Abstract
A quantitative assay based on high-performance liquid chromatography analysis of bile salts and bacterial protein determination was established for investigating bile salt hydrolase (BSH) activity in bacteria isolated from the small intestine of chickens. Bacteria were isolated using various media and were subsequently grouped according to cell morphology, fermentation profile, and 16S ribosomal DNA sequence. Representative isolates from each bacterial group were assayed for BSH activity. The isolates differed in BSH activity with respect to the state of growth and preculturing with and without taurochenodeoxycholate. The highest levels of BSH activity were found with Enterococcus faecium and Clostridium perfringens.
Several gram-positive bacteria inhabiting the small intestine of chickens are capable of hydrolyzing the amide bond of conjugated bile salts, liberating free bile salts with markedly lower detergent properties in the emulsification of fat (4, 12). The main target for antibiotic growth promoters in broiler feed is the gram-positive microflora, and a reduction of the bile salt hydrolase (BSH)-producing microorganisms is one of the suggested mechanisms by which antibiotics elicit their beneficial effect (8). However, little is known about the extent of BSH activity expressed by individual bacteria colonizing the chicken gut.
An in vitro method, based on high-performance liquid chromatography (HPLC) analysis of bile salts and bacterial protein determination, was established for quantifying BSH activity in bacteria isolated from the small intestine of chickens.
Animals and sampling.
Ten male broiler chickens (Ross 208, Fællesrugeriet DK-8900 Randers), reared and fed according to standard recommendations for Danish broiler production, were randomly selected at the age of 35 days from the production plant of the Danish Institute of Agricultural Sciences (Research Centre Foulum, DK-8830 Tjele). The broilers were killed by cervical dislocation, the small intestine was excised, and the intestinal content was collected and pooled for bacteriological analysis. The experiment complied with the guidelines of the Danish Ministry of Justice with respect to animal experimentation and care of animals under study.
Isolation of bacteria from small intestine content.
Isolation of total anaerobes (CFE agar), lactic acid bacteria (MRS agar), lactobacilli (ROG agar), enterococci (SLA agar), and Clostridium perfringens (TSC agar) from small intestinal contents was performed as described by Engberg et al. (7). The selective medium used for isolation of bifidobacteria was MW agar (24). An average of 20 colonies were picked randomly from the highest dilution of each medium and transferred aseptically and under anaerobic conditions (10:10:80; CO2-H2-N2 atmosphere) into Hungate tubes containing reinforced clostridial bouillon (MERCK 5411) with an added 0.005 g of hemin liter−1. After incubation of the cells overnight at 37°C, the cell morphology was investigated by phase-contrast microscopy. Glycerol (20%) was added to the remaining overnight cultures before storage at −80°C.
Fermentation test of the isolates.
The isolates were subcultured (0.2 ml of inoculum of an overnight culture) in anaerobic (N2 atmosphere) roll tubes containing 9 ml of the prereduced sterilized peptone yeast glucose medium described by Holdeman et al. (14). After incubation at 37°C for 48 h, the concentrations of fermentation products in terms of organic acids (16) and gas (15) were measured by gas chromatography.
DNA extraction and PCR amplification.
The nucleic acid extraction from isolates cultured overnight at 37°C in reinforced clostridial bouillon (MERCK 5411) with an added 0.005 g of hemin, and the subsequent PCR amplification of 16S ribosomal DNA (rDNA), were performed as described by Knarreborg (18).
Sequencing of 16S rDNA.
The 16S rDNA nucleotide sequences of all isolates were sequenced at the 3′-terminal end of the molecule using a single primer as described by Leser et al. (19). This partial determination provided sequences of approximately 530 bp, which together with the phenotypic characterization of the isolates were used for provisional grouping of the isolates. Based on the grouping, representative isolates were selected and subjected to near-full-length 16S rDNA sequencing according to the procedure outlined by Leser et al. (19). To determine the closest relatives of the partial and near-full-length 16S rDNA sequences retrieved, searches were conducted in GenBank using the BLAST algorithm (1).
Nucleotide sequence accession numbers.
The near-full-length sequences of the representative isolates AK21, AK51, AK61, AK68, AK89, and AK113 have been deposited in GenBank under accession numbers AY098491, AY098488, AY098492, AY098489, AY098486, and AY098490, respectively.
BSH assay: isolates, growth conditions, and sampling.
