Abstract
Genetic studies with Burkholderia cepacia complex isolates are hampered by the limited availability of cloning vectors and by the inherent resistance of these isolates to the most common antibiotics used for genetic selection. Also, some of the promoters widely employed for gene expression in Escherichia coli are inefficient in B. cepacia. In this study, we have utilized the backbone of the vector pME6000, a derivative of the pBBR1 plasmid that was originally isolated from Bordetella bronchiseptica, to construct a set of vectors useful for gene expression in B. cepacia. These vectors contain either the constitutive promoter of the S7 ribosomal protein gene from Burkholderia sp. strain LB400 or the arabinose-inducible PBAD promoter from E. coli. Promoter sequences were placed immediately upstream of multiple cloning sites in combination with the minimal sequence of pME6000 required for plasmid maintenance and mobilization. The functionality of both vectors was assessed by cloning the enhanced green fluorescent protein gene (e-gfp) and determining the levels of enhanced green fluorescent protein expression and fluorescence emission for a variety of clinical and environmental isolates of the B. cepacia complex. We also demonstrate that B. cepacia carrying these constructs can readily be detected intracellularly by fluorescence microscopy following the infection of Acanthamoeba polyphaga.
Burkholderia cepacia was first described as the causative agent of soft rot in onions (7). This microorganism can degrade many chemical compounds (16, 27) and produce a number of substances that antagonize soilborne plant pathogens (5, 22, 25, 26, 28). These properties have generated considerable interest in the use of B. cepacia as a bioremediation agent and a biopesticide (19, 23). In the past 20 years, B. cepacia has also emerged as a multidrug-resistant opportunistic pathogen, particularly in patients with cystic fibrosis (17, 18). Chronic pulmonary infections in cystic fibrosis patients that are caused by B. cepacia cannot be eradicated with antimicrobial agents, and they are often associated with increased rates of morbidity and mortality (24).
In the past few years, the taxonomy of B. cepacia has been reexamined (for a recent review, see reference 9). Discrete genomic species that are phenotypically similar, originally classified as B. cepacia, have been reclassified into distinct genomovars that form what is collectively known as the B. cepacia complex (9, 41). Members of the B. cepacia complex have also been isolated from environmental sources. Currently it is difficult to differentiate between clinical and environmental isolates, since characteristic virulence factors have not yet been clearly defined (4). The genome sequencing of three members of the Burkholderia genus, including the environmental Burkholderia sp. isolate LB400 (http://www.jgi.doe.gov/JGI_microbial/html/burkholderia/burk_mainpage.html) and the clinical isolates of Burkholderia pseudomallei K96243 and B. cepacia genomovar III J2315 (http://www.sanger.ac.uk/Projects/B_cepacia/Sanger), will facilitate the genetic analysis of these microorganisms. This includes the identification of genes encoding virulence traits as well as other properties related to the ability of Burkholderia species to survive and thrive in various ecological niches. Unfortunately, genetic studies with B. cepacia complex isolates are difficult, in part because of the limited availability of cloning vectors and also because of the inherent resistance of these isolates to most of the antibiotics commonly employed for genetic selection.
The plasmid pME6000 is a derivative of the pBBR1 plasmid originally isolated from Bordetella bronchiseptica (3), which is maintained at roughly 20 to 30 copies per cell. The vector encodes a replication protein (Rep) that shares sequence homology with replication proteins in gram-negative bacteria. The Rep protein is essential for stable plasmid maintenance even in the absence of antibiotic selection, since the original pBBR1 plasmid was naturally maintained in B. bronchiseptica while lacking any known selectable markers. pBBR1 also carries a mobilization (mob) region encoding a putative protein that shares sequence homology to Mob/Pre proteins of gram-positive plasmids and is required to mediate the transfer of the plasmid when transfer functions are supplied in trans (3). Compatibility studies involving pBBR1 suggest that this plasmid is not a member of the broad-host-range IncC, IncP, IncQ, or IncW plasmids, and it may therefore represent a novel incompatibility group.
pME6000 carries a copy of the PLAC promoter to drive expression of cloned genes. Unpublished studies in our laboratory suggest that this promoter does not function in any of the clinical isolates examined that belong to the genomovars II, III, IV, and V. In this study, we report the construction of a set of expression vectors for use in B. cepacia which contain either the constitutive promoter of the S7 ribosomal protein gene from Burkholderia sp. LB400 or the arabinose-inducible BAD promoter from Escherichia coli. Promoter sequences were placed immediately upstream of a multiple cloning site in combination with the minimal sequence of pME6000 required for plasmid maintenance and mobilization. The functionality of both vectors was assessed by cloning the enhanced green fluorescent protein (eGFP) gene (e-gfp) and determining the level of protein expression and fluorescence emission in various clinical and environmental isolates of the B. cepacia complex. In previous studies, we have shown that representative members of B. cepacia genomovars can survive intracellularly within amoebae (31) and macrophages (36). Thus, we also tested the utility of our vectors by determining microscopically the fluorescence emission of intracellular bacteria following experimental infections of Acanthamoeba polyphaga. Our results suggest that these newly constructed constitutive and inducible expression vectors will improve genetic manipulations of B. cepacia complex strains as well as other Burkholderia isolates.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains and plasmids used in this study are listed in Table 1. Bacteria were grown in Luria-Bertani (LB) (Sigma Chemical Co., St. Louis, Mo.) and SOB (2% tryptone, 0.5% yeast extract, 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, 10 mM MgSO4) media. For B. cepacia, trimethoprim and tetracycline were each used at final concentrations of 100 μg/ml. Antibiotics for E. coli were used at the following concentrations: trimethoprim, 50 μg/ml; tetracycline, 20 μg/ml; ampicillin, 100 μg/ml; and kanamycin, 40 μg/ml. l-arabinose was added to final concentrations ranging from 0.02% (wt/vol) to 4.0% as required.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| E. coli | ||
| DH5α | F−, φ80 lacZΔM15 endA1 recA1 hsdR17(rK− mK+) supE44 thi-1 ΔgyrA96 (ΔlacZYA-argF)U169 relA1 | Lab stocks |
| Burkholderia sp. | ||
| ATCC 25416 | B. cepacia, genomovar I, environmental isolate | D. P. Speert |
| CEP509 | B. cepacia, genomovar I, clinical isolate | D. P. Speert |
| Cep484 | B. multivorans, clinical isolate | D. P. Speert |
| C5424 | B. cepacia, genomovar III, clinical isolate | D. P. Speert |
| C4455 | B. cepacia, genomovar III, clinical isolate | D. P. Speert |
| K56-2 | B. cepacia, genomovar III, clinical isolate | D. P. Speert |
| CEP559 | B. stabilis, clinical isolate | D. P. Speert |
| CEP040 | B. vietnamiensis, clinical isolate | D. P. Speert |
| CEP021 | B. cepacia, genomovar VI, clinical isolate | D. P. Speert |
| LB400 | Burkholderia sp., environmental isolate | S. Sehay |
| Plasmids | ||
| pME6000 | pBBR1 ori, TetrlacZ mob+ | S. Heeb |
| pML50 | pME6000, PS7-dhfr | This study |
| pML51 | pME6000, promoterless dhfr | This study |
| pML52 | pME6000, araC-PBAD-dhfr | This study |
| pBAD24 | AraC AmprcolE1 | D.E. Heinrichs |
| pRK2013 | RK2 derivative, Kmrmob+tra+ ColE1 | 15 |
| pTnMod-OTp′ | Plasposon, Tpr | 13 |
| pMLS7 | pBBR1 ori, PS7 promoter, Tprmob+ | This study |
| pMLBAD | pBBR1 ori, araC-PBAD Tprmob+ | This study |
| pMLS7-eGFP | pMLS7, e-gfp | This study |
| pMLBAD-eGFP | pMLBAD, e-gfp | This study |
| pML53 | pMLBAD-eGFP, PDHFR-araE | This study |
| pIJ8668 | e-gfp Aprr | 40 |
Ampr, ampicillin resistance; Aprr, apramycin resistance; Kmr, kanamycin resistance; Tetr, tetracycline resistance; Tpr, trimethoprim resistance.
