Abstract
Thorough assessments of fungal diversity are currently hindered by technological limitations. Here we describe a new method for identifying fungi, oligonucleotide fingerprinting of rRNA genes (OFRG). ORFG sorts arrayed rRNA gene (ribosomal DNA [rDNA]) clones into taxonomic clusters through a series of hybridization experiments, each using a single oligonucleotide probe. A simulated annealing algorithm was used to design an OFRG probe set for fungal rDNA. Analysis of 1,536 fungal rDNA clones derived from soil generated 455 clusters. A pairwise sequence analysis showed that clones with average sequence identities of 99.2% were grouped into the same cluster. To examine the accuracy of the taxonomic identities produced by this OFRG experiment, we determined the nucleotide sequences for 117 clones distributed throughout the tree. For all but two of these clones, the taxonomic identities generated by this OFRG experiment were consistent with those generated by a nucleotide sequence analysis. Eighty-eight percent of the clones were affiliated with Ascomycota, while 12% belonged to Basidiomycota. A large fraction of the clones were affiliated with the genera Fusarium (404 clones) and Raciborskiomyces (176 clones). Smaller assemblages of clones had high sequence identities to the Alternaria, Ascobolus, Chaetomium, Cryptococcus, and Rhizoctonia clades.
Fungi are important components of ecosystems and human civilization. They play vital roles in processes such as soil formation, nutrient cycling, nutrient transportation to plant roots, and the transformation of waste materials into useful commodities such as compost (9, 23). Fungi also represent a source of food, pharmaceuticals, and biological control agents (8). Yet despite their importance, there still are no efficient methods for describing the considerable diversity of fungi inhabiting most environments.
Traditional methods for examining fungal diversity include isolation on culture media and analysis of fruiting bodies (7). However, since not all fungi readily grow on culture media (12) and the diversity of extant fungi is considerable (12, 13), new approaches for describing these organisms are needed. The development of rRNA gene (ribosomal DNA [rDNA])-based strategies, which have led to the discovery of thousands of new prokaryotic phylotypes (1, 3, 11, 20, 27), should also provide a valuable means to analyze fungal communities.
Several rDNA-based approaches have been devised for analysis of fungal community composition. When developing such strategies, the first requirement is to obtain PCR primers that selectively amplify fungal rDNA from the sample of interest. These primers must have high specificity, because fungal DNA will likely constitute only a minor fraction of the total DNA isolated from most samples. Investigators examining a variety of environments, including plant roots, soil, and human tissues, have developed primer sets for this purpose (2, 4, 10, 14, 15, 18, 19, 21, 22, 24, 26). After the rDNA genes are amplified, they can be analyzed by several methods, including cloning and sequencing (26) or by separating them through processes such as denaturing gradient gel electrophoresis (DGGE) (17, 19, 22, 24). A new and potentially more effective approach, however, may come from array-based technologies, which offer the necessary capability for thorough analysis of fungal community composition.
Oligonucleotide fingerprinting of rRNA genes (OFRG) is an array-based method that allows extensive analysis of microbial community composition (25). OFRG works by sorting arrayed rDNA clones into taxonomic clusters through a series of hybridization experiments, each using a single oligonucleotide probe. Although this approach was originally developed to examine bacterial community composition, a simulated annealing algorithm can be used to design probe sets that discriminate any clone type (5). In this work, we developed an OFRG probe set for fungal rDNA and then demonstrated its utility by examining the fungal community composition of an agricultural soil.
MATERIALS AND METHODS
The methods used in this work are based on previously described protocols (25). Several significant modifications have been made to improve the reliability and accuracy of the OFRG process.
Soil treatments and DNA extraction.