The representative isolates were tested quantitatively for their BSH activity. Escherichia coli strain AK108 (GenBank accession number AY098487), previously isolated in our lab from the chicken gut and characterized according to the procedure described above, was used as a negative control in the assay for BSH activity (4, 12). Overnight cultures of the isolates were prepared in appropriate media using MRS broth (MERCK 0661) for culturing isolates identified as Lactobacillus strains and using reinforced clostridial broth (MERCK 5411) with an added 0.005 g of hemin liter−1 for the remaining isolates. Taurochenodeoxycholate (TCDC), which is the major bile salt present in avian bile, was used in the BSH assay and was purchased from Calbiochem (Darmstadt, Germany). Batches (50 ml each) containing the appropriate culture medium without TCDC and with addition of 2 mM TCDC were prepared anaerobically (N2 atmosphere) in 125-ml sterile serum bottles with butyl rubber stoppers. In a pilot study, we found that autoclaving did not affect the concentration of TCDC; hence, bile salt was added and the pH was adjusted to 6.8 prior to autoclaving. Inoculates from each overnight culture of the representative isolates (2% [vol/vol]) were transferred aseptically into the two culture media and incubated for 24 h in a shaking water bath at 39°C. Aliquots of samples (1.0 ml) from each culture medium were removed with sterile injection syringes at 0, 2, 4, 6, 8, and 24 h for measurement of pH and growth and analysis of BSH activity. In addition, a sample (100 μl) was collected from the culture medium containing TCDC for determination of the bile salt concentration. Corresponding volumes were removed and discarded from the culture medium without TCDC. Immediately after collection, the samples for HPLC analysis of bile salt concentration were diluted 50-fold in an extraction mixture containing 20% acetonitrile (super gradient; LAB-SCAN, Dublin, Ireland), 70% H2O, and 10% NaOH, where ursodeoxycholate (Sigma, St. Louis, Mo.) was added as an internal standard to a final concentration of 40 μM. Samples (1.0 ml each) for determination of growth were centrifuged at 5,000 × g for 10 min, and the pellet stored at −20°C until analysis for bacterial protein. To assay BSH activity in the isolates at different times during the 24-h growth period, samples (1.0 ml each) from the two culture media were transferred into 25-ml sterile serum bottles containing 9 ml of anaerobe (N2 atmosphere) TCDC-phosphate buffer (0.1 M NaH2PO4, 2 mM TCDC [pH 6.5]). This suspension was incubated for 2 h at 39°C in a shaking water bath. Samples (100 μl) for bile salt determination by HPLC were removed at 0, 1, and 2 h and immediately transferred to the extraction mixture as described above. An unaltered pH of the buffer measured at the end of the 2-h incubation period confirmed that a constant cell concentration was maintained during incubation.
Measurement of bacterial protein.
Bacterial growth was assessed by measuring the concentrations of protein in the two culture media according to the procedure described by Knarreborg (18).
HPLC analysis of conjugated and free bile salts.
Conjugated and free bile salts were quantified by reversed-phase HPLC with pulsed amperometric detection as outlined by Dekker et al. (6). The preextracted samples were mixed using a Shaker VXR vibrax (IKA-Werke, Staufen, Germany) at 1,500 rpm for 30 s and subsequently centrifuged at 5,000 × g for 10 min. The supernatant (1.0 ml) was passed through a 0.20-μm-pore-size nylon syringe filter membrane (Cameo 17N-DDR02T17NB) prior to injection onto the HPLC. The chromatographic conditions used are reported by Knarreborg (18).
Calculation of BSH activity.
Calculation of BSH activity was based on the release of chenodeoxycholate (CDC) from the bacterial hydrolysis of the amide bond of TCDC. The free bile salt CDC was released linearly during the 2-h incubation period (data not shown). The slope of the linear regression was related to the parallel determination of bacterial protein, and the BSH activity was expressed as nanomoles of CDC produced per microgram of bacterial protein per hour.
BSH activity in isolates from the small intestine of chickens.
