Abstract
Bile is one of many barriers that Listeria monocytogenes must overcome in the human gastrointestinal tract in order to infect and cause disease. We demonstrated that stationary-phase cultures of L. monocytogenes LO28 were able to tolerate concentrations of bovine, porcine, and human bile and bile acids well in excess of those encountered in vivo. Strain LO28 was relatively bile resistant compared with other clinical isolates of L. monocytogenes, as well as with Listeria innocua, Salmonella enterica serovar Typhimurium LT2, and Lactobacillus sakei. While exponential-phase L. monocytogenes LO28 cells were exquisitely sensitive to unconjugated bile acids, prior adaptation to sublethal levels of bile acids or heterologous stresses, such as acid, heat, salt, or sodium dodecyl sulfate (SDS), significantly enhanced bile resistance. This adaptive response was independent of protein synthesis, and in the cases of bile and SDS adaptation, occurred in seconds. In order to identify genetic loci involved in the bile tolerance phenotype of L. monocytogenes LO28, transposon (Tn917) and plasmid (pORI19) integration banks were screened for bile-sensitive mutants. The disrupted genes included a homologue of the capA locus required for capsule formation in Bacillus anthracis; a gene encoding the transcriptional regulator ZurR; a homologue of an Escherichia coli gene, lytB, involved in isoprenoid biosynthesis; a gene encoding a homologue of the Bacillus subtilis membrane protein YxiO; and a gene encoding an amino acid transporter with a putative role in pH homeostasis, gadE. Interestingly, all of the identified loci play putative roles in maintenance of the cell envelope or in stress responses.
Listeria monocytogenes is responsible for 27.6% of food-related deaths in the United States annually (37). The ability of the pathogen to occupy a wide variety of environmental niches reflects an ability to sense, respond, and adapt to the many stressful conditions encountered during the passage from the food-processing environment to the host.
Following consumption, the acidity of the stomach is the first major barrier encountered by the pathogen. Recent studies have shown that L. monocytogenes possesses many acid resistance systems that enable the bacterium to combat low-pH environments (6, 7, 8, 12, 21). Bacteria that survive and transit the stomach enter the small intestine, where they encounter stresses associated with volatile fatty acids, variations in pH, low oxygen, competition with normal flora, and elevated osmolarity (5). As the liver secretes up to a liter of bile into the intestinal tract each day (24), exposure to bile also represents a major challenge. The ability of pathogens to tolerate bile is likely to be important for survival and colonization. For example, the commensal intestinal microbiota is inherently bile resistant, as are common gram-negative intestinal pathogens (39, 40, 41, 46, 48). Indeed, bile salts are often used in the selective enrichment of such organisms.
Bile is a digestive secretion that plays a major role in the emulsification and solubilization of lipids (49). It is synthesized in the liver but is stored and concentrated in the gallbladder during the fasting state. Bile is primarily composed of bile acids (12% by weight), which are found as sodium salts under physiological conditions. These “biological detergents” are synthesized from cholesterol, where the steroid nucleus is conjugated with either glycine or taurine (23). Forming part of the body's physicochemical defense system, bile salts possess potent antimicrobial activity and have the ability to dissolve the phospholipids, cholesterol, and proteins of cell membranes, causing cells to lyse (24).
L. monocytogenes is capable of colonizing the human gallbladder (1, 2), indicating an inherent ability to tolerate high concentrations of bile. However, little information is available concerning the levels of bile tolerance or factors contributing to bile responses in this pathogen. In this report, we describe the physiological response of L. monocytogenes LO28 to bile and bile acid stress. We examine the relationship between resistance to bile stress and heterologous sublethal conditions. We also initiate an investigation into the molecular mechanisms underlying bile resistance in this important human pathogen.
MATERIALS AND METHODS
Media, chemicals, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Listeria strains were grown in brain heart infusion (BHI) broth or tryptic soy broth supplemented with 0.6% yeast extract (TSB-YE). Escherichia coli was grown in Luria-Bertani medium. Erythromycin and chloramphenicol were made up as concentrated stocks and added to media at the required levels. For solid media, agar was added to 1.5%. Bile and bile acids were solubilized in water, and filter-sterilized stocks were added to autoclaved media. Plates supplemented with bile or bile acids were inverted and incubated anaerobically for at least 48 h before use. Sodium dodecyl sulfate (SDS) was solubilized in water to obtain a concentration of 10% (wt/vol).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant properties | Source or referencea |
|---|---|---|
| Strains | ||
| E. coli EC101 | E. coli JM101 with repA from pWV01 integrated in the chromosome | 30 |
| Serovar Typhimurium LT2 | UCC Culture Collection | |
| L. sakei | UCC Culture Collection | |
| L. monocytogenes | ||
| LO28 | Serotype 1/2c | P. Cossart, Institut Pasteur |
| ScottA | Serotype 4b | T. Abee |
| EGDe | Serotype 1/2a | W. Goebel |
| DPC4591 (ATCC 19118) | Serotype 4c | Teagasc; Moorepark |
| DPC 4593 (ATCC 19117) | Serotype 4d | Teagasc; Moorepark |
| DPC 4594 (ATCC 15313) | Serotype 1/2c | Teagasc; Moorepark |
| DPC 4609 (ATCC 19115) | Serotype 3a | Teagasc; Moorepark |
| L. innocua | ||
| DPC1770 | Teagasc; Moorepark | |
| FH 2097 | Food isolate | St. Finbarrs Hospital, Cork, Ireland |
| FH2327 | Food isolate | St. Finbarrs Hospital, Cork, Ireland |
| Plasmids | ||
| pORI19 | Emr Ori+ RepA− derivative of pORI28 | 30 |
| pVE6007 | Cmr; temperature-sensitive derivative of pWV01 | 35 |
| pTV1-OK | Kmr Emr; temperature-sensitive derivative of pWV01 | 20 |
UCC, University College Cork.
Growth of cultures in the presence of bile or bile acids.
