Abstract
Wood-boring bivalves of the family Teredinidae (commonly called shipworms) are known to harbor dense populations of gram-negative bacteria within specialized cells (bacteriocytes) in their gills. These symbionts are thought to provide enzymes, e.g., cellulase and dinitrogenase, which assist the host in utilizing wood as a primary food source. A cellulolytic, dinitrogen-fixing bacterium, Teredinibacter turnerae, has been isolated from the gill tissues of numerous teredinid bivalves and has been proposed to constitute the sole or predominant symbiont of this bivalve family. Here we demonstrate that one teredinid species, Lyrodus pedicellatus, contains at least four distinct bacterial 16S rRNA types within its gill bacteriocytes, one of which is identical to that of T. turnerae. Phylogenetic analyses indicate that the three newly detected ribotypes are derived from gamma proteobacteria that are related to but distinct (>6.5% sequence divergence) from T. turnerae. In situ hybridizations with 16S rRNA-directed probes demonstrated that the pattern of occurrence of symbiont ribotypes within bacteriocytes was predictable and specific, with some bacteriocytes containing two symbiont ribotypes. However, only two of the six possible pairwise combinations of the four ribotypes were observed to cooccur within the same host cells. The results presented here are consistent with the existence of a complex multiple symbiosis in this shipworm species.
Lyrodus pedicellatus is a member of the family Teredinidae, a group of worm-like marine bivalves that burrow into and consume floating or submerged wood with the aid of endosymbiotic bacteria. Adult specimens range from a few millimeters to more than 2 m in length and up to several centimeters in diameter. These bivalves, commonly called shipworms, cause severe damage to ships, piers, fishing equipment, and other wooden structures worldwide. Shipworms also play an important ecological role as the principle agents of mineralization of cellulosic plant materials in shallow (<150 m) marine and brackish environments (6). Wooden substrates serve as both food and shelter for shipworms, which are the only marine animals known to be capable of normal growth and reproduction with wood as a sole food source (14).
The mechanism by which shipworms digest wood is thought to involve gram-negative bacterial endosymbionts found within the gills. These bacteria were first observed by transmission electron microscopy in the shipworm Bankia australis (32) and were proposed to synthesize essential amino acids lacking in the shipworm's wood-based diet (31). However, the axenic cultivation of a cellulolytic, dinitrogen-fixing bacterium from the gills of several shipworm species suggests that the symbionts play a more direct role in wood feeding (41). This symbiotic bacterium, recently named Teredinibacter turnerae (10), was shown to reside within specialized epithelial cells (bacteriocytes) in the shipworm L. pedicellatus (9) in a gill region previously referred to as the Gland of Deshayes (36). Enzymes produced by this symbiont were proposed to facilitate the digestion of wood and to provide an internal source of combined nitrogen to supplement the host's protein-deficient diet (16, 41). Phylogenetic analysis, based on 16S rRNA sequences, showed T. turnerae to be a member of the gamma subdivision of the proteobacteria (9, 10).
As in most intracellular bacterial endosymbioses, the symbiont population L. pedicellatus has been presumed to be a monoculture (9, 37, 41). However, in a few cases, multiple intracellular endosymbiont species have been observed within a single host. For example, the gill bacteriocytes of certain mussel species from hydrothermal vents and cold seeps (8, 12) contain two coexisting symbiont species. Multiple endosymbioses have also been reported in other invertebrate taxa; however, in these cases, the symbionts either were extracellular (29, 30), were localized to separate tissues (28), or were pathogens that infect only a portion of the host population (34). The purpose of this investigation was to explore the diversity and phylogenetic composition of symbiont populations in the gills of L. pedicellatus. The results provide evidence for an intracellular bacterial consortium potentially more complex than others previously described.
MATERIALS AND METHODS
Cultivation of L. pedicellatus.
Adult specimens of L. pedicellatus were obtained from cultures maintained at Woods Hole Oceanographic Institute, Woods Hole, Mass., and were kept in the laboratory at room temperature in seawater aquaria. Larvae from spawning adults were collected on pine blocks (1 by 1 by 15 cm) suspended vertically above the aquaria with the lower 2 cm submerged. After 1 to 2 days, the blocks were transferred to new aquaria. Fresh seawater was supplied every 14 to 16 days. No food source other than the wood substrate was provided.
Isolation of T. turnerae.
T. turnerae was isolated from the gill tissue of lab-reared L. pedicellatus as described previously (41) by serial dilution (101 to 1010) of gill homogenate in shipworm basal medium (SBM) containing 0.2% agar with cellulose (Sigmacell, 2 g/liter; Sigma Chemical) added as a carbon source and with no added source of combined nitrogen. Axenic stock cultures were maintained in the same medium. SBM liquid cultures, used for nucleic acid extractions, contained SBM supplemented with Sigmacell (2 g/liter) and ammonium chloride (5 mM).
Nucleic acid preparation, PCR amplification, and cloning.
DNA was extracted from ∼100 mg of fresh gill tissue from each adult specimen of L. pedicellatus as described previously (7). DNA extraction from T. turnerae cultures was done as described previously (10). Bacterial 16S rRNA genes were amplified by PCR (35) from bulk nucleic acids extracted from L. pedicellatus gill tissue (two adult specimens) and cultivated T. turnerae cells. Nearly complete 16S rRNA gene sequences were amplified from each DNA preparation independently by using bacterial-domain-specific primers 27f and 1492r (23). PCRs were performed in 50-μl volumes as described previously (7) with 20 or 35 cycles and either 1 ng of gill DNA/μl or 0.2 ng of bacterial genomic DNA/μl.
PCR amplification products from L. pedicellatus gill tissue were cloned into Escherichia coli by using the pGEM-T vector system (Promega) following the manufacturer's recommended protocol. Plasmid DNAs were recovered by using the Wizard Plus SV Miniprep DNA purification system (Promega) as directed by the manufacturer and were screened for the presence of insert DNA by agarose gel electrophoresis. A combined total of 43 clones containing inserts of the appropriate size (∼1.5 kb as determined by agarose gel electrophoresis) were selected and screened by sequencing both strands of a region corresponding to E. coli positions 357 to 907 (∼500 bp) (23).
Sequencing.