Recombinant DNA methods.
DNA ligations, restriction endonuclease digestions, and agarose gel electrophoresis were performed according to standard techniques (37). Restriction enzymes, T4 DNA polymerase, and T4 DNA ligase were purchased from Roche Diagnostics (Dorval, Quebec, Canada). Polynucleotide kinase was obtained from MBI Fermentas Inc. (Burlington, Ontario, Canada). DNA transformation experiments with E. coli were carried out by the calcium chloride method (10). Transfer of plasmids into B. cepacia was conducted by triparental mating (12) using the pRK2013 helper plasmid (15).
Oligonucleotides.
The following synthetic oligonucleotides were used for vector constructions and cloning experiments: P6000NT, 5′-CACTCACTACAGCAGAGCCATTTAAACAACATCCCC-3′ (forward); P6000CT, 5′-ATTGGTCGACTTAAACGCCTGGTGCTACGCC-3′ (reverse); DHFRNT, 5′-TCTACGGGGTCTGACGCTCAGTGGAACG-3′ (forward); DHFRCT, 5′-AGGGATCCTAAGATATCGCTTAGGCCACACGTTCAAG-3′ (reverse) (underline indicates restriction endonuclease sites); S7NT, 5′-TATACAGCTAGCCGAATTGCTGGAAGTCGTCGC-3′ (forward); S7CT, 5′-CGTAGAATTCGTTTCTTCCTTTAACTT TTCAGTTGGAGC-3′ (reverse); PBADNT, 5′-TTAAGATCTCCCATCGGTGATGTCGGCGATATAGGC-3′ (forward); PBADCT, 5′-AAGAGTTTGTAGAAACGCAAAAAGGCCATCCGTCAGG-3′ (reverse); PGFPNT, 5′-GATAGAATTCCATGGTGAGCAAGGGCGA-3′ (forward); PGFPCT, 5′-GATCGAATTCGCGGCCGCTTTACTTGTA-3′ (reverse). PCRs were carried out using the PTC-0200 DNA engine (MJ Research, Incline Village, Nev.). Pwo polymerase (Roche Diagnostics) was used for PCRs expected to produce products of 3 kb or less. All other reactions were performed with the EXPAND High Fidelity PCR system (Roche Diagnostics). PCR templates were always denatured at 94°C for 3 min prior to cycle initiation. The remaining conditions for amplification were optimized for each primer pair, and they are described below. PCR products were separated on 0.7 to 1.0% (wt/vol) agarose gels, and the bands were purified with the QiaQuick gel extraction system (Qiagen, Valencia, Calif.).
Vector construction.
pME6000 is a low-copy-number, broad-host-range cloning vector kindly provided by S. Heeb, Laboratoire de Biologie Microbienne, Université de Lausanne (Lausanne, Switzerland). pME6000 was constructed by replacing the cat gene of pBBR1MCS (30) with the tetRA genes of pVK100 (29). The sequence carrying the replication and mob regions of pME6000 was used as the backbone for the construction of the new vectors (Fig. 1A). This 4.2-kb fragment was generated by PCR amplification with the primers P6000NT and P6000CT under the following conditions: 10 cycles of 1 min at 94°C, 1 min at 55°C, and 4 min at 68°C each, an additional 20 cycles of 1 min at 94°C, 1 min at 55°C, and 4 min plus 5 s per cycle at 68°C, and a final extension at 68°C for 8 min.
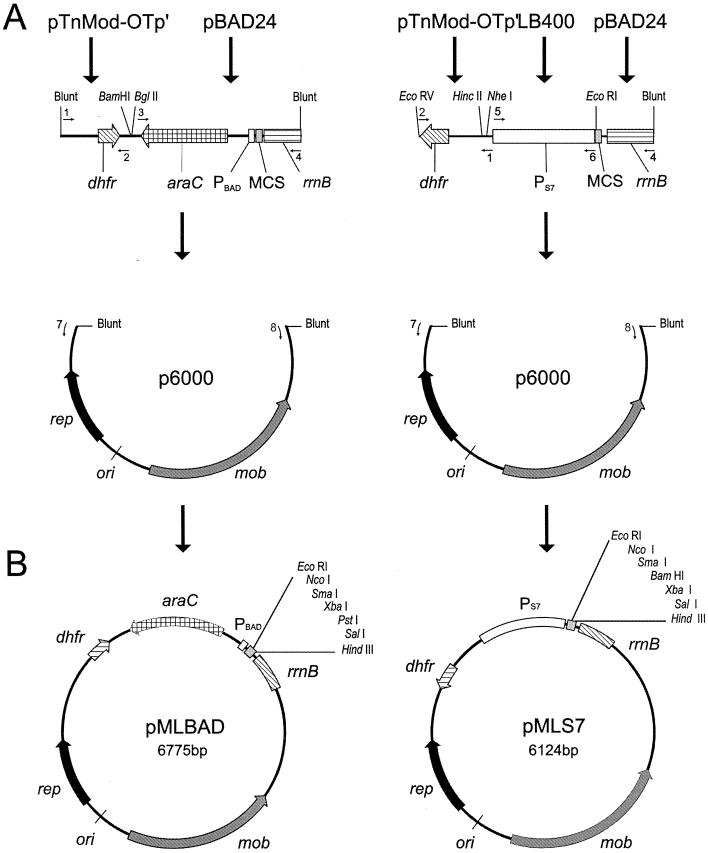
FIG. 1.
pMLBAD and pMLS7 expression vector maps. (A) Construction of pMLBAD and pMLS7 vectors. Sources of DNA fragments described in Materials and Methods are indicated by arrows above each construct. Primers used in plasmid constructions are indicated by the numbers above or below the small arrows: 1, DHFRNT; 2, DHFRCT; 3, PBADNT; 4, PBADCT; 5, S7NT; 6, S7CT; 7, P6000NT; and 8, P6000CT. Blunt, end of the fragment cloned directly from the PCR product; dhfr, dihydrofolate reductase gene encoding trimethoprim resistance; PBAD, arabinose-inducible promoter; araC, transcriptional regulator gene; rrnB, transcriptional terminator; MCS, multiple cloning site; PS7, region carrying the S7 ribosomal protein promoter from Burkholderia sp. strain LB400; p6000, backbone sequence derived from broad-host-range vector pME6000; mob, gene required for conjugal transfer of plasmid; rep, replication protein gene; ori, origin of replication. (B) Map of pMLBAD and pMLS7. Unique restriction endonuclease sites in the MCS of both plasmids are indicated. Plasmids are not drawn to scale.