Cultures were screened for growth on BHI agar plates supplemented with various concentrations of bovine bile (B-8381; Sigma Chemical Co. Ltd., Poole, United Kingdom), oxgall (B-3883; Sigma), and porcine bile (B-8631; Sigma). Growth on BHI containing individual bile acids was also examined. Sodium salts of the following bile acids (Sigma) were used: taurocholic acid (TCA), glycocholic acid (GCA), taurodeoxycholic acid (TDCA), glycodeoxycholic acid (GDCA), taurochenodeoxycholic acid, and glycochenodeoxycholic acid. The plates were incubated at 37°C under anaerobic conditions using BBL anaerobic jars and the Merck Anaerocult A Gas Pak system. For broth assays, overnight cultures were inoculated (3%) into BHI broth containing various concentrations of bile or bile acids. The cultures were incubated anaerobically at 37°C without shaking. Cell growth was measured either spectrophotometrically by determining the optical density at 600 nm or by performing viable cell counts by diluting cultures in one-quarter-strength Ringer's solution and enumeration on BHI. Before optical density determinations, the medium was adjusted to pH 7 to dissolve precipitated bile salts. Human bile was obtained from gallbladders at laparoscopic cholecystectomy and stored at −70°C. Samples were thawed and sterilized at 80°C for 10 min before addition to sterilized cooled media.
pH dependency of bile salt toxicity.
The effects of 5 mM GDCA and TDCA on LO28 cultures was investigated in BHI broth set at initial pH values ranging from 4.5 to 7.5 with 3 M lactic acid. The broth was inoculated (3%) with cells from an overnight culture and incubated anaerobically. Viable-cell counts were performed after 18 h by serial dilution in one-quarter-strength Ringer's solution and enumeration on BHI. The pH was not controlled during the experiment.
Bile challenge and adaptation assays.
Cultures were grown to an A600 of 0.15 (log phase). The cells were centrifuged (12,000 × g for 6 min) and adapted to bile by resuspending them in BHI containing sublethal levels of bile salts (sodium cholate-sodium deoxycholate [1:1]) or were left unadapted (BHI alone). The cells were centrifuged and challenged with BHI containing 0.3% (wt/vol) bile salts. After the 5-s adaptation period, the challenge concentration was obtained by adding the remaining bile salts to the medium to reach a final concentration of 0.3%. When necessary, chloramphenicol (10 μg/ml) was added during adaptation. Viable-cell counts were performed at intervals. Where results are presented as percent survival, this was calculated as viable-cell counts after challenge expressed as a percentage of viable-cell counts at time zero, i.e., immediately prior to treatment.
Stress cross-protection assays.
Log-phase cells were harvested by centrifugation and adapted to sublethal concentrations of various stressors (BHI supplemented with 0.08% bile salts for bile adaptation, BHI adjusted to pH 5.5 with 3 M lactic acid for acid adaptation, BHI prewarmed at 42°C for heat adaptation, or BHI supplemented with 5% NaCl for salt adaptation). Nonadapted cultures were resuspended in fresh BHI. After 1 h at 37°C, the cultures were centrifuged and the cells were resuspended in BHI containing 0.3% bile salts. Viable-cell counts were performed at intervals. For experiments involving SDS, 0.01% was used for adaptation and the challenge concentration was 0.2%.
Creation of a pORI19 integration bank in L. monocytogenes LO28.
An L. monocytogenes LO28 pORI19 integration bank was created as previously described (44) using a modified version of the procedure outlined by Law et al. (30). Briefly, genomic DNA was partially digested with Sau3A to generate fragments with an average size of 500 bp and ligated to plasmid pORI19, which had been digested with BamHI and dephosphorylated with shrimp alkaline phosphatase. The resulting recombinant plasmids were transformed into E. coli EC101, and colonies were selected on Luria-Bertani plates containing erythromycin (250 μg/ml), IPTG (isopropyl-1-β-d-thiogalactopyranoside) (1 mM), and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (40 μg/ml). Plasmids were extracted from pooled transformants and used to transform LO28 harboring pVE6007. Recombinants were selected on BHI with 5 μg of erythromycin/ml at 30°C. The transformants were harvested from the plates and stocked in BHI containing 40% glycerol at −20°C. To force plasmid integration, 50 μl of the bank was used to inoculate 10 ml of prewarmed BHI broth at 42°C overnight, followed by plating on prewarmed BHI with 5 μg of erythromycin/ml at 42°C. Loss of pVE6007 was determined by replica plating onto BHI plates containing 10 μg of chloramphenicol/ml.
Construction of a Tn917 mutant bank.
An L. monocytogenes LO28 Tn917 bank was created using the temperature-sensitive plasmid pTV1-OK (20). Transformants were grown overnight in TSB-YE containing kanamycin (50 μg/ml) at 30°C and subcultured (1%) into TSB-YE containing erythromycin (0.04 μg/ml) at 42°C, and kanamycin-sensitive Tn917 integrants were selected for on TSA-YE containing 10 μg of erythromycin/ml.
Isolation of mutants with diminished bile tolerance and identification of disrupted genes.
Individual mutants were grown overnight in BHI broth in 96-well plates. Aliquots were transferred to fresh BHI broth supplemented with 30% oxgall in 96-well plates. Following 5 h of incubation at 37°C, the cultures were serially diluted in Ringer's solution and enumerated on BHI plates. Disrupted genes were identified in mutants with altered bile tolerance. An inverse-PCR approach was used to elucidate the point of insertion of transposon Tn917. Genomic DNA was digested with HindIII. Following self-ligation, PCRs were performed using primers 4292 (5′-CAAAGCCTAGTAATGCGGTCATTCC-3′) and 4293 (5′-CTTGGAGAGTATAAATTTGACTTG-3′). The PCR products were purified with the QiaexII gel extraction kit (Qiagen, Hilden, Federal Republic of Germany) and cloned into the pCR2.1 vector using the TOPO TA cloning kit (Invitrogen, San Diego, Calif.). Plasmids were isolated with the QIAprep Spin Miniprep kit. Nucleotide sequence determination was performed on an ABI 373 automated sequencer. Restriction enzymes, T4 DNA ligase, and PCR reagents were purchased from Boehinger Mannhein GmbH, Mannheim, Germany, and used according to the manufacturer's recommendations. The bile-sensitive pORI19 mutant was electroporated with the RepA+ helper plasmid, pVE6007, and recovered at 30°C on BHI plates containing erythromycin and chloramphenicol. A transformant was passaged in BHI broth containing erythromycin and chloramphenicol at 30°C. Inserts on the rescued plasmids were amplified by PCR using primers M13F (5′-GTTTTCCCAGTCACGAC-3′) and M13R (5′-CAGGAAACAGCTATGAC-3′).
