Abstract
A novel haloarchaeal strain, Haloarcula sp. strain D1, grew aerobically on 4-hydroxybenzoic acid (4HBA) as a sole carbon and energy source and is the first member of the domain Archaea reported to do so. Unusually, D1 metabolized 4HBA via gentisic acid rather than via protocatechuic acid, hydroquinone, or catechol. Gentisate was detected in 4HBA-grown cultures, and gentisate 1,2-dioxygenase activity was induced in 4HBA-grown cells. Stoichiometric accumulation of gentisate from 4HBA was demonstrated in 4HBA-grown cell suspensions containing 2,2′-dipyridyl (which strongly inhibits gentisate 1,2-dioxygenase). To establish whether initial 1-hydroxylation of 4HBA with concomitant 1,2-carboxyl group migration to yield gentisate occurred, 2,6-dideutero-4HBA was synthesized and used as a substrate. Deuterated gentisate was recovered from cell suspensions and identified as 3-deutero-gentisate, using gas chromatography-mass spectrometry and proton nuclear magnetic resonance spectroscopy. This structural isomer would be expected only if a 1,2-carboxyl group migration had taken place, and it provides compelling evidence that the 4HBA pathway in Haloarcula sp. strain D1 involves a hydroxylation-induced intramolecular migration. To our knowledge, this is the first report of a pathway which involves such a transformation (called an NIH shift) in the domain Archaea.
Biodegradation of aromatic compounds has historically been one of the most intensively studied aspects of microbial catabolism in both bacteria and eukaryotic microorganisms. In contrast, little is known about the metabolism of aromatic compounds by microorganisms within the domain Archaea. This is despite growing evidence that the Archaea are a very diverse group, the members of which inhabit a broad range of environmental niches (5, 8, 21, 47) and are intimately involved in many important and poorly understood biogeochemical processes (43). Aromatic degradation pathways in the Archaea are of considerable interest in evolutionary terms. Also, it has been suggested that biodegradative enzymes from Archaea (and other extremophilic microorganisms) may be exploited to remove pollutants from industrial effluents or as novel biocatalysts (44, 54, 70).
Aerobic degradation of aromatic compounds occurs in some members of the extremely halophilic Archaea, or haloarchaea (order Halobacteriales)—the major group of aerobic Archaea (67). These organisms are obligate and extreme halophiles, often growing optimally in near-saturated brines (∼5.2 M NaCl). Unlike many halophilic bacteria that use compatible organic solutes (31), haloarchaea actively accumulate very high cytoplasmic concentrations of KCl to avoid osmotic stress (51, 67). Consequently, as many parts of their cellular machinery are highly adapted to function in the presence of very high salt concentrations (20, 53), enzymes from haloarchaea are of considerable biotechnological interest (44). To date, aerobic metabolism of a limited range of aromatic compounds has been demonstrated in a single haloarchaeal isolate from the nutritionally versatile genus Haloferax (24, 29, 30). Haloferax sp. strain D1227 metabolizes benzoic acid (BA), 3-hydroxybenzoic acid (3HBA), 3-phenylpropionic acid, and cinnamic acid via a gentisate pathway. The only ring cleavage dioxygenase enzyme described in any archaeon to date (gentisate 1,2-dioxygenase [GDO]) has been purified, and the encoding gene has been cloned from this strain (29). As yet, there have been no reports describing metabolism of aromatic compounds in other haloarchaeal genera. In particular, almost nothing is known about the initial hydroxylation reactions that occur during metabolism of aromatic compounds by Haloferax sp. strain D1227 or by any other member of the domain Archaea.
During the course of work to investigate the metabolism of aromatic compounds in hypersaline environments, a novel haloarchaeal strain (Haloarcula sp. strain D1) was isolated. The strain grew on 4-hydroxybenzoic acid (4HBA) as a sole carbon and energy source and is the first member of the domain Archaea reported to do so. Although not a priority pollutant in the wider environment, 4HBA is present as a contaminant in certain highly saline industrial effluents, such as wastewater from olive oil manufacture (7, 22). Thus, the biodegradation of 4HBA by halophilic microorganisms is of interest in terms of effluent bioremediation. 4HBA is also a common intermediate in the biodegradation of lignin (14) and is therefore an important compound in the short-term cycling of environmental carbon.
In order to compare 4HBA degradation pathways in different evolutionary domains, experimental work was undertaken to establish which pathway Haloarcula sp. strain D1 utilizes to assimilate 4HBA. Specifically, we tested the hypothesis that Haloarcula sp. strain D1 metabolized 4HBA by a somewhat unusual metabolic route, involving 2,5-dihydroxybenzoic acid (gentisic acid [GA]) as an intermediate. Using selectively deuterated 4HBA and proton nuclear magnetic resonance (1H-NMR) spectroscopy, convincing evidence was obtained that an intramolecular migration (called an NIH shift) of a carboxyl group, or possibly a thioester derivative, occurred during metabolism of 4HBA to GA by Haloarcula sp. strain D1. Several alternative mechanisms that might account for this unusual NIH shift are considered here.
MATERIALS AND METHODS
Microbial strains.
Haloferax sp. strain D1227 (ATCC 51408) (24) was used as a reference strain throughout this study. Although unable to grow on 4HBA, Haloferax sp. strain D1227 can grow aerobically on a limited range of other aromatic acids as sole carbon and energy sources: BA, 3HBA, phenylpropionic acid, and cinnamic acid. Haloferax sp. strain D1227 appears to metabolize all of these compounds via a previously characterized gentisate pathway (29, 30). Haloarcula sp. strain D1 was isolated from a high-salt enrichment culture containing BA.
Media and growth conditions.
Both strains were maintained on agar plates containing 1.5% (wt/vol) ultrapure agarose (Bethesda Research Laboratories) dissolved in a halophile growth medium (HGM) which contained the following basal salts: MgCl2 · 6H2O (19.5 g · liter−1), MgSO4 · 7H2O (29.0 g · liter−1), CaCl2 · 2H2O (1.1 g · liter−1), KCl (6.0 g · liter−1), NaBr (0.5 g · liter−1), (NH4)2SO4 (10.0 g · liter−1), and NaCl (174.0 g · liter−1). The medium was adjusted to pH 7.5 (5 M KOH), and Tris-HCl buffer (10 mM) was routinely added to maintain the pH at 7.5. Agar plates also contained yeast extract (1.5 g · liter−1; Oxoid Ltd.) and Casamino Acids (1.5 g · liter−1; Difco Ltd.). To prevent precipitation of medium components, a stock solution of yeast extract and Casamino Acids was prepared, autoclaved separately, and aseptically added to molten HGM agar when it had cooled below 50°C. After inoculation, the plates were incubated at 40°C in sealed polyethylene bags until colonies were visible (1 to 2 weeks) and were then stored at room temperature in sealed bags.
Single colonies were inoculated into liquid HGM starter cultures (5 ml) and incubated in 25-ml universal bottles in a rotary shaker (40°C; 200 rpm). When late logarithmic growth phase was reached (typically after 1 to 3 days), they were used to inoculate subsequent cultures. Starter cultures contained both yeast extract (1.5 g · liter−1) and Casamino Acids (1.5 g · liter−1), aseptically added to autoclaved HGM from a sterile stock solution as described above. Otherwise, liquid HGM cultures contained yeast extract (100 mg · liter−1; added from an autoclaved stock solution) and either pyruvic acid, BA, 3HBA, or 4HBA (all sodium salts) as growth substrates, added to autoclaved HGM as filter-sterilized stock solutions. The substrate concentrations used for growth were either 4 (aromatic acids) or 10 mM (pyruvic acid). Liquid cultures (25 ml) were incubated in 150-ml bottles. When required, larger-scale cultivation was performed using cultures (750 ml) grown in 2-liter conical flasks or a 10-liter fermentor (Biolab; B. Braun Biotech Ltd.). Fermentor cultures contained 7 liters of HGM (40°C; 200-rpm agitation; airflow, 10 liters · min−1; air outlet condenser fitted) and were inoculated with three 25-ml starter cultures. Fed-batch growth was employed to maximize the yield of cells from larger-scale cultures grown on aromatic acids. As the cultures reached the late logarithmic or early stationary phase of growth (as determined by measurement of the optical density at 550 nm [OD550]), additional growth substrate (4 mM) and additional yeast extract (100 mg · liter−1) were aseptically added to the growth medium. The cells were harvested 24 h later by centrifugation (8,000 × g; 30 min; 4°C).
Growth experiments.
Triplicate cultures were incubated in 150-ml bottles along with sterile (uninoculated) and substrate-free controls. Duplicate samples (0.5 ml) were aseptically removed at intervals and used for determination of the OD550 before being centrifuged (15,800 × g; 5 min). Aliquots of the supernatant (0.2 ml) were diluted 1:1 with appropriate double-strength isocratic high-performance liquid chromatography (HPLC) buffer or solvent (see below), and the diluted samples were sealed in crimp vials with butyl rubber-polytetrafluoroethylene caps (Supelco Ltd.). Samples were stored in the dark at 4°C until they were analyzed.
