nishina, and Y. Izumi. 2001. Purification, characterization, and overexpression of flavin reductase involved in dibenzothiophene desulfurization by Rhodococcus erythropolis D-1. Appl. Environ. Microbiol. 67:1179-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miles, J. R. W., and P. Moy. 1979. Degradation of endosulfan and its metabolites by a mixed culture of soil microorganisms. Bull. Environ. Contam. Toxicol. 23:13-19. [DOI] [PubMed] [Google Scholar]
- 21.Parry, J. R., and W. Li. 1997. An NADPH:FAD oxidoreductase from valanimycin producer Streptomyces viridifaciens. J. Biol. Chem. 272:23303-23311. [DOI] [PubMed] [Google Scholar]
- 22.Piddington, C. S., B. R. Kovacevich, and J. Rambosek. 1995. Sequence and molecular characterization of a DNA region encoding dibenzothiophene desulfurization operon of Rhodococcus sp. strain IGTS8. Appl. Environ. Microbiol. 61:468-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Sutherland, T. D., I. Horne, M. J. Lacey, R. L. Harcourt, R. J. Russell, and J. G. Oakeshott. 2000. Enrichment of an endosulfan-degrading mixed bacterial culture. Appl. Environ. Microbiol. 66:2822-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sutherland, T. D., I. Horne, R. L. Harcourt, R. J. Russell, and J. G. Oakeshott. Isolation of an endosulfan-degrading Mycobacterium species. J. Appl. Microbiol., in press.
- 26.Thibaut, D., N. Tatet, D. Bisch, D. Faucher, L. Debussche, and F. Blanche. 1995. Purification of the two-enzyme system catalyzing the oxidation of the d-proline residue of pristinamycin IIB during the last step of pristinamycin IIA biosynthesis. J. Bacteriol. 177:5199-5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Woerden, H. F. 1963. Organic sulfites. Chem. Rev. 63:557-571. [Google Scholar]
- 28.Xi, L., C. H. Squires, D. J. Monticello, and J. D. Childs. 1997. A flavin reductase stimulates DszA and DszC proteins of Rhodococcus erythropolis IGTS8 in vitro. Biochem. Biophys. Res. Commun. 230:73-75. [DOI] [PubMed] [Google Scholar]