Abstract
Obligate bacterial endosymbionts of paramecia able to form refractile inclusion bodies (R bodies), thereby conferring a killer trait upon their ciliate hosts, have traditionally been grouped into the genus Caedibacter. Of the six species described to date, only the Paramecium caudatum symbiont Caedibacter caryophilus has been phylogenetically characterized by its 16S rRNA gene sequence, and it was found to be a member of the Alphaproteobacteria related to the Rickettsiales. In this study, the Caedibacter taeniospiralis type strain, an R-body-producing cytoplasmatic symbiont of Paramecium tetraurelia strain 51k, was investigated by comparative 16S rRNA sequence analysis and fluorescence in situ hybridization with specific oligonucleotide probes. C. taeniospiralis is not closely related to C. caryophilus (80% 16S rRNA sequence similarity) but forms a novel evolutionary lineage within the Gammaproteobacteria with the family Francisellaceae as a sister group (87% 16S rRNA sequence similarity). These findings demonstrate that the genus Caedibacter is polyphyletic and comprises at least two phylogenetically different bacterial species belonging to two different classes of the Proteobacteria. Comparative phylogenetic analysis of C. caryophilus, five closely related Acanthamoeba endosymbionts (including one previously uncharacterized amoebal symbiont identified in this study), and their hosts suggests that the progenitor of the alphaproteobacterial C. caryophilus lived within acanthamoebae prior to the infection of paramecia.
The ability of certain paramecia to kill other paramecia was first described in 1938 by Sonneborn, who observed that sensitive paramecia exhibit distinct morphological symptoms upon ingestion of toxic “particles” released by the killer strain and ultimately die (52). In 1958, electron microscopy studies revealed that these heritable cytoplasmic particles are gram-negative rod-shaped prokaryotes (16) which eluded cultivation using standard laboratory media (30, 32) and that the toxic effect is associated with proteinaceous, refractile inclusion bodies (R bodies) found inside the bacterial endosymbionts (30, 32). Subsequently, all R-body-producing obligate intracellular symbionts of paramecia were combined into the genus Caedibacter and classified according to morphological, functional, and phenotypic properties (17, 32, 35). Within the genus Caedibacter, the six species Caedibacter caryophilus, Caedibacter varicaedens, Caedibacter taeniospiralis, Caedibacter pseudomutans, Caedibacter paraconjugatus, and Caedibacter macronucleorum (11, 38, 47) have been recognized. C. caryophilus, the only member of the genus Caedibacter that has been characterized by its 16S rRNA gene sequence, was found to be affiliated with the Alphaproteobacteria (57). Within the Alphaproteobacteria, C. caryophilus clusters together with obligate endosymbionts of acanthamoebae (6, 20) and the paramecium endosymbionts Holospora obtusa and Holospora elegans (3, 14).
In this study, the phylogenetic affiliation of the Paramecium tetraurelia symbiont C. taeniospiralis strain 51k (initially described by Beale et al. [5], named by Preer et al. [32], and validated by Preer and Preer [34]) was investigated by the full-cycle rRNA approach, including comparative 16S rRNA gene sequence analysis and detection of endosymbionts within their host cells by fluorescence in situ hybridization (FISH) using specific 16S rRNA-targeted oligonucleotide probes. These analyses showed that despite several consistent morphological and functional features of its members, the genus Caedibacter contains at least two only distantly related bacteria which are affiliated with different classes of the Proteobacteria. In addition, a previously uncharacterized Acanthamoeba endosymbiont was included in this study, as it was found to hybridize with a C. caryophilus-specific oligonucleotide probe, indicating close phylogenetic relatedness. Comparative analysis of the phylogeny of these alphaproteobacterial symbionts and their hosts suggested that the progenitor of C. caryophilus lived and coevolved within acanthamoebae prior to the infection of paramecia.
MATERIALS AND METHODS
Isolation and maintenance of protozoa.
P. tetraurelia strain 51k containing endosymbiotic C. taeniospiralis was isolated previously in Spencer, Ind. (ATCC 30632). The morphology and fine structure of the symbiont, at that time designated the kappa particle, was described by Beale et al. (5), and the symbiont was later classified as the type strain of C. taeniospiralis (17). The paramecia were maintained over the years in lettuce medium or in a decoction of cereal leaves supplemented with living Enterobacter aerogenes cells at 23°C as described elsewhere (54). Acanthamoeba sp. strain TUMK-23 harboring rod-shaped endosymbionts was recovered in this study from activated sludge of a wastewater treatment plant connected to a rendering plant (Kraftisried, Germany), using the technique described by Visvesvara (58). Amoebal cultures were axenized as described previously (20) and maintained in trypticase-soy-yeast extract broth at 20°C.
Electron microscopy.
Acanthamoeba sp. strain TUMK-23 was examined by electron microscopy using a modification of a previously published method (15). Briefly, aliquots of amoebae in broth were fixed with 2% glutaraldehyde in 0.1 M cacodylate. The fixed amoebae were then pelleted in agar and embedded. Thin sections were stained with uranyl acetate and lead citrate and examined with a Phillips CM-10 electron microscope. C. taeniospiralis was examined by electron microscopy as described for C. caryophilus by Schmidt et al. (47). Briefly, symbiont-bearing paramecia were fixed with glutaraldehyde, postfixed with OsO4, and embedded in epoxid resin. Sections were stained with uranyl acetate and lead citrate and investigated with a Zeiss 10 electron microscope.
Orcein staining.
Paramecia were fixed on a slide with osmium vapor for 5 to 10 s, postfixed with a drop of a mixture of ethanol and acetic acid (3:1), and stained with a drop of orcein dissolved in acetic and lactic acid as described by Beale and Jurand (4).
Killer tests.
To determine whether C. taeniospiralis-bearing paramecia display killer activity, symbiont-free paramecia of the same strain were incubated together with symbiont-bearing cells in small wells of depression slides as described by Sonneborn (54). The wells were observed for deformed, spinning, or dying cells for up to 48 h (the descriptions of different killer traits are summarized in reference 32).
DNA isolation, PCR, cloning, and sequencing.
In order to separate extracellular bacteria from the C. taeniospiralis-bearing paramecium cells, 2 ml of a dense xenically grown paramecium culture was filtered through a 0.45-μm-pore-size syringe filter (PALL Gelman Laboratory, Ann Arbor, Mich.) and washed twice with distilled water. Paramecia were recovered from the filter disk by reverse filtration with 2 ml of distilled water and subsequently harvested by centrifugation (21,910 × g; 3 min). Simultaneous DNA isolation from P. tetraurelia and its intracellular bacteria was performed using the DNeasy tissue kit (Qiagen, Hilden, Germany) according to the instructions of the manufacturer. Simultaneous isolation of DNA from Acanthamoeba sp. strain TUMK-23 and its endosymbionts was performed using a modified UNSET procedure (22) as described by Horn et al. (20).
PCR amplification of near-full-length bacterial 16S rRNA gene fragments was performed using primers targeting signature regions of the 16S rRNA gene that are highly conserved within the domain Bacteria. The nucleotide sequences of forward and reverse primers were 5′-AGAGTTTGATYMTGGCTCAG-3′ (Escherichia coli 16S ribosomal DNA [rDNA] positions 8 to 27 [7, 59]) and 5′-GGYTACCTTGTTACGACT-3′ (E. coli 16S rDNA positions 1492 to 1511) or 5′-CAKAAAGGAGGTGATCC-3′ (E. coli 16S rDNA positions 1529 to 1546). Amplification of near-full-length paramecium and amoeba 18S rRNA genes was carried out using primers SSU1 (5′-AACCTGGTTGATCCTGCCAG-3′) and SSU2 (5′-GATCCTTCTGCAGGTTCACCTAT-3′), complementary to conserved target regions at both ends of the 18S rRNA gene (12). 16S rRNA and 18S rRNA gene amplification reactions were performed separately using annealing temperatures of 54 (16S rRNA gene) and 55°C (18S rRNA gene), respectively. Negative controls without a DNA template were included in all PCRs. The presence and sizes of the amplification products were determined by agarose gel electrophoresis and ethidium bromide staining.
Amplified products were cloned into E. coli using the TOPO TA cloning kit (Invitrogen, Carlsbad, Calif.) following the instructions of the manufacturer. Nucleotide sequences of cloned rRNA gene fragments were determined by the dideoxynucleotide method (45) by cycle sequencing of purified plasmid preparations (Qiagen) using the Thermo Sequenase cycle-sequencing kit (Amersham Life Science, Little Chalfont, England) and an infrared automated DNA sequencer (Li-Cor Inc., Lincoln, Nebr.) under conditions recommended by the manufacturers. For sequencing, the dye-labeled vector-specific primers M13/pUC V (5′-GTAAAACGACGGCCAGT-3′) and M13/pUC R (5′-GAAACAGCTATGACCATG-3′) were applied. Primer Ac1138 (5′-CTCTAAGAAGCACGGACG-3′ [20]) was used to complete the 18S rRNA gene sequence of Acanthamoeba sp. strain TUMK-23.
Comparative sequence analysis.
Sequence homology searches within the public databases DDBJ-EMBL-GenBank were performed using the BLASTn service available at the National Center for Biotechnology Information website (2). The 16S and 18S rRNA sequences obtained were added to the rRNA sequence database of the Technische Universität München (encompassing about 15,000 published and unpublished homologous small-subunit rRNA primary structures) by use of the program package ARB (available at http://www.arb-home.de). Alignment of retrieved the rRNA sequences was performed by using the ARB automated alignment tool (Fast Aligner version 1.03). The alignments were refined by visual inspection and by secondary-structure analysis (26). Phylogenetic analyses were performed by applying the distance matrix, parsimony, and maximum-likelihood methods implemented in ARB to different data sets. Bootstrap analysis (1,000 resamplings) was performed for the parsimony trees using the Phylip program package (10). To determine the robustness of the phylogenetic trees, analyses were performed with and without the application of filter sets excluding highly variable positions. In detail, the filters included only those positions that are conserved in at least 50% of all bacteria, all Alphaproteobacteria, and all Gammaproteobacteria within the 16S rRNA database. For the phylogenetic analysis of the protozoa, filters were constructed which considered only those 18S rRNA positions that are conserved in at least 50% of all eukaryotic or Acanthamoeba sequences present in the 18S rRNA database. Unless otherwise noted in the text, the bacterial nomenclature proposed in the taxonomic outline (release 1; April 2001) of the second edition of Bergey's Manual of Systematic Bacteriology (http://www.cme.msu.edu/bergeys/) was used.
Oligonucleotide probes.
The oligonucleotide probes used in this study are listed in Table 1. New oligonucleotide probes were designed using the Probedesign and Probematch tools implemented in the ARB software package. In order to ensure probe specificity, all available rRNA sequences included in the ARB database were checked for the presence of the probe target sites. The oligonucleotides were synthesized and directly labeled with 5(6)-carboxyfluorescein-N-hydroxysuccinimide ester (FLUOS) or the hydrophilic sulfoindocyanine fluorescent dye Cy3 or Cy5 (Interactiva, Ulm, Germany). Newly designed oligonucleotide probe sequences were deposited at the oligonucleotide probe database probeBase (http://www.probebase.net).
TABLE 1.