Resting cell experiments.
Cells were harvested from 750-ml or 7-liter cultures by centrifugation as described above and washed once (in buffered HGM), before being resuspended in buffered HGM to give an OD550 of approximately 8.0. Aliquots (5 ml) were dispensed into sterile universal tubes, test substrates (2 mM) were added, time zero samples (0.5 ml) were withdrawn for HPLC analysis, and the tubes were incubated at 40°C. The tubes were incubated on their sides in an orbital shaker (200 rpm). Further samples (0.5 ml) were withdrawn at intervals (typically every 2 to 4 h) and were processed and stored as described above prior to HPLC analysis. Control tubes contained either cells and no added substrate or buffered HGM and the test substrate only. In some experiments, cell suspensions also contained 2,2′-dipyridyl (1 mM), which was dissolved in HGM prior to resuspension of the harvested cells.
Preparation of cell extracts.
Cells from two 750-ml cultures were harvested, washed (in HGM), and resuspended in 20 ml of 100 mM Tris-HCl buffer (pH 7.5) containing 3.0 M KCl, 50 mM MgCl2 · 6H2O, and 5 mM sodium ascorbate (29). The cells were lysed by sonication (six times for 30 s each time; the lysate was cooled on ice between bursts), and the lysate was clarified by centrifugation (25,000 × g; 60 min). The filtered (0.2-μm pore size) supernatant was referred to as cell-free extract (CFE) and was diluted (using high-salt buffer; see above) as necessary prior to enzyme assays. Protein concentrations in CFE samples were estimated using the BCA Protein Assay kit (Pierce Chemicals Ltd.) and were typically between 2 and 5 mg · ml of protein−1 when undiluted.
Enzyme assays.
Ring cleavage dioxygenase activity, using a range of di- and trihydroxyaromatic substrates, was assayed in cell extracts by detection of substrate-dependent oxygen uptake (polarography), using a Clarke-type oxygen electrode (Rank Brothers, Cambridge, United Kingdom). Oxygen uptake rates were measured at 30°C following the addition of test substrates to aliquots of diluted CFE, essentially as described previously (41).
Rates of substrate-induced oxygen uptake were determined in duplicate samples of diluted CFE (2 ml), and specific activities (nanomoles of O2 consumed per minute per milligram of protein) were calculated. For negative controls, aliquots of distilled H2O (dH2O) were added to cell extracts, substrates were added to buffer containing no CFE, or solid 2,2′-dipyridyl (1 mM) was added directly to cell extracts prior to enzyme assays. Assays were performed in the presence and absence of ferrous ammonium sulfate [Fe(NH4)2(SO4)2; 50 μM], which has been shown to activate Fe(II)-containing ring cleavage dioxygenases in some instances (27, 40, 41, 52, 63). A total of 12 aromatic compounds were tested as possible ring cleavage dioxygenase substrates (Table 1). For calculation of oxygen uptake rates, dH2O which was saturated with oxygen at 30°C was assumed to contain 250 μM O2 (0.5 μmol in a 2-ml cell volume) (32).
TABLE 1.
GDO activity in cell extractsa
| Strain | Growth substrate | Sp act (nmol of O2/min/mg of (protein)b |
|---|---|---|
| Haloferax sp. strain D1227 | BA | 427 ± 32 |
| Pyruvic acid | ND | |
| Haloarcula sp. strain D1 | BA | 3.7 ± 0.1 |
| 4-HBA | 2.6 ± 0.1 | |
| Pyruvic acid | ND |
GDO activity in crude cell extracts was assayed by measuring the rate of gentisate-dependent oxygen uptake using a Clarke-type oxygen electrode, as described in the text.
The specific activities shown are the means of duplicate assays (± standard deviation) performed in buffer containing 3 M KCl; ND, no activity was detected. No GDO activity remained in control extracts which were preincubated with 2,2′-dipyridyl (1 mM) for 15 min. No substrate-dependent oxygen uptake was measured in any extract in response to the addition of the following compounds: catechol (1,2-dihydroxybenzene); protocatechuate (3,4-dihydroxybenzoic acid); 2,3-, 2,4-, 2,6-, or 3,5-dihydroxybenzoic acid; 2,3,4- or 2,4,6-trihydroxybenzoic acid; or hydroquinone (1,4-dihydroxybenzene). Addition of either hydroxyhydroquinone (HHQ) or 3,4,5-trihydroxybenzoic acid to cell extracts resulted in detectable increases in O2 uptake rates. However, as these compounds also induced equivalent or higher levels of O2 uptake in buffer-only control assays (up to 60 nmol of O2/min in the case of HHQ), this was attributed to their rapid autoxidation in the oxygen electrode cell.
Synthesis of 2,6-dideutero-4HBA.
4HBA was selectively deuterated in the presence of a rhodium catalyst, using a method similar to that used for the synthesis of 2,6-dideuterobenzoic acid (11, 42). Dry sodium 4-hydroxybenzoate (2.5 g; 15.6 mmol) and rhodium(III) chloride (0.5 g; 2.4 mmol) were dissolved in a mixture of anhydrous dimethylformamide (50 ml) and D2O (25 ml). The reaction mixture was refluxed under a dry nitrogen atmosphere for 18 h; the pH of the reaction mixture was then adjusted to 2.0 by dropwise addition of concentrated H2SO4. It was extracted with diethyl ether (three times with 50 ml each time), and the ether extracts were combined and washed with saturated NaCl solution (50 ml) followed by dH2O (50 ml). The organic phase was dried over anhydrous Na2SO4, and the ether was removed under reduced pressure to yield the deuterated product, 2,6-dideutero-4HBA (see Fig. 4, structure I), as an off-white crystalline solid. The deuterated compound was dried in a desiccator under vacuum and then stored under dry nitrogen. No further purification of the product was considered necessary. Samples of the deuterated compound and of authentic 4HBA were analyzed by mass spectrometry (MS) and by 1H-NMR spectroscopy to estimate the yield of deuterated product and for assignment of the incorporated deuterium atoms (see Results).
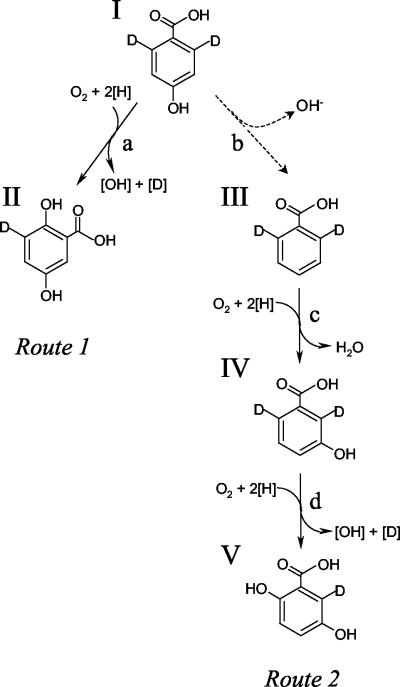
FIG. 4.
Alternative metabolic routes which could account for biotransformation of 4HBA to 2,5-dihydroxybenzoic acid (GA). Route 1 (I → II), 1-hydroxylation with 1,2-migration of COOH group; route 2 (I → III → IV → V), dehydroxylation and upper benzoate pathway. The structures shown are deuterated isomers of 4HBA (I), BA (III), 3HBA (IV), and GA (II and V). The putative enzyme activities shown are 4HBA 1-hydroxylase (a), 4HBA 4-dehydroxylase (b), BA 3-hydroxylase (c), and 3HBA 6-hydroxylase (d). Note that 4-dehydroxylation (b, broken arrow) of 4HBA is an entirely hypothetical transformation.
Recovery of deuterated 2,5-dihydroxybenzoic acid.
Deuterated GA was recovered from the combined supernatants of resting cell experiments as follows. 2,2′-Dipyridyl was complexed by the addition of solid Fe(II)(SO4)2 (1 g), as it forms a highly water-soluble complex with Fe2+ ions. The resulting deep-red solution was acidified (to pH 3) by dropwise addition of concentrated H2SO4, and the GA was extracted using diethyl ether (three times with 50 ml each time). Optimal recovery of GA was confirmed using HPLC before the aqueous phase was discarded. The combined organic extracts were dried over anhydrous Na2SO4, and the ether was removed under reduced pressure to yield a mixture of recovered dideuterated substrate and deuterated GA product; these were separated by preparative layer chromatography (PLC). In order to maximize the recovery of separated acids from PLC plates (and to facilitate analysis by gas chromatography [GC]), the mixture was first methylated with diazomethane (CH2N2). Diazomethane was synthesized from Diazald (N-methyl-N-nitroso-p-toluenesulfonamide) in a Mini Diazald apparatus (Sigma Aldrich Ltd.), used according to the manufacturer's instructions.
The compounds to be methylated were redissolved in a minimum volume (∼1 ml) of diethyl ether, and an ethereal solution of freshly prepared diazomethane was added, dropwise and with swirling, until the solution remained yellow. The reaction mixture was kept at 0°C for 60 min, after which the excess diazomethane was destroyed by dropwise addition of glacial acetic acid. The solvents were then removed under vacuum to leave the methylated products, which were separated by PLC.