RESULTS
Growth in the presence of bovine, porcine, and human bile.
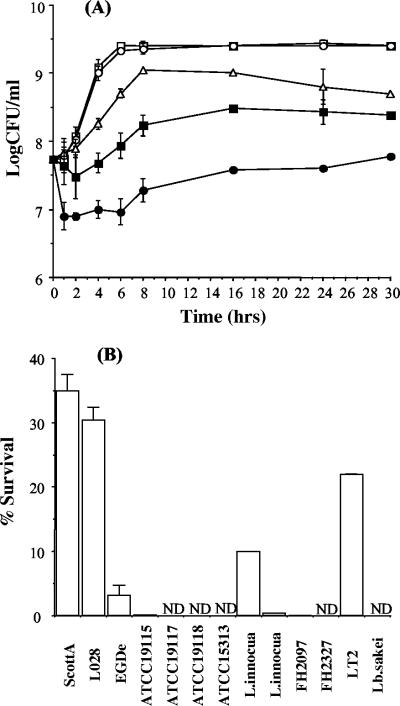
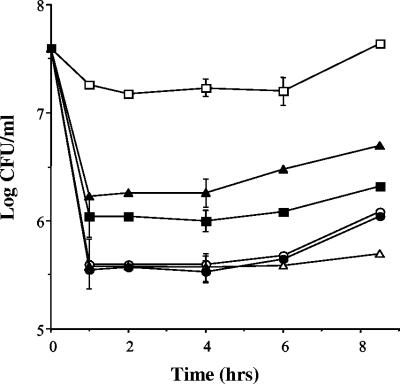
L. monocytogenes LO28 was capable of growth in the presence of all types of bile examined. All experiments in this study were performed anaerobically to mimic in vivo conditions. Confluent growth was observed on agar plates supplemented with the highest concentrations tested (15% oxgall, 15% bovine bile, or 2% porcine bile). We examined the ability of LO28 to tolerate oxgall in BHI broth (Fig. 1A). The growth of cultures was unaffected by the addition of 0.3% oxgall, which is considered an approximation of bile levels encountered in vivo (15, 17). At higher concentrations (10 to 30%), there was a noticeable effect on growth, but even at 30% oxgall, strain LO28 was capable of growth after an initial kill and with an extended lag phase. Generally, Lactobacillus spp. cannot grow in bile levels higher than 0.3% (18, 26, 27), and although Salmonella enterica serovar Typhimurium survives initial exposure to 30% bile, cultures fail to survive beyond 10 h of exposure (48). Importantly, L. monocytogenes LO28 grew to confluence on plates containing physiological concentrations of 0.3% (vol/vol) human bile, and addition of 10% human bile to the broth did not inhibit growth. Therefore, these results indicate the extreme tolerance of L. monocytogenes LO28 to concentrations of bovine, porcine, and, importantly, human bile that exceed levels which would be expected in vivo.
FIG. 1.
(A) Survival of L. monocytogenes LO28 in bovine bile (oxgall). Overnight cultures were inoculated (3%) into BHI broth containing 0.3 (○), 10 (▵), 20 (▪), or 30% (•) oxgall or BHI alone (□). The cultures were incubated anaerobically at 37°C. Viable-cell counts were performed at intervals by serial dilution in one-quarter-strength Ringer's solution and enumeration on BHI. (B) Comparative bile tolerances of various bacteria. Overnight cultures were inoculated into broth containing 30% oxgall. Viable-cell counts (% survival) were performed after 5 min and are expressed as percentages of counts obtained for control cultures, i.e., cells grown in broth without bile. The error bars represent standard deviations of triplicate experiments. ND, not detected.
Comparative bile tolerances of various strains.
We compared the survival of L. monocytogenes LO28 after 5 min in 30% oxgall to the survival of various other bacteria (Fig. 1B). This level was selected because previous experiments had shown that LO28 numbers were reduced by almost one-half log unit after 5 min at this concentration (Fig. 1A). Significant strain variation was observed in the ability to tolerate these high levels of bile. L. monocytogenes ScottA and LO28 showed similarly high tolerances, but strain EGDe, which has recently been sequenced by the Listeria Genome Consortium (16), was significantly more sensitive. For three other L. monocytogenes strains examined (ATCC 19117, ATCC 19118, and ATCC 15313), no survivors were recovered after 5 min. Similar variations were observed among Listeria innocua strains. As expected, the Salmonella strain tested was extremely tolerant and Lactobacillus sakei was not able to survive this high concentration of bile.
Effect of bile salts on growth of L. monocytogenes LO28.