Sequencing reactions were performed with an ABI PRISM Dye Terminator cycle sequencing ready reaction kit by using AmpliTaq DNA polymerase FS on an ABI 373A DNA sequencer. Approximately 500 bp of the double-strand sequence was determined for each clone by using primers 357f and 907r (23). Nearly complete double-strand sequences of 16S rRNA genes (1,390 to 1,441 bp) were determined for selected clones and PCR products by using primers 27f, 357f, 704f, 690r, 907r, 1101r, and 1492r (23).
Fluorescent in situ hybridization. (i) Tissue and cell preparation.
Gill tissues were rinsed in sterile seawater and fixed in Davidson's fixative (10% glycerol, 8% formaldehyde, 28.5% ethanol, 10% acetic acid, 30% seawater) (20) overnight at room temperature. Tissues were then dehydrated through a series of graded ethanol wash steps (50% once, 70% once, 80% once, 95% twice, and 100% thrice for 30 min each), followed by two washes in xylene at room temperature (45 min each). Tissues were infiltrated with paraffin (three changes, 30 min, 60°C). Consecutive tissue sections (5-μm thickness) were cut and mounted on APTS (3-aminopropyltriethoxysilane; Sigma)-treated slides. Slides were then baked at 57°C for 1 h. Paraffin was removed by three washes in xylene (15 min) and rehydrated through a graded ethanol series (inverse of that described above). Slides were washed in 0.2 M HCl and rinsed in 0.02 M Tris-HCl, pH 8.2. Following a 5-min treatment with proteinase K (0.5 μl/ml), slides were rinsed as described above, postfixed in 3.7% formaldehyde in 0.02 M Tris-HCl, and rinsed again. After drying, slides were treated with triethanolamine-acetic anhydride (24). T. turnerae cells grown in vitro and symbiont cells from gill tissue homogenates were fixed for 1 h to overnight in 3.7% formaldehyde in 90% methanol. Cells were dotted on APTS-coated slides and air dried. Slides were postfixed in 90% methanol-3.7% formaldehyde for 20 min at room temperature (5) and then rinsed in distilled water (dH2O) for 1 min. Slides were then treated in triethanolamine-acetic anhydride, and hybridizations were performed as previously described.
(ii) Hybridization conditions.
In situ hybridizations were performed on gill tissue sections and gill tissue homogenates from four adult shipworms sacrificed on 31 October 1996, 6 November 1996, 17 August 1997, and 14 October 1997. Hybridization buffer (750 mM NaCl, 5 mM EDTA, 100 mM Tris-HCl [pH 7.8], 0.025% sodium dodecyl sulfate) was added to each section along with 50 ng of each probe. Sections were covered with coverslips cut from Parafilm (American National Can) to prevent evaporation and were incubated in a humid chamber at 50°C for 3 to 24 h. Slides were washed three times (5 min) in 0.2× SET (30 mM NaCl, 200 nM EDTA, 4 mM Tris-HCl [pH 7.8]) at 50°C (∼11 to 14°C below melting temperature). Slides were then air dried and mounted with Vectashield mounting medium (Vector Laboratories).
(iii) Oligonucleotide design, synthesis, and labeling.
Oligonucleotide (DNA) test probes were designed as described previously (9) to hybridize specifically with a variable region (E. coli positions 637 to 662) of the RNA-like strand of the consensus sequence of clone groups LP1 through LP6. These clone-specific probes were labeled with either a red or green fluorochrome (Texas Red or fluorescein isothiocyanate [FITC], respectively) and were designated lpx-r or lpx-g, respectively, where x is a number designating the clone consensus group (1 through 6) for which the probe was designed, r is red fluorochrome, and g is green fluorochrome. Negative-control probes were labeled similarly and designated lpxn-r or lpxn-g. Each negative-control probe contained two differences (G→C) from its target sequence at E. coli positions 648 and 656. A bacterial-domain-specific probe corresponding to E. coli positions 338 to 355 (2) was labeled with FITC and designated bac-g. Probe sequences are listed in Table 1.
TABLE 1.
Probe sequences
| Probe | Positions | Sequencea | Melting temp specificityb |
|---|---|---|---|
| lp1 | 637-662 | GCAATACTCTAGCAACCCAGTTCTG | 65 LP1 |
| lp1n | 637-662 | GCAATACTCTACCAACCCACTTCTG | 65 NA |
| lp2 | 637-662 | GCTGTACTCTAGCTATACAGTTCTA | 61 LP2 |
| lp2n | 637-662 | GCTGTACTCTACCTATACACTTCTA | 61 NA |
| lp3 | 637-662 | GCAATACTCTAGTAATCCAGTTCTG | 61 LP3 |
| lp3n | 637-662 | GCAATACTCTACTAATCCACTTCTG | 61 NA |
| lp4 | 637-662 | GCTGTACTCAAGTTACCCAGTTCTA | 63 LP4 |
| lp4n | 637-662 | GCTGTACTCAACTTACCCACTTCTA | 63 NA |
| bac | 338-355 | LGCTGCCTCCCGTGTAGGAGT | 64 Bacteria |
Mismatches to target sequences are in bold.
NA, not applicable.
Oligonucleotides were synthesized commercially (Operon Technologies Inc., Alameda, Calif.) with incorporation of an amino linker (Amino Modifier C6) at the 5′ end of each oligonucleotide. Fluorochrome labels were added postsynthesis. Reaction cocktails contained 100 μl of oligonucleotide, 50 μl of 500 mM carbonate buffer (50 mM Na2CO3, 450 mM NaHCO3 [pH 9.2]), 60 μl of dH2O, and 40 μl of either carboxyfluorescein or Texas Red-X succinimidyl esters (10 mg/ml each; Molecular Probes) in dimethyl formamide. Labeling reactions were conducted in the dark at room temperature for 12 h, after which the unincorporated label was removed by chromatography (10 mM Tris-HCl [pH 7.5 to 8.0]) on a Sephadex G-25 column. Reaction products were vacuum dried and redissolved in dH2O to a final volume of 50 to 70 μl. Labeled probes were purified by thin-layer chromatography on silica gel 60 F254 thin-layer chromatography plates (EM Separations Technology) with 100 ml of solvent (55 ml of n-propanol, 35 ml of NH4OH, 10 ml of dH2O) for 4 to 5 h. Fluorochrome-conjugated probe was visualized by UV light (254 nm), scraped from the plate, and eluted in 300 to 600 μl of dH2O. Silica gel was removed by centrifugation (19,000 × g, 10 min), and probe concentrations were determined spectrophotometrically. Dual-probe in situ hybridizations were performed as described above except that a red-labeled and a green-labeled probe of different specificities were applied simultaneously in equal concentrations. In most cases, a red-labeled ribotype-specific probe was added in combination with the bacterial-domain-specific bac-g probe that served as an internal positive control for bacterial staining. Under these conditions, hybridization of a single probe to a given target resulted in red or green primary color staining while combined hybridization of two probes produced secondary colors ranging from yellow to orange.