For construction of pMLS7 (Fig. 1B), the promoter region of the S7 ribosomal protein gene from Burkholderia sp. strain LB400 was amplified by PCR using the primers S7NT and S7CT under the followings conditions: 5 cycles of 45 s at 94°C, 45 s at 50°C, and 1 min at 72°C each, an additional 25 cycles of 45 s at 94°C, 45 s at 54.5°C, and 1 min at 72°C, and a final extension of 5 min at 72°C. The resulting 847-bp fragment was digested with NheI and EcoRI, which cut at the 3′ and 5′ ends, respectively. The vector pBAD24 was also digested with NheI and EcoRI and ligated overnight with the 847-bp promoter fragment. In this manner, the multiple cloning site and rrnB terminator sequence of pBAD24 was placed directly downstream of the PS7 promoter. A 1,320-bp fragment was PCR amplified from the ligation mixture using the primers S7NT and PBADCT under the following conditions: 25 cycles of 45 s at 94°C, 45 s at 54°C, and 1.5 min at 72°C each, and a final extension of 7 min at 72°C. The 1,320-bp amplicon was treated with polynucleotide kinase to facilitate blunt-end cloning. In a parallel experiment, the trimethoprim resistance gene (dhfr) was amplified by PCR from the plasposon pTnMod-OTp′ (13) using the DHFRNT and DHFRCT primers. The conditions for PCR amplification were as follows: 5 cycles of 45 s at 94°C, 45 s at 53°C, and 55 s at 72°C each, 22 cycles of 45 s at 94°C, 45 s at 55°C, and 55 s at 72°C, and a final extension cycle of 4 min at 72°C. The 665-bp product was digested with HincII and EcoRV, cutting at the 5′ and 3′ ends, respectively, to generate blunt ends suitable for cloning. The digested PCR product containing the coding region of the dhfr gene was ligated with the 1,320-bp fragment carrying the P7S promoter and the pBAD24 multiple cloning site followed by the rrnB transcriptional terminator. The ligated fragments served as templates for PCR using the DHFRCT and PBADCT primers (Fig. 1A). The conditions of this reaction were as follows: 5 cycles of 45 s at 94°C, 45 s at 45°C, and 2 min at 72°C each, 22 cycles of 45 s at 94°C, 45 s at 55°C, and 2 min plus 5 s per cycle at 72°C, and a final extension cycle of 7 min at 72°C. The 1,981-bp product was treated with polynucleotide kinase and digested with EcoRV to generate blunt ends suitable for ligation with the pME6000 backbone amplicon described above. The ligation mix was transformed into E. coli DH5α, and transformants were plated on LB agar plates plus 50 ìg of trimethoprim/ml. Resistant colonies were isolated and screened for the presence of plasmid. The resulting 6,124-bp plasmid was designated pMLS7.
For construction of pMLBAD (Fig. 1B), the region of pBAD24 containing the araC gene, PBAD promoter, the multiple cloning site, and the rrnB terminator region was amplified by PCR using the PBADNT and PBADCT primers and the following conditions: 25 cycles of 45 s at 94°C, 45 s at 55°C each, 2 min plus 5 s per cycle at 70°C, and a final extension cycle of 7 min at 70°C. The resulting 1,912-bp product was treated with polynucleotide kinase and digested with BglII. The BglII recognition site was included within the PBADNT primer. The dhfr gene cassette described above was digested with BamHI to generate a 5′-end overhang compatible for ligation to the BglII end of the fragment carrying the araC-PBAD promoter region. After ligation, the mixture was amplified by PCR using the DHFRCT and PBADCT primers (Fig. 1A). The conditions for this reaction were the following: 5 cycles of 45 s at 94°C, 45 s at 55°C, and 2 min 40 s at 72°C each, 25 cycles of 45 s at 94°C, 45 s at 55°C, and 2 min 40 s plus 5 s per cycle at 72°C, and a final extension cycle of 7 min at 72°C. The resulting 2,584-bp product was treated with polynucleotide kinase and ligated to the p6000 backbone as described previously for the construction of pML7S. The ligation mix was transformed into E. coli DH5α cells, and the transformants were plated on LB agar plates containing 50 μg of trimethoprim/ml. Plasmid DNA was isolated from these colonies and examined by restriction endonuclease mapping to confirm the correct orientation of cloned DNA. The resulting 6,775-bp plasmid was named pMLBAD.
Plasmid stability.
To assess the stability of the constructed plasmids in the absence of antibiotic selection cultures of B. cepacia genomovar III strain C5424, Burkholderia vietnamiensis (formerly B. cepacia genomovar V) strain CEP040, and E. coli DH5α harboring either pMLS7 or pMLBAD were grown to the mid-logarithmic phase and used to inoculate tubes of LB broth without antibiotics at an initial optical density at 600 nm (OD600) of 0.02. Aliquots were removed at an OD600 of 0.32 (approximately corresponding to four generations) and plated onto LB plates with and without trimethoprim. A third aliquot was transferred to fresh LB broth and incubation continued as before to generate data for the subsequent generation points until a set of 20 generations was completed. Plates were incubated at 37°C overnight, and colonies were counted. The percent plasmid stability for each strain over the course of 20 generations was calculated by dividing the number of colonies in the LB plates supplemented with trimethoprim by the number of colonies in the LB plates without antibiotic.
Cloning of the e-gfp gene.
The coding region of e-gfp was amplified by PCR using the PEGFPNT and PEGFPCT primers and pIJ8668 DNA template according to the following conditions: 25 cycles of 45 s at 94°C, 45 s at 50°C, and 1 min at 72°C each, and a final extension cycle of 5 min at 72°C. The 800-bp fragment was treated with polynucleotide kinase and digested with NcoI, which cuts at the 5′ end of the e-gfp gene. This fragment was ligated into both pML7S and pMLBAD, which were digested with both NcoI and SmaI. The respective ligation mixtures were transformed into E. coli DH5α cells and plated on LB agar plates containing 50 μg of trimethoprim/ml. These experiments resulted in the isolation of the plasmids pML7S-eGFP and pMLBAD-eGFP containing the e-gfp gene under the control of the PS7 and PBAD promoters, respectively.