PLC.
Normal-phase PLC was performed using silica gel G-UV254 PLC plates. Samples of the methylated products were applied to the origin of each plate in a horizontal line ∼15 cm in length, and the plates were developed twice in a solvent system containing 10% (vol/vol) diethyl ether and 90% (vol/vol) n-hexane. In this solvent system, methylated 4HBA (methyl-4-hydroxybenzoate) had Rf values of 0.264 and 0.4 (first and second development, respectively), and methylated GA (methyl-2,5-dihydroxybenzoate or methylgentisate) had Rf values of 0.04 and 0.067 (first and second developments, respectively). Development of PLC plates was performed under a nitrogen atmosphere to minimize autoxidation of potentially unstable dihydroxyaromatic compounds. Bands on the plates were visualized under short-wavelength UV light, their positions were marked, and they were carefully scraped off the plates and extracted using diethyl ether (three times with 5 ml each time). The extracts were combined and dried over anhydrous NaSO4, and the ether was removed under a stream of dry nitrogen gas to yield the isolated compounds, which were stored under nitrogen to prevent autoxidation.
Analytical techniques.
Aromatic acids were routinely detected and quantified using a System Gold HPLC system (Beckman Instruments Ltd.), which comprised a model AS507E autosampler, a model 128 pump module, and a model 168 UV photodiode array detector. Analyses were performed in reverse-phase mode at room temperature, using isocratic elution (flow rate, 1 ml · min−1) in a solvent containing ammonium acetate buffer (100 mM; pH 4.2) and either 20 or 40% (vol/vol) methanol. Under these conditions, BA typically eluted in 4.72 min (40% MeOH), 3HBA typically eluted in 5.58 min (20% MeOH), 4HBA typically eluted in 4.97 min (20% MeOH), and GA typically eluted in 2.05 min (20% MeOH). Aromatic acids were quantified by reference to appropriate external standards, which were detected at the following wavelengths: 234 (BA), 236 (3HBA), 253 (4HBA), and 320 nm (GA). These analytes had the following UV spectral characteristics [λmax (intensity)]: BA, 234 nm (100%) and 272 nm (13%); 3HBA, 236 nm (100%) and 292 nm (44%); 4HBA, 253 nm (100%); and GA, 234 nm (100%) and 320 nm (94%). All HPLC separations were performed using a Beckman Ultrasphere ODS (C18) column (5 μm; 4.7 by 150 mm).
Mass spectra were obtained at 70 eV (electron impact [EI+] mode) using a VG Autospec X series mass spectrometer (Fisons Instruments Ltd.). GC-MS was performed using a model 8065 gas chromatograph (Fisons Instruments Ltd.) interfaced to a mass selective detector (VG Quattro Dual Sector mass spectrometer; Fisons Instruments Ltd.). Ion currents between m/z 30 and 650 were monitored.
1H-NMR spectroscopy was performed using either a 300- (Avance 300; Bruker Instruments Ltd.) or a 500-MHz (DRX500; Bruker Instruments Ltd.) NMR spectrometer. Samples were dissolved in 500-μl aliquots of either deuterated trichloromethane (CDCl3) or deuterated acetone (CD3COCD3), and proton chemical shifts (δH) were reported in parts per million downfield from a tetramethylsilane [Si(CH3)4] external standard.
Chemicals.
All chemicals, reagents, and solvents used were of analytical grade and were purchased from commercial sources.
RESULTS
Aerobic growth of Haloarcula sp. strain D1 on 4HBA.
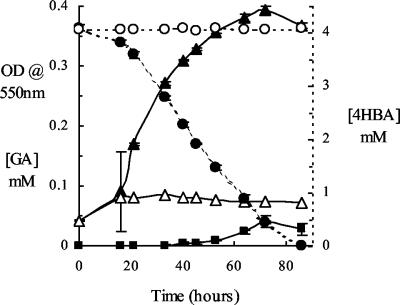
To confirm that Haloarcula sp. strain D1 was able to grow aerobically on 4HBA as a sole carbon and energy source, time course growth experiments were undertaken and culture samples were withdrawn and analyzed at intervals using reverse-phase HPLC. 4HBA-dependent growth and complete removal of the substrate from the growth medium over an 86-h period were demonstrated (Fig. 1). As the growth medium contained a trace of yeast extract (100 mg · liter−1), some background growth was seen in the no-4HBA control cultures (which reached a maximum OD550 of 0.085). A compound which was tentatively identified as GA (on the basis of its very distinctive UV spectrum and chromatographic properties) was detected in 4HBA-grown cultures during the late logarithmic-early stationary phase of growth (reaching a maximum concentration of 3.9 μM). GA was detected during growth of Haloferax sp. strain D1227 on both BA and 3HBA (data not shown), which was consistent with results obtained previously with this strain (29, 30). It was also confirmed that Haloferax sp. strain D1227 is unable to grow on 4HBA, as noted earlier (24).
FIG. 1.
Growth of Haloarcula sp. strain D1 on 4HBA. OD550s in cultures containing 4HBA (▴) and in controls containing no added substrate (▵; background growth on 100 mg of yeast extract liter−1 in medium), along with substrate concentrations in cultures (•) and sterile controls (○), are shown. Low concentrations of GA (▪) were detected during the late logarithmic and early stationary phases of growth. The data shown are the means of triplicate determinations; the error bars represent ±1 standard deviation.
Growth of Haloarcula sp. strain D1 on 4HBA occurred only under aerobic conditions; the strain was unable to grow on 4HBA under anaerobic conditions, even in the presence of alternative respiratory electron acceptors, such as nitrate, dimethyl sulfoxide, and trimethylamine-N-oxide (data not shown). To our knowledge, 4HBA degradation in growth media containing >20% (wt/vol) dissolved salts has not been described previously, and Haloarcula sp. strain D1 is the first member of the domain Archaea that has been reported to grow on 4HBA. The suggestion that this isolate possessed a somewhat unusual 4HBA degradation pathway (in which GA might be a metabolic intermediate) was particularly interesting and was the subject of further experimental work.
Accumulation of GA in resting cell suspensions containing 2,2′-dipyridyl.
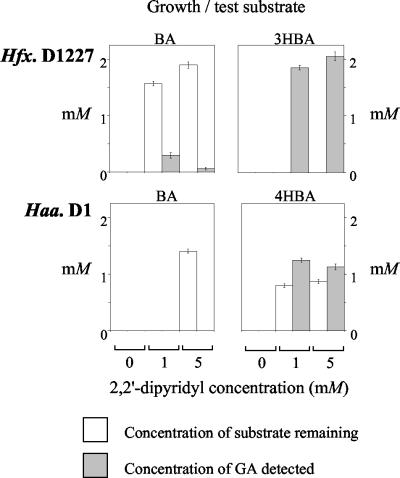
Cultures of Haloferax sp. strain D1227 and Haloarcula sp. strain D1 were grown on various substrates (pyruvic acid, BA, 3HBA, or 4HBA as appropriate), and then washed cell suspensions were incubated with the appropriate growth substrates (2 mM) for 18 h in the presence and absence of 2,2′-dipyridyl (0, 1, and 5 mM). Clear accumulation of GA was demonstrated in some cell suspensions (Fig. 2). The pattern of GA accumulation in suspensions of Haloferax sp. strain D1227 was consistent with previous work on this strain (29, 30). Significantly, stoichiometric accumulation of GA (up to 1.25 mM) was observed in suspensions of 4HBA-grown Haloarcula sp. strain D1 which were incubated with 4HBA in the presence of 2,2′-dipyridyl. This again suggested that GA may be an intermediate in the 4HBA degradation pathway expressed by this strain.
FIG. 2.
Accumulation of 2,5-dihydroxybenzoic acid (GA) in resting cell suspensions. Each histogram represents one strain grown on the substrate indicated; washed cells were incubated with the growth substrate (2 mM) in the presence or absence of 2,2′-dipyridyl (0, 1, or 5 mM). The bars represent the concentrations of substrate remaining and the concentrations of GA detected (if any) after incubation for 18 h at 40°C. The data shown represent the means of triplicate determinations; the error bars represent ±1 standard deviation. Hfx., Haloferax; Haa., Haloarcula.