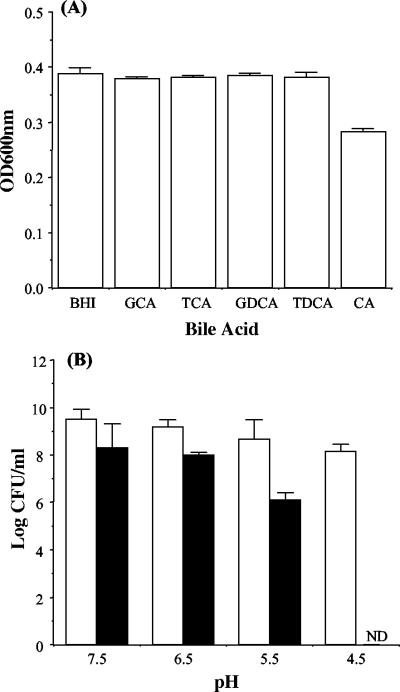
As human bile is primarily composed of conjugated bile acids, the influence of 5 mM selected acids (TCA, GCA, TDCA, GDCA, taurochenodeoxycholic acid, and glycochenodeoxycholic acid) on growth was examined; 5 mM is considered the average concentration prevailing in the intestinal tract (10). It was observed that the growth of L. monocytogenes LO28 was not inhibited on agar containing any of these bile acids. The growth of cultures in broth was also unaffected. After 18 h of incubation, optical density readings were comparable to the reading for L. monocytogenes LO28 grown in BHI alone (Fig. 2A). However, unlike the conjugated bile acids tested, the only unconjugated bile acid tested (cholic acid) significantly suppressed the growth of L. monocytogenes LO28. To approximate in vivo intraluminal conditions under which a wide range of pHs may be encountered (4, 28), we investigated the effect of 5 mM GDCA or TDCA on LO28 in BHI broth adjusted to different pHs. Cultures were unable to grow in BHI at pH 4.5 supplemented with 5 mM GDCA (plated after 18 h of anaerobic incubation), indicating that inhibition is strongly pH dependent for this glycoconjugated bile acid (Fig. 2B). Resistance against the tauroconjugated bile acid TDCA was significantly higher at all pHs tested (as determined by the Student t test).
FIG. 2.
(A) Effects of individual bile acids on growth. Overnight cultures were inoculated into BHI supplemented with 5 mM GCA, TCA, GDCA, TDCA, or cholic acid (CA). The cultures were incubated anaerobically for 16 h at 37°C without shaking. Cell growth was measured spectrophotometrically by determining the optical density at 600 nm (OD600nm). (B) pH dependency of TDCA (open bars) and GDCA (solid bars) toxicities. Viable-plate counts were performed after 18 h of anaerobic incubation in BHI broth set at different initial pHs and containing 5 mM TDCA or GDCA. The error bars represent standard deviations of triplicate experiments. ND, not detected.
Bile challenge and adaptation assays.
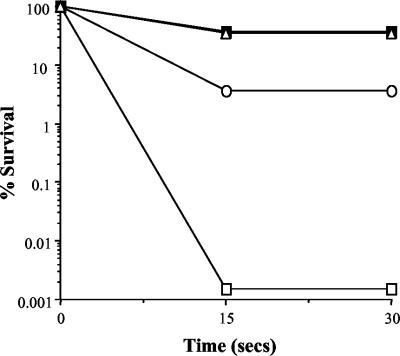
The response of log-phase L. monocytogenes LO28 to lethal concentrations of unconjugated bile salts (sodium cholate-sodium deoxycholate [1:1]) was examined. This mix was chosen because the cells proved to be resistant to the high concentrations of bovine bile and individual conjugated bile acids tested. It was observed that killing was nearly instantaneous in that there was a 5-log-unit reduction in numbers after only 15 s of exposure to 0.3% bile (Fig. 3). Significantly, rapid adaptation to low levels of bile provided substantial protection against challenge with lethal levels. Exposing cells to 0.08% bile salts for as little as 5 s resulted in 37% survival after a normally lethal challenge; this represents a 4.5-log-unit increase over unadapted cells. As expected, addition of chloramphenicol during the brief adaptation period did not prevent the development of acquired bile salt tolerance, confirming that de novo protein synthesis is not required for unconjugated bile salt adaptation (Fig. 3). Although stationary-phase cells showed increased resistance to lethal levels of unconjugated bile salts, adaptation to sublethal levels resulted in the same degree of protection as log-phase cells (37%), and again, addition of chloramphenicol did not affect acquired tolerance (Fig. 3). Given that the adaptive mechanisms are so rapid and are independent of protein synthesis, they most probably result from changes in membrane architecture and composition.
FIG. 3.
Survival of stationary-phase (○) and exponential-phase (□) L. monocytogenes in BHI broth supplemented with 0.3% bile salts (sodium cholate-sodium deoxycholate [1:1]). The cultures were adapted to 0.08% bile for 30 min in the absence (▪) or presence (▵) of chloramphenicol (10 μg/ml).
The relationship between adaptation to bile and other stresses was examined (Table 2). Adaptation to both heat (42°C) and low pH (5.5) resulted in significant cross-protection against lethal levels of bile salts (a 3-log-unit increase in survival). Adaptation to either NaCl or the detergent SDS conferred even greater protection (4-log-unit increased survival). The response to SDS was interesting in that it closely mimicked the response to bile (rapid killing and “flash” adaptation; data not shown) and probably reflects a similar mechanistic basis. Bile adaptation provided a degree of cross-protection against heat and SDS but actually resulted in sensitization to acid challenge (lactic acid [pH 3.5]) (data not shown).
TABLE 2.
Effects of adaptation to sublethal stresses on survival in lethal concentrations of bile saltsa
| Adaptation | % Survival after bile salt challenge | Log increase in survival |
|---|---|---|
| Control | 0.001 ± 0 | |
| Bile salts (0.08%) | 37.0 ± 1.1 | 4.57 |
| Acid (pH 3.5) | 1.30 ± 0.05 | 3.11 |
| Heat (42°C) | 1.00 ± 0.03 | 3.00 |
| NaCl (5%) | 13.38 ± 0.80 | 4.13 |
| SDS (0.01%) | 14.54 ± 1.30 | 4.16 |
Cultures were grown to log phase, adapted to the appropriate stress, and challenged with 0.3% (wt/vol) sodium cholate-sodium deoxycholate (1:1). The results shown are averages of triplicate experiments ± standard deviations.
Isolation and analysis of mutants with diminished bile tolerance.