Expression analysis.
eGFP expression by pML7S-eGFP and pMLBAD-eGFP in B. cepacia complex isolates was assessed by Western blotting as described elsewhere (2). Both plasmids were mobilized by triparental mating to the Burkholderia sp. isolates indicated in Table 1. Cultures were grown overnight in LB broth alone or supplemented with increasing concentrations of l-arabinose or glucose as appropriate. Samples were first calibrated based on total wet weight of bacterial pellets following centrifugation and then boiled for 10 min in the presence of Laemmli Tris glycine denaturing reducing sample buffer (Novex, San Diego, Calif.). Equal volumes of each sample were loaded into a sodium dodecyl sulfate-14% polyacrylamide gel and separated at 200 V for 1 h. Samples were transferred to nitrocellulose membranes at 4°C for 1.5 h at 100 V. Membranes were blocked overnight using a 5% skim milk-TBST (16 mM Tris, 135 mM NaCL, 0.2% Tween 20) solution at 4°C and then incubated with rabbit anti-eGFP primary antibodies (Chemicon International, Temecula, Calif.), at a 1:5,000 dilution in TBST-5% skim milk for 1 h at room temperature. After a wash, membranes were incubated for 1 h at room temperature with donkey anti-rabbit serum conjugated to horseradish peroxidase (Amersham Biosciences, Baie d'Urfé, Quebec, Canada), which was diluted to 1:3,000 in TBST-5% skim milk. The blot was developed with the ECL chemiluminescent substrate (Roche Diagnostics) as recommended by the supplier.
The intensity of fluorescence emitted by strains carrying both the constitutive and the arabinose-inducible eGFP constructs was measured using a Microfluor1 fluorescence spectrometer (Dynex, Chantilly, Va.). Strains were grown overnight in LB broth alone or supplemented with l-arabinose or glucose. Each culture was calibrated to an OD600 of 0.2 prior to examination and aliquoted into a white Microfluor1 96-well plate (Dynex). The samples were analyzed under the following conditions: excitation wavelength of 485 nm, emission wavelength of 510 nm, 1-s interval, 37°C incubation temperature, 5-s agitation, and scale 100. To ensure that values recorded were due to expression of eGFP, cultures of untransformed strains were examined for autofluorescence and also served as the appropriate blanks for calculation of the relative units of fluorescence.
Fluorescence microscopy.
Amoeba infection assays were conducted using A. polyphaga strain JAC/S2 (ATCC 50372) as described in an earlier study (31). All infections were conducted with the genomovar III strain C5424 carrying pML7S-eGFP and the B. vietnamiensis CEP40 strains carrying either pML7S-eGFP or pMLBAD-eGFP. Amoebae were maintained axenically at 25°C as a monolayer in 25-cm2 flasks using a medium similar to that described by Page (35) except that glucose was replaced by glycerol. l-arabinose at a 1% final concentration was added to the axenic growth media and the nonnutrient Acanthamoeba buffer (35) to maintain promoter induction in the experiments involving pMLBAD-eGFP. Cells were washed in Acanthamoeba buffer, and approximately 1,000 amoeba cells were transferred to each well of a four-well Lab Tek II chamber slide (Nalge Nunc International, Rochester, N.Y.). Bacteria washed in Acanthamoeba buffer were added at a multiplicity of infection ranging from 1:1 to 1:100. Bacteria and amoebae were centrifuged for 2 min at 1,000 × g and incubated at 25°C. Slides were analyzed by light and fluorescence microscopy at 4, 12, and 24 h postinfection. Fluorescence and differential interference contrast images were acquired using a Qimaging (Burnaby, British Columbia, Canada) cooled charged-coupled-device camera on an Axioscope II (Carl Zeiss, Thornwood, N.Y.) microscope with a ×100/1.3 numerical aperture Plan-Neofluor objective, 150-W xenon lamp, and an endow GFP bandpass emission filter set (Chroma Technology, Brattleboro, Vt.) with a 470 ± 20 nm excitation range and a 525 ± 25 emission range. Images were digitally processed using Northern Eclipse version 6.0 (Empix Imaging, Mississauga, Ontario, Canada) imaging analysis software.
Nucleotide sequence accession numbers.
The nucleotide sequences of pMLBAD and pMLS7 have been deposited in GenBank under accession no. AY112733 and AY112734, respectively.
RESULTS AND DISCUSSION
Construction of a constitutive expression system for B. cepacia.
Previously constructed broad-host-range plasmid vectors derived from IncP and IncW incompatibility groups can in principle replicate in B. cepacia (1, 33, 38). However, these vectors lack appropriate antibiotic resistance gene markers that would be useful for genetic manipulations of clinical isolates from the B. cepacia complex, since the majority of these strains are highly resistant to ampicillin, tetracycline, and the aminoglycosides kanamycin and gentamicin (see below). The plasmid pBBR1 is a relatively small broad-host-range plasmid from B. bronchiseptica (3), and its derivatives have been used as cloning and expression vectors in nonenteric bacteria (14, 39). Thus, to construct vectors suitable for gene expression in B. cepacia, we decided to use the backbone of the cloning vector pME6000, a derivative from pBBR1 that is similar to pBBR1MCS (30) except that it encodes tetracycline resistance. Initial experiments based on a deletion analysis of pME6000 identified a 4.2-kb fragment containing the minimum sequence required for stable maintenance and plasmid transfer functions in both E. coli and B. cepacia (data not shown). This fragment was amplified using the p6000NT and p6000CT primers and served as the backbone for the construction of both the constitutive and inducible expression vectors described in this work (Fig. 1). We chose the dhfr gene, encoding trimethoprim resistance, as the selectable marker for our vectors, since our studies have shown that 4 out of 38 clinical isolates of B. cepacia (collected from diverse geographic and clinical sources) and all of the 8 environmental strains examined are sensitive to trimethoprim at a concentration of 100 μg/ml. These results are in agreement with a recent report showing that trimethoprim was more effective than both chloramphenicol and tetracycline against B. cepacia complex isolates (34). In fact, the MIC of trimethoprim was comparable to that of tobramycin, ciprofloxacin, and ceftazidime, which are used clinically to treat B. cepacia infections (34). Therefore, in contrast to the dhfr resistance marker, the use of resistance markers against tobramycin, ciprofloxacin, and ceftazidime for vector constructions would not be ethical. The dhfr resistance gene corresponds to a small open reading frame, which makes it desirable for use in an expression vector as it will limit the construct size while minimizing redundancy of restriction sites. The dhfr gene was amplified from the plasposon pTnMod-OTp′ as described in Materials and Methods.