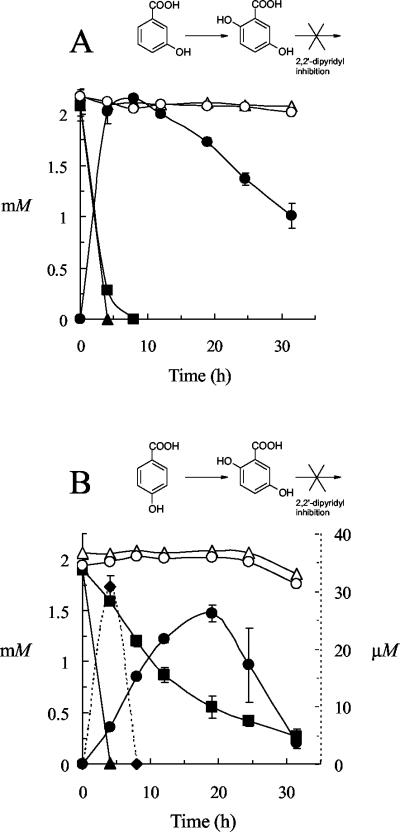
Additional time course experiments were conducted (using cell suspensions containing 1 mM 2,2′-dipyridyl) in order to investigate further and to confirm that transient accumulation of other intermediates had not been overlooked. As expected, 3HBA rapidly disappeared from suspensions of Haloferax sp. strain D1227 cells which had been grown on 3HBA, and stoichiometric accumulation of GA was observed in suspensions which contained both 3HBA and 2,2′-dipyridyl (Fig. 3A). No GA was detected in the absence of the inhibitor. In contrast, transient accumulation of GA (33 μM maximum) was detected as 4HBA disappeared from suspensions of 4HBA-grown Haloarcula sp. strain D1, which contained no 2,2′-dipyridyl. Furthermore, stoichiometric accumulation of GA from 4HBA was demonstrated (Fig. 3B). Only the added aromatic substrates (3HBA and 4HBA) and GA were detected in these experiments, and no other aromatic metabolites or intermediates were detected at any time. Thus, it appeared likely that GA was indeed an intermediate in the 4HBA pathway in Haloarcula sp. strain D1, which would also account for the presence of low concentrations of GA in 4HBA-grown cultures (Fig. 1).
FIG. 3.
Accumulation of GA in resting cell suspensions of Haloferax sp. strain D1227 (A) and Haloarcula sp. strain D1 (B). The concentrations of substrate (3HBA in panel A; 4HBA in panel B) in the presence (▪) and absence (▴) of 2,2′-dipyridyl and in sterile controls in the presence (○) and absence (▵) of 2,2′-dipyridyl are shown. The concentrations of GA in the presence (•) and absence (♦) of 2,2′-dipyridyl are also shown. The data shown represent the means of triplicate determinations; the error bars represent ±1 standard deviation.
Ring cleavage dioxygenase activity in cell extracts.
In order to further investigate the metabolism of 4HBA via GA, enzyme assays to detect ring cleavage dioxygenase activity in Haloarcula sp. strain D1 cell extracts were carried out. Cell extracts from Haloferax sp. strain D1227 were also assayed for comparative purposes. Induced GDO activity was detected in extracts prepared from Haloarcula sp. strain D1 cells grown on either BA or 4HBA but not on pyruvic acid (Table 1). Addition of 50 μM Fe2+ (as ferrous ammonium sulfate) did not increase the levels of GDO activity detected, and despite exhaustive efforts, no other ring cleavage dioxygenase activity was detected in any cell extract at any time (either in the presence or absence of Fe2+). In particular, no catechol or protocatechuate dioxygenase activity could be detected, which provided further indirect support for the hypothesis that Haloarcula sp. strain D1 metabolized 4HBA via a gentisate pathway. Addition of 1 mM 2,2′-dipyridyl completely inhibited any GDO activity which was present in the extracts tested.
Synthesis and biotransformation of deuterated 4HBA.
In view of the results described above (particularly the stoichiometric conversion of 4HBA into GA in the presence of 2,2′-dipyridyl), it was hypothesized that the 4HBA degradation pathway in Haloarcula sp. strain D1 in fact involved C-1 hydroxylation of 4HBA with a concomitant 1,2-carboxyl group migration (called an NIH shift [18]), to yield GA (Fig. 4, route 1). Like Crawford (16), we considered that 4-dehydroxylation of 4HBA to yield BA (Fig. 4, route 2) was unlikely, as enzymatic dehydroxylation of 4HBA is unprecedented. However to completely exclude this possibility, an experiment using selectively deuterated 4HBA as a biotransformation substrate was devised. Using 2,6-dideutero-4HBA as a substrate, the experiment illustrated in Fig. 3B was essentially repeated on a larger scale, and enough deuterated GA was recovered for analysis using both GC-MS and 1H-NMR methods. Under these conditions, the two metabolic routes outlined in Fig. 4 would be expected to yield deuterated GA products with different substitution patterns, which could be easily distinguished using 1H-NMR.
The product from a pathway involving a 1,2-carboxyl group shift (Fig. 4, route 1) would be 3-deutero-2,5-dihydroxybenzoic acid (Fig. 4, structure II). The 1H-NMR spectrum of this compound would be expected to show two single, uncoupled peaks, each assigned to protons para (H-4) and ortho (H-6) to the carboxyl group. In contrast, the product from a pathway initially involving 4-dehydroxylation and then metabolism by the upper benzoate pathway (Fig. 4, route 2) would be 6-deutero-2,5-dihydroxybenzoic acid (Fig. 4, structure V). The 1H-NMR spectrum of this compound would be expected to show two vicinally coupled (split) peaks, each assigned to protons meta (H-3) and para (H-4) to the carboxyl group, and the peak corresponding to a proton ortho to the carboxyl group would be absent.
The synthesis of 2,6-dideutero-4HBA (Fig. 4, structure I) was achieved with an overall yield of approximately 40% (wt/wt). The retention of two deuterium atoms in the product was confirmed using GC-MS. Authentic 4HBA gave major mass ions at m/z 138 (M+; 89%), 121 (M+-OH; 100%), and 93 (M+-COOH; 52%). The deuterated 4HBA product gave major mass ions at m/z 138 (M+; undeuterated; 3%), 140 (M+; dideuterated; 39%), 123 (M+-OH; dideuterated; 42%), and 95 (M+-COOH; dideuterated; 11%), which indicated 92% (±3%) incorporation of D2. Peaks in the 1H-NMR spectrum of the dideuterated 4HBA product [δH (300 MHz, CDCl3) 6.83 ppm (s, 2H, 3-H, and 5-H), 7.88 ppm (d, J = 9Hz, 0.16H, 2-H, and 6-H)] were assigned to protons in the structure by comparison to a reference 1H-NMR spectrum of authentic 4HBA [δH (300 MHz, CDCl3) 6.83 ppm (d, J = 9Hz, 2H, 3-H, and 5-H), 7.84 ppm (d, J = 9Hz, 2H, 2-H, and 6-H)] and to the published 1H-NMR spectrum (Aldrich Technical Library). The 1H-NMR data indicated a high degree of incorporation of deuterium (92% D2) at positions C-2 and C-6 of the aromatic ring in the dideuterated product.
Recovery and analysis of deuterated GA.
Deuterated GA was extracted from the combined supernatants of 50 5-ml cell suspensions as the GA concentration peaked (as determined by HPLC analysis of samples from a control tube containing undeuterated 4HBA). The deuterated GA was recovered along with residual 2,6-dideutero-4HBA. Both compounds were methylated and then separated by PLC before analysis using GC-MS and 1H-NMR methods.
A compound corresponding to methylgentisate (the methyl ester derivative of GA) eluted from the GC column with a retention time (Rt) of 15.69 min compared to a sample of authentic methylgentisate, which eluted with an Rt of 15.62 min. The mass spectra obtained from these compounds were broadly similar and were consistent with the recovered product being a monodeuterated isomer of methylgentisate (Fig. 4, structure II or V, and data not shown). Data from MS alone cannot permit the unambiguous identification of even authentic methylgentisate as the methyl ester of GA, because the mass spectra of methyl esters derived from different dihydroxybenzoic acids exhibit almost identical fragmentation patterns. Nevertheless, taken in combination with the near-identical GC and HPLC retention times and the very distinctive UV absorbance spectrum of the recovered compound (which, prior to methylation, was identical to that of authentic GA), the MS data did support the identification of the recovered compound as deuterated methylgentisate. This identification was confirmed by 1H-NMR data. A minor GC peak with an Rt of 14.54 min was also seen and indicated the presence of some 2,6-dideutero-methyl-4-hydroxybenzoate (Fig. 4, methyl ester of structure I) [major mass ions at m/z 154 (M+; 12%), 123 (M+-OCH3), 124 (100%), 95 (M+-CO2CH3)], derived from residual substrate.