This greenhouse experiment was part of another project targeted at identifying the microorganisms involved in soil suppressiveness against the plant-parasitic nematode Heterodera schachtii. Soil (top 10 cm) was collected from the 9E field at the Agriculture Research Station at the University of California, Riverside (28). A portion of the soil was fumigated with methyl iodide as previously described (28). Three days after fumigation, all soils were mixed 10:1 with silica sand. Six-inch pots were filled with methyl iodide-fumigated and nonfumigated soils in two ratios: 99.9:0.1 (0.1% suppressive soil treatment) and 90:10 (10% suppressive soil treatment). There were five replicate pots per treatment. Five seeds of mustard greens (Brassica juncea cv. Florida broadleaf; Lockhart Seed, Inc., Stockton, Calif.) were planted per pot. All pots were placed in a greenhouse under natural light at 23 ±3°C. After emergence, the seedlings were thinned to one per pot. Four weeks after seeding, each pot was infested with 10,000 second-stage juveniles of the plant-parasitic nematode Heterodera schachtii. Soil was collected 11 weeks later. The samples were dried by overnight incubation at 30°C and then stored at −20°C. DNA was extracted from each soil sample (0.5 g) by using the FastDNA Spin Kit for Soil (Bio101, Vista, Calif.) (6). DNA from the five replicate samples was pooled for analysis of fungal composition.
rDNA library construction.
Fungal small-subunit rDNA samples were PCR amplified in 10-μl glass capillary tubes by using a 1002 RapidCycler (Idaho Technologies, Idaho Falls) from gel-purified soil DNA. One-hundred-microliter PCR mixtures contained 50 mM Tris (pH 8.3), 500 μg of bovine serum albumin per ml, 2.5 mM MgCl2, 250 μM each deoxynucleoside triphosphate (dNTP), 400 nM each of the fungal small-subunit rDNA primers 463 (TCAAGTTAGCATGGAATAATRRAATAGGA) and 464 (AACTCATTGCAATGCYCTATCCCCA), 5 U of AmpliTaq DNA polymerase (ABI) and 10 μl of soil DNA, composed of equal volumes of DNA from each of the replicate soil samples. The primers are derivatives of nu-SSU-0817-5′ and nu-SSU-1536-3′ (4). The cycling parameters were as follows: 94°C for 2 min; 20 cycles of 94°C for 10 s, 55°C for 15 s, and 72°C for 4 min; followed by 72°C for 2 min. PCR products were gel isolated and purified with QIAquick PCR purification kit (Qiagen, Chatsworth, Calif.), ligated into pOFRG (an unpublished T-cloning vector), transformed into competent Escherichia coli DH5α (Gibco-BRL), and plated on Luria-Bertani (LB) agar plates containing 100 μg of ampicillin per ml, which were surface spread with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) and isopropyl-β-d-thiogalactopyranoside (IPTG); other T-cloning vectors (25) can be used instead of pOFRG, but note that the array construction PCR primer or primers (see below) must match the vector. For each soil treatment, 768 white colonies were randomly picked into 384-well culture plates, with each well containing 30 μl of LB agar (100 μg of ampicillin per ml), except for the perimeter wells, which were filled with 60 μl of LB agar to prevent drying. For array construction (see below), the culture plates were incubated for 7 to 9 h at 37°C; these plates were then transferred to a HiGro shaking incubator (GeneMachines, Genomic Instrumentation Services, Inc.) and shaken (500 rpm) overnight at 37°C with an airflow setting of 0.5 standard liters per min. For long-term storage, the culture plates were incubated overnight at 37°C without shaking in an open plastic bag, with each well containing 30 μl of LB medium (100 μg of ampicillin per ml). The next day, the plates were stored at −70°C after addition of 30 μl of LB medium supplemented with 30% glycerol.
Array construction.