To ensure efficient expression in Burkholderia strains, we chose a ribosomal protein promoter sequence from Burkholderia sp. strain LB400, which was formerly believed to be a member of the B. cepacia complex. The S7 ribosomal protein promoter from LB400 was amplified by PCR. The functionality of this promoter in B. cepacia was assessed by constructing a fragment encoding the dhfr gene under the control of PS7. This fragment was ligated into pME6000, resulting in the plasmid pML50 (Table 1). E. coli DH5α cells transformed with pML50 did not grow in the presence of trimethoprim, indicating that PS7 does not function in E. coli. In contrast, B. cepacia cells conjugated with pML50 were resistant to 100 μg of trimethoprim/ml. As a control, B. cepacia conjugated with pML51, which carries a promoterless dhfr gene, did not survive in the presence of antibiotic selection (data not shown). These data indicate that PS7 is functional only in B. cepacia, and therefore it could serve to drive gene expression in these microorganisms. The construction of the constitutive expression vector pMLS7 was then completed by placing the PS7 promoter region upstream from the multiple cloning sites derived from the pBAD24 vector, as outlined in Fig. 1. This strategy provided for convenient restriction sites placed immediately downstream of the promoter region to facilitate gene cloning. The strong ribosomal rrnB transcriptional terminator from pBAD24, located downstream of the multiple cloning site, was also included to prevent readthrough transcription into the vector backbone.
Construction of an inducible expression system for B. cepacia.
Past experience in our laboratory has indicated that the PLAC and PTAC promoters, widely employed for regulated gene expression in E. coli, do not function in the B. cepacia isolates from genomovars II, III, and V that we have examined. Since the E. coli araC-PBAD system has been used in nonenteric bacteria (39), we decided to evaluate its functionality in B. cepacia. This system is based on the regulatory protein AraC, which in the presence of l-arabinose activates transcription from the PBAD promoter (20). In contrast, in the presence of glucose, AraC functions as a transcriptional repressor, thereby preventing transcription from the PBAD promoter, thus providing a tightly regulated system for gene expression. We tested the araC-PBAD system in B. cepacia by cloning the dhfr gene under the control of the PBAD promoter in a manner similar to that described above for the construction of pML50. The resulting plasmid, pML52, was mobilized by triparental mating into B. cepacia isolates and shown to confer resistance to 100-μg/ml trimethoprim in the presence of l-arabinose. No bacterial growth was observed on plates containing glucose and trimethoprim, indicating that the regulation of the PBAD promoter functions effectively in B. cepacia. These data demonstrated that the araC-PBAD system functions in B. cepacia, and it was therefore used to construct the l-arabinose-inducible vector pMLBAD (Fig. 1) as described in Materials and Methods.
Stability of pMLS7 and pMLBAD.
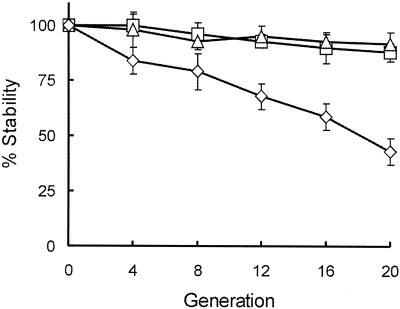
The stability of the constructed plasmids was investigated in both E. coli and Burkholderia sp. backgrounds. Two B. cepacia complex strains were used in these experiments: the genomovar III isolate C5424 and the B. vietnamiensis strain CEP040. We were especially interested in these two isolates because they are used in our laboratory for studies on the intracellular survival of B. cepacia within amoebae and macrophages. Bacteria harboring pMLS7 and pMLBAD were grown in the absence of trimethoprim for up to 20 generations. Aliquots were plated after every 4 generations on LB agar plates with or without trimethoprim, and the plasmid stability was expressed as a percent value from the number of colonies obtained with antibiotic selection relative to the number of colonies obtained in the absence of selection. Figure 2 shows that after 20 generations in the absence of antibiotic selection, the stability of pMLS7 in B. cepacia C5424 (88%) is comparable to that in E. coli DH5α (92%). In contrast, pMLS7 is less stable in B. vietnamiensis CEP040, exhibiting 44% stability following 20 generations without antibiotic selection pressure. These results suggest that there are strain variations regarding plasmid stability in Burkholderia species. However, even in the case of B. vietnamiensis CEP040, the plasmid remains relatively stable for a number of generations in the absence of selection. As expected, similar results were obtained with pMLBAD (data not shown). The lower level of stability of these plasmids in B. vietnamiensis could be due to decreased expression levels of the Rep protein, host-specific effects on plasmid partition, or incompatibility with indigenous plasmids. Further studies are required to examine these possibilities.
FIG. 2.
Analysis of pMLS7 stability in the absence of antibiotic selection. Cultures of Burkholderia strains C5424 (□) and CEP040 (⋄) and E. coli DH5α (trio), all containing pMLS7, were grown in LB without trimethoprim for 20 generations. Aliquots were removed after every four generations and plated on LB agar with or without trimethoprim. The percent stability of pMLS7 in each strain was determined by dividing the number of colonies obtained on LB-trimethoprim plates by the number of colonies obtained on LB plates with no trimethoprim. The data points represent the means and standard deviations from the experiments completed in triplicate.
Expression of GFP in B. cepacia.
The green fluorescent protein (GFP) of the jellyfish Aequorea victoria is a valuable tool for examining individual bacterial cells under nondestructive conditions in both laboratory model systems and natural environments (6). Since B. cepacia strains are present in the rhizosphere (4) and can also be found in association with amoebae (31) and eukaryotic cells (8, 32, 36), the availability of GFP-labeled bacteria would facilitate studying bacterium-host cell interactions. Earlier attempts in our laboratory employed GFP mutant forms that were optimized for bacterial use (11) as reporters for gene expression in B. cepacia complex strains. These experiments failed to reveal fluorescence in most cases or showed very poor yellow fluorescence in some isolates. In contrast, the same reporter constructs revealed intense green fluorescence when expressed in E. coli DH5α (data not shown). We interpreted the poor fluorescence in B. cepacia as a consequence of the relatively AT-rich nature of the gfp gene (38 mol% G+C compared to the estimated 67 mol% G+C of B. cepacia), which would determine a marked difference in codon usage between gfp and its bacterial host. The eGFP encoded by a mutated form of the gfp gene has recently been successfully expressed in Streptomyces coelicolor (40), a microorganism that has a mol% G+C content higher than that of B. cepacia. The coding region of e-gfp contains many silent nucleotide replacements (21), resulting in a 61 mol% G+C and an altered codon usage that corresponds much more closely to that of B. cepacia genes than the codon usage of the other gfp genes. Therefore, we evaluated e-gfp expression in B. cepacia by cloning this gene into both expression vectors as described in Material and Methods. At the same time, these experiments allowed us to evaluate the usefulness of our constitutive and inducible expression plasmids.