Optimal hybridization and wash conditions for each probe were determined empirically by in situ hybridization to fixed and mounted cells under successively increasing stringencies until fluorescence due to probe hybridization was undetectable. Hybridization specificity was also confirmed for each probe by competitive hybridization with unlabeled exact-match probes. Specimens were observed by using a Nikon Labophot 2 epifluoresence microscope with Texas Red, fluorescein, and Texas Red, fluorescein, dual-wavelength filter sets (Chroma). Photomicrographs were taken with a Nikon FX-35DX camera with Kodak Gold (200 ASA) color print film or Kodak Elite II (200 ASA) slide film. Exposure times ranged from 30 to 524 s. Exposure times for negative-control probes were doubled compared to the corresponding exposure times for experimental probes to rule out the possibility of faint, nonspecific staining.
Phylogenetic analysis.
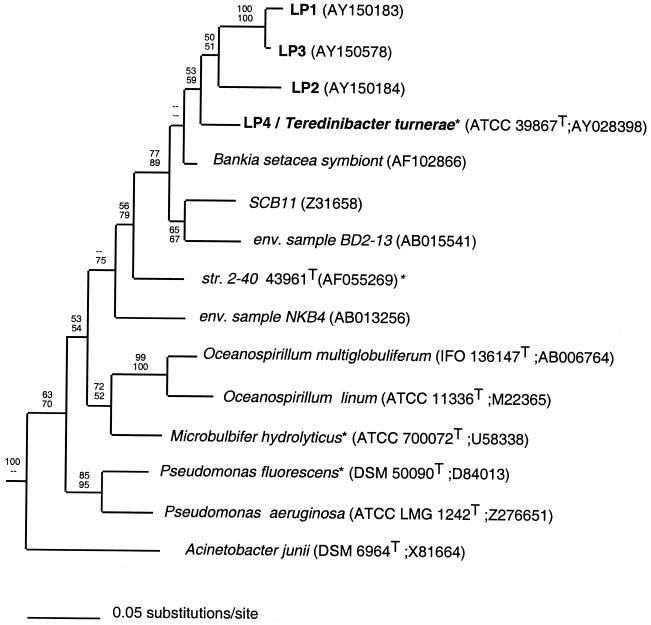
Sequences of 16S rRNA gene clone groups LP1 (1,390 bp, three pooled clones), LP2 (1,397 bp, two pooled clones), LP3 (1,441 bp, one clone), and LP4(Tt) (1,352 bp, one clone) and the direct-PCR sequence of the T. turnerae 16S rRNA gene were determined bidirectionally and aligned with 16S rRNA genes from selected gamma proteobacteria and environmental samples contained in the GenBank and Ribosomal Database Project databases. Database and culture collection accession numbers are reported in Fig. 1. Reference taxa were selected from among members of the gamma subdivision of the proteobacteria most closely related to T. turnerae, including taxa with known cellulolytic activity and the gill symbionts of other bivalves, as described previously (10).
FIG. 1.
A phylogenetic tree for T. turnerae, ribotypes LP1 through LP4, and representative gamma proteobacteria. The tree shown is extracted from a single best tree inferred by maximum likelihood analysis with 16S rRNA sequences. The maximum likelihood tree is identical to one of four best trees inferred by maximum parsimony. Bootstrap proportions are indicated at each node (maximum likelihood above, maximum parsimony below) as a percentage of 1,000 replicates. Dashes indicate bootstrap proportions of <50%. Type cultures and taxa with known cellulolytic capabilities are indicated by the superscripts T and *, respectively. Culture collection and sequence database accession numbers are in given in parentheses. Taxa included in the analyses but not shown in the tree are Aeromonas salmonicida (CCM 4103T; AT009859), Bathymodiolus puteoserpentis chemoautotrophic symbiont (U29163), Bathymodiolus puteoserpentis methanotrophic symbiont (U29164), Erwinia carotovora* (ATCC 15713T; M59149), Lucina nassula symbiont (X95299), Serratia marcescens* (ATCC 13880T; M59160), Vibrio harveyi (ATCC 14126T; X56578), Xanthomonas campestris* (LMG 568T; X95917), and Xanthomonas albilinians* (LMG 494T; X95918).
Alignments maximized agreement with the proposed secondary structure for E. coli (17). Agreement with this model was verified in new sequences by the occurrence of compensatory substitutions that preserve secondary structure in conserved internal helices. Gaps were inserted to compensate for length variation in identified loops and helices or to accommodate missing data. Additional gaps were not added to increase primary sequence similarity among taxa. Alignments were edited by using SeqLab (version 10.1; GCG). Sites within regions of uncertain alignment were identified and eliminated from further analyses, leaving a final character set of 1,381 nucleotide positions, 387 of which were parsimony informative.
Phylogenetic analyses were performed using algorithms contained in PAUP* (Phylogenetic Analysis Using Parsimony, version 4.0b10) (D. L. Swofford, PAUP* 4.0. Sinauer, Sunderland, Mass., 1997). Maximum likelihood trees were inferred by using the HKY85 substitution model (18) with empirical base frequencies, equal rates for all sites, and a transition/transversion ratio of 2. Maximum parsimony analyses were performed with character states optimized by accelerated transformation (ACCTRAN). In all analyses, branch swapping was by tree bisection-reconnection, with characters weighted equally (weight = 1) and gaps treated as missing data. Bootstrap analyses were performed by using the full heuristic search option with 1,000 replicates. Bootstrap analyses under maximum likelihood were limited to a reduced taxon set (20 taxa) to accommodate the greater computational demands of this technique.
RESULTS AND DISCUSSION
Analysis of 16S rRNA gene sequences.
Each of the 43 examined clones was assigned to one of six consensus groups (designated LP1 through LP6) based on their sequence identity. Clones were placed in a given group if they displayed a ≥99% sequence identity to the consensus sequence for that group over the ∼500 bp of sequence determined. Of the 43 clones, 31 were included in group LP1 (72%), 4 in group LP5 (9%), 2 in group LP2 (5%), and 1 each in groups LP3, LP4, and LP6 (2%). Consensus sequence LP4 was identical to the sequence determined for pure-cultured cells of T. turnerae and is hereafter referred to as LP4(Tt). Inserts contained in three remaining clones (3W16, 3W49, and 3W59) were determined to be probable chimeras and so were not assigned to consensus groups. All three putative chimeras were recovered from PCR 1 (35 cycles), consistent with the observation that chimera formation may increase with an increasing number of cycles during PCR amplification (26, 38, 39).
Sequence variability within consensus groups.
Sequence variability was observed within consensus groups; however, the frequency and pattern of most nucleotide substitutions were consistent with that expected due to polymerase error. For example, the 31 clones corresponding to the largest consensus group (LP1) contained a total of 22 divergences from the group consensus sequence, distributed over 18 positions (an average of <1 difference per 700 bp). At 15 (83%) of these variable positions, a difference from the consensus was observed in only 1 of the 31 clones. Only three positions (0.6% of all positions) contained differences from the consensus in two or more clones. In only two of these positions did differences covary (two clones shared the same difference [A→G] at E. coli position 498, and four clones shared the same difference [T→G] at E. coli position 610). The total number of differences observed among the 31 clones could be explained by a nucleotide misincorporation rate of about 5 × 10−5 incorporation errors/bp/cycle. This is within the range of values (2 × 10−4 to 7.2 × 10−5 errors/bp/cycle) reported for Taq polymerase under similar PCR conditions (11, 21, 27). Furthermore, none of the differences that occurred in stems were matched by compensating changes in the complementary strand, and five of these differences resulted in mismatches that could destabilize conserved helices. These results indicate that sequence polymorphism in the symbiont population represented by this consensus group is low by comparison to between-group variation (∼1.5 to 8.1%). Therefore, in this investigation, each consensus group sequence was treated as a unique rRNA type, or ribotype. Each ribotype was considered to represent a proxy for a putative symbiont phylogenetic type, or phylotype.
Phylogenetic analyses of ribotypes.
The phylogenetic analyses presented here include only those ribotypes (LP1 through LP4) that were detected in tissue and tissue homogenates by in situ hybridization (see below). Pairwise sequence identity (calculated as 1-Kimura 2-parameter distance) between these ribotypes ranged from 91.9 to 93.4%, with the exception of LP1 versus LP3, which shared a 98.5% sequence identity.
Comparison to published sequences (Ribosomal Database Project, GenBank) demonstrated that the L. pedicellatus putative symbiont ribotypes are derived from gamma proteobacteria. The sequence most similar to ribotypes LP1 through LP4 (93.3 to 95.3%) is that reported for an uncultivated symbiont from the shipworm Bankia setacea (37). Ribotypes LP5 and LP6 were distantly related to those detected in tissue, the former showing the greatest similarity (∼90%) to epsilon proteobacteria, while the latter was weakly associated with alpha proteobacteria.
A heuristic search by maximum likelihood analysis retained a single best tree with an ln likelihood of 11,037.2 (Fig. 1). Maximum parsimony retained the four best trees of 1,732 steps (consistency index = 0.4330, homoplasy index = 0.5670, retention index = 0.5290, and rescaled consistency index = 0.2291). Trees inferred by both methods were topologically identical with respect to all nodes receiving significant (>70%) (19) bootstrap support.
In these analyses, the T. turnerae sequence (identical to ribotype LP4) and ribotypes LP1, LP2, and LP3 were nested within a single clade that also included the 16S rRNA genes of an uncultivated symbiont from the shipworm B. setacea (37); an uncultivated bacterium, environmental sample BD2-13 (GenBank accession no. AB015541), from deep-sea sediments (1,521 m) in Sugura Bay, Japan (1,521 m; 34°55′N, 138°39′E) (25); and an undescribed marine heterotrophic bacterium cultivated from seawater collected along the coast of La Jolla, Calif. (SCB11, GenBank accession no. Z31658) (33). This clade receives significant bootstrap support (19) in both maximum parsimony (89%) and maximum likelihood (77%) analyses and is hereafter referred to informally as the Teredinibacter clade.
No strong support was observed for any specific relationship between the Teredinibacter clade and other named genera. Analyses described here suggest a peripheral relationship with either Pseudomonas spp. (sensu stricto) or Oceanospirillum spp., although with weak to insignificant bootstrap support. Consistent with previous reports, a possible relationship is suggested between the Teredinibacter clade and strain 2-40, an unnamed bacterium isolated from a marine sea grass bed in the Chesapeake Bay (15).
These results indicate that ribotypes LP1 through LP3 are derived from bacterial species that are candidates for classification within the genus Teredinibacter, the only formally described taxon in this lineage (10). Though no convention has been established to correlate degree of 16S rRNA sequence identity with taxonomic standing, most taxa that are considered to be separate species within the same genus differ by at least 1.5% (13). By this criterion, ribotypes LP1, LP2, and LP3, which differ from that of T. turnerae by >6.5%, may represent one or more new species within this genus. However, taxonomic classification must await more complete characterization of the putative symbionts and related free-living bacteria.
Fluorescent in situ hybridization.(i) Single-probe hybridizations.
When applied to tissue, probes lp1-r through lp4(Tt)-r [specific for ribotypes LP1 through LP4(Tt)] resulted in fluorescent staining of the symbiont-containing vesicles in sectioned bacteriocytes and individual bacterial cells in gill homogenates (single-probe data not shown). Staining and appearance of symbionts within bacteriocytes was similar to that observed in a previous study (9). Fluorescent staining by the bacterial-domain-specific positive-control probe bac-g was also observed in the same regions. No staining of gill tissue was detected by using ribotype-specific probes lp5-r or lp6-r or negative-control probes lp1n-r through lp4(Tt)n-r. These results indicate that ribotypes LP1 through LP4(Tt) derive from symbionts that reside within gill bacteriocytes. Only these symbiont ribotypes were examined further in this investigation.