The arrays were constructed by applying spots of PCR-amplified rDNA onto nylon membranes. Thirty-five-microliter PCR mixtures contained 50 mM Tris (pH 8.3), 500 μg of BSA per ml, 2.5 mM MgCl2, 250 μM each dNTP, and 800 nM ArrayPCR primer (GTCGTGCGTGGGACACCAGTAG), which anneals to the multiple cloning site of the vector, as well as 1.75 U of Taq DNA polymerase. The reagents (35 μl per well) were added to 384-well PCR plates (Marsh Bio Products, Rochester, N.Y.). (Note that it is important to avoid air bubbles at the bottom of the wells.) Freshly grown, overnight cultures (0.5 μl of each) of the rDNA clones (described in the previous paragraph) were added to the PCR reagents with a 384-pin solid pin replicator (V & P Scientific, Inc., San Diego, Calif.). The plates were sealed with Thermo-Seal foil (Marsh Bio Products) by using a preheated Combi Thermo-sealer (ABgene, Epsom, United Kingdom) for 4 s. PCR was then performed by alternately submerging the PCR plates in two water baths. The cycling parameters were as follows: 94°C for 10 min; 35 cycles of 94°C for 1 min and 72°C for 2 min; and finally 72°C for 5 min. The PCR products were applied as spots with a surfactant-coated 0.5-μl slot pin replicator onto dry 11- by 8-cm Hybond N+ membranes by using a Multi-Print replication registration device (V&P Scientific, San Diego, Calif.) (Amersham Pharmacia Biotech). One microliter of each PCR product was delivered to the membrane by two sequential spotting applications. For each spotting application, the membranes were placed on two sheets of 0.35-mm chromatography paper (with the paper changed for each membrane), and the replicator was firmly pressed against the membrane for 5 s. The Multi-Print device allows the contents of four different 384-well plates to be printed onto a single 11- by 8-cm membrane, resulting in an array of 1,536 clones.
Array hybridization.
The nylon membranes containing the rDNA fragments, which we now call “arrays,” were fixed by UV cross-linking (70 mJ). Immediately before hybridization, the arrays were denatured with 0.5 N NaOH-1.5 M NaCl (two times for 5 min each on chromatography paper), neutralized with 50 mM Na phosphate (pH 7.2) (three times for 3 min each on chromatography paper), covered with boiling 0.1% sodium dodecyl sulfate (SDS), and allowed to cool for 10 min. The arrays were then prehybridized in bottles containing 5 ml of hybridization solution (5% sarcosyl, 0.2 M Na phosphate [pH 7.2]) with rotation for 30 min at 12°C. Hybridizations were performed by adding 10 μl of a 33P-labeled DNA oligonucleotide probe (5 μl for probes 14 and 27) to each bottle and rotating the bottles overnight at 12°C. DNA oligonucleotides were end labeled with T4 polynucleotide kinase (T4 PNK) (New England Biolabs); the 10-μl reaction mixtures contained 2 μM oligonucleotide, 15 μCi of [γ-33P]ATP, 1 μl of 10× T4 PNK buffer, and 6.5 U of T4 PNK and were incubated at 37°C for 30 min. Following the overnight hybridization, the hybridization solution was collected and saved for the second hybridization, and the arrays were washed twice in 0.1 to 4× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 15 to 30 min at 12°C (Table 1). After being washed, the arrays were briefly placed on chromatography paper to remove excess fluid and then enclosed with plastic wrap to prevent drying. The membranes were exposed to an Imaging Screen (Bio-Rad) for 16 h and then scanned with a Personal Molecular Imager FX (Bio-Rad). The signal intensities with background correction were obtained with ImaGene 4.0 software (Biodiscovery). The arrays were used two to four times. To remove the probe from the arrays, the arrays were covered with boiling stripping buffer (1× SSC, 0.1% SDS, 200 mM Tris [pH 7.5]), allowed to cool for 15 min, and then either put in hybridization bottles for a second hybridization or dried for 30 min for long-term storage.
TABLE 1.
Array washing conditions for each OFRG probea
| Probe no. | Washing buffer (× SSC) | Washing time (min) |
|---|---|---|
| 1 | 1 | 30 |
| 2 | 1 | 30 |
| 3 | 1 | 30 |
| 4 | 4 | 15 |
| 5 | 0.1 | 30 |
| 6 | 0.1 | 30 |
| 7 | 4 | 15 |
| 8 | 1 | 30 |
| 9 | 1 | 30 |
| 10 | 1 | 30 |
| 11 | 0.1 | 30 |
| 12 | 0.1 | 30 |
| 13 | 1 | 15 |
| 14 | 0.1 | 30 |
| 15 | 1 | 30 |
| 16 | 0.1 | 30 |
| 17 | 1 | 15 |
| 18 | 4 | 15 |
| 19 | 1 | 30 |
| 20 | 0.1 | 30 |
| 21 | 1 | 15 |
| 22 | 1 | 15 |
| 23 | 1 | 15 |
| 24 | 4 | 15 |
| 25 | 1 | 15 |
| 26 | 4 | 15 |
| 27 | 1 | 30 |
After hybridization with the 33P-labeled probes, arrays were washed twice with the buffers and for the times indicated
Oligonucleotide probes.