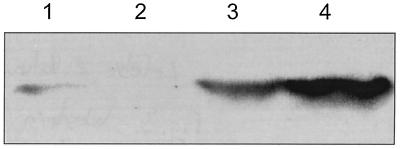
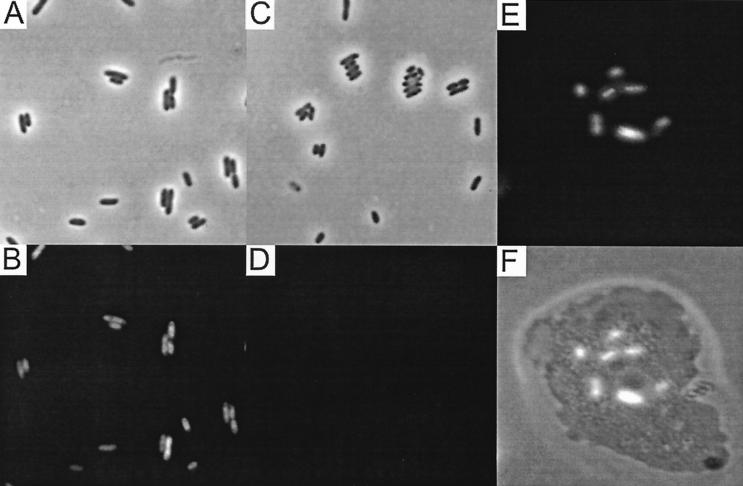
The expression of the eGFP protein was first assessed for B. vietnamiensis CEP040 by Western blot analysis. CEP040 was selected based on previous studies that have demonstrated that it readily survives intracellularly within epithelial cells, amoebae, and macrophages (8, 31, 36) and thus would be a suitable isolate for fluorescence microscopy studies. Figure 3 shows that the amount of eGFP produced by CEP040(pMLBAD-eGFP) increased as a function of the l-arabinose concentration in the culture medium. Maximal eGFP expression was reached with 2% l-arabinose (Fig. 3, lane 4). The band intensity in samples prepared from cells grown at 4% l-arabinose decreased, and also these cultures did not grow as well as the other samples (data not shown). This is likely due to increased osmolarity by the high solute concentrations and not due to overexpression of eGFP, as both the parent strain and strains carrying the expression plasmids showed the same growth defect when incubated in 4% l-arabinose. The sample obtained from CEP040(pMLBAD-eGFP) cells grown in the absence of arabinose shows a weak signal (Fig. 3, lane 1), suggesting that a small amount of protein is produced. As expected, no band was visible in lysates from cells grown in 0.2% glucose (Fig. 3, lane 2), indicating that promoter activity was fully repressed. Consistent with the protein expression results, a strong fluorescence was observed in bacterial cells carrying pMLBAD-eGFP under induction with 2% l-arabinose (Fig. 4A and B), while bacterial cells grown in the presence of glucose did not fluoresce (Fig. 4C and D). Overall, data from the protein expression experiments and fluorescence microscopy conclusively demonstrate that the eGFP protein is abundantly expressed and also that the PBAD promoter is functional in strain CEP040. Similar results in regard to protein expression and fluorescence microscopy of bacterial cells were obtained with pMLS7-eGFP, although in this case fluorescence was detected without induction (data not shown).
FIG. 3.
Western blot analysis of arabinose-induced eGFP expression from CEP040(pMLBAD-eGFP) cultures. Cultures were grown in LB alone or with the addition of various concentrations of arabinose and glucose. Samples were processed as described in Materials and Methods. Lanes: 1, LB alone; 2, 0.2% glucose; 3, 1% arabinose; 4, 2% arabinose.
FIG. 4.
eGFP expression in B. cepacia cells in culture and following the infection of A. polyphaga. (A and B) Images representing the same field of view of CEP040(pMLBAD-eGFP) cells grown in the presence of 2% arabinose visualized by differential interference phase contrast and fluorescence microscopy, respectively. (C and D) Images representing the same field of view of CEP040(pMLBAD-eGFP) cells grown in the presence of 1% glucose visualized by differential interference phase contrast and fluorescence microscopy, respectively. (E) Image of an A. polyphaga cell following 12 h of infection with C5424(pMLBAD-eGFP) visualized by fluorescence microscopy. (F) Overlay of a phase contrast image with the fluorescent image in panel E. All images were obtained at magnification ×1,000.
Functional eGFP expression determined by relative fluorescence measurements.
To assess functional eGFP expression associated with both the constitutive vector and the inducible vector, the relative fluorescence emitted by eGFP was quantified using a spectrofluorometer. The numbers of relative fluorescence units (rFU) displayed by each strain under various growth conditions are shown in Table 2. The values reveal that pMLS7-eGFP functioned well in members of the B. cepacia genomovars I through VI, albeit with varying levels of expression. More importantly, the level of fluorescence associated with the E. coli strain DH5α ranged from 5- to 21-fold lower than that observed for the set of Burkholderia strains examined. These data demonstrate that eGFP emits strong fluorescence in B. cepacia strains and also confirm our previous conclusion that the PS7 promoter has greater expression efficiency in B. cepacia than in E. coli.
TABLE 2.
Comparison of fluorescence levels for various strains of the B. cepacia complex associated with pMLS7-eGFP
| Strain | Genomovar or species | eGFP expression (rFU)a |
|---|---|---|
| CEP509 | I | 193 ± 16 |
| ATCC25416 | I | 126 ± 17 |
| CEP484 | II | 76 ± 6 |
| K56-2 | III | 337 ± 9 |
| C4455 | III | 75 ± 10 |
| C5424 | III | 260 ± 18 |
| CEP40 | V | 405 ± 13 |
| CEP21 | VI | 207 ± 9 |
| LB400 | Burkholderia sp. | 127 ± 15 |
| DH5α | E. coli | 16 ± 16 |
Values reflect the mean ± standard deviation of eight determinations. Measurements with strain CEP559 (genomovar IV) were not included since this strain exhibited a high level of autofluorescence that interfered with the detection of eGFP-specific fluorescence.
In agreement with the Western blot analysis of the strains carrying the pMLBAD-eGFP construct, the rFU values increased with the arabinose concentration (Table 3 and data not shown). However, the repression of eGFP expression by the addition of glucose was not as obvious. In all cases, the rFU values in cultures grown in the presence of glucose were intermediate compared to the rFU values of cells grown in LB alone and those of cells grown in 0.2% arabinose. This phenotype is likely due to changes in metabolic functions of the cells in the presence or absence of the various carbon sources, since wild-type cells alone grown in the presence of glucose showed significantly higher levels of autofluorescence than cells grown in LB. The absence of a band detectable by Western blot analysis in the cultures grown in the presence of glucose (Fig. 3, lane 1) supports this interpretation. Although there was some variability in the total number of fluorescence units associated with the different strains under the same growth conditions, the positive correlation between arabinose concentration and fluorescence intensity was constant. The relatively low rFU values measured for E. coli strain DH5α is likely a consequence of the extended induction times used in these experiments. Protocols using this promoter in enteric bacteria commonly use much less arabinose and shorter induction intervals (20), and therefore the conditions used in these experiments may not have been optimal for protein overexpression in E. coli.
TABLE 3.
Comparison of fluorescence levels associated with pMLBAD-eGFP in the presence of various concentrations of arabinose
| Strain | Genomovar or species | Growth conditiona | eGFP expression (rFU)b |
|---|---|---|---|
| 5424 | III | No arabinose | 24 ± 23 |
| 0.2% arabinose | 111 ± 24 | ||
| 2.0% arabinose | 380 ± 24 | ||
| CEP40 | V | No arabinose | 13 ± 12 |
| 0.2% arabinose | 433 ± 32 | ||
| 2.0% arabinose | 1705 ± 12 | ||
| DH5 | E. coli | No arabinose | 77 ± 29.12 |
| 0.2% arabinose | 103 ± 12.63 | ||
| 2.0% arabinose | 174 ± 21.00 |
Bacteria were grown in LB broth with the appropriate supplement of arabinose.
Values reflect the mean ± standard deviation of eight determinations.
The elevated concentrations of arabinose required for protein expression in B. cepacia possibly reflect poor transport of the sugar into the cell, resulting in low levels of intracellular pools being available for interaction with the AraC protein. To examine this possibility, we cloned the E. coli araE gene encoding an l-arabinose permease under control of the weak constitutive dfhr promoter into pMLBAD-eGFP, resulting in the plasmid pML53. In this construct, the expression of the permease is independent of the l-arabinose concentration in the medium. B. cepacia cells carrying pML53 displayed increased levels of eGFP production as determined by rFU values and by direct observation with fluorescence microscopy using l-arabinose concentrations as low as 0.02% (data not shown). These results suggest that B. cepacia isolates are relatively impermeable to l-arabinose and that this phenotype can be overcome by supplying the AraE l-arabinose permease. Altogether, our data show that at least in B. cepacia complex strains of genomovars I, II, III, IV, V, and V, eGFP is efficiently expressed in both of our constitutive and inducible promoter expression vectors.
In vivo expression of eGFP in intracellular B. cepacia.
Previous studies in our laboratory have shown that certain strains of the B. cepacia complex can survive within amoebae for extended periods of time (31). To test the functionality of our vectors in vivo and to confirm the expression of eGFP as a reporter for intracellular B. cepacia, A. polyphaga cells were infected with strains CEP040 and C5424 harboring pMLBAD-eGFP or pMLS7-eGFP. Amoebae were observed by light and fluorescence microscopy at 6, 12, and 24 h postinfection. The results show that fluorescent bacteria were clearly visible within amoeba cells (Fig. 4E to F). In experiments conducted using the regulatable vectors, the maintenance of arabinose in the minimal salt Acanthamoeba buffer at a concentration of 1% enhanced the level of bacterial fluorescence. It was also found that the substitution of glucose in the axenic medium with glycerol improved the intensity of the fluorescent bacteria. Therefore, the constitutive and inducible expression of eGFP by these plasmids will provide a valuable tool for investigating the characteristics of intracellular infection of amoebae and other host cells by B. cepacia complex strains.
Conclusions.
The results from this study demonstrate the usefulness of the constitutive and inducible expression vectors for gene expression in B. cepacia complex strains. We have also established that the arabinose-inducible promoter functions appropriately in the Burkholderia background and is fully repressed in the presence of glucose. In addition, we have shown a robust expression of eGFP under the control of the constitutive and inducible promoters in the vectors constructed in this study. We conclude that these vectors will function as suitable genetic complementation systems for characterizing gene knockout mutants in B. cepacia, especially those which display defects in tolerance to various stress conditions, including lack of intracellular survival within host cells as well as poor, or negligible, survival in plant and animal models of infection.
Acknowledgments
We thank the colleagues mentioned or referenced in Table 1 for providing strains and plasmids used in this study. We also thank Cristina L. Marolda for technical help, G. Lazarovits for help with the spectrofluorometric determinations, and D. E. Heinrichs and T. A. Hunt for critical reading of the manuscript.
The Prokaryotic Microscopy Facility was supported by grants from the Academic Development Fund, University of Western Ontario, and the Canadian Institutes of Health Research. M.D.L. was supported by a Doctoral Research Award from the Canadian Institutes of Health Research. This study was supported by operating grants from the Canadian Institutes of Health Research and the Canadian Cystic Fibrosis Foundation. M.A.V. holds a Canada Research Chair in Infectious Diseases and Microbial Pathogenesis.
REFERENCES
- 1.Abe, M., M. Tsuda, M. Kimoto, S. Inouye, A. Nakazawa, and T. Nakazawa. 1996. A genetic analysis system of Burkholderia cepacia: construction of mobilizable transposons and a cloning vector. Gene 174:191-194. [DOI] [PubMed] [Google Scholar]
- 2.Amer, A. O., and M. A. Valvano. 2002. Conserved aspartic acids are essential for the enzymatic activity of the WecA protein initiating the biosynthesis of O-specific lipopolysaccharide and enterobacterial common antigen in Escherichia coli. Microbiology 148:571-582. [DOI] [PubMed] [Google Scholar]
- 3.Antoine, R., and C. Locht. 1992. Isolation and molecular characterization of a novel broad-host-range plasmid from Bordetella bronchiseptica with sequence similarities to plasmids from gram-positive organisms. Mol. Microbiol. 6:1785-1799. [DOI] [PubMed] [Google Scholar]
- 4.Bevivino, A., C. Dalmastri, S. Tabacchioni, L. Chiarini, M. L. Belli, S. Piana, A. Materazzo, P. Vandamme, and G. Manno. 2002. Burkholderia cepacia complex bacteria from clinical and environmental sources in Italy: genomovar status and distribution of traits related to virulence and transmissibility. J. Clin. Microbiol. 40:846-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bevivino, A., S. Sarrocco, C. Dalmastri, S. Tabacchioni, C. Cantale, and L. Chiarini. 1998. Characterization of a free-living maize-rhizosphere population of Burkholderia cepacia: effect of seed treatment on disease suppression and growth promotion in maize. FEMS Microbiol. Ecol. 27:225-237. [Google Scholar]
- 6.Bloemberg, G. V., G. A. O'Toole, B. J. J. Lugtenberg, and R. Kolter. 1997. Green fluorescent protein as a marker for Pseudomonas spp. Appl. Environ. Microbiol. 63:4543-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burkholder, W. H. 1950. Sour skin, a bacterial rot of onion bulbs. Phytopathology 40:115-117. [Google Scholar]
- 8.Burns, J. L., M. Jonas, E. Y. Chi, D. K. Clark, A. Berger, and A. Griffith. 1996. Invasion of respiratory epithelial cells by Burkholderia (Pseudomonas) cepacia. Infect. Immun. 64:4054-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coenye, T., P. Vandamme, J. R. W. Govan, and J. J. LiPuma. 2001. Taxonomy and identification of the Burkholderia cepacia complex. J. Clin. Microbiol. 39:3427-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen, S. N., A. C. Chang, and L. Hsu. 1972. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc. Natl. Acad. Sci. USA 69:2110-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 12.Craig, F. F., J. G. Coote, R. Parton, J. H. Freer, and N. J. Gilmour. 1989. A plasmid which can be transferred between Escherichia coli and Pasteurella haemolytica by electroporation and conjugation. J. Gen. Microbiol. 135:2885-2890. [DOI] [PubMed] [Google Scholar]
- 13.Dennis, J. J., and G. J. Zylstra. 1998. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Appl. Environ. Microbiol. 64:2710-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elzer, P. H., M. E. Kovach, R. W. Phillips, G. T. Robertson, K. M. Peterson, and R. M. Roop II. 1995. In vivo and in vitro stability of the broad-host-range cloning vector pBBR1MCS in six Brucella species. Plasmid 33:51-57. [DOI] [PubMed] [Google Scholar]
- 15.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Folsom, B. R., P. J. Chapman, and P. H. Pritchard. 1990. Phenol and trichloroethylene degradation by Pseudomonas cepacia G4: kinetics and interactions between substrates. Appl. Environ. Microbiol. 56:1279-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Govan, J. R., J. E. Hughes, and P. Vandamme. 1996. Burkholderia cepacia: Medical, taxonomic and ecological issues. J. Med. Microbiol. 45:395-407. [DOI] [PubMed] [Google Scholar]
- 18.Govan, J. R. W., and V. Deretic. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Govan, J. R. W., and P. Vandamme. 1998. Agricultural and medical microbiology: a time for bridging gaps. Microbiology 144:2373-2375. [DOI] [PubMed] [Google Scholar]
- 20.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haas, J., E. C. Park, and B. Seed. 1996. Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr. Biol. 6:315-324. [DOI] [PubMed] [Google Scholar]
- 22.Hebbar, K. P., M. H. Martel, and T. Heulin. 1998. Suppression of pre- and postemergence damping-off in corn by Burkholderia cepacia. Eur. J. Plant Pathol. 104:29-36. [Google Scholar]
- 23.Holmes, A., J. Govan, and R. Goldstein. 1998. Agricultural use of Burkholderia (Pseudomonas) cepacia: a threat to human health? Emerg. Infect. Dis. 4:221-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Isles, A., I. Maclusky, M. Corey, R. Gold, C. Prober, P. Fleming, and H. Levison. 1984. Pseudomonas cepacia infection in cystic fibrosis: an emerging problem. J. Pediatr. 104:206-210. [DOI] [PubMed] [Google Scholar]
- 25.Jayaswal, R. K., M. Fernandez, R. S. Upadhyay, L. Visintin, M. Kurz, J. Webb, and K. Rinehart. 1993. Antagonism of Pseudomonas cepacia against phytopathogenic fungi. Curr. Microbiol. 26:17-22. [DOI] [PubMed] [Google Scholar]
- 26.Kang, Y., R. Carlson, W. Tharpe, and M. A. Schell. 1998. Characterization of genes involved in biosynthesis of a novel antibiotic from Burkholderia cepacia BC11 and their role in biological control of Rhizoctonia solani. Appl. Environ. Microbiol. 64:3939-3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kilbane, J. J., D. K. Chatterjee, and A. M. Chakrabarty. 1983. Detoxification of 2,4,5-trichlorophenoxyacetic acid from contaminated soil by Pseudomonas cepacia. Appl. Environ. Microbiol. 45:1697-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.King, E., and J. Parke. 1996. Population density of the biocontrol agent Burkholderia cepacia AMMDR1 on four pea cultivars. Soil Biol. Biochem. 28:306-312. [Google Scholar]
- 29.Knauf, V. C., and E. W. Nester. 1982. Wide host range cloning vectors: a cosmid clone bank of an Agrobacterium Ti plasmid. Plasmid 8:45-54. [DOI] [PubMed] [Google Scholar]
- 30.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. Roop, and K. M. Peterson. 1994. pBBR1MCS: a broad-host-range cloning vector. BioTechniques 16:800-802. [PubMed] [Google Scholar]
- 31.Marolda, C. L., B. Hauröder, M. A. John, R. Michel, and M. A. Valvano. 1999. Intracellular survival and saprophytic growth of isolates from the Burkholderia cepacia complex in free-living amoebae. Microbiology 145:1509-1517. [DOI] [PubMed] [Google Scholar]
- 32.Martin, D. W., and C. D. Mohr. 2000. Invasion and intracellular survival of Burkholderia cepacia. Infect. Immun. 68:24-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morales, V. M., A. Backman, and M. Bagdasarian. 1991. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97:39-47. [DOI] [PubMed] [Google Scholar]
- 34.Nzula, S., P. Vandamme, and J. R. W. Govan. 2002. Influence of taxonomic status on the in vitro antimicrobial susceptibility of the Burkholderia cepacia complex. J. Antimicrob. Chemother. 50:265-269. [DOI] [PubMed] [Google Scholar]
- 35.Page, F. C. 1976. An illustrated key to freshwater and soil amoebae. The Ferry House, Ambleside Cumbria, United Kingdom.
- 36.Saini, L., S. Galsworthy, M. John, and M. A. Valvano. 1999. Intracellular survival of Burkholderia cepacia complex isolates in the presence of macrophage cell activation. Microbiology 145:3465-3475. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1990. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 38.Santos, P. M., I. Di Bartolo, J. M. Blatny, E. Zennaro, and S. Valla. 2001. New broad-host-range promoter probe vectors based on the plasmid RK2 replicon. FEMS Microbiol. Lett. 195:91-96. [DOI] [PubMed] [Google Scholar]
- 39.Sukchawalit, R., P. Vattanaviboon, R. Sallabhan, and S. Mongkolsuk. 1999. Construction and characterization of regulated L-arabinose-inducible broad host range expression vectors in Xanthomonas. FEMS Microbiol. Lett. 181:217-223. [DOI] [PubMed] [Google Scholar]
- 40.Sun, J., G. H. Kelemen, J. M. Fernandez-Abalos, and M. J. Bibb. 1999. Green fluorescent protein as a reporter for spatial and temporal gene expression in Streptomyces coelicolor A3(2). Microbiology 145:2221-2227. [DOI] [PubMed] [Google Scholar]
- 41.Vandamme, P., B. Holmes, M. Vancanneyt, T. Coenye, B. Hoste, R. Coopman, H. Hevets, S. Lauwers, M. Gillis, K. Kersters, and J. R. W. Govan. 1997. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int. J. Syst. Bacteriol. 47:1188-1200. [DOI] [PubMed] [Google Scholar]