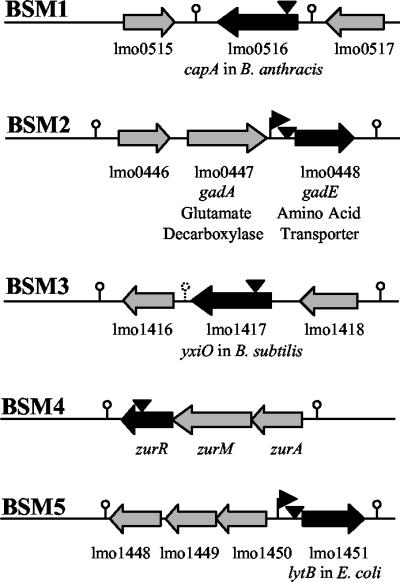
In order to identify the genes involved in the resistance of L. monocytogenes LO28 to bile, we screened independent pORI19 and Tn917 integration banks for mutants unable to cope with these high levels (30% oxgall). Five hundred individual mutants were examined. Of these, five demonstrated a reproducible increase in bile sensitivity compared to the parent. The sites of insertion of the plasmid (one mutant) or the transposon (four mutants) were identified either by plasmid rescue or by inverse PCR (as outlined in Materials and Methods). Southern hybridization confirmed a single insertion event for Tn917 in each of the four mutants (data not shown). The sequence at the insertion point was used to identify the disrupted gene in each case (Fig. 4). For each gene, the National Center for Biotechnology Information annotation number for the corresponding gene in L. monocytogenes EGDe is given. The figure also shows adjacent open reading frames in L. monocytogenes EGDe and putative transcriptional terminators.
FIG. 4.
Sites (vertical arrowheads) of pORI19 (BSM1) or Tn917 (BSM2 to -5) insertion in L. monocytogenes LO28 bile-sensitive mutants. For all genes, the National Center for Biotechnology Information annotation number for the corresponding gene in L. monocytogenes EGDe is given. Genes disrupted in LO28 are indicated by solid arrows. Adjacent open reading frames in EGDe are represented by shaded arrows. Horizontal arrowheads represent putative promoter regions. Putative terminator regions are depicted by lollipops.
In bile-sensitive mutant 1 (BSM1), pORI19 has been inserted at the coding site for amino acid 394 of lmo0516, a homologue of Bacillus anthracis capA, a gene involved in capsule synthesis in that organism. In BSM2, Tn917 has been inserted between the putative promoter region and the ribosomal binding site of the gadE gene (lmo0448), which encodes a putative transporter of the glutamate decarboxylase acid resistance system. In BSM3, the transposon has been inserted into lmo1417 at the coding site for amino acid 115. This locus is homologous to the Bacillus subtilis yxiO gene, which encodes a putative major facilitator superfamily transporter that has significant homologies with putative efflux pumps. The Tn917 insertion in BSM4 was mapped to the coding site for amino acid 116 of lmo1445, which encodes ZurR, a member of the Fur family of transcriptional regulators. Finally, in BSM5, the transposon disrupted the putative promoter region of lmo1451, a homologue of E. coli lytB, a gene involved in isoprenoid biosynthesis.
All five mutants displayed growth rates similar to that of the wild type (as assayed by doubling time) when grown in BHI (pH 7) at 37°C, indicating that the disrupted genes were not necessary for growth under normal physiological conditions (data not shown). On initial exposure to 30% oxgall, there is the expected half-log-unit decrease in survival of the parent, while the numbers of the mutants are significantly lower (as determined by the Student t test), with BSM1 and BSM2 showing ∼1.5-log-unit reductions and BSM3, BSM4, and BSM5 showing ∼2-log-unit reductions (Fig. 5).
FIG. 5.
Survival of wild-type L. monocytogenes LO28 (□) and BSM1 (capA; ▴), BSM2 (gadE; ▪), BSM3 (yxiO; ○), BSM4 (zurR; ▵), and BSM5 (lytB; •) in high levels of bile. Overnight cultures were inoculated (3%) into BHI broth supplemented with 30% (wt/vol) oxgall. Viable-cell counts were performed at intervals by serial dilution in one-quarter-strength Ringer's solution and enumeration on BHI.
For two of the four transposon mutants (gadE and yxiO), an independently created knockout was available from our laboratory culture collection. These were used to confirm an identical reduction in bile tolerance, as displayed by the equivalent transposon mutant. The pORI19::capA mutant was recreated by a second insertional integration into the same gene, and once again an identical phenotype was observed. These results suggest that the observed phenotypes are specific for each gene under investigation and not the results of independent secondary mutations.
DISCUSSION
To survive in the human intestinal tract and subsequently cause disease, food-borne pathogens must resist the action of bile, ∼500 to 1,000 ml of which is secreted daily from the gallbladder into the upper small intestine (24). In this study, we have investigated the ability of L. monocytogenes LO28 to tolerate bile, we have confirmed the ability of LO28 to adapt to sublethal bile concentrations, and we have examined the relationships between bile stress and other environmental signals. Finally, we have identified a number of genetic loci involved in bile resistance in this organism.
L. monocytogenes LO28 was capable of growth in all types of bile examined and at levels that greatly exceed those expected in vivo. In probiotic research, where bile tolerance is considered of primary importance in the selection of strains, resistance is currently measured by the ability of a strain to grow in the presence of 0.3% bovine bile (oxgall) (15, 17). L. monocytogenes LO28 is not affected at this level, and cultures can tolerate and even grow in broth containing levels as high as 30% bovine bile. Importantly, L. monocytogenes grows to confluence at the physiological concentration of 0.3% (vol/vol) human bile, and addition of 10% (vol/vol) human bile to broth does not affect growth.
Human bile is composed of individual conjugated and unconjugated bile acids that are present in the small intestine at average concentrations of 5 mM (10, 22). L. monocytogenes LO28 is very tolerant of individual conjugated bile acids at concentrations up to 20 mM. However, while L. monocytogenes is resistant to the tauroconjugated bile acid TDCA (pKa, 1) at a wide pH range, some sensitivity is demonstrated to the glycoconjugated bile acid GDCA (pKa, 3.9), especially at low pH. This most likely reflects toxicity by intracellular acidification, with the effects of glycoconjugated bile acids manifesting in a manner similar to those of weak inorganic acids (11). Furthermore, stationary-phase L. monocytogenes LO28 was relatively sensitive to the effects of the unconjugated bile salts examined. Such unconjugated bile acids may be encountered in the intestine, where conjugated bile acids are hydrolyzed by the resident microflora (10, 11). The data suggest that L. monocytogenes LO28 is extremely resistant to the effects of human gallbladder bile, which consists primarily of conjugated bile salts. However, it is likely that the growth of the pathogen may be inhibited in transient microenvironments within the intestinal lumen, where glycoconjugated and unconjugated bile acids may be present at low pH.