The following 26 discriminating oligonucleotide probes were used: 1, ATAGGGATAG; 2, CTGGCTTCTT; 3, GTCTTTGGGT; 4, GATTTGTCTG; 5, AGGGATCGGG; 6, GCTACACTGA; 7, AAATAGCCCG; 8, CGGTTCTATT; 9, TGATAGCTCT; 10, CGCGCGCTAC; 11, GTTGGTGGAG; 12, CTGGGTAATC; 13, AATCAAAGTC; 14, GCCGTTCTTA; 15, GGCTTCTTAG; 16, CAGAGCCAGC; 17, CAGACATAAC; 18, TTTGAGGCAA; 19, GCACCTTACG; 20, CCAGACACAA; 21, TATGCCGACT; 22, CTTAACCTGC; 23, TTGATAGCTC; 24, AAATTCTTGG; 25, TACTGCGAAA; and 26, TCAAAGTCTT. The reference probe (no. 27) was GGTGAGTTTCCC; this probe is expected to hybridize to all fungal rDNA clones and is derived from the PCR primer nu-SSU-1196-3′ (4). These probes were designed with a previously described simulated annealing algorithm (5). Simulated annealing is a popular heuristic method for efficiently solving difficult optimization problems (16). The original goal of our probe set design was to construct a probe set that could discriminate 616 fungal small-subunit rDNA sequences obtained from GenBank. However, since some of the probes provided by the simulated annealing algorithm did not hybridize in a consistent and predictable manner in the actual experiments, the probes used in this study were a collection of oligonucleotides from three different probe sets that produced strong signal intensities and hybridized to the control clones in the expected manner. Even though this probe set was generated through suboptimal means, it was still able to produce high-resolution results (see Results and Discussion section). Future refinements of the probe selection algorithm allowing replacement of ineffective probes should increase the resolution of this approach.
Data analysis.
The signal intensities used for this OFRG analysis were averaged values obtained from two replicate hybridization experiments. The averaged values were classified as 0, 1, or N, according to the intensity values of control clones. For this experiment, 1,536 clones were arrayed, 30 of which had defined nucleotide sequences and served as control clones for each hybridization experiment. For most probes, the control clones expected not to hybridize with the probe (negative controls) had intensity values less than the control clones expected to hybridize with the probe (positive controls). Conversely, the intensity values from the positive clones were higher that those from the negative controls. For these probes, clones with intensity values less than or equal to x were given a 0 classification, where x is the highest intensity value generated by a negative control. Clones with intensity values greater than or equal to y were given a 1 classification, where y is the lowest value generated by a positive control. All other clones were given an N classification. For some probes, not all of the control clones performed in the predicted manner. For example, some positive controls had intensity values that were lower than some of the negative control values and vice versa. For these probes, clones with intensity values less than x were given a 0 classification, where x is the lowest intensity value generated from a positive control. Clones with intensity values greater than y were given a 1 classification, where y is the highest value generated by a negative control. All other clones were given an N classification. The process created a hybridization fingerprint for each clone, which is a vector of values resulting from its hybridizations with all probes. The fingerprints were clustered by UPGMA (unweighted pair group method with arithmetic mean) from PAUP 4.0 beta 10, with default parameters. Each cluster was defined as a group of clones with the same fingerprint (with N classifications consistently resolved). Twenty-nine clones did not hybridize to the reference probe and were excluded from this analysis.
Sequence analysis.