A total of 105 isolates were obtained from the small intestinal contents of 35-day-old chickens, and the distribution of recovery from the different media is given together with the viable counts in Table 1. Provisional grouping, based on phenotypic (morphology and fermentation profile) and genotypic (partial sequencing of 16S rDNA) characterization, yielded six groups belonging to the genera Lactobacillus (L. salivarius and L. aviarius), Enterococcus (E. faecalis and E. faecium), Clostridium (C. perfringens), and Streptococcus (S. alactolyticus). Consistently, previous studies have found a relatively low level of microbial diversity in the small intestine of chickens, mainly comprising lactobacilli, enterococci, streptococci, C. perfringens, and enterobacteria (2, 7, 25, 26). From each group, representative isolates were subjected to near-full-length 16S rDNA sequence analysis (Table 1) and were tested for BSH activity (Fig. 1). Growth curves and BSH activities measured for the two lactobacillus isolates (L. salivarius AK21 and L. aviarius AK113) were similar, and hence, only results for L. salivarius AK21 are presented in Fig. 1. A decrease in pH from approximately 6.8 to 5.0 was registered in cultures of isolates grown in the absence and presence of TCDC as a result of organic acids produced during the 24-h incubation period (data not shown).
TABLE 1.
Grouping, based on partial 16S rDNA sequence analysis, cell morphology, and product profiles from glucose fermentation, of isolates from small intestine content of 35-day-old chickens
| Identity by 16S rDNA | No. of isolates recovered fromd:
|
Morphology | Production of organic acidse
|
Amt of gas (mM)
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Isolatea | % Similarb | No. of bpc | CFE | MRS | ROG | SLA | TSC | MW | Total concn (mM) | Molar ratio (%) of:
|
||||||||
| F | A | P | B | L | S | H2 | CO2 | |||||||||||
| L. aviarius AK113 | 98 | 1,091 | 6 | 4 | Rod | 43.0 | 0.2 | 3.6 | 0.0 | 0.0 | 96.0 | 0.2 | 0.0 | 18.3 | ||||
| L. salivarius AK21 | 98 | 1,354 | 2 | 11 | 6 | Rod | 43.3 | 1.8 | 2.7 | 0.0 | 0.0 | 95.0 | 0.5 | 0.0 | 20.3 | |||
| E. faecalis AK89 | 99 | 943 | 5 | Coccus | 49.6 | 7.0 | 8.7 | 0.2 | 0.0 | 84.0 | 0.1 | 0.0 | 7.0 | |||||
| E. faecium AK61 | 99 | 1,349 | 10 | 8 | 12 | 1 | 3 | Coccus | 44.8 | 1.6 | 1.8 | 0.0 | 0.0 | 96.6 | 0.0 | 0.0 | 11.5 | |
| C. perfring. AK51 | 98 | 1,065 | 16 | 16 | Rod | 33.6 | 0.0 | 54.7 | 5.1 | 25.7 | 11.8 | 2.7 | 61.3 | 52.1 | ||||
| S. alactolyt. AK68 | 99 | 1,334 | 4 | 1 | Coccus | 32.7 | 3.2 | 0.6 | 0.2 | 0.0 | 95.5 | 0.5 | 0.0 | 10.6 | ||||
Representative isolates were identified by near-full-length 16S rDNA sequence analysis. Abbreviations: C. perfring., C. perfringens; S. alactolyt., S. alactolyticus.
Percentage sequence similarity obtained by comparison to database sequences in GenBank.
Length of the near-full-length 16S rDNA sequence.
Number of pure isolates recovered from each medium. Total log10 CFU of bacteria/g of medium were as follows: from CFE, 8.52; from MRS, 7.86; from ROG, 7.07; from SLA, 6.95; from TSC, 5.38; and from MW, 4.88.
F, formic acid; A, acetic acid; P, propionic acid; B, butyric acid; L, lactic acid; S, succinic acid.
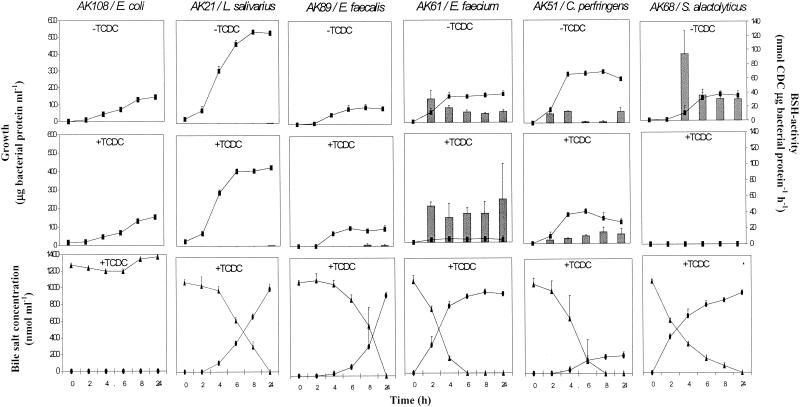
FIG. 1.