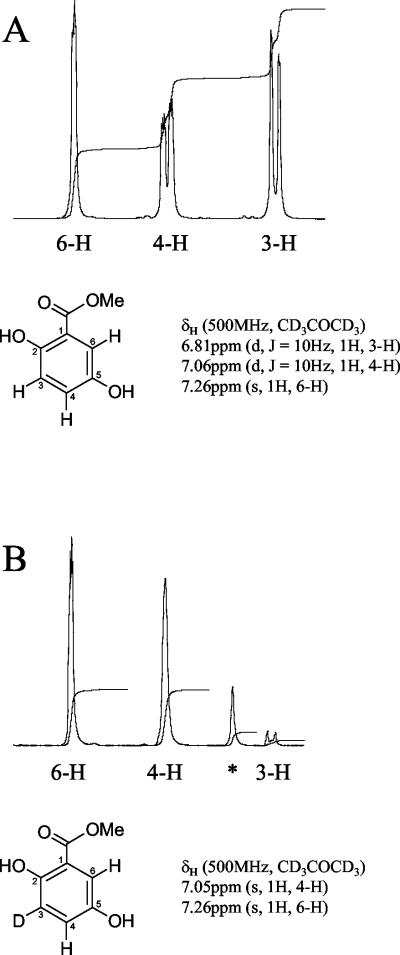
The results of 1H-NMR analyses of both the recovered deuterated methylgentisate and authentic methylgentisate are presented in Fig. 5, along with details from the 1H-NMR spectrum of each compound (the region between 6.7 and 7.3 ppm is enlarged). The 1H-NMR spectra of the deuterated products that would be expected from a pathway involving a 1,2-carboxyl group migration and an alternative (hypothetical) pathway involving dehydroxylation to BA were explained previously. The 1H-NMR spectrum obtained from the deuterated methylgentisate (Fig. 5B) supports the unambiguous identification of this compound as 3-deutero-methylgentisate (Fig. 4, structure II) and not 6-deutero-methylgentisate (Fig. 4, structure V). The absence of vicinal coupling of the peak assigned to proton 4-H and the absence of a signal of comparable intensity corresponding to proton 3-H indicate that the deuterated methylgentisate which was isolated is the structural isomer shown in Fig. 5B. As this is the compound expected if a 1,2-carboxyl group migration (Fig. 4, route 1) had occurred, this is strong evidence that the pathway responsible for the conversion of 4HBA to GA proceeds by this route. The unassigned peak was attributed to the presence of residual methyl-2,6-dideutero-4-hydroxybenzoate [Fig. 4, methyl ester of structure I; δH (500 MHz, CD3COCD3) 6.89 ppm (s, 3-H and 5-H)], which was also detected by GC-MS (see above).
FIG. 5.
1H-NMR spectra. (A) Authentic methyl 2,5-dihydroxybenzoate (methylgentisate). (B) Deuterated methylgentisate. Peaks in the 1H-NMR spectra were assigned to individual protons in the positions indicated. The peak marked with an asterisk was attributed to the presence of a trace of the methylated substrate, methyl-2,6-dideutero-4-hydroxybenzoate [δH (500 MHz, CD3COCD3) 6.89 ppm (s, 3-H and 5-H)]. Only data from the chemical shift range between 6.7 and 7.3 ppm are shown.
In the unlikely event that 4HBA was actually being metabolized by initial 4-dehydroxylation to BA (Fig. 4, route 2), then the 1H-NMR spectrum of the deuterated methylgentisate product, 6-deutero-methylgentisate (Fig. 4, structure V), would be essentially identical to the authentic methylgentisate spectrum, except that the single peak assigned to proton 6-H (7.26 ppm) would be absent. The vicinal coupling between protons 4-H and 3-H would still be clearly apparent as splitting of the peaks assigned to these protons.
DISCUSSION
We show here that aerobic 4HBA metabolism in Haloarcula sp. strain D1 proceeds via gentisate. Induction of GDO was demonstrated in extracts from 4HBA-grown cells, along with stoichiometric accumulation of GA from 4HBA in biotransformation experiments. In contrast, most bacteria aerobically metabolize 4HBA by 3-hydroxylation to yield protocatechuate, which serves as the ring cleavage substrate (25, 28, 34, 35, 38, 39, 55, 60, 62, 64). Metabolism of 4HBA by decarboxylation to phenol, followed by ortho hydroxylation to catechol, has also been suggested in Rhodococcus opacus (35). In eucarya, such as yeasts and fungi, three 4HBA pathways are known. Like many bacteria, some fungi also metabolize 4HBA via 3-hydroxylation to protocatechuate, which serves as the ring cleavage substrate (56). Others 1-hydroxylate 4HBA, with concomitant decarboxylation to yield hydroquinone, which is further metabolized by ortho hydroxylation to hydroxyhydroquinone (26, 68). Metabolism of 4HBA to protocatechuate, followed by 1-hydroxylation and decarboxylation to hydroxyhydroquinone, has also been reported (4, 56, 66). Hydroxyhydroquinone serves as the ring cleavage substrate in these pathways.
Two feasible routes for the conversion of 4HBA to GA are outlined in Fig. 3, and the hypothesis that the Haloarcula pathway involves 1-hydroxylation of 4HBA with a concomitant 1,2-carboxyl group migration (Fig. 4, route 1) is strongly supported by the experimental data. An alternative explanation, involving successive 4-dehydroxylation, 3-hydroxylation, and 6-hydroxylation (Fig. 4, route 2), can be ruled out. Reductive dehydroxylation of 4-hydroxybenzoyl-coenzyme A (CoA) to benzoyl-CoA under anaerobic conditions has been reported (33, 48). However, there are no reports in the literature to suggest that 4HBA is aerobically metabolized by dehydroxylation to BA in any group of organisms.
Although unusual, the haloarchaeal pathway described here is not without precedent. Evidence for the aerobic metabolism of 4HBA via gentisate (based initially on patterns of ring cleavage dioxygenase induction) has been reported in the gram-positive genus Bacillus (13, 16). In these early studies, no isotopic evidence of an intramolecular carboxyl group migration was presented. However, a further thermophilic Bacillus stearothermophilus strain which grew on 4HBA was subsequently isolated (49), and it was confirmed that this strain metabolized 4HBA by 1-hydroxylation and 1,2-carboxyl group migration, yielding GA. GC-MS analysis of deuterated GA which was recovered following 2,2′-dipyridyl-inhibited biotransformation of 2- or 3-deutero-4HBA by 4HBA-grown cells indicated 50% loss of label in GA derived from 2-deutero-4HBA but negligible loss in GA derived from 3-deutero-4HBA. This provided strong evidence that a 1,2-carboxyl group migration was occurring in this case. Thus, on the basis of work on 4HBA metabolism which has been reported to date, it appears that this unusual pathway may be present only in some Bacillus strains and in the haloarchaeal genus Haloarcula.
Intramolecular migrations during monooxygenase-catalyzed aryl hydroxylation reactions are certainly not without precedent. Hydroxylation-induced intramolecular migrations of this type (typically involving deuterium, tritium, halogen, or methyl substituents) are called NIH shifts (18, 36), and numerous examples (>100) have been reported in both mammalian and bacterial systems. NIH shifts have traditionally been attributed to the involvement of arene oxides as transient intermediates in monooxygenase-mediated hydroxylation reactions (19, 37), although there have been suggestions that some observed NIH shifts occur as the result of alternative mechanisms of hydroxylation (57, 69), including dioxygenase-catalyzed cis dihydroxylation (6).
Monooxygenase-catalyzed hydroxylations which involve arene oxide intermediates appear to proceed by the initial enzyme-catalyzed epoxidation of the aromatic ring (by introduction of a single oxygen atom derived from dioxygen) to yield an arene oxide (oxirane). Benzenoid arene oxides exist in equilibrium with valence tautomeric oxepines, with the equilibrium favoring either the arene oxide or the oxepine form in different compounds (10). The oxirane ring in the arene oxide form (but not the oxepine) can be opened to produce a transient hydroxyl-substituted cationoid intermediate, which is stabilized by either migration or elimination of the atom or group that was initially present at the site of hydroxylation. The intermediate finally aromatizes to form a phenol (10, 19). Stable arene oxides of naphthalene, quinoline, and methylbenzoate have been isolated as metabolic intermediates during enzyme-catalyzed hydroxylation of these compounds (1, 11, 45, 46). Consequently, as well as providing a mechanistic explanation for the NIH shift, there is direct evidence that arene oxides are intermediates in the enzymatic hydroxylation of at least some aromatic compounds containing one or more aromatic rings. However, it is rearrangement of the carbocation intermediate which arises following opening of the oxirane ring which accounts for the NIH shift itself.
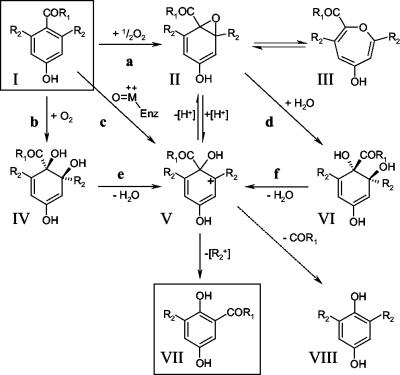
The conversion of 2,6-dideutero-4HBA (Fig. 6, structure I; R1 = OH, R2 = D) to 3-deuterogentisic acid (Fig. 6, structure VII; R1 = OH, R2 = D) can therefore be explained by invoking the involvement of a carbocation intermediate (Fig. 6, structure V; R1 = OH, R2 = D), which could in principle arise by four distinct routes: (i) C2—O opening of the oxirane ring in 2,6-dideutero-4HBA 1,2-oxide (Fig. 6, structure II; R1 = OH, R2 = D); (ii) direct electrophilic addition of an enzyme-bound activated oxygen species at position 1 of the aromatic ring of 2,6-dideutero-4HBA (Fig. 6, structure I; R1 = OH, R2 = D); (iii) dehydration of a trans-1,2-diol (Fig. 6, structure VI; R1 = OH, R2 = D), arising indirectly from a 1,2-arene oxide (Fig. 6, structure II; R1 = OH, R2 = D) as the product of an epoxide hydrolase; (iv) dehydration of a cis-1,2-diol (Fig. 6, structure IV; R1 = OH, R2 = D), produced from an arene (Fig. 6, structure I; R1 = OH, R2 = D) by a ring-hydroxylating dioxygenase.
FIG. 6.
Alternative mechanisms which could account for the observed 1,2-carboxyl group migration. The structures shown (R1 = OH, R2 = H) are 4HBA (I), 4HBA 1,2-epoxide (II), valence tautomer of arene oxide II (III), 4-HBA cis-1,2-diol (IV), intermediate carbocation (V), 4HBA trans-1,2-diol (VI), GA (VII), and hydroquinone (VIII). The putative enzyme activities shown (R1 = OH, R2 = H) are 4HBA monooxygenase (a), 4HBA 1,2-dioxygenase (b), reaction with an enzyme-bound activated oxygen species (c), 4HBA 1,2-epoxide hydrolase (d), 4HBA cis-1,2-diol dehydratase (e), and 4HBA trans-1,2-diol dehydratase (f). Dotted arrow, decarboxylation.
Rearrangement of a carbocation, arising by any of these routes, could then account for the observed 1,2-carboxyl migration. However, there are significant problems with this explanation. First, the presence of an ionizable para-hydroxyl substituent on the aromatic ring of 4HBA suggests the possible formation of an unstable para-benzoquinone from an intermediate cation of type V (Fig. 6). This species would readily decarboxylate (Fig. 6), ultimately yielding hydroquinone (Fig. 6, structure VIII; R1 = OH, R2 = H or D), essentially as described by Daly et al. (18). No hydroquinone (or any other metabolites which would suggest that decarboxylation was occurring) was detected at any time during the course of the work described here. Second, it has been shown that the carboxyl group itself does not migrate, even in the absence of other ionizable ring substituents. Boyd and Berchtold (9) reported that chemically synthesized arene 1,2-oxides of o-toluic acid and p-toluic acid underwent exclusively C2—O oxirane opening, with hydroxylation at position 1. However, this resulted only in decarboxylation to o-cresol and p-cresol, respectively, and no 1,2-migration of the carboxyl group (or ortho hydroxylation, following C1—O opening) was observed. Similarly, while the 1,2-oxide of BA was reported to undergo both C2—O oxirane opening (resulting in 1-hydroxylation) and C1—O opening (ortho hydroxylation, yielding 2-hydroxybenzoic acid), 1,2-migration of the carboxyl group was never observed. C2—O opening and 1-hydroxylation of BA were always accompanied by decarboxylation, yielding phenol. There is therefore compelling chemical evidence that 1,2-migration of carboxyl groups does not occur following rearrangement of cations of type V (Fig. 6), derived from aromatic carboxylic acids, such as 4HBA (as opposed to aromatic methylene-carboxylic acids, such as 4-hydroxyphenylacetic acid).
In view of the above observations, the 1,2-carboxyl group migration reported here (during conversion of 4HBA to GA) and that reported by Keenan and Chapman (49) are chemically rather surprising and would not be expected to occur. It should be noted that the original report suggesting a carboxyl group migration (49) appeared before the synthesis of a 1,2-arene oxide of BA and its aromatization without evidence of carboxyl group migration (9). Significantly, however, intramolecular 1,2-migration of derivatized carboxylic acids (such as methyl esters and amides) has been observed (9, 12). The unequivocal 1,2-migration of a carbomethoxy group has recently been demonstrated in fungal biotransformation experiments using substituted methylbenzoates as substrates and, during the acid-catalyzed aromatization of methylbenzoate 1,2-oxide, isolated as a metabolic intermediate (11, 42). Since the ester and amide derivatives of carboxylic acids have been shown to migrate during aromatization of arene-1,2-oxides, it is probable that thioesters would behave in a similar manner. Indeed, Dagli et al. (17) reported that thioesters have very high “migratory aptitudes” in reactions involving the nonenzymatic rearrangement of epoxides, and intramolecular migration of a CoA-thioester group (COSCoA) during enzyme-mediated conversion of methylmalonyl-CoA to succinyl-CoA has also been demonstrated (61). With this in mind, it is postulated that the substrate for the 1-hydroxylase enzyme implicated in the 4HBA pathway in Haloarcula sp. strain D1 (and in Bacillus strains which appear to possess the same pathway) is 4-hydroxybenzoyl-CoA (Fig. 6, structure I; R1 = SCoA, R2 = H). The involvement of a thioester could account for the apparent intramolecular migration of a carboxyl group, which is otherwise chemically unprecedented, and the phenolic thioester product of the transformation could subsequently be hydrolyzed to yield GA (Fig. 6, structure VII; R1 = OH, R2 = H). Given the apparent involvement of CoA thioesters in the metabolism of both BA and 3HBA in Haloferax sp. strain D1227 (30), a mechanism involving thioester formation, migration, and hydrolysis appears feasible. It is notable that during the earlier studies using Bacillus strains, repeated attempts were made to demonstrate 1-hydroxylation of 4HBA to GA in cell extracts without success (16, 49). Crawford (16) attempted to detect cell-free hydroxylation in the presence of “various combinations of cofactors.” However, it was not realized until much more recently that some monooxygenase-mediated (aerobic) aromatic degradation pathways involve CoA thioesterification (2, 3, 29, 30, 50, 59). Consequently, CoA and ATP were probably not included as potential cofactors in Crawford's experiments. There is still some uncertainty about the precise biochemical role of CoA in these pathways, and recent work has suggested that an entirely novel type of monooxygenase may be involved (23).
There are four postulated routes to the intermediate carbocation (Fig. 6, structure V), although we consider the involvement of an arene oxide (Fig. 6, structure II) to be most likely. There is evidence that carbocation intermediates of type V (Fig. 6) which undergo NIH shifts can arise from cis-1,2-dihydrodiols (Fig. 6, structure IV) without the involvement of arene oxides (6). However, metabolism of 4HBA by a pathway involving a ring-hydroxylating dioxygenase has never been reported. In addition, known ring-hydroxylating 1,2-dioxygenases which hydroxylate aromatic acids are multicomponent enzyme systems which contain Fe(II) as a cofactor in one or more subunits (58, 71, 72, 73), which would make them extremely sensitive to inhibition by 2,2′-dipyridyl. Whether a trans-diol intermediate (Fig. 6, structure VI) is involved is still unknown. Many fungi and some bacteria (including cyanobacteria) can partially oxidize polycyclic aromatic hydrocarbon compounds to 1,2-trans-dihydrodiols, apparently via epoxide hydrolase-catalyzed hydration of 1,2-arene oxides (15, 65). Although these pathways generally appear to be cometabolic “dead ends” in that the substrates are not fully metabolized or mineralized (65), trans-dihydrodiols can also be dehydrated to form carbocations of type V (Fig. 6), which can undergo an NIH shift. Barr et al. (6) demonstrated that the NIH shift of a deuterium atom occurred following nonenzymatic dehydration of both naphthalene and quinoline 1,2-trans-dihydrodiols.
To our knowledge, this is the first report of an NIH shift within the domain Archaea. Efforts are under way to identify and clone the genes which encode enzymes in the haloarchaeal 4HBA pathway, and in similar pathways in Bacillus spp., so that comparison at the molecular genetic level will be possible.
Acknowledgments
We thank Manus Carey and Robert Boyd for assistance with GC-MS and EI+-MS analyses, respectively, and Richard Murphy for assistance with 1H-NMR analysis.
This study was funded by a Cooperative Award for Science and Technology grant awarded by the Department of Education (Northern Ireland) in collaboration with ICI Technology.
REFERENCES
- 1.Agarwal, S. K., D. R. Boyd, H. P. Porter, W. B. Jennings, S. J. Grossman, and D. M. Jerina. 1986. Arene oxides and trans-dihydrodiols of quinoline. Tetrahedron Lett. 27:4253-4256. [Google Scholar]
- 2.Altenschmidt, U., B. Oswald, and G. Fuchs. 1991. Purification and characterization of benzoate-coenzyme A ligase and 2-aminobenzoate-coenzyme A ligases from a denitrifying Pseudomonas sp. J. Bacteriol. 173:5494-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altenschmidt, U., B. Oswald, E. Steiner, H. Herrmann, and G. Fuchs. 1993. New aerobic benzoate oxidation pathway via benzoyl-coenzyme A and 3-hydroxybenzoyl-coenzyme A in a denitrifying Pseudomonas sp. J. Bacteriol. 175:4851-4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson, J. J., and S. Dagley. 1980. Catabolism of aromatic acids in Trichosporon cutaneum. J. Bacteriol. 141:534-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barns, S. M., C. F. Delwiche, J. D. Palmer, and N. R. Pace. 1996. Perspectives on Archaeal diversity, thermophily and monophyly from environmental rRNA sequences. Proc. Natl. Acad. Sci. USA 93:9188-9193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barr, S. A., N. Bowers, D. R. Boyd, N. D. Sharma, L. Hamilton, R. Austin, S. McMordie, and H. Dalton. 1998. The potential role of cis-dihydrodiol intermediates in bacterial aromatic hydroxylation and the NIH shift. J. Chem. Soc. Perkin 1 20:3443-3451.
- 7.Benitez, F. J., J. Beltran-Heredia, J. A. Peres, and J. R. Dominguez. 2000. Kinetics of p-hydroxybenzoic acid photodecomposition and ozonation in a batch reactor. J. Hazard. Mater. 73:161-178. [DOI] [PubMed] [Google Scholar]
- 8.Bintrim, S. B., T. J. Donohue, J. Haldelsman, G. P. Roberts, and R. M. Goodman. 1997. Molecular phylogeny of Archaea from soil. Proc. Natl. Acad. Sci. USA 94:277-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyd, D. R., and G. A. Berchtold. 1979. Aromatization of arene 1,2-oxides. 1-carboxy- and 1-carboalkoxybenzene oxides. J. Am. Chem. Soc. 101:2470-2474. [Google Scholar]
- 10.Boyd, D. R., and N. D. Sharma. 1996. The changing face of arene oxide-oxepine chemistry. Chem. Soc. Rev. 25:289-296. [Google Scholar]
- 11.Boyd, D. R., J. T. G. Hamilton, N. D. Sharma, J. S. Harrison, W. C. McRoberts, and D. B. Harper. 2000. Isolation of stable benzene oxide from a fungal biotransformation and evidence for an ‘NIH shift' of the carbomethoxy group during hydroxylation of methyl benzoates. Chem. Commun. 16:1481-1482.
- 12.Busch, F. R., and G. A. Berchold. 1985. Aromatization of benzamide 1,2-oxide and N,N-dimethylbenzamide 1,2-oxide. J. Org. Chem. 50:1590-1592. [Google Scholar]
- 13.Buswell, J. A., and J. S. Clark. 1976. Oxidation of aromatic acids by a facultative thermophilic Bacillus sp. J. Gen. Microbiol. 96:209-213. [DOI] [PubMed] [Google Scholar]
- 14.Cain, R. B., R. F. Bilton, and J. A. Darrah. 1968. The metabolism of aromatic acids by microorganisms. Metabolic pathways in the fungi. Biochem. J. 108:797-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cerniglia, C. E. 1992. Biodegradation of polycyclic aromatic hydrocarbons. Biodegradation 3:351-368. [Google Scholar]
- 16.Crawford, R. L. 1976. Pathways of 4-hydroxybenzoate degradation among species of Bacillus. J. Bacteriol. 127:204-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dagli, D. J., R. A. Gorski, and J. Wemple. 1975. Synthesis and rearrangement of glycidic thiol esters; migratory aptitudes. J. Org. Chem. 40:1741-1745. [Google Scholar]
- 18.Daly, J., D. Jerina, and B. Witkop. 1968. Migration of deuterium during hydroxylation of aromatic substrates by liver microsomes. Arch. Biochem. Biophys. 128:517-527. [DOI] [PubMed] [Google Scholar]
- 19.Daly, J. W., D. M. Jerina, and B. Witkop. 1972. Arene oxides and the NIH shift: the metabolism, toxicity and carcinogenicity of aromatic compounds. Experientia 28:1129-1264. [DOI] [PubMed] [Google Scholar]
- 20.Danson, M. J., and D. W. Hough. 1997. The structural basis of protein halophilicity. Comp. Biochem. Physiol. A 117:307-312. [Google Scholar]
- 21.DeLong, E. F., K. Y. Wu, B. B. Prezelin, and R. V. M. Jovine. 1994. High abundance of Archaea in Antarctic marine picoplankton. Nature 371:695-697. [DOI] [PubMed] [Google Scholar]
- 22.Di Gioia, D., L. Bertin, F. Fava, and L. Marchetti. 2001. Biodegradation of hydroxylated and methoxylated benzoic, phenylacetic and phenylpropionic acids present in olive mill wastewaters by two bacterial strains. Res. Microbiol. 152:83-93. [DOI] [PubMed] [Google Scholar]
- 23.El-Said Mohamed, M., A. Zaar, C. Ebenau-Jehle, and G. Fuchs. 2001. Reinvestigation of a new type of aerobic benzoate metabolism in the proteobacterium Azoarcus evansii. J. Bacteriol. 183:1899-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Emerson, D., S. Chauhan, P. Oriel, and J. A. Breznak. 1994. Haloferax sp. D1227, a halophilic archaeon capable of growth on aromatic compounds. Arch. Microbiol. 161:445-452. [Google Scholar]
- 25.Engelhardt, G., H. G. Rast, and P. R. Wallnöfer. 1979. Degradation of aromatic carboxylic acids by Nocardia spec. DSM 43251. FEMS Microbiol. Lett. 5:245-251. [Google Scholar]
- 26.Eppink, M. H. M., S. A. Boeren, J. Vervoort, and W. J. H. Vanberkel. 1997. Purification and properties of 4-hydroxybenzoate 1-hydroxylase (decarboxylating), a novel flavin adenine dinucleotide-dependent monooxygenase from Candida parapsilosis CBS604. J. Bacteriol. 179:6680-6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng, Y., H. E. Khoo, and C. L. Poh. 1999. Purification and characterization of gentisate 1,2-dioxygenases from Pseudomonas alcaligenes NCIB 9867 and Pseudomonas putida NCIB 9869. Appl. Environ. Microbiol. 65:946-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fernandez, J., A. A. Dimarco, L. N. Ornston, and S. Harayama. 1995. Purification and characterization of Acinetobacter calcoaceticus 4-hydroxybenzoate 3-hydroxylase after its overexpression in Escherichia coli. J. Biochem. 117:1261-1266. [DOI] [PubMed] [Google Scholar]
- 29.Fu, W. J., and P. Oriel. 1998. Gentisate 1,2-dioxygenase from Haloferax sp. D1227. Extremophiles 2:439-446. [DOI] [PubMed] [Google Scholar]
- 30.Fu, W. J., and P. Oriel. 1999. Degradation of 3-phenylpropionic acid by Haloferax sp. D1227. Extremophiles 3:45-53. [DOI] [PubMed] [Google Scholar]
- 31.Galinski, E. A. 1995. Osmoadaptation in bacteria. Adv. Microb. Physiol. 37:272-328. [PubMed] [Google Scholar]
- 32.Gevantman, L. H. 2000. Solubility of selected gases in water, p. 8-86. In D. R. Lide (ed.), CRC handbook of chemistry and physics, 80th ed. CRC Press, London, United Kingdom.
- 33.Glöckler, R., A. Tschech, and G. Fuchs. 1989. Reductive dehydroxylation of 4-hydroxybenzoyl-CoA to benzoyl-CoA in a denitrifying Pseudomonas species. FEBS Lett. 251:237-240. [DOI] [PubMed] [Google Scholar]
- 34.Grund, E., C. Knorr, and R. Eichenlaub. 1990. Catabolism of benzoate and monohydroxylated benzoates by Amycolatopsis and Streptomyces spp. Appl. Environ. Microbiol. 56:1459-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grund, E., and H. J. Kutzner. 1998. Utilisation of quinate and p-hydroxybenzoate by actinomycetes: key enzymes and taxonomic relevance. J. Basic Microbiol. 38:241-255. [DOI] [PubMed] [Google Scholar]
- 36.Guroff, G., J. W. Daly, D. M. Jerina, J. Renson, B. Witkop, and S. Udenfriend. 1967. Hydroxylation-induced migration: the NIH shift. Science 157:1524-1530. [DOI] [PubMed] [Google Scholar]
- 37.Hamilton, G. A. 1974. Chemical models and mechanisms for oxygenases, p. 405-452. In O. Hayaishi (ed.), Molecular mechanisms of oxygen activation. Academic Press, New York, N.Y.
- 38.Hammann, R., and H. J. Kutzner. 1998. Key enzymes for the degradation of benzoate, m- and p-hydroxybenzoate by some members of the order Actinomycetales. J. Basic Microbiol. 38:207-220. [PubMed] [Google Scholar]
- 39.Haribabu, B., A. V. Kamath, and C. S. Vaidyanathan. 1984. Degradation of substituted benzoic acids by a Micrococcus species. FEMS Microbiol. Lett. 21:197-200. [Google Scholar]
- 40.Harpel, M. R., and J. D. Lipscomb. 1990. Gentisate 1,2-dioxygenase from Pseudomonas-substrate coordination to active-site Fe2+ and mechanism of turnover. J. Biol. Chem. 265:22187-22196. [PubMed] [Google Scholar]
- 41.Harpel, M. R., and J. D. Lipscomb. 1990. Gentisate 1,2-dioxygenase from Pseudomonas acidovorans. Methods Enzymol. 188:101-107. [DOI] [PubMed] [Google Scholar]
- 42.Harrison, J. 1999. Fungal oxidation of arene compounds. Ph.D. thesis. The Queen's University of Belfast, Belfast, Northern Ireland.
- 43.Hinrichs, K. U., J. M. Hayes, S. P. Sylva, P. G. Brewer, and E. F. DeLong. 1999. Methane-consuming archaebacteria in marine sediments. Nature 398:802-805. [DOI] [PubMed] [Google Scholar]
- 44.Hough, D. W., and M. J. Danson. 1999. Extremozymes. Curr. Opin. Chem. Biol. 3:39-46. [DOI] [PubMed] [Google Scholar]
- 45.Jerina, D. M., J. W. Daly, B. Witkop, P. Zaltzman-Nirenberg, and S. Udenfriend. 1968. The role of arene oxide-oxepin systems in the metabolism of aromatic substrates. III. Formation of 1,2-naphthalene oxide from naphthalene by liver microsomes. J. Am. Chem. Soc. 90:6525-6527. [DOI] [PubMed] [Google Scholar]
- 46.Jerina, D. M., J. W. Daly, B. Witkop, P. Zaltzman-Nirenberg, and S. Udenfriend. 1970. 1,2-Naphthalene oxide as an intermediate in the microsomal hydroxylation of naphthalene. Biochemistry 9:147-155. [DOI] [PubMed] [Google Scholar]
- 47.Jurgens, G., K. Lindström, and A. Saano. 1997. Novel group within the kingdom Crenarchaeota from boreal forest soil. Appl. Environ. Microbiol. 63:803-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kasmi, A. E., R. Brachmann, G. Fuchs, and S. W. Ragsdale. 1995. Hydroxybenzoyl-CoA reductase: coupling kinetics and electrochemistry to derive enzyme mechanisms. Biochemistry 34:11668-11677. [DOI] [PubMed] [Google Scholar]
- 49.Keenan, S. L., and P. J. Chapman. 1978. Carboxy-migration facilitated by bacterial hydroxylation of 4-hydroxybenzoate. J. Chem. Soc. Chem. Commun. 17:731-732.
- 50.Kiemer, P., B. Tshisuaka, S. Fetzner, and F. Lingens. 1996. Degradation of benzoate via benzoyl-coenzyme A and gentisate by Bacillus stearothermophilus PK1, and purification of gentisate 1,2-dioxygenase. Biol. Fertil. Soils 23:307-313. [Google Scholar]
- 51.Kushner, D. J. 1986. Molecular adaptation of enzymes, metabolic systems and transport systems in halophilic bacteria. FEMS Microbiol. Rev. 39:121-127. [Google Scholar]
- 52.Lipscomb, J. D., and A. M. Orville. 1992. Mechanistic aspects of dihydroxybenzoate dioxygenases. Met. Ions Biol. Syst. 28:243-298. [Google Scholar]
- 53.Madern, D., C. Ebel, and G. Zaccai. 2000. Halophilic adaptation of enzymes. Extremophiles 4:91-98. [DOI] [PubMed] [Google Scholar]
- 54.Margesin, R., and F. Schinner. 2001. Potential of halotolerant and halophilic microorganisms for biotechnology. Extremophiles 5:73-83. [DOI] [PubMed] [Google Scholar]
- 55.Middlehoven, W. J., A. Coenen, B. Kraakman, and M. D. Sollewijn Gelpke. 1992. Degradation of some phenols and hydroxybenzoates by the imperfect ascomycetous yeasts Candida parapsolisis and Arxula adeninivorans: evidence for an operative gentisate pathway. Antonie Leeuwenhoek 62:181-187. [DOI] [PubMed] [Google Scholar]
- 56.Middlehoven, W. J. 1993. Catabolism of benzene compounds by ascomycetous and basidiomycetous yeasts and yeastlike fungi. Antonie Leeuwenhoek 63:125-144. [DOI] [PubMed] [Google Scholar]
- 57.Nasir, M. S., B. I. Cohen, and K. D. Karlin. 1992. Mechanism of aromatic hydroxylation in a copper monooxygenase model system—1,2-methyl migrations and the NIH shift in copper chemistry. J. Am. Chem. Soc. 114:2482-2494. [Google Scholar]
- 58.Neidle, E. L., C. Hartnett, L. N. Ornston, A. Bairoch, M. Rekik, and S. Harayama. 1991. Nucleotide sequences of the Acinetobacter calcoaceticus benABC genes for benzoate 1,2-dioxygenase reveal evolutionary relationships among multicomponent oxygenases. J. Bacteriol. 173:5385-5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Niemetz, R., U. Altenschmidt, S. Brucker, and G. Fuchs. 1995. Benzoyl-CoA 3-monooxygenase, a flavin-dependent hydroxylase: purification, some properties and its role in aerobic benzoate metabolism via gentisate in a denitrifying bacterium. Eur. J. Biochem. 227:161-168. [DOI] [PubMed] [Google Scholar]
- 60.Seibold, B., M. Matthes, M. H. Eppink, F. Lingens, W. J. H. Vanberkel, and R. Müller. 1996. 4-hydroxybenzoate hydroxylase from Pseudomonas sp. CBS3—purification, characterization, gene cloning, sequence analysis and assignment of structural features determining the coenzyme specificity. Eur. J. Biochem. 239:469-478. [DOI] [PubMed] [Google Scholar]
- 61.Sprecher, M., M. J. Clark, and D. B. Sprinson. 1966. The absolute configuration of methylmalonyl coenzyme A and stereochemistry of the methylmalonyl coenzyme A mutase reaction. J. Biol. Chem. 241:872-877. [PubMed] [Google Scholar]
- 62.Sterjiades, R. 1993. Properties of NADH/NADPH-dependent p-hydroxybenzoate hydroxylase from Moraxella sp. Biotechnol. Appl. Biochem. 17:77-90. [Google Scholar]
- 63.Suemori, A., R. Kurane, and N. Tomizuka. 1993. Purification and properties of gentisate 1,2-dioxygenase from Rhodococcus erythropolis S-1. Biosci. Biotechnol. Biochem. 57:1781-1783. [Google Scholar]
- 64.Suemori, A., K. Nakajima, R. Kurane, and Y. Nakamura. 1995. o-, m- and p-hydroxybenzoate degradative pathways in Rhodococcus erythropolis. FEMS Microbiol. Lett. 125:31-36. [DOI] [PubMed] [Google Scholar]
- 65.Sutherland, J. B., F. Rafii, A. A. Khan, and C. E. Cerniglia. 1995. Mechanisms of polycyclic aromatic hydrocarbon degradation, p. 269-306. In L. Y. Young and C. E. Cerniglia (ed.), Microbial transformation and degradation of toxic organic chemicals. Wiley-Liss, New York, N.Y.
- 66.Sze, I. S., and S. Dagley. 1984. Properties of salicylate hydroxylase and hydroxyquinol 1,2-dioxygenase purified from Trichosporon cutaneum. J. Bacteriol. 159:353-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tindall, B. J. 1992. The family Halobacteriaceae, p. 768-808. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K. H. Schleifer (ed.), The prokaryotes, 2nd ed., volume I. Springer Verlag, New York, N.Y.
- 68.Vanberkel, W. J. H., H. M. Eppink, W. J. Middelhoven, J. Vervoort, and I. M. C. M. Rietjens. 1994. Catabolism of 4-hydroxybenzoate in Candida parapsilosis proceeds through initial oxidative decarboxylation by a FAD-dependent 4-hydroxybenzoate 1-hydroxylase. FEMS Microbiol. Lett. 121:207-215. [DOI] [PubMed] [Google Scholar]
- 69.Vannelli, T., and A. B. Hooper. 1995. NIH shift in the hydroxylation of aromatic-compounds by the ammonia-oxidizing bacterium Nitrosomonas europaea—evidence against an arene oxide intermediate. Biochemistry 34:11743-11749. [DOI] [PubMed] [Google Scholar]
- 70.Ventosa, A., and J. J. Nieto. 1995. Biotechnological applications and potentialities of halophilic microorganisms. World J. Microbiol. Biotechnol. 11:85-94. [DOI] [PubMed] [Google Scholar]
- 71.Yamaguchi, M., and H. Fujisawa. 1980. Purification and characterization of an oxygenase component in benzoate 1,2-dioxygenase system from Pseudomonas arvilla C-1. J. Biol. Chem. 255:5058-5063. [PubMed] [Google Scholar]
- 72.Yamaguchi, M., and H. Fujisawa. 1981. Reconstitution of iron-sulfur cluster of NADH-cytochrome C reductase, a component of benzoate 1,2-dioxygenase system from Pseudomonas arvilla C-1. J. Biol. Chem. 256:6783-6787. [PubMed] [Google Scholar]
- 73.Yamaguchi, M., and H. Fujisawa. 1982. Subunit structure of oxygenase component in benzoate-1,2-dioxygenase system from Pseudomonas arvilla C-1. J. Biol. Chem. 257:12497-12502. [PubMed] [Google Scholar]