The nucleotide sequences of 117 rDNA clones were determined by using the ABI PRISM BigDye Terminators v3.0 cycle sequencing kit and an ABI 3100 genetic analyzer. These sequences were used to examine the resolution and accuracy of this OFRG experiment. For the resolution analysis, 37 sequences from seven randomly chosen clusters were determined. For the accuracy analysis, we used the sequences from the resolution analysis plus 80 additional sequences that were distributed throughout the tree. Plasmid DNA was extracted with a QIAprep Spin Miniprep kit (Qiagen, Chatsworth, Calif.). The sequencing primers used were T725 and SP650 (GGCCCGACGTCGCATGCTC and TGGTCGACCTGCAGGCGGC, respectively). rDNA sequences were assembled with ContigExpress (Vector NTI). Sequence identities were determined with BLAST (National Center for Biotechnology Information) and Align X (Vector NTI).
Nucleotide sequence accession numbers.
The nucleotide sequences of the following rDNA clones from Fig. 1B have been deposited in the GenBank database (accession numbers in parentheses): 1011-2 (AF515307), 111-1 (AF515315), 1152-1 (AF515316), 1183-1 (AF515317), 1388-1 (AF515331), 15-1 (AF515340), 21-1 (AF515353), 33-1 (AF515363), 336-1 (AF515364), 432-1 (AF515376), 496-1 (AF515380), 621-1 (AF515388), 67-2 (AF515392), 720-1 (AF515398), 852-2 (AF515407), 864-2 (AF515408), 960-2 (AF515416). The accession numbers for the other nucleotide sequences used in this study were AF515305, AF515306, AF515308 to AF515314, AF515318 to AF515330, AF515332 to AF515339, AF515341 to AF515352, AF515354 to AF515362, AF515365 to AF515375, AF515377 to AF515379, AF515381 to AF515387, AF515389 to AF515391, AF515393 to AF515397, AF515399 to AF515406, and AF515409 to AF515415.
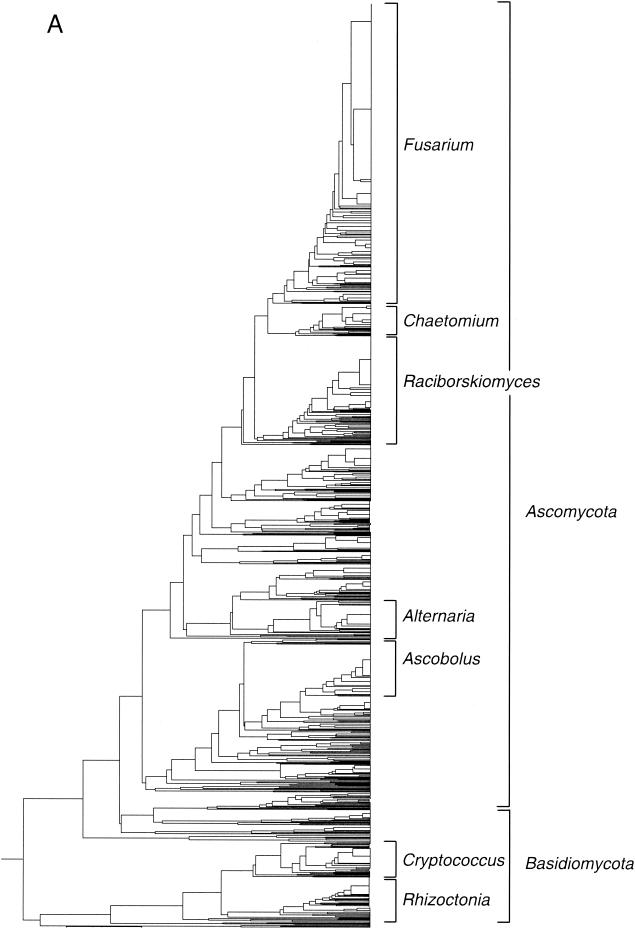
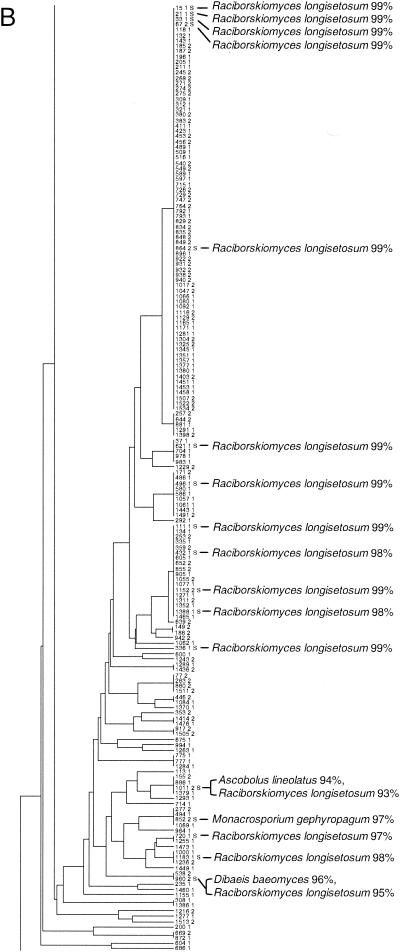
FIG. 1.