In order to study the lethality of bile salts in L. monocytogenes, we subjected exponential-phase cells to a concentrated mixture of unconjugated bile salts, a procedure that resulted in a significant and rapid bactericidal effect. However, exposure to sublethal levels of unconjugated bile acids conferred an increased ability to survive subsequent lethal challenges. This adaptation was extremely rapid and occurred independently of protein synthesis. A similar flash adaptation to bile has been previously reported for Enterococcus faecalis (13). Furthermore, L. monocytogenes had a similar physiological response to the anionic detergent SDS, and a significant degree of cross-protection between this detergent and bile was observed. Industrial detergents have previously been shown to form mixed micelles with lipids, resulting in rapid solubilization and disaggregation (31). In our experiments, it is possible that low levels of bile salts or SDS rapidly intercalate with membrane lipids, rendering the subsequent mixed membranes resistant to further detergent effects. Overall, the data suggest that exposure of L. monocytogenes LO28 to sublethal levels of industrial detergents in the food-processing environment, or to low levels of bile in the small intestine during fasting, could potentially influence the bile tolerance of exponential-phase cells. Indeed, any environmental or physiological stimulus capable of altering membrane characteristics, such as acid adaptation (45, 47), increased osmolarity (43), or entry into stationary phase (25, 33), was seen to increase resistance to unconjugated bile salts in L. monocytogenes LO28.
Despite the fact that protein synthesis is not required for physiological adaptation to bile, it is likely that the extremely high level of bile resistance exhibited by L. monocytogenes LO28 is a function of a wide array of proteins which govern cell envelope architecture or enable the maintenance of intracellular homeostasis during exposure to bile. Relatively little information is available concerning genes involved in bile resistance in gram-positive organisms. In a study of bile adaptation in E. faecalis, Flahaut et al. (13) demonstrated the induction of 45 proteins by bile, including the heat shock proteins GroEL. Increased transcription of the groESL operon in response to bile has also been demonstrated in L. monocytogenes (14). Genes that have been shown to play a role in the bile resistance of gram-negative microorganisms include the Tol efflux pump, genes for porins, regulatory genes, and genes involved in lipopolysaccharide synthesis (41, 46, 48). Of the aforementioned genes, analysis of the recently published L. monocytogenes EGDe genome (16) could only identify homologues of the efflux pumps. However, the presence of three genes possibly involved in the degradation of bile salts was noted previously (16), and these are under investigation in our laboratory.
In this study, we identify five genes that, to our knowledge, represent the first indication of loci contributing to bile tolerance in a gram-positive organism. Interestingly, all of the identified loci can be postulated to play putative roles in maintenance of the cell envelope or in stress responses. One of the genes identified is homologous to B. anthracis capA, a locus encoding a membrane-associated enzyme which mediates the polymerization of d-glutamic acid and is essential for encapsulation (36). Analysis of the entire EGDe genome shows the presence of a second capA homologue, lmo0117, but no apparent homologues of the remaining genes in the Bacillus capsule synthesis operon could be identified. It is possible that the capA gene products are involved in cell envelope biogenesis in L. monocytogenes. Another mutation resulted in the disruption of gadE, a gene encoding a putative glutamate transporter and most likely a component of the glutamate decarboxylase acid resistance system (8). This system may play a role in combating the low-pH stress arising from intracellular dissociation of bile salts. Another bile-sensitive mutant contained a disruption in a homologue of B. subtilis yxiO, encoding a putative major facilitator superfamily membrane transporter that shows significant homologies to antibiotic efflux pumps. Such pumps play a significant role in bile resistance in gram-negative bacteria by expelling bile salts that subvert the barrier imposed by the outer membrane (29, 32, 34, 38, 46). Interestingly, we have recently isolated an identical yxiO transposon mutant during a screen for acid-sensitive mutants in L. monocytogenes. We have shown that this locus plays a role in the maintenance of homeostasis under a variety of suboptimal environmental stresses (M. Begley et al., unpublished data). Another disrupted gene, zurR, encodes a member of the Fur family of transcriptional regulators. Although this protein has been shown to play a role in zinc homeostasis (9), the entire regulon it controls has not been identified. It is clear from our data, however, that genes under the influence of ZurR play a role in the tolerance of bile. Analysis of the final mutant identified by our screening procedure revealed an insertion into a homologue of lytB, a gene in E. coli that plays a role in the nonmelvonate pathway of isoprenoid biosynthesis (42). As isoprenoids are involved in peptidoglycan metabolism, disruption of lytB most likely alters cell wall synthesis and could thereby affect resistance to bile. Mutation of lytB has been reported to confer tolerance to penicillin in E. coli (19) and to render Burkholderia pseudomallei more susceptible to polymyxin B (3). Interestingly, L. innocua does not possess an ortholog of the gene disrupted in L. monocytogenes (16).
In conclusion, L. monocytogenes LO28 can tolerate the average concentrations of bile and bile acids encountered in vivo. Furthermore, the data demonstrate that prior adaptation of the bacterium to sublethal levels of bile or other stresses encountered during food processing and sanitizing treatments (acid, heat, salt, and SDS) may alter cellular physiology to enhance bile resistance. Finally, we uncovered some of the molecular mechanisms that confer an extremely high bile tolerance on this strain.
Acknowledgments
We thank C. Dunne for sourcing human bile and H. Cowman for supplying Listeria strains.
This work was funded by the Irish government under the National Development Plan 2000-2006.