(ii) Dual-probe hybridizations in individual symbiont cells.
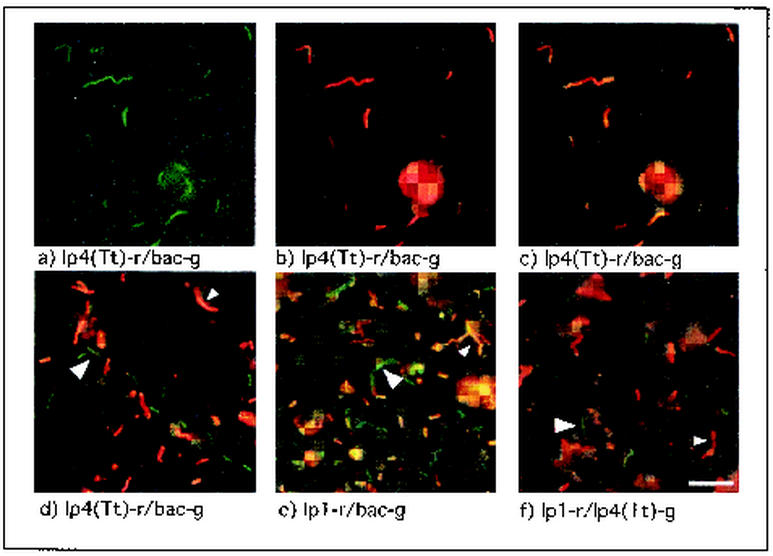
Dual-probe hybridizations, performed on individual symbiont cells released from bacteriocytes by gentle homogenization, demonstrated that no single ribotype-specific probe stained all symbiont cells visible in a given homogenate. When any red-labeled ribotype-specific probe [lp1-r through lp4(Tt)-r] was applied in combination with the green-labeled bacterial-domain-specific probe, both green-stained cells and yellow- to orange-stained cells were observed (Fig. 2d and e). These results indicate that, although each ribotype-specific target sequence could be detected in some symbiont cells, no single ribotype could account for all bacteria present in the gill homogenates. This is consistent with the possibility that the four ribotypes correspond to distinct symbiont phylotypes that coexist in the gill tissue.
FIG. 2.
Dual-probe in situ hybridizations with ribotype-specific and bacterial-domain-specific probes to label individual bacterial cells from T. turnerae pure culture and L. pedicellatus gill homogenate. Pure-cultured T. turnerae (a-c) and symbionts from L. pedicellatus gill homogenate (d-f) stained with probe combinations indicated below the corresponding panel and viewed with the green (a), red (b), or red and green dual-wavelength filter sets (c-f) are shown. Pure cultures of the symbiont T. turnerae stained uniformly with ribotype- and domain-specific probes, while symbionts in shipworm gill homogenate were heterogeneous with respect to staining with ribotype-specific probes. Large arrows indicate cells stained green by a single domain-specific (d and e) or ribotype-specific (f) probe, while small arrows indicate cells stained yellow to orange due to the combination of red- and green-labeled probes (d and e) or red due to staining with a single ribotype-specific probe (f). Similar results were observed for probe combinations lp3-r-lp4(Tt)-g and lp3-r-bac-g (data not shown). Scale bar, 5 μm.
Alternatively, these results could be explained by expression of multiple divergent 16S rRNA gene copies present in the genome(s) of one (or fewer than four) symbiont phylotypes. This possibility, however, is not supported by dual-probe in situ hybridization results determined by using two ribotype-specific probes. When either the red-labeled lp1-r (Fig. 2f) or lp3-r (data not shown) probe was applied to gill homogenates in combination with the green-labeled lp4(Tt)-g probe, both red- and green-labeled symbiont cells were observed. These results suggest that ribotypes LP1 and LP3 are expressed in a different subset of symbiont cells than is ribotype LP4. It was not possible, however, to examine all possible pairwise combinations of ribotype-specific probes in gill homogenates. This was due to the low detection efficiency of the FITC-labeled versions of probes lp1 and lp3 in individual symbiont cells and to the low abundance of cells staining with probe lp2 in gill homogenates. Furthermore, faint secondary colors (yellow to orange) are difficult to distinguish from pure primary colors (red and green) in individual bacterial cells in tissue homogenates. Therefore, it is possible that some cells express a second 16S rRNA type at levels below the detection limits of this system.
Although these results cannot rule out the possibility that some symbiont cells may express multiple ribotypes, hybridization results indicate that the cells of T. turnerae express only one of the four ribotypes when grown in pure culture. Only one of the four ribotype-specific probes, probe lp4(Tt)-r, was observed to stain pure-cultured log-phase cells of T. turnerae. Furthermore, in dual-probe experiments with probes lp4-r and bac-g, all cells were stained by both probes (Fig. 2a-c), indicating a homogeneous bacterial population expressing ribotype LP4.
(iii) Dual-probe hybridizations in bacteriocytes.