Oligonucleotide probes used for FISHa
| Probe name | Designationb | Sequence (5′-3′) | Target rRNA | Specificity | % FAc | Reference |
|---|---|---|---|---|---|---|
| Ctaenio998 | S-S-Ctaenio-998-a-A-18 | CTCTCTCGTCTTCTATGG | 16S | C. taeniospiralis 51k | 60 | This study |
| Ctaenio129 | S-S-Ctaenio-129-a-A-18 | CCCTCTGTACGGCAGATT | 16S | C. taeniospiralis 51k | 70 | This study |
| Ctaenio86 | S-S-Ctaenio-86-a-A-18 | GAAAGTGGAAGTCGAACC | 16S | C. taeniospiralis 51k | ND | This study |
| EUB338 | GCTGCCTCCCGTAGGAGT | 16S | Almost all bacteria | 0-50 | 3 | |
| EUB338-II | GCAGCCACCCGTAGGTGT | 16S | Planctomycetales | 0-50 | 8 | |
| EUB338-III | GCTGCCACCCGTAGGTGT | 16S | Verrucomicrobiales | 0-50 | 8 | |
| ALF1B | CGTTCGYTCTGAGCCAG | 16S | Alphaproteobacteria | 20 | 28 | |
| BET42a | GCCTTCCCACTTCGTTT | 23S | Betaproteobacteria | 35 | 28 | |
| GAM42a | GCCTTCCCACATCGTTT | 23S | Gammaproteobacteria | 35 | 28 | |
| CC23a | TTCCACTTTCCTCTCTCG | 16S | C. caryophilus | ND | 58 |
Difference alignments are available at probeBase (http://www.probebase.net).
According to Alm et al. (1).
Optimal formamide (FA) concentration in the hybridization buffer. ND, not determined.
FISH.
For in situ hybridization, 10 μl of dense protozoal cultures was spotted on glass slides and dried at room temperature. Lysis of the paramecia was prevented by using a constant airflow to speed up the drying process. The immobilized specimens were fixed with 4% formaldehyde for 30 to 60 min at room temperature, washed twice with sterile water or Page's saline, and subsequently dehydrated in 50, 80, and 98% ethanol for 5 min each. The optimal hybridization and washing conditions for the endosymbiont-specific probes S-S-Ctaenio-998-a-A-18 and S-S-Ctaenio-129-a-A-18 were determined using the hybridization and washing buffers of Manz et al. (27) following procedures described previously (20). The intensity of the fluorescence signal remained the same with up to 60 (S-S-Ctaenio-998-a-A-18) or 70% (S-S-Ctaenio-129-a-A-18) formamide in the hybridization buffer. Both probes were subsequently used at 35% formamide in the hybridization buffer. In some experiments, the DNA-binding dye DAPI (4′,6-diamino-2-phenylindole; Sigma, Buchs, Switzerland) was used for the visualization of intracellular bacteria. Subsequent to FISH, the specimens were covered with 5 μl of DAPI solution (1 μg/μl), incubated for 5 min in the dark, washed with sterile water, and air dried.
Slides were examined using a laser scanning confocal microscope (LSM 510; Carl Zeiss, Oberkochen, Germany). Images were recorded and processed using the standard software package delivered with the instrument (version 2.01).
Nucleotide sequence accession numbers.
The recovered 16S and 18S rRNA gene sequences have been submitted to the DDBJ-EMBL-GenBank databases under accession numbers AY102612 (16S rRNA gene of C. taeniospiralis strain 51k), AY102613 (18S rRNA gene of P. tetraurelia strain 51k), AY102614 (16S rRNA gene of the endosymbiont of Acanthamoeba sp. strain TUMK-23), and AY102615 (18S rRNA gene of Acanthamoeba sp. strain TUMK-23).
RESULTS
Morphology, phylogeny, and in situ identification of C. taeniospiralis 51k.
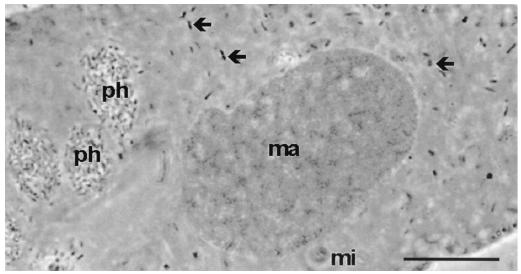
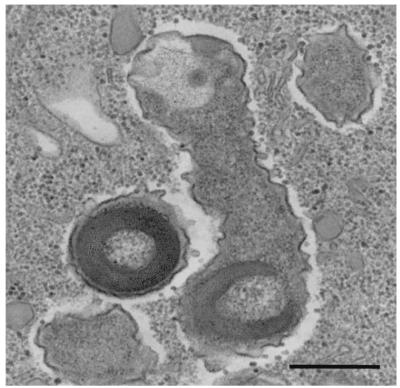
Bacterial endosymbionts were readily visualized within P. tetraurelia by orcein staining and phase-contrast light microscopy (Fig. 1). C. taeniospiralis 51k was formerly described as a cytoplasmic endosymbiont of P. tetraurelia able to produce type 51 R bodies (with R-body genes encoded on a plasmid) conferring a hump killer trait upon its paramecium host (5, 17). Consistent with this description, electron microscopy revealed that the rod-shaped endosymbionts of the investigated P. tetraurelia 51k measured 0.4 to 0.7 by 1.0 to 2.5 μm and possessed a gram-negative-type cell wall (Fig. 2). The bacteria were equally distributed within the cytoplasm and were surrounded by an electron-translucent layer (Fig. 2). Type 51 R bodies were observed within the investigated endosymbionts and were shown to confer a hump killer trait upon their P. tetraurelia hosts (9, 32, 56), inducing the formation of aboral blisters in sensitive (symbiont-free) paramecia.
FIG. 1.
Phase-contrast micrograph of P. tetraurelia strain 51k stained with orcein. Numerous symbionts (arrows) in the cytoplasm and bacteria in phagosomes (ph) are visible. ma, macronucleus; mi, micronucleus. Bar, 10 μm.
FIG. 2.
Ultrastructure of C. taeniospiralis harboring type 51 R bodies within the cytoplasm of its host, P. tetraurelia. Bar, 0.5 μm.
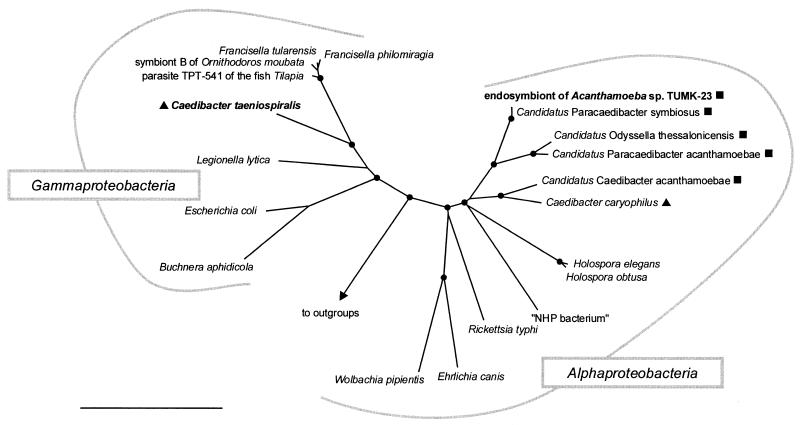
Near-full-length 16S rRNA gene sequences (1,544 bp) of intracellular bacteria of P. tetraurelia were amplified, cloned, and sequenced. Out of 10 16S rRNA clones analyzed, 7 were found to have identical sequences. This sequence was considered to represent the endosymbionts of P. teraurelia, while the other three sequences were assumed to be derived from bacteria that were present in the food vacuoles of the xenically grown paramecium culture. This 16S rRNA gene sequence was novel and showed moderate 16S rRNA sequence similarity with members of the Gammaproteobacteria. Within the Gammaproteobacteria, the highest 16S rRNA similarity values were obtained with representatives of the Francisella group (86.5 to 87.3%). A significantly lower 16S rRNA similarity (80%) was observed with the alphaproteobacterial symbiont C. caryophilus. All applied treeing methods consistently confirmed the affiliation of the retrieved 16S rRNA gene sequences with the Gammaproteobacteria and demonstrated that they form a novel evolutionary lineage within this subgroup with the Francisella cluster as a sister group (Fig. 3).
FIG. 3.
16S rRNA-based neighbor-joining tree showing (i) the phylogenetic affiliation of C. taeniospiralis 51k (endosymbiont of P. tetraurelia) with representative members of the Gammaproteobacteria and (ii) the relationship of the endosymbiont of Acanthamoeba sp. strain TUMK-23 with the alphaproteobacterial C. caryophilus (endosymbiont of P. caudatum) and other representative members of the Alphaproteobacteria. “Candidatus Caedibacter acanthamoebae,” “Candidatus Paracaedibacter acanthamoebae,” “Candidatus Paracaedibacter symbiosus,” and “Candidatus Odyssella thessalonicensis” were recently described as endosymbionts of acanthamoebae (5, 18). The respective eukaryotic hosts are indicated by symbols: ▴, Paramecium sp.; ▪, Acanthamoeba sp. Parsimony bootstrap values (1,000 resamplings) of >97% are indicated as solid circles. “NHP bacterium,” shrimp pathogen causing necrotizing hepatopancreatitis. Bar, 10% estimated evolutionary distance.
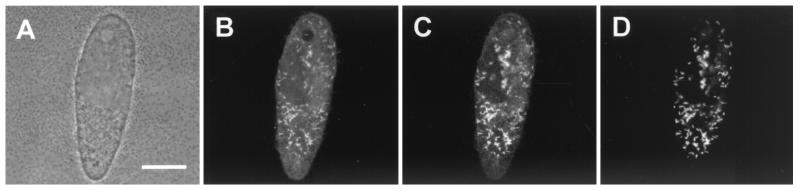
Consistent with these findings, C. taeniospiralis was readily visualized within its Paramecium host cells by FISH using the probe GAM42a (targeting a signature region on the 23S rRNA of the Gammaproteobacteria) (Fig. 4), while no fluorescence was detected with the probes ALF1B and CC23a (specific for the Alphaproteobacteria and C. caryophilus, respectively; data not shown). Application of the newly designed probes S-S-Ctaenio-998-a-A-18 and S-S-Ctaenio-129-a-A-18, complementary to signature regions on the retrieved Francisella-related 16S rRNA sequence, demonstrated that this sequence originated from the intracytoplasmatic P. tetraurelia endosymbiont C. taeniospiralis (Fig. 4). Simultaneous hybridization with the probes S-S-Ctaenio-998-a-A-18 and S-S-Ctaenio-129-a-A-18 and the bacterial probe EUB338 labeled with different dyes illustrated that all detectable bacteria within the cytoplasm of P. tetraurelia were stained by all three probes, demonstrating the absence of phylogenetically different symbiotic bacteria within the host cells (Fig. 4).
FIG. 4.
In situ identification of C. taeniospiralis 51k within its host, P. tetraurelia. Identical microscopic fields are depicted. The fluorescence images (B to D) show a median section through the paramecium cell seen in the simulated phase-contrast image (A). Shown is FISH using the FLUOS-labeled oligonucleotide probe EUB338 (B), the Cy3-labeled endosymbiont-specific probe S-S-Ctaenio-129-a-A-18 (C), and the Cy5-labeled endosymbiont-specific probe S-S-Ctaenio-998-a-A-18 (D). Bar, 20 μm
The 16S rRNA gene of C. taeniospiralis contained a stretch of 97 additional base pairs replacing the tetraloop corresponding to helix 6 of the E. coli rRNA secondary structure (positions 82 to 87), which did not show significant similarity to any rRNA gene sequence deposited in public databases. This putative intervening sequence is able to form a stable stem-loop structure, as indicated by secondary-structure prediction using the software RNAstructure version 3.5 (28). The presence of such intervening sequences within rRNA genes has been reported previously for C. caryophilus and other intracellular bacteria (21, 25, 46, 57). However, a search within the publicly available 16S rRNA gene sequences revealed that <20 only distantly related sequences also possess additional base pairs in this region (for example, members of the Firmicutes, the Actinobacteria, the Planctomycetes, and the Epsilonproteobacteria). FISH using the probe S-S-Ctaenio-86-a-A-18, which targets the putative intervening sequence of C. taeniospiralis, resulted in no detectable signals, suggesting that this sequence stretch is either excised during maturation of the RNA or is simply not accessible due to the formation of a stable secondary structure.
Phylogeny and in situ identification of a C. caryophilus-related endosymbiont of Acanthamoeba sp.
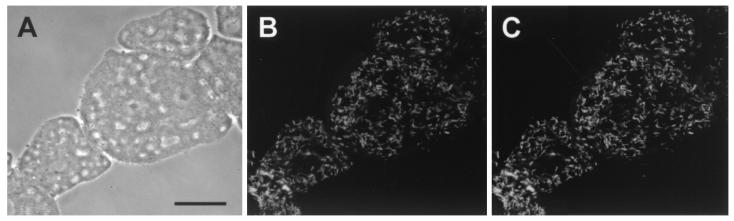
Previous studies demonstrated that bacteria closely related to the alphaproteobacterial Paramecium caudatum symbiont C. caryophilus also occur as endosmybionts of free-living amoebae (provisionally designated “Candidatus Paracaedibacter” and “Candidatus Odyssella” [6, 20]). Therefore, the Acanthamoeba isolate TUMK-23, which was found to contain bacterial endosymbionts that hybridize with the C. caryophilus-specific oligonucleotide probe CC23a (indicating a close relationship with C. caryophilus), was included in this study (Fig. 5). Electron microscopy revealed that this rod-shaped endosymbiont measured 0.2 to 0.3 by 1.3 to 1.7 μm and possessed a gram-negative-type cell wall (Fig. 6). The bacteria were equally distributed within the cytoplasm and were surrounded by an electron-translucent layer (Fig. 6). Consistent with previous descriptions of C. caryophilus-related symbionts of amoebae, no R-body-like structures were observed within these symbionts.
FIG. 5.
In situ identification of the bacterial endosymbiont of Acanthamoeba sp. strain TUMK-23. Identical microscopic fields are depicted. (A) Phase-contrast image. (B and C) FISH using FLUOS-labeled oligonucleotide probe EUB338 (B) and Cy3-labeled C. caryophilus-specific probe CC23a (C). Bar, 10 μm.
FIG. 6.
Ultrastructure of the C. caryophilus-related endosymbiont (solid arrows) within the cytoplasm of its host, Acanthamoeba sp. strain TUMK-23. cm, amoebal cell membrane; n, nucleus. The open arrows indicate mitochondria. Bar, 0.5 μm.
Near-full-length 16S rRNA gene sequences (1,464 bp) of the bacterial symbiont of Acanthamoeba sp. strain TUMK-23 were amplified, cloned, and sequenced. Comparative sequence analysis demonstrated that this endosymbiont shows the highest 16S rRNA sequence similarities to members of the Alphaproteobacteria and that it is closely affiliated with the C. caryophilus-related amoebal symbiont “Candidatus Paracaedibacter symbiosus” (99% 16S rRNA sequence similarity) (Fig. 3).
Phylogeny of the protozoan hosts of Caedibacter endosymbionts.
Comparative analysis of 18S rRNA gene fragments (1,701 and 2,239 bp) retrieved from the P. tetraurelia and the Acanthamoeba sp. hosts revealed the highest sequence similarities to members of the genus Paramecium (98% with P. tetraurelia [50]) and Acanthamoeba (98% with Acanthamoeba sp. strain UWE39 [20]), respectively, and thus confirmed the morphology-based classification of these protozoa.
DISCUSSION
Ever since the cytoplasmic heritable killer trait of paramecia has been associated with endosymbiotic bacteria, several models for classification of these bacteria have been proposed (17, 32, 37, 38, 55). Initially, these endosymbionts were designated by Greek letters (for example, as kappa or mu particles) (32, 33, 53). While this codification is still in use, a binominal nomenclature and the genus name Caedibacter (earlier designated Caedobacter [36]) were introduced in the 1970s (32). In brief, there is general agreement that the presence of R bodies, the host specificity of the symbionts, and the cell compartment in which the symbiont multiplies (macronucleus or cytoplasm) are the key taxonomic properties for classification of Caedibacter species. In addition, the different types of killing that the symbionts confer upon their hosts, the morphology of R bodies, the potential association with bacteriophages or bacteriophage-like structures, and the existence of different types of extrachromosomal elements were used for subclassification of the genus Caedibacter (30). However, taxonomic systems that are based on phenotypic properties do not necessarily reflect evolutionary history (29, 44, 61, 62). With respect to the Caedibacter endosymbionts of paramecia, significant differences in G+C content and low DNA-DNA hybridization values between different Caedibacter species provided initial indications that (i) this genus might contain genetically diverse bacteria and (ii) their common characteristic phenotype might be determined by genetically related extrachromosomal elements (40, 42, 43). The 16S rRNA-based phylogenetic analysis presented in this study substantiated this hypothesis and demonstrated that the genus Caedibacter is actually polyphyletic and comprises bacteria belonging to two different classes of the Proteobacteria. C. taeniospiralis forms a novel evolutionary lineage within the Gammaproteobacteria which groups together with other intracellular bacteria, namely, a symbiont of the tick Ornithodorus mobuta, a fish parasite, and the human pathogen Francisella tularensis. In contrast, C. caryophilus belongs to the alphaproteobacterial family Holosporaceae and is most closely related to amoebal endosymbionts (e.g., the endosymbiont of Acanthamoeba sp. strain TUMK-23 analyzed in this study) and the parasitic paramecium symbionts H. obtusa and H. elegans (3, 20).
One traditional key taxonomic criterion of the genus Caedibacter is the production of R bodies (13, 23, 30, 48). The findings reported here indicate that R-body production and the associated killer trait either resulted from convergent evolution (within different evolutionary lineages of the Proteobacteria) or evolved only once and were subsequently passed on by horizontal gene transfer. Supportive evidence for the latter hypothesis was provided by the discovery that some R-body proteins are encoded by transposable genetic elements like phages or plasmids (24, 31, 39, 40, 41). This scenario might also explain the otherwise peculiar existence of R-body-like structures in nonsymbiotic bacteria, like Pseudomonas avenae and Pseudomonas taeniospiralis, and their absence in the Caedibacter-related Acanthamoeba endosymbionts (20, 23, 24, 60). While comparative analysis of the genes necessary for R-body production (the genes necessary for type 51 R-body synthesis were cloned and heterologically expressed by Quackenbush and Burbach [19, 40]) may help to clarify their evolutionary history, our data clearly demonstrate that the presence of R bodies alone must not be used as a phylogenetically meaningful taxonomic marker.