Growth (▪) of the representative isolates and the BSH-negative control (E. coli AK108) in culture medium without TCDC (−TCDC) and in culture medium with addition of TCDC (+TCDC). The BSH activity (bars) for isolates was determined at intervals during the 24-h growth period. The concentrations of TCDC (▴) and CDC (•) were determined during growth in the TCDC-supplemented culture medium. Values for the representative isolates are expressed as means ± standard deviations (n = 3).
The negative control, E. coli AK108, showed no BSH activity, as demonstrated by the lack of CDC production during 2-h incubations of cells preincubated in the two culture media. In addition, no hydrolysis of TCDC was detected in the TCDC-supplemented medium during the 24-h incubation period (Fig. 1). As shown by the bottom plots in Fig. 1, all of the representative isolates were capable of hydrolyzing TCDC, where the total amount of TCDC was completely transformed within the 24-h incubation period. BSH has been demonstrated to be synthesized constitutively in the majority of bacteria (11, 21, 22). Accordingly, this study showed that BSH activity in C. perfringens AK51, E. faecium AK61, and S. alactolyticus AK68 was expressed independently of whether or not the cells were precultured with bile salts. Although at very low levels, BSH activities were also detected in cells of the two Lactobacilli isolates (L. salivarius AK21 and L. aviarius AK113) from both preincubation conditions. In contrast, cells of E. faecalis AK89 precultured in the absence of TCDC showed no activity, whereas cells preincubated with TCDC expressed low BSH activity. This suggested that BSH activity in E. faecalis AK89 was inducible by TCDC. Whether BSH is synthesized constitutively or not, the enzyme must be considered as activated in the small intestine, where the bacteria are constantly exposed to bile salts.
BSH activity was detected only in stationary-phase cells of the lactobacillus isolates (L. salivarius AK21 and L. aviarius AK113) and E. faecalis AK89. This has been demonstrated previously in Lactobacillus johnsonii (21) and Lactobacillus reuteri (28). In contrast, E. faecium AK61, C. perfringens AK51, and S. alactolyticus AK68 expressed BSH activities throughout the growth period. A higher level of BSH activity was found in E. faecium AK61 preincubated with TCDC, suggesting that some BSH-enhancing factors were involved (21). The BSH activity in cells of C. perfringens AK51 that were precultured with TCDC followed the course of growth, whereas BSH activity in cells preincubated in the absence of TCDC fluctuated and showed no clear relation to the state of growth. The BSH enzyme is located mainly intracellularly in C. perfringens (10), as reported for the majority of bacteria (3, 11, 21); however, extracellular BSH activity has also been demonstrated for C. perfringens (17, 22). Masuda (22) showed that the extracellular BSH activity in C. perfringens gradually increased during growth, which is comparable to the present observations on BSH activities in cells of C. perfringens AK51 preincubated in the presence of TCDC. In addition, studies have provided evidence for multiple BSH activities for C. perfringens (5, 23). Hence, it is likely that different hydrolase systems were activated in response to growth and growth conditions. The total amount of bile salts in the TCDC-containing medium inoculated with C. perfringens AK51 decreased during the 24-h incubation period. This was not due to precipitation of bile salts, as verified by the negative control and by the cultures of the remaining isolates, where CDC was formed in stoichiometric amounts compared with TCDC disappearance in the solution during the 24-h incubation period. Further, a disappearance of total bile salts was not noted when the BSH activities were determined. C. perfringens bacteria possess several bile salt transformation activities other than deconjugation, including 7-dehydroxylation of primary bile salts into secondary bile salts and further conversion of secondary bile salts into 3-keto bile salts (13, 22, 29). Hence, it is likely that growing cells of C. perfringens AK51 further transformed CDC into a compound which was undetectable by the applied HPLC analysis; however, investigating the fate of CDC was beyond the scope of this study.