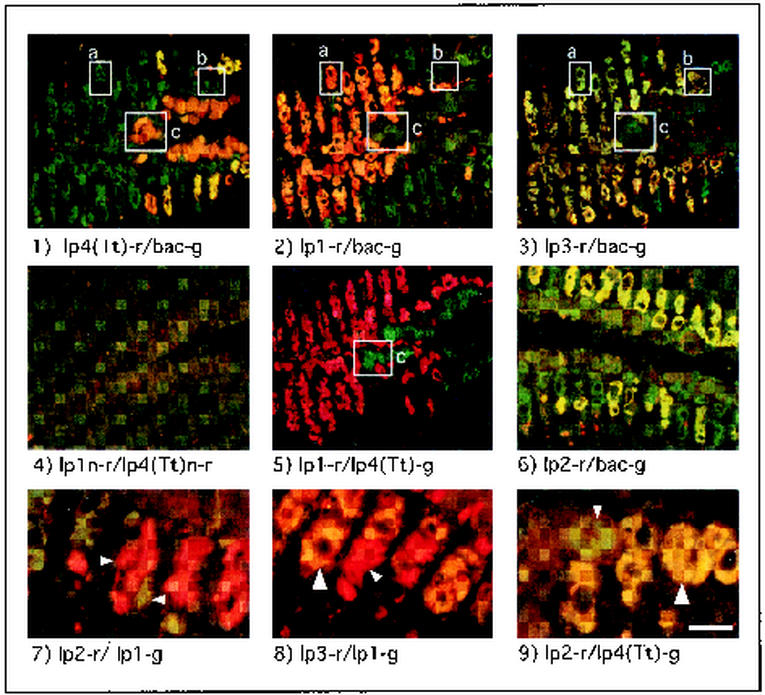
Dual-probe hybridizations performed in sectioned gill tissue produced distinct and specific staining patterns for each ribotype. This is evident in Fig. 3, panels 1 to 6, which shows a series of tissue sections (panels 1 to 5 are consecutive) from the gill of a single specimen of L. pedicellatus. For example, in section 1, treated with probes lp4(Tt)-r and bac-g, approximately one-third of the bacteriocytes were stained orange to yellow, indicating hybridization of both probes while the remaining bacteriocytes were stained green, indicating hybridization of only the bacterial-domain-specific probe bac-g. However, a different and nearly inverse staining pattern is shown for probe lp1-r in section 2. That is, cells that stained green in panel 1 appear orange to yellow in panel 2 and vice versa. This can be seen clearly in the boxed regions in Fig. 3, which indicate bacteriocytes that can be identified by shape, size, and position in several sections. Bacteriocyte a was stained by probe lp1-r (panel 2) but not by probe lp4(Tt)-r (panel 1), while the inverse is shown for bacteriocyte c. These results indicate that the symbiont ribotypes LP1 and LP4(Tt) occur in separate and mutually exclusive subsets of bacteriocytes. This conclusion is further supported by the section shown in panel 5, in which ribotype-specific probes lp1-r and lp4(Tt)-g show patterns of staining consistent with those found in panels 1 and 2 but stained only with the primary colors red and green.
FIG. 3.
Dual-probe in situ hybridizations with ribotype-specific and bacteria-domain-specific probes to label symbiont cells in sectioned gill tissue from L. pedicellatus. Panels 1 to 5 show consecutive sections from a single specimen of L. pedicellatus. Probe combinations applied to each section are indicated below the corresponding panel. Boxes a to c delineate bacteriocytes that could be identified by size, shape, and position in several sections. Small arrows in panels 7 to 9 delineate individual bacteriocytes stained by a single red or green ribotype-specific probe; large arrows delineate yellow- to orange-stained bacteriocytes, indicating the colocalization of two ribotype-specific probes. Note that symbiont ribotypes LP1 and LP3 are localized in a different subset of bacteriocytes than ribotypes LP2 and LP4. Scale bar, 75 μm (panels 1 to 6) or 25 μm (panels 7 to 9).
In contrast, probe lp3-r stained bacteriocytes with a pattern similar to that observed for lp1-r in panel 2. That is, many but not all bacteriocytes that stained with probe lp1-r also stained with probe lp3-r (see bacteriocytes a and b in panels 2 and 3). Moreover, bacteriocytes that stained with lp4(Tt)-r failed to stain with either lp1-r or lp3-r (box c, panels 1 to 3). These results indicate that ribotypes LP1 and LP3 cooccur in some but not all bacteriocytes, though neither cooccurs with LP4(Tt). Finally, though staining by this probe was observed in relatively few bacteriocytes, lp2-r stained with a pattern similar to that of lp4(Tt)-r (section 6). That is, some but not all bacteriocytes that stained with probe lp4(Tt)-g also stained with probe lp2-r (Fig. 3, panel 9). Staining with probe lp2-r, however, was not observed in bacteriocytes that stained with probes lp1-g (Fig. 3, panel 7) or lp3-g (data not shown).
Distribution of ribotypes.
Staining by ribotype-specific probes also showed consistent patterns of distribution with respect to gill structure. Bacteriocytes labeled by the lp4(Tt) and lp2 probes were distributed symmetrically in the more dorsal and medial regions of the gill, closely adjoining the central axis dividing the right and left gill demibranchs. This axis runs approximately along the horizontal midline of panels 1 to 6 in Fig. 3. Probes lp1 and lp3 were widely distributed in the more lateral and ventral regions of the gill filaments.
Cooccurrence of ribotypes.
There are six possible pairwise staining combinations that can be generated with a group of four probes (1:2, 1:3, 1:4, 2:3, 2:4, and 3:4). To date, simultaneous staining by only two of these six possible pairwise combinations (1:3 and 2:4) has been observed in single bacteriocytes (Fig. 3, panels 8 and 9). This pattern of colocalization suggests that these ribotypes correspond to four distinct symbiont phylotypes that may be paired in specific combinations within bacteriocytes. This might indicate that symbiotic associations also occur between bacteria within bacteriocytes, i.e., bacteria-bacteria symbioses may exist within the larger animal-bacteria symbiosis.
Again, the observed pattern of cooccurring ribotypes could be also be explained by the differential expression of multiple divergent 16S rRNA gene copies present in the genome(s) of fewer than four symbiont phylotypes. However, pairwise evolutionary distances among the four ribotypes are larger than those typically observed among operons in a single genome. Multiple rRNA operons are common in bacteria, and sequence variability between 16S rRNA gene copies as high as 6.4% (1, 40, 42) has been reported. However, such large differences appear to be unusual (3). A survey of 14 complete bacterial-genome sequences (representing 55 rRNA operons) revealed a range of inter-operon variation in 16S rRNA genes from 0 to 1.23%, with over half of all taxa showing no inter-operon differences and nearly 80% (11 of 14) showing a less than 0.2% sequence difference among 16S rRNA gene copies (22). In comparison, the sequence divergences observed among consensus groups LP1 through LP4(Tt) are greater than 6.5% for all pairwise comparisons, except that of LP1 versus LP3, which differs by 1.5%. Thus, these differences are larger than those typically observed between rRNA operons in a single genome and, in fact, with the exception of the LP1 versus LP3 comparison, exceed any inter-operon differences yet reported.
PCR amplification and cloning results are also inconsistent with multiple divergent operons, at least for ribotypes LP1 and LP4. In amplifications performed by using bacterial-domain-specific primers with genomic DNA extracted from pure cultures of T. turnerae, a single sequence identical to the LP4 consensus sequence was detected. In contrast, the LP1 consensus sequence predominates in similar amplifications from L. pedicellatus gill DNA. It is difficult to explain how such differences could arise if both sample types contain a monoculture of the same bacterial phylotype.
Although the data presented here support the existence of multiple symbionts in L. pedicellatus, the functional significance of this multiple symbiosis remains unknown. In nature, wood degradation is a complex process that is rarely accomplished by a single microbial species (4). Thus, it is possible that each symbiont type in L. pedicellatus contributes different enzymes to a complex cellulolytic system. Alternatively, multiple symbiont phylotypes could provide flexibility to the host in adapting to a variety of wood types or environmental conditions. In either case, it may be revealing to determine whether multiple endosymbiosis is unique to this species or is a more widespread phenomenon. Unfortunately, only a small number of adult specimens have been examined in this study and these have been collected and reared in similar locations and under similar conditions. Therefore, although uniform results were observed in these specimens, it remains to be determined whether the composition of symbiont populations is consistent among all members of this species or whether variation may occur with the differing condition, environment, and developmental stage of the hosts. Finally, although T. turnerae (LP4) has been isolated from the gills of several shipworm species (10), it remains uncertain whether the other putative symbionts detected here are distributed widely among shipworm species. Further investigations are presently under way in this laboratory with a broader range of host specimens and taxa and more quantitative molecular and histological methods to better describe the diversity and complexity of symbiont populations in teredinid bivalves.
In summary, (i) four distinct but related symbiont ribotypes have been detected in the gills of L. pedicellatus, (ii) no single ribotype was detected in all symbiont cells in gill homogenates or in all bacteriocytes in gill tissue, (iii) the 16S rRNA gene sequence of one ribotype (LP4) is identical to that determined for T. turnerae grown in pure culture, (iv) ribotypes LP1 through LP3 have not been detected in cultivated cells of T. turnerae, (v) two ribotypes were detected in some but not all bacteriocytes, and (vi) within individual bacteriocytes, the ribotype combinations LP1 and LP3 and LP2 and LP4 have been observed but other possible pairwise combinations have not. Taken together, these observations provide strong evidence that at least two symbiont phylotypes (represented by ribotypes LP1 and LP3 and LP2 and LP4, respectively) and as many as four distinct symbiont phylotypes coexist in the gills of L. pedicellatus.
Acknowledgments
This work was supported by grants from the National Science Foundation (DEB-9420051 and IBN-9982982), the Maine Science and Technology Foundation's Center for Innovation in Biotechnology, and the University of Maine's Faculty Research program.
We express our thanks to Scott Gallager (Woods Hole Oceanographic Institution) for providing the breeding stock of L. pedicellatus and Martin Polz (MIT) for thoughtful discussion of the manuscript.
REFERENCES
- 1.Amann, G., K. O. Stetter, E. Llobet-Brossa, R. Amann, and J. Anton. 2000. Direct proof for the presence and expression of two 5% different 16S rRNA genes in individual cells of Haloarcula marismortui. Extremophiles 4:373-376. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R. I., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clayton, R. A., G. Sutton, P. S. Hinkle, Jr., C. Bult, and C. Fields. 1995. Intraspecific variation in small-subunit rRNA sequences in GenBank: why single sequences may not adequately represent prokaryotic taxa. Int. J. Syst. Bacteriol. 45:595-599. [DOI] [PubMed] [Google Scholar]
- 4.Coughlan, M. P., and F. Mayer. 1992. The cellulose-decomposing bacteria and their enzyme systems, p. 451-516. In A. Ballows, H. Truper, W. Harder, and K. H. Schleifer (ed.), vol. 4. Springer-Verlag, Berlin, Germany.
- 5.DeLong, E. F., G. S. Wickham, and N. R. Pace. 1989. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science 243:1360-1363. [DOI] [PubMed] [Google Scholar]
- 6.Distel, D. L. The biology of wood-boring bivalves and their bacterial endosymbionts. In T. Schulz, B. Goodell, and D. Nicholas (ed.), Current knowledge of wood deterioration mechanisms and its impact on biotechnology and wood preservation, in press. American Chemical Society, Washington, D.C.
- 7.Distel, D. L. 2000. Phylogenetic relationships among Mytilidae (Bivalvia): 18S rRNA data suggest convergence in mytilid body plans. Mol. Phylogenet. Evol. 15:25-33. [DOI] [PubMed] [Google Scholar]
- 8.Distel, D. L., and C. M. Cavanaugh. 1995. Intracellular coexistence of methano- and thioautotrophic bacteria in a hydrothermal vent mussel. Proc. Natl. Acad. Sci. USA 92:9598-9602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Distel, D. L., E. F. DeLong, and J. B. Waterbury. 1991. Phylogenetic characterization and in situ localization of the bacterial symbiont of shipworms (Teredinidae: Bivalvia) by using 16S rRNA sequence analysis and oligodeoxynucleotide probe hybridization. Appl. Environ. Microbiol. 57:2376-2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Distel, D. L., W. Morrill, N. MacLaren-Toussaint, D. Franks, and J. Waterbury. Teredinibacter turnerae, gen. nov., sp. nov., a dinitrogen fixing, cellulolytic, endosymbiotic gammaproteobacterium isolated from the gills of wood-boring mollusks (Bivalvia; Teredinidae). Int. J. Syst. Evol. Microbiol., in press. [DOI] [PubMed]
- 11.Dunning, A. M., P. Talmud, and S. E. Humphries. 1988. Errors in polymerase chain reaction. Nucleic Acids Res. 16:10393.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fisher, C. R., J. M. Brooks, J. S. Vodenicher, J. M. Zande, J. J. Childress, and R. A. Burke, Jr. 1993. The co-occurrence of methanotrophic and chemoautotrophic sulfur-oxidizing bacterial symbionts in a deep-sea mussel. PSZNI Microb. Ecol. 14:277-289. [Google Scholar]
- 13.Fox, G. E., J. D. Wistozkey, and P. J. Jurtshuck. 1992. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Intl. J. Syst. Bacteriol. 42:166-170. [DOI] [PubMed] [Google Scholar]
- 14.Gallager, S. M., R. D. Turner, and C. J. Berg. 1981. Physiological aspects of wood consumption, growth, and reproduction in the shipworm Lyrodus pedicellatus quatrefages. J. Exp. Mar. Biol. Ecol. 52:63-77. [Google Scholar]
- 15.Gonzalez, J. M., and R. M. Weiner. 2000. Phylogenetic characterization of marine bacterium strain 2-40, a degrader of complex polysaccharides. Int. J. Syst. Evol. Microbiol. 50:831-834. [DOI] [PubMed] [Google Scholar]
- 16.Greene, R. V., H. L. Griffin, and S. N. Freer. 1988. Purification and characterization of an extracellular endoglucanase from the marine shipworm bacterium. Arch. Biochem. Biophys. 1:334-341. [DOI] [PubMed] [Google Scholar]
- 17.Gutell, R. R. 1994. Collection of small subunit (16S- and 16S-like) ribosomal RNA structures: 1994. Nucleic Acids Res. 22:3502-3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hasegawa, M., Y. Iida, T. Yano, F. Takaiwa, and M. Iwabuchi. 1985. Phylogenetic relationships among eukaryotic kingdoms inferred from ribosomal RNA sequences. J. Mol. Evol. 22:32-38. [DOI] [PubMed] [Google Scholar]
- 19.Hillis, D. M., and J. J. Bull. 1993. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 42:182-192. [Google Scholar]
- 20.Humason, G. L. 1967. Animal tissue techniques. W. H. Freeman and Co., San Francisco, Calif.
- 21.Keohavong, P., and W. G. Thilly. 1989. Fidelity of DNA polymerases in DNA amplification. Proc. Natl. Acad. Sci. USA 86:9253-9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klappenbach, J. A., P. R. Saxman, J. R. Cole, and T. M. Schmidt. 2001. rrndb: the ribosomal RNA operon copy number database. Nucleic Acids Res. 29:181-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lane, D. J. 1991. 16S/23S sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, New York, N.Y.
- 24.Leitch, A. R. 1994. In situ hybridization, vol. 27. Bios Scientific Publications, Ltd., Oxford, United Kingdom.
- 25.Li, L., C. Kato, and K. Horikoshi. 1999. Bacterial diversity in deep-sea sediments from different depths. Biodivers. Conserv.
- 26.Liesack, W., H. Weyland, and E. Stackebrandt. 1991. Potential risks of gene amplification by PCR as determined by 16S rDNA analysis of a mixed-culture of strict barophilic bacteria. Microb. Ecol. 21:191-198. [DOI] [PubMed] [Google Scholar]
- 27.Ling, L. L., C. Keohavong, C. Dias, and W. G. Thilly. 1991. Optimization of the polymerase chain reaction with regard to fidelity: modified T7, Taq, and Vent polymerases. PCR Methods Appl. 1:63-69. [DOI] [PubMed] [Google Scholar]
- 28.Moran, N. A., and P. Baumann. 2000. Bacterial endosymbionts in animals. Curr. Opin. Microbiol. 3:270-275. [DOI] [PubMed] [Google Scholar]
- 29.Nishiguchi, M. K. 2000. Temperature affects species distribution in symbiotic populations of Vibrio spp. Appl. Environ. Microbiol. 66:3550-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishiguchi, M. K., E. G. Ruby, and M. J. McFall-Ngai. 1998. Competitive dominance among strains of luminous bacteria provides an unusual form of evidence for parallel evolution in sepiolid squid-vibrio symbioses. Appl. Environ. Microbiol. 64:3209-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Popham, J. D. 1974. Further observations of the Gland of Deshayes in the Teredo Bankia australis. Veliger 18:55-59. [Google Scholar]
- 32.Popham, J. D., and M. R. Dickson. 1973. Bacterial associations in the Teredo Bankia australis (Lamellibranchia, Mollusca). Mar. Biol. 19:338-340. [Google Scholar]
- 33.Rehnstam, A.-S., S. Bachman, D. C. Smith, F. Azam, and A. Hagstrom. 1993. Blooms of sequence-specific culturable bacteria in the sea. FEMS Microbiol. Ecol. 102:161-166. [Google Scholar]
- 34.Rousset, F., H. Braig, and S. L. O'Neill. 1999. A stable triple Wolbachia infection in Drosophila with nearly additive incompatibility effects. Heredity 82:620-667. [DOI] [PubMed] [Google Scholar]
- 35.Saiki, R., D. H. Gelfand, S. Stoffel, S. J. Scharf, R. Higuchi, G. T. Horn, K. B. Mullis, and H. A. Erlich. 1987. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 239:487-491. [DOI] [PubMed] [Google Scholar]
- 36.Sigerfoos, C. P. 1907. Natural history, organization and late development of the Teredinidae or shipworms. Bull. Bureau Fish. 37:191-231. [Google Scholar]
- 37.Sipe, A. R., A. E. Wilbur, and S. C. Cary. 2000. Bacterial symbiont transmission in the wood-boring shipworm Bankia setacea (Bivalvia: Teredinidae). Appl. Environ. Microbiol. 66:1685-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang, G. C., and Y. Wang. 1996. The frequency of chimeric molecules as a consequence of PCR co-amplification of16S. rRNA genes from different bacterial species. Microbiology 142:1107-1114. [DOI] [PubMed] [Google Scholar]
- 39.Wang, G. C., and Y. Wang. 1997. Frequency of formation of chimeric molecules as a consequence of PCR coamplification of 16S rRNA genes from mixed bacterial genomes. Appl. Environ. Microbiol. 63:4645-4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, Y., Z. Zhang, and N. Ramanan. 1997. The actinomycete Thermobispora bispora contains two distinct types of transcriptionally active 16S rRNA genes. J. Bacteriol. 179:3270-3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Waterbury, J. B., C. B. Calloway, and R. D. Turner. 1983. A cellulolytic-nitrogen fixing bacterium cultured from the Gland of Deshayes in shipworms (Bivalvia: Teredinidae). Science 221:1401-1403. [DOI] [PubMed] [Google Scholar]
- 42.Yap, W. H., Z. Zhang, and Y. Wang. 1999. Distinct types of rRNA operons exist in the genome of the actinomycete Thermomonospora chromogena and evidence for horizontal transfer of an entire rRNA operon. J. Bacteriol. 181:5201-5209. [DOI] [PMC free article] [PubMed] [Google Scholar]