REFERENCES
- 1.Allerberger, F., B. Langer, O. Hirsch, M. P. Dierich, and H. P. Seeliger. 1989. Listeria monocytogenes cholecystitis. Z. Gastroenterol. 27:145-147. [PubMed] [Google Scholar]
- 2.Briones, V., M. M. Blanco, A. Marco, N. Prats, J. F. Fernandez-Garayzabal, G. Suarez, M. Domingo, and L. Dominguez. 1992. Biliary excretion as possible origin of Listeria monocytogenes in fecal carriers. Am. J. Vet. Res. 53:191-193. [PubMed] [Google Scholar]
- 3.Burtnick, M. N., and D. E. Woods. 1999. Isolation of polymyxin B-susceptible mutants of Burkholderia pseudomallei and molecular characterization of genetic loci involved in polymyxin resistance. Antimicrob. Agents Chemother. 43:2648-2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carey, M. C., D. M. Small, and C. M. Bliss. 1983. Lipid digestion and absorption. Annu. Rev. Physiol. 45:651-677. [DOI] [PubMed] [Google Scholar]
- 5.Chowdhury, R., G. K. Sahu, and J. Das. 1996. Stress response in pathogenic bacteria. J. Biosci. 21:149-160. [Google Scholar]
- 6.Cotter, P. D., N. Emerson, C. G. M. Gahan, and C. Hill. 1999. Identification and disruption of lisRK, a genetic locus encoding a two-component signal transduction system involved in stress tolerance and virulence in Listeria monocytogenes. J. Bacteriol. 181:6840-6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cotter, P. D., C. G. M. Gahan, and C. Hill. 2001. A glutamate decarboxylase system protects Listeria monocytogenes in gastric fluid. Mol. Microbiol. 40:465-475. [DOI] [PubMed] [Google Scholar]
- 8.Cotter, P. D., C. G. M. Gahan, and C. Hill. 2000. Analysis of the role of the Listeria monocytogenes F0F1-ATPase operon in the acid tolerance response. Int. J. Food Microbiol. 60:137-146. [DOI] [PubMed] [Google Scholar]
- 9.Dalet, K., E. Gouin, Y. Cenatiempo, P. Cossart, and Y. Héchard. 1999. Characterisation of a new operon encoding a Zur-like protein and an associated ABC zinc permease in Listeria monocytogenes. FEMS Microbiol. Lett. 174:111-116. [DOI] [PubMed] [Google Scholar]
- 10.De Boever, P., and W. Verstraete. 1999. Bile salt deconjugation by Lactobacillus plantarum 80 and its implication for bacterial toxicity. J. Appl. Microbiol. 87:345-352. [DOI] [PubMed] [Google Scholar]
- 11.De Smet, I., L. Van Hoorde, M. Vande Woestyne, H. Christiaens, and W. Verstraete. 1995. Significance of bile salt hydrolytic activities of Lactobacilli. J. Appl. Bacteriol. 79:292-301. [DOI] [PubMed] [Google Scholar]
- 12.Ferreira, A., C. P. O'Byrne, and K. J. Boor. 2001. Role of σB in heat, ethanol, acid, and oxidative stress resistance and during carbon starvation in Listeria monocytogenes. Appl. Environ. Microbiol. 67:4454-4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flahaut, S., J. Frere, P. Boutibonnes, and Y. Auffray. 1996. Comparison of bile salts and sodium dodecyl sulfate stress responses in Enterococcus faecalis. Appl. Environ. Microbiol. 62:2416-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gahan, C. G. M., J. O'Mahony, and C. Hill. 2001. Characterization of the groESL operon in Listeria monocytogenes: utilization of two reporter systems (gfp and hly) for evaluating in vivo expression. Infect. Immun. 69:3924-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilliland, S. E., T. E. Stanley, and L. J. Bush. 1984. Importance of bile tolerance of Lactobacillus acidophilus used as a dietary adjunct. J. Dairy Sci. 67:3045-3051. [DOI] [PubMed] [Google Scholar]
- 16.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. Garcia-Del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. Mata Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Pérez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vázquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 17.Goldin, B. R., and S. L. Gorbach. 1992. Probiotics for humans, p. 355-376. In R. Fuller (ed.), Probiotics, the scientific basis. Chapman and Hall, London, United Kingdom.
- 18.Gupta, P. K., B. K. Mital, and S. K. Garg. 1996. Characterization of Lactobacillus acidophilus strains for use as dietary adjunct. Int. J. Food Microbiol. 29:105-109. [DOI] [PubMed] [Google Scholar]
- 19.Gustafson, C. E., S. Kaul, and E. E. Ishiguro. 1993. Identification of the Escherichia coli lytB gene, which is involved in penicillin tolerance and control of the stringent response. J. Bacteriol. 175:1203-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gutierrez, J. A., P. J. Crowley, D. P. Brown, J. D. Hillan, P. Youngman, and A. S. Bleiweis. 1996. Insertional mutagenesis and recovery of interrupted genes of Streptococcus mutans by using transposon Tn917: preliminary characterization of mutants displaying acid sensitivity and nutritional requirements. J. Bacteriol. 178:4166-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hill, C., P. D. Cotter, R. D. Sleator, and C. G. M. Gahan. 2002. Bacterial stress response in Listeria monocytogenes: jumping the hurdles imposed by minimal processing. Int. Dairy J. 12:273-283. [Google Scholar]
- 22.Hofmann, A. F., G. Molino, M. Milanese, and G. Belforte. 1983. Description and stimulation of a physiological pharmacokinetic model for the metabolism and enterohepatic circulation of bile acids in man. J. Clin. Investig. 71:1003-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hofmann, A. F., and K. J. Mysels. 1992. Bile acid solubility and precipitation in vitro and in vivo: the role of conjugation, pH and Ca2+ ions. J. Lipid Res. 33:617-626. [PubMed] [Google Scholar]
- 24.Hofmann, A. F. 1994. Bile acids, p. 677-718. In I. M. Arias, J. L. Boyer, N. Fausto, W. B. Jackoby, D. A. Schachter, and D. A. Shafritz (ed.), The liver: biology and pathobiology. Raven Press, New York, N.Y.