S. alactolyticus AK68 that was preincubated in the absence of TCDC exhibited a high level of BSH activity. However, growth of S. alactolyticus AK68 was completely inhibited at the physiological concentration of bile salts, and consequently no BSH activity was detected when cells were preincubated in the TCDC-containing medium. Still, deconjugation did occur, as shown by the increase in CDC in the culture medium. An explanation could be that the inoculum alone, which was at too low a level to monitor by measurement of protein, was responsible for the rapid formation of CDC in the culture medium. The strong bactericidal effect on S. alactolyticus AK68 and the growth-inhibiting effect on the remaining isolates when they were cultured in the TCDC-containing medium were very likely due to accumulation of the more toxic compound CDC as a result of deconjugation (27). Hence, a reverse relationship was found between growth and BSH activity. Accordingly, growth of the BSH-negative E. coli AK108 was not affected by TCDC.
Thus, it was demonstrated that E. faecium AK61 and C. perfringens AK51 expressed high levels of BSH activity at the physiological concentration of bile salts and, although showing depressed growth, were tolerant to the presence of CDC. Even though contributions from bacteria other than the ones isolated cannot be excluded, E. faecium and C. perfringens were found to be dominant among common bacteria isolated from the small intestine of chickens with respect to BSH activity. Of these two species, E. faecium is considered to be the major contributor to BSH activity, since it accounted for an essential part of the indigenous microflora in the small intestine of chickens (Table 1). Hence, this study provided evidence in favor of previous theories suggesting that E. faecium (9) and C. perfringens (20, 26) are responsible for depression of growth in chickens.
Acknowledgments
We thank Trine Poulsen and Mona Dinesen for excellent help with collection of data and Karin Durup for excellent technical assistance with the sequence analysis. We address special thanks to Thomas Rebsdorf for his skillful technical assistance with the HPLC analysis.
We are grateful to the Danish Ministry of Food, Agriculture and Fisheries for financial support.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Barnes, E. M., G. C. Mead, and D. A. Barnum. 1972. The intestinal flora of the chicken in the period 2 to 6 weeks of age, with particular reference to the anaerobic bacteria. Br. Poult. Sci. 13:311-326. [DOI] [PubMed] [Google Scholar]
- 3.Christiaens, H., and W. Verstraete. 1992. Conjugated bile acid hydrolysis by intestinal bacteria. A mini-review of the literature. Med. Fac. Landbouww. Univ. Gent. 57/4b:1973-1979. [Google Scholar]
- 4.Cole, C. B., and R. Fuller. 1984. Bile acid deconjugation and attachment of chicken gut bacteria: their possible role in growth depression. Br. Poult. Sci. 25:227-231. [DOI] [PubMed] [Google Scholar]
- 5.Coleman, J. P., and L. L. Hudson. 1995. Cloning and characterisation of a conjugated bile acid hydrolase gene from Clostridium perfringens. Appl. Environ. Microbiol. 61:2514-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dekker, R., R. van der Meer, and C. Olieman. 1991. Sensitive pulsed amperometric detection of free and conjugated bile acids in combination with gradient reversed-phase HPLC. Chromatography 31:549-553. [Google Scholar]
- 7.Engberg, R. M., M. S. Hedemann, T. D. Leser, and B. B. Jensen. 2000. Effect of zinc bacitracin and salinomycin on intestinal microflora and performance of broilers. Poult. Sci. 79:1311-1319. [DOI] [PubMed] [Google Scholar]
- 8.Feighner, S. D., and M. P. Dashkevicz. 1987. Subtherapeutic levels of antibiotics in poultry feeds and their effects on weight gain, feed efficiency, and bacterial cholyltaurine hydrolase activity. Appl. Environ. Microbiol. 53:331-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuller, R., C. B. Cole, and M. E. Coates. 1984. The role of Streptococcus faecium in antibiotic-relieved growth depression of chickens, p. 395-403. In M. Woodbine, (ed.), Antimicrobials and agriculture. Buttenwoods, London, United Kingdom.
- 10.Gopal-Srivastava, R., and P. B. Hylemon. 1988. Purification and characterization of bile salt hydrolase from Clostridium perfringens. J. Lipid Res. 29:1079-1085. [PubMed] [Google Scholar]
- 11.Grill, J. P., F. Schneider, J. Crociani, and J. Ballongue. 1995. Purification and characterization of conjugated bile salt hydrolase from Bifidobacterium longum BB536. Appl. Environ. Microbiol. 61:2577-2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hill, M. J. 1995. Bacteria and fat digestion, p. 131-142. In M. J. Hill (ed.), Role of gut bacteria in human toxicology and pharmacology. Taylor and Francis Inc., London, United Kingdom.
- 13.Hirano, S., N. Masuda, H. Oda, and H. Mukai. 1981. Transformation of bile acids by Clostridium perfringens. Appl. Environ. Microbiol. 42:394-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holdeman, K. A. L., E. P. Cato, and E. C. Moore. 1977. Anaerobe laboratory manual. Virginia Polytechnic Institute and State University, Blacksburg, Va.
- 15.Jensen, B. B., and H. Jørgensen. 1994. Effect of dietary fiber on microbial activity and microbial gas production in various regions of the gastrointestinal tract of pigs. Appl. Environ. Microbiol. 60:1897-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jensen, M. T., R. P. Cox, and B. B. Jensen. 1995. Microbial production of skatole in the hind gut of pigs given different diets and its relation to skatole deposition in backfat. Anim. Sci. 61:293-304. [Google Scholar]
- 17.Kishinaka, M., A. Umeda, and S. Kuroki. 1994. High concentrations of conjugated bile acids inhibit bacterial growth of Clostridium perfringens and induce its extracellular cholylglycine hydrolase. Steroids 59:485-489. [DOI] [PubMed] [Google Scholar]
- 18.Knarreborg, A. 2002. The impact of microbial deconjugation of bile salts on fat digestion in broiler chickens. Ph.D. thesis. ISBN87-88976-58-0. The Royal Veterinary and Agricultural University, Frederiksberg, Denmark.
- 19.Leser, T. D., J. Z. Amenuvor, T. K. Jensen, R. H. Lindecrona, M. Boye, and K. Møller. 2002. Culture-independent analysis of gut bacteria: the pig gastrointestinal tract microbiota revisited. Appl. Environ. Microbiol. 68:673-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lev, M., and M. Forbes. 1959. Growth response to dietary penicillin of germ-free chicks and chicks with a defined intestinal flora. Br. J. Nutr. 13:78-84. [DOI] [PubMed] [Google Scholar]
- 21.Lundeen, S. G., and D. C. Savage. 1990. Characterization and purification of bile salt hydrolase from Lactobacillus sp. strain 100-100. J. Bacteriol. 172:4171-4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masuda, N. 1981. Deconjugation of bile salts by Bacteroides and Clostridium. Microbiol. Immunol. 25:1-11. [DOI] [PubMed] [Google Scholar]
- 23.Nair, P. P., M. Gordon, S. Gordon, J. Reback, and A. I. Mendeloff. 1965. The cleavage of bile acid conjugates by cell-free extracts from Clostridium perfringens. Life Sci. 4:1887-1892. [DOI] [PubMed] [Google Scholar]
- 24.Rada, V., K. Sirotek, and J. Petr. 1999. Evaluation of selective media for bifidobacteria in poultry and rabbit caecal samples. J. Vet. Med. B 46:369-373. [DOI] [PubMed] [Google Scholar]
- 25.Salanitro. J. P., I. G. Blake, P. A. Muirhead, M. Maglio, and J. R. Goodman. 1978. Bacteria isolated from the duodenum, ileum, and cecum of young chicks. Appl. Environ. Microbiol. 35:782-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stutz, M. W., S. L. Jonhson, and F. R. Judith. 1983. Effects of diet and bacitracin on growth, feed efficiency, and populations of Clostridium perfringens in the intestine of broiler chicks. Poult. Sci. 62:1619-1625. [DOI] [PubMed] [Google Scholar]
- 27.Tannock, G. W., J. M. Bateup, and H. F. Jenkinson. 1997. Effect of sodium taurocholate on the in vitro growth of lactobacilli. Microb. Ecol. 33:163-167. [DOI] [PubMed] [Google Scholar]
- 28.Taranto, M. P., F. Sesma, and G. F. Valdez. 1999. Localization and primary characterisation of bile salt hydrolase activity from Lactobacillus reuteri. Biotechnol. Lett. 21:935-938. [Google Scholar]
- 29.Wells, J. E., and P. B. Hylemon. 2000. Identification and characterization of a bile acid 7α-dehydroxylation operon in Clostridium sp. strain TO-931, a highly active 7α-dehydroxylating strain isolated from human feces. Appl. Environ. Microbiol. 66:1107-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]