Without considering R bodies as a key criterion for Caedibacter taxonomy, the alphaproteobacterial C. caryophilus phenotypically seems to differ significantly from all other validly described Caedibacter species. C. caryophilus is, for example, the only species that multiplies within P. caudatum, while nearly all other Caedibacter species, including C. taeniospiralis, thrive within members of the Paramecium aurelia species complex (11, 35, 47). The only exeption, C. macronucleorum, resides in Paramecium duboscqui (11). In addition, C. caryophilus and the phylogenetically uncharacterized C. macronucleorum live within the macronuclei of their hosts, whereas all other Caedibacter species are located directly within the host cytoplasm (11, 35, 47). In addition, it has been shown that C. caryophilus can lose its ability to produce R bodies (47, 49, 51) and switch from symbiotic to parasitic behavior under certain growth conditions (47). In summary, the phylogeny and the phenotypic properties of C. caryophilus are more similar to those of the closely related paramecium symbionts Holospora sp. and clearly separate C. caryophilus from other members of the genus Caedibacter.
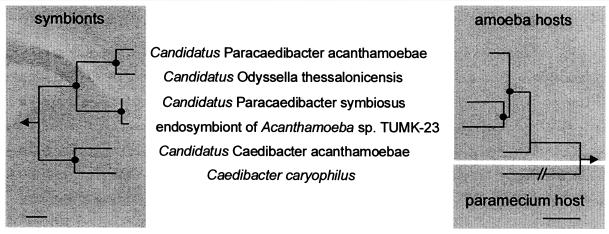
Comparison of the evolutionary history of the alphaproteobacterial C. caryophilus, the closely related endosymbionts of acanthamoebae, and their respective eukaryotic host cells reveals a striking congruency of the branching patterns of their rRNA-based dendrograms (Fig. 7). This finding indicates coevolution between the symbiosis partners and suggests that the ancestor of the C. caryophilus-related endosymbionts lived within an amoebal progenitor and coevolved with its hosts during their diversification into the different Acanthamoeba sublineages. According to this scenario, C. caryophilus-related bacteria originally thrived in amoebal hosts and were relatively recently laterally transferred into a Paramecium host. Since the Acanthamoeba endosymbionts do not carry R bodies, the uptake of R-body-coding mobile genetic elements by the C. caryophilus symbiont might have taken place after the transfer into the new Paramecium host cell, for example, by coinfection of the host with different Caedibacter species (17). This hypothesis is also supported by the observation that R-body production is not essential for the Caedibacter-Paramecium symbiosis (47). A more robust inference of coevolution between alphaproteobacterial Caedibacter symbionts and their amoebal hosts, however, must await the isolation and comparative sequence analysis of further amoebae carrying C. caryophilus-related endosymbionts.
FIG. 7.
Comparison of 16S and 18S rRNA-based neighbor-joining dendrograms of alphaproteobacterial C. caryophilus-related endosymbionts (left) and their Acanthamoeba or Paramecium host organisms (right) suggesting (i) coevolution between bacterial symbionts and Acanthamoeba hosts and (ii) relatively recent transfer of a C. caryophilus-like symbiont from Acanthamoeba to Paramecium. The 18S rRNA gene sequence of the amoebal host of “Candidatus Odyssella thessalonicensis” is not available (6). Parsimony bootstrap values (1,000 resamplings) higher than 99% are indicated as solid circles. The arrow points to outgroups. Bars, 10% estimated evolutionary distance.
Clearly, additional rRNA sequence information from the other four validly described Caedibacter species and their host paramecia is necessary for a more detailed understanding of the phylogeny of these unique bacteria and the evolution of the Caedibacter-Paramecium symbiosis. We obtained several Paramecium strains from the American Type Culture Collection which had been described as carrying Caedibacter endosymbionts (P. tetraurelia stock 51m1k carrying C. pseudomutans ATCC 30633, Paramecium biaurelia stock 7K carrying C. varicaedens ATCC 30637, and P. biaurelia stock 570 carrying C. paraconjugatus ATCC 30638). Unfortunately, DAPI staining and FISH analysis revealed that those cultures had lost their Caedibacter endosymbionts, as had the stock cultures at the American Type Culture Collection (data not shown and N. Hetrick, personal communication). Consequently, these Caedibacter-type strains are no longer publicly available. Joint efforts of protozoologists and bacteriologists are required to reisolate paramecia carrying these Caedibacter species for a more comprehensive phylogenetic analysis. A comprehensive knowledge of the evolutionary history of Caedibacter species will help us to understand the evolution of this complex, unique symbiotic system in which plasmids or phages induce bacterial hosts to produce R bodies and associated toxins (30, 36) that provide their eukaryotic host cells a selective advantage against closely related ciliates.
Acknowledgments
This study was supported by Deutsche Forschungsgemeinschaft grant WA 1047/2-2 to M.W.
REFERENCES
- 1.Alm, E. W., D. B. Oerther, M. Larsen, D. A. Stahl, and L. Raskin. 1996. The oligonucleotide probe database. Appl. Environ. Microbiol. 62:3557-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R., N. Springer, W. Ludwig, H. D. Görtz, and K. H. Schleifer. 1991. Identification in situ and phylogeny of uncultured bacterial endosymbionts. Nature 351:161-164. [DOI] [PubMed] [Google Scholar]
- 4.Beale, G. H., and A. Jurand. 1966. Three different types of mate-killer (mu) particles in Paramecium aurelia, stock 540. J. Gen. Microbiol. 23:243-252. [DOI] [PubMed] [Google Scholar]
- 5.Beale, G. H., A. Jurand, and J. R. Preer. 1969. The classes of endosymbionts of Paramecium aurelia. J. Cell Sci. 5:65-91. [DOI] [PubMed] [Google Scholar]
- 6.Birtles, R. J., T. J. Rowbotham, R. Michel, D. G. Pitcher, B. Lascola, S. Alexiou-Daniel, and D. Raoult. 2000. ′Candidatus Odyssella thessalonicensis' gen. nov., sp. nov., an obligate intracellular parasite of Acanthamoeba species. Int. J. Syst. Evol. Microbiol. 50:63-72. [DOI] [PubMed] [Google Scholar]
- 7.Brosius, J., T. J. Dull, D. D. Sleeter, and H. F. Noller. 1981. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J. Mol. Biol. 148:107-127. [DOI] [PubMed] [Google Scholar]
- 8.Daims, H., A. Brühl, R. Amann, K.-H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 9.Dippell, R. V. 1958. The fine structure of kappa in killer stock 51 of Paramecium aurelia. Preliminary observations. J. Biophys. Biochem. Cytol. 4:125-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Felsenstein, J. 1988. Phylogenies from molecular sequences: inference and reliability. Annu. Rev. Genet. 22:521-565. [DOI] [PubMed] [Google Scholar]
- 11.Fokin, S., and H.-D. Görtz. 1993. Caedibacter macronucleorum sp. nov., a bacterium inhabiting the macronucleus of Paramecium duboscqui. Arch. Protistenkd. 143:319-324. [Google Scholar]
- 12.Gast, R. J., P. A. Fuerst, and T. J. Byers. 1994. Discovery of group I introns in the nuclear small subunit ribosomal RNA genes of Acanthamoeba. Nucleic Acids Res. 22:592-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Görtz, H.-D., and T. Brigge. 1998. Intracellular bacteria in protozoa. Naturwissenschaften 85:359-368. [DOI] [PubMed] [Google Scholar]
- 14.Görtz, H.-D., H. W. Kuhlmann, M. Mollenbeck, A. Tiedtke, J. Kusch, H. J. Schmidt, and A. Miyake. 1999. Intra- and intercellular communication systems in ciliates. Naturwissenschaften 86:422-434. [DOI] [PubMed] [Google Scholar]
- 15.Hall, J., and H. Voelz. 1985. Bacterial endosymbionts of Acanthamoeba sp. J. Parasitol. 71:89-95. [PubMed] [Google Scholar]
- 16.Hamilton, L. D., and M. E. Gettner. 1958. Fine structure of kappa in Paramecium aurelia. J. Biophys. Biochem. Cytol. 4:122-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heckmann, K., and H.-D. Görtz. 1992. Prokaryotic symbionts of ciliates, p. 3865-3890. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes, 2nd ed. Springer, New York, N.Y.
- 18.Hentschel, U., M. Steinert, and J. Hacker. 2000. Common molecular mechanisms of symbiosis and pathogenesis. Trends Microbiol. 8:226-231. [DOI] [PubMed] [Google Scholar]
- 19.Heruth, D. P., F. R. Pond, J. A. Dilts, and R. L. Quackenbush. 1994. Characterization of genetic determinants for R-body synthesis and assembly in Caedibacter taeniospiralis 47 and 116. J. Bacteriol. 176:3559-3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horn, M., T. R. Fritsche, R. K. Gautom, K.-H. Schleifer, and M. Wagner. 1999. Novel bacterial endosymbionts of Acanthamoeba spp. related to the Paramecium caudatum symbiont Caedibacter caryophilus. Environ. Microbiol. 1:357-367. [DOI] [PubMed] [Google Scholar]
- 21.Horn, M., T. R. Fritsche, T. Linner, R. K. Gautom, M. D. Harzenetter, and M. Wagner. 2002. Obligate bacterial endosymbionts of Acanthamoeba spp. related to the beta-Proteobacteria: proposal of ′Candidatus Procabacter acanthamoebae' gen. nov., sp. nov. Int. J. Syst. E vol. Microbiol. 52:599-605. [DOI] [PubMed] [Google Scholar]
- 22.Hugo, E. R., R. J. Gast, T. J. Byers, and V. J. Stewart. 1992. Purification of amoeba mtDNA using the unset procedure, p. D7.1-D7.2. In J. J. Lee and A. T. Solldo (ed.), Protocols in protozoology. Allen Press, Lawrence, Kans.
- 23.Kanabrocki, J. A., J. Lalucat, B. J. Cox, and R. L. Quackenbush. 1986. Comparative study of refractile (R) bodies and their genetic determinants: relationship of type 51 R bodies to R bodies produced by Pseudomonas taeniospiralis. J. Bacteriol. 168:1019-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lalucat, L., O. Meyer, F. Mayer, R. Pares, and H.-G. Schlegel. 1979. R bodies in newly isolated free-living hydrogen-oxydizing bacteria. Arch. Microbiol. 121:9-15. [Google Scholar]
- 25.Linton, D., J. P. Clewley, A. Burnens, R. J. Owen, and J. Stanley. 1994. An intervening sequence (IVS) in the 16S rRNA gene of the eubacterium Helicobacter canis. Nucleic Acids Res. 22:1954-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ludwig, W., and K.-H. Schleifer. 1994. Bacterial phylogeny based on 16S and 23S rRNA sequence analysis. FEMS Microbiol. Rev. 15:155-173. [DOI] [PubMed] [Google Scholar]
- 27.Manz, W., R. Amann, W. Ludwig, M. Vancanneyt, and K.-H. Schleifer. 1992. Phylogenetic oligonucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst. Appl. Microbiol. 142:1097-1106. [Google Scholar]
- 28.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 29.Palleroni, N. J. 1984. Pseudomonaceae, vol. 1. Williams & Wilkins, Baltimore, Md.
- 30.Pond, F. R., I. Gibson, J. Lalucat, and R. L. Quackenbush. 1989. R-body-producing bacteria. Microbiol. Rev. 53:25-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Preer, J. R., Jr., L. B. Preer, B. Rudman, and A. Jurand. 1971. Isolation and composition of bacteriophage-like particles from kappa of killer paramecia. Mol. Gen. Genet. 111:202-208. [DOI] [PubMed] [Google Scholar]
- 32.Preer, J. R., Jr., L. B. Preer, and A. Jurand. 1974. Kappa and other endosymbionts in Paramecium aurelia. Bacteriol. Rev. 38:113-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Preer, J. R., Jr. 1975. The hereditary symbionts of Paramecium aurelia. Symp. Soc. Exp. Biol. 29:125-144. [PubMed] [Google Scholar]
- 34.Preer, J. R., and L. B. Preer. 1982. Revival of names of protozoan endosymbionts and proposal of Holospora caryophila nom. nov. Int. J. Syst. Bacteriol. 32:140-141. [Google Scholar]
- 35.Preer, J. R., and L. B. Preer,. 1984. Endosymbionts of protozoa, p. 795-813. In N. R. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 1. Willliams & Wilkins, Baltimore, Md..
- 36.Preer, L. B., A. Jurand, J. R. Preer, Jr., and B. M. Rudman. 1972. The classes of kappa in Paramecium aurelia. J. Cell Sci. 11:581-600. [DOI] [PubMed] [Google Scholar]
- 37.Quackenbush, R. L. 1977. Phylogenetic relationships of bacterial endosymbionts of Paramecium aurelia: polynucleotide sequence relationships of 51 kappa and its mutants. J. Bacteriol. 129:895-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quackenbush, R. L. 1978. Genetic relationships among bacterial endosymbionts of Paramecium aurelia: deoxyribonucleotide sequence relationships among members of Caedobacter. J. Gen. Microbiol. 108:181-187. [Google Scholar]
- 39.Quackenbush, R. L., J. A. Dilts, and R. L. Maser. 1982. Physical map of a plasmid from Caedibacter taeniospiralis 51. J. Bacteriol. 152:939-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quackenbush, R. L. 1983. Plasmids from bacterial endosymbionts of hump-killer paramecia. Plasmid 9:298-306. [DOI] [PubMed] [Google Scholar]
- 41.Quackenbush, R. L., and J. A. Burbach. 1983. Cloning and expression of DNA sequences associated with the killer trait of Paramecium tetraurelia stock 47. Proc. Natl. Acad. Sci. USA 80:250-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quackenbush, R. L., J. A. Dilts, and B. J. Cox. 1986. Transposonlike elements in Caedibacter taeniospiralis. J. Bacteriol. 166:349-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quackenbush, R. L., B. J. Cox, and J. A. Kanabrocki. 1986. Extrachromosomal elements of extrachromosomal elements of Paramecium and their extrachromosomal elements. Basic Life Sci. 40:265-278. [DOI] [PubMed] [Google Scholar]
- 44.Rosello-Mora, R. A., L. Wolfgang, and K.-H. Schleifer. 1993. Zoogloea ranugera: a phylogenetically diverse species. FEMS Microbiol. Lett. 114:129-134. [Google Scholar]
- 45.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sauer, C., E. Stackebrandt, J. Gadau, B. Holldobler, and R. Gross. 2000. Systematic relationships and cospeciation of bacterial endosymbionts and their carpenter ant host species: proposal of the new taxon Candidatus Blochmannia gen. nov. Int. J. Syst. Evol. Microbiol. 50:1877-1886. [DOI] [PubMed] [Google Scholar]
- 47.Schmidt, H. J., H.-D. Görtz, and R. L. Quackenbush. 1987. Caedibacter caryophila sp. nov., a killer symbiont inhabiting the macronucleus of Paramecium caudatum. Int. J. Syst. Bacteriol. 37:459-462. [Google Scholar]
- 48.Schmidt, F. R. H. J., Pond, and H.-D. Görtz. 1987. Refractile bodies (R bodies) from the macronuclear killer particle Caedibacter caryophila. J. Cell Sci. 88:177-184. [Google Scholar]
- 49.Schmidt, H. J., H. D. Görtz, F. R. Pond, and R. L. Quackenbush. 1988. Characterization of Caedibacter endonucleobionts from the macronucleus of Paramecium caudatum and the identification of a mutant with blocked R-body synthesis. Exp. Cell Res. 174:49-57. [DOI] [PubMed] [Google Scholar]
- 50.Sogin, M. L., and H. J. Elwood. 1986. Primary structure of the Paramecium tetraurelia small-subunit rRNA coding region: phylogenetic relationships within the Ciliophora. J. Mol. Evol. 23:53-60. [DOI] [PubMed] [Google Scholar]
- 51.Soldo, A. T., G. A. Gustavo, and W. J. van Wagendonk. 1966. Growth of particle-bearing and particle-free Paramecium aurelia in axenic culture. J. Protozool. 13:492-497. [Google Scholar]
- 52.Sonneborn, T. M. 1938. Mating types in P. aurelia: diverse conditions for mating in different stocks; occurrence, number and interrelation of the types. Proc. Am. Phil. Soc. 79:411-434. [Google Scholar]
- 53.Sonneborn, T. M. 1943. Gene and cytoplasm. I. The determination and inheritance of the killer character in variety 4 of P. aurelia. Proc. Natl. Acad. Sci. USA 29:329-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sonneborn, T. M. 1950. Methods in the general biology and genetics of Paramecium aurelia. J. Exp. Zool. 113:87-143. [Google Scholar]
- 55.Sonneborn, T. M. 1959. Kappa and related particles in Paramecium. Adv. Virus Res. 6:229-356. [Google Scholar]
- 56.Sonneborn, T. M. 1965. The metagon: RNA and cytoplasmic inheritance. Am. Nat. 99:279-307. [Google Scholar]
- 57.Springer, N., W. Ludwig, R. Amann, H. J. Schmidt, H. D. Görtz, and K. H. Schleifer. 1993. Occurrence of fragmented 16S rRNA in an obligate bacterial endosymbiont of Paramecium caudatum. Proc. Natl. Acad. Sci. USA 90:9892-9895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Visvesvara, G. S.,. 1995. Pathogenic and opportunistic free-living amebae, p. 1196-1203. In P. R. Murray (ed.), Manual of clinical microbiology, 6th ed. American Society for Microbiology, Washington, D.C.
- 59.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wells, B., and R. W. Horne. 1983. The ultrastructure of Pseudomonas avenae. II. Intracellular refractile (R-body) structure. Micron Microsc. Acta 14:329-344. [Google Scholar]
- 61.Wheelis, M. L., O. Kandler, and C. R. Woese. 1992. On the nature of global classification. Proc. Natl. Acad. Sci. USA 89:2930-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]