- 25.Huisman, G. W., D. A. Siegele, M. M. Zambrano, and R. Kolter. 1996. Morphological and physiological changes during stationary phase, p. 1672-1682. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, D.C.
- 26.Hyronimus, B., C. Le Marrec, A. Hadj Sassi, and A. Deschamps. 2000. Acid and bile tolerance of spore-forming lactic acid bacteria. Int. J. Food Microbiol. 61:193-197. [DOI] [PubMed] [Google Scholar]
- 27.Jacobsen, C. N., V. Rosenfeldt Nielsen, A. E. Hayford, P. L. Møller, K. F. Michaelsen, A. Paerregaard, B. Sandström, M. Tvede, and M. Jakobsen. 1999. Screening of probiotic activities of 47 strains of Lactobacillus spp. by in vitro techniques and evaluation of the colonization ability of 5 selected strains in humans. Appl. Environ. Microbiol. 65:4949-4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kitagawa, K., A. Nishigori, N. Murata, K. Nishimoto, and H. Takada. 1966. Radiotelemetry of the pH of the gastrointestinal tract by glass electrode. Gastroenterology 51:368-372. [PubMed] [Google Scholar]
- 29.Lacroix, F. J., A. Cloeckaert, O. Grepinet, C. Pinault, M. Y. Popoff, H. Waxin, and P. Pardon. 1996. Salmonella typhimurium acrB-like gene: identification and role in resistance to biliary salts and detergents and role in murine infection. FEMS Microbiol. Lett. 135:161-167. [DOI] [PubMed] [Google Scholar]
- 30.Law, J., G. Buist, A. Haandrikman, J. Kok, G. Venema, and K. Leenhouts. 1995. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J. Bacteriol. 177:7011-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le Maire, M., P. Champeil, and J. V. Møller. 2000. Interaction of membrane proteins and lipids with solubilizing detergents. Biochim. Biophys. Acta 1508:86-111. [DOI] [PubMed] [Google Scholar]
- 32.Lomovskaya, O., and K. Lewis. 1992. Emr, an Escherichia coli locus for multidrug resistance. Proc. Natl. Acad. Sci. USA 89:8938-8942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lou, Y., and A. E. Yousef. 1996. Resistance of Listeria monocytogenes to heat after adaptation to environmental stresses. J. Food Prot. 59:465-471. [DOI] [PubMed] [Google Scholar]
- 34.Ma, D., D. N. Cook, M. Alberti, N. G. Pon, H. Nikaido, and J. E. Hearst. 1995. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol. Microbiol. 16:45-55. [DOI] [PubMed] [Google Scholar]
- 35.Maguin, E., P. Duwat, T. Hege, D. Ehrlich, and A. Gruss. 1992. New thermosensitive plasmid for gram-positive bacteria. J. Bacteriol. 174:5633-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Makino, S., I. Uchida., N. Terakado, C. Sasakawa, and M. Yoshikawa. 1989. Molecular characterization and protein analysis of the cap region, which is essential for encapsulation in Bacillus anthracis. J. Bacteriol. 171:722-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morita, Y., K. Kodama, S. Shiota, T. Mine, A. Kataoka, T. Mizushima, and T. Tsuchiya. 1998. NorM, a putative multidrug efflux protein of Vibrio parahaemolyticus and its homolog in Escherichia coli. Antimicrob. Agents Chemother. 42:1778-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pace, J. L., T. J. Chai, H. A. Rossi, and X. Jiang. 1997. Effect of bile on Vibrio parahaemolyticus. Appl. Environ. Microbiol. 63:2372-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pope, L. M., K. E. Reed, and S. M. Payne. 1995. Increased protein secretion and adherence to HeLa cells by Shigella spp. following growth in the presence of bile salts. Infect. Immun. 63:3642-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prouty, A. M., J. C. Van Velkhinburgh, and J. S. Gunn. 2002. Salmonella enterica serovar Typhimurium resistance to bile: identification and characterization of the tolQRA cluster. J. Bacteriol. 184:1270-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rohdich, F., S. Hecht, K. Gärtner, P. Adam, C. Krieger, S. Amslinger, D. Arigoni, A. Bacher, and W. Eisenreich. 2002. Studies on the nonmevalonate terpene biosynthetic pathway: metabolic role of IspH (LytB) protein. Proc. Natl. Acad. Sci. USA 99:1158-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Russell, N. J., R. I. Evans, P. F. ter Steeg, J. Hellemons, A. Verheul, and T. Abee. 1995. Membranes as a target for stress adaptation. Int. J. Food Microbiol. 28:255-261. [DOI] [PubMed] [Google Scholar]
- 44.Sleator, R. D., J. Wouters, C. G. M. Gahan, T. Abee, and C. Hill. 2001. Analysis of the role of opuC, an osmolyte transport system, in salt tolerance and virulence potential of Listeria monocytogenes. Appl. Environ. Microbiol. 67:2692-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Slonczewski, J. L., and J. W. Foster. 1996. pH-regulated genes and survival at extreme pH, p.1539-1549. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, D.C.
- 46.Thanassi, D. G., L. W. Cheng, and H. Nikaido. 1997. Active efflux of bile salts by Escherichia coli. J. Bacteriol. 179:2512-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Schaik, W., C. G. M. Gahan, and C. Hill. 1999. Acid-adapted Listeria monocytogenes displays enhanced tolerance against the lantibiotics nisin and lacticin 3147. J. Food. Prot. 62:536-539. [DOI] [PubMed] [Google Scholar]
- 48.Van Velkinburgh, J. C., and J. S. Gunn. 1999. PhoP-PhoQ-regulated loci are required for enhanced bile resistance in Salmonella spp. Infect. Immun. 67:1614-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vlahcevic, Z. R., D. M. Heuman, and P. B. Hylemon. 1990. Physiology and pathophysiology of enterohepatic circulation of bile acids, p. 341-377. In D. Zakim and T. D. Boyer (ed.), Hepatology. A textbook of liver disease. W. B. Saunders Company, Philadelphia, Pa.