Taxonomic depiction of soil fungi produced by OFRG. The UPGMA tree was constructed from hybridization fingerprints of rDNA clones. (A) Complete UPGMA tree. (B) Detailed depiction of the Raciborskiomyces clade. rDNA clones are designated by a number followed by a space and then either 1 or 2: 1 indicates the clones from the 0.1% suppressive soil treatment, and 2 indicates the clones from the 10% suppressive soil treatment. Clones whose nucleotide sequences were determined are indicated by the suffix S; identities to GenBank sequences are indicated. Clusters are designated by vertical lines adjacent to the clone numbers. For example, the first cluster contains the clones 15 1 S and 1534 2 and all of the clones in between. Each cluster was defined as a group of clones with the same fingerprint. The full-length tree can be obtained from the corresponding author.
RESULTS AND DISCUSSION
This report describes an array-based approach (OFRG) for analysis of fungal community composition. OFRG permits extensive analysis of fungal community composition by sorting rDNA clones into taxonomic clusters. Clone libraries are constructed with PCR primers designed to selectively amplify fungal rRNA genes from DNA isolated from environmental samples (4). The cloned rDNA fragments are arrayed on nylon membranes and then subjected to a series of hybridization experiments, each using a single DNA oligonucleotide probe. For every hybridization experiment, the signal intensities are transformed into three discrete values: 0, 1, and N, where 0 and 1, respectively, specify negative and positive hybridization events and N designates an uncertain assignment. This process creates a hybridization fingerprint for each clone, which is a vector of values resulting from its hybridizations with all probes. The clones are identified by clustering their hybridization fingerprints with those of known sequences and by nucleotide sequence analyses of representative clones within a cluster.
To demonstrate this approach, we analyzed 1,536 fungal rDNA clones by using an OFRG probe set comprised of 26 oligonucleotides, each 10 nucleotides in length. The rDNA clones were derived from two treatments of the same agricultural soil. Since no considerable differences in community composition between these treatments were observed, we refer to these clones in this manuscript as simply “soil clones,” without reference to their treatment origin. UPGMA analysis of the hybridization fingerprints produced a tree comprised of 455 clusters (Fig. 1 and Table 2); each cluster was defined as a group of clones with the same fingerprint. Eighty-eight percent of the clones were affiliated with Ascomycota, while 12% belonged to Basidiomycota. No Chytridiomycota or Zygomycota clones were identified. The most predominant group of clones was affiliated with the Fusarium clade (Fig. 1 and Table 2). The second largest group of clones had high sequence identity with Raciborskiomyces longisetosum, which belongs to the family Pseudoperisporiaceae. The Raciborskiomyces clade is shown in greater detail to demonstrate a typical assemblage within this fungal rDNA tree (Fig. 1B). A BLAST analysis of the nucleotide sequences of clones distributed throughout this group showed that most had high sequence identity (≥98%) to Raciborskiomyces longisetosum. At the end of this assemblage, where the branch lengths are longer, indicating larger differences in the hybridization fingerprints, the sequenced clones had relatively low sequence identity (≤98%) to R. longisetosum or other fungi. Other smaller groups of clones within the tree belonged to the Alternaria, Ascobolus, Chaetomium, Cryptococcus, and Rhizoctonia clades. Two clones with high sequence identity to nematode rDNA were also found.
TABLE 2.
Taxonomic distribution of fungal rDNA clones obtained from soila
| Taxon | No. of clones in soil |
|---|---|
| Alternaria | 63 |
| Ascobolus | 82 |
| Chaetomium | 53 |
| Cryptococcus | 58 |
| Fusarium | 404 |
| Raciborskiomyces | 176 |
| Rhizoctonia | 69 |
The data were obtained by adding the numbers of clones in the well-defined taxonomic groups from the UPGMA tree (Fig. 1). Clones not counted in this analysis were from regions of the tree that contained relatively few redundant fingerprints.
The resolution of this OFRG experiment was evaluated by a nucleotide sequence analysis of clones within selected clusters distributed throughout the UPGMA tree. The nucleotide sequences from at least four clones from seven clusters were determined. For each cluster, a pairwise sequence analysis showed that clones with an average sequence identity of 99.2% were grouped into the same cluster; the range of identities was 98.2 to 99.9%. This result demonstrates that this OFRG probe set is capable of discriminating fungal rDNA clones with high sequence identities.
The accuracy of the taxonomic identities produced by this OFRG experiment was evaluated by a sequence analysis of rDNA clones distributed throughout the UPGMA tree. OFRG allows identification of rDNA clones through their association with fingerprints from known sequences within the tree or by sequence analysis of representative clones within a cluster. To evaluate this OFRG experiment, we compared the taxonomic identities obtained from the UPGMA tree with those obtained by a BLAST (National Center for Biotechnology Information) analysis of the nucleotide sequences of 117 clones. This analysis showed that the taxonomic identities produced by this OFRG experiment were consistent with those generated by a nucleotide sequence analysis for 115 of the 117 rDNA clones examined (data not shown). One of the misidentified or misplaced clones was grouped in an undefined assemblage within Ascomycota, yet its best match from a BLAST analysis was a Basidiomycete sequence (95% identity). The other clone had 97% sequence identity to Ascobolus lineolatus, yet it did not cluster with the other Ascobolus clones in the UPGMA tree. The misplacement of these two clones may have been caused by a variety of factors, including experimental error, probe design, or the limited number of sequences in the databases.
Several areas should be addressed in future studies to increase the accuracy and resolution of this analysis. For probe design, our original goal for this project was to develop a probe set that could discriminate 616 fungal small-subunit rDNA sequences obtained from GenBank. However, since some of the probes provided by the simulated annealing algorithm did not hybridize in a consistent and predictable manner in the actual experiments, the probes used in this study were a collection of oligonucleotides from three different probe sets that produced strong signal intensities and hybridized to the control clones in the expected manner. To enable more efficient probe design, we have recently developed a new iterative algorithm that will address this issue by allowing unsuccessful probes to be replaced (Della Vedova et al., unpublished algorithm). Another factor limiting the OFRG approach is the number and breadth of rDNA sequences in the databases. As with other comparative sequence analyses, OFRG's ability to accurately identify rDNA sequences will be enhanced by more comprehensive databases. Increasing the numbers of available sequences will also lead to better probe design. Other approaches for increasing the resolution and accuracy of a fungal OFRG include examination of either a larger portion of the small-subunit gene or the more variable internal transcribed spacer (ITS) region.
In summary, OFRG provides an efficient and relatively inexpensive means to analyze fungal diversity. It allows more thorough analysis of community composition than methods such as DGGE and is less costly than nucleotide sequence analysis of clone libraries. For comparison, this OFRG analysis cost approximately $1,720 in supplies and sequencing charges, while a nucleotide sequence analysis of 1,536 rDNA clones would have cost approximately $15,360 (two sequencing reactions per clone at $5 per reaction). In addition, this particular OFRG experiment was relatively expensive, because we were examining the resolution and accuracy of the analysis, which involved obtaining the nucleotide sequences of 112 clones ($1,120). Furthermore, if a larger clone library were examined with a higher-density array, the cost of the OFRG analysis would stay approximately the same, while the cost of the sequence analysis would increase considerably. We anticipate that OFRG will facilitate extensive examinations of fungal diversity, which should lead to the discovery of new phylotypes and to a better understanding of the relationships between community composition and function. Although PCR-based rDNA methods do not generate quantitative depictions of community composition, they do provide an excellent starting point for further investigations. OFRG has been used to identify differences in bacterial and fungal community composition that correlate with pathogen suppressiveness in soil; these differences have been subsequently verified by quantitative PCR analyses, confirming the usefulness of this approach (J. Borneman, unpublished data).
Acknowledgments
We thank Alexandra J. Scupham for critical review of the manuscript.
This research was funded in part by grants from the NSF BDI Program (J.B. and T.J.), the UC Biotechnology Research & Training program (J.B.), and the NSF ITR program (G.D.V. and T.J). L.V. was supported by Vaddia-BARD Postdoctoral Award FI-306-00 from BARD, The United States-Israel Binational Agricultural Research and Development Fund. G.D.V. was supported by MURST grant “Bioinformatica e Ricerca Genomica.”
REFERENCES
- 1.Barns, S. M., R. E. Fundyga, M. W. Jeffries, and N. R. Pace. 1994. Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc. Natl. Acad. Sci. USA 91:1609-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bock, M., M. Maiwald, R. Kappe, P. Nickel, and H. Naeher. 1994. Polymerase chain reaction-based detection of dermatophyte DNA with a fungus-specific primer system. Mycoses 37:79-84. [DOI] [PubMed] [Google Scholar]
- 3.Borneman, J., and E. W. Triplett. 1997. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl. Environ. Microbiol. 63:2647-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borneman, J., and R. J. Hartin. 2000. PCR primers that amplify fungal rRNA genes from environmental samples. Appl. Environ. Microbiol. 66:4356-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borneman, J., M. Chrobak, G. Della Vedova, A. Figueroa, and T. Jiang. 2001. Probe selection algorithms with applications in the analysis of microbial communities. Bioinformatics 17(Suppl. 1):S39-S48. [DOI] [PubMed] [Google Scholar]
- 6.Borneman, J., P. W. Skroch, K. M. O'Sullivan, J. A. Palus, N. G. Rumjanek, J. L. Jansen, J. Nienhuis, and E. W. Triplett. 1996. Molecular microbial diversity of an agricultural soil in Wisconsin. Appl. Environ. Microbiol. 62:1935-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bridge, P., and B. Spooner. 2001. Soil fungi: diversity and detection. Plant Soil 232:147-154. [Google Scholar]
- 8.Chapela, I. H. 1997. Bioprospecting: myths, realities and potential impact on sustainable development, p. 238-256. In M. E. Palm and I. H. Chapela (ed.) Mycology in sustainable development: expanding concepts, vanishing borders. Parkway Publishers, Inc., Boone, N.C.
- 9.Christensen, M. 1989. A view of fungal ecology. Mycologia 81:1-19. [Google Scholar]
- 10.Gardes, M., and T. D. Bruns. 1993. ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Mol. Ecol. 2:113-118. [DOI] [PubMed] [Google Scholar]
- 11.Giovannoni, S. J., T. B. Britschgi, C. L. Moyer, and K. G. Field. 1990. Genetic diversity in Sargasso Sea bacterioplankton. Nature 345:60-63. [DOI] [PubMed] [Google Scholar]
- 12.Hawksworth, D. L. 1991. The fungal dimension of biodiversity: magnitude, significance, and conservation. Mycol. Res. 95:641-655. [Google Scholar]
- 13.Hawksworth, D. L., and A. Y. Rossman. 1997. Where are all the undescribed fungi? Phytopathology 87:888-891. [DOI] [PubMed] [Google Scholar]
- 14.Henson, J. M., and R. French. 1993. The polymerase chain reaction and plant disease diagnosis. Annu. Rev. Phytopathol. 31:81-109. [DOI] [PubMed] [Google Scholar]
- 15.Kappe, R., C. Fauser, C. N. Okeke, and M. Maiwald. 1996. Universal fungus-specific primer systems and group-specific hybridization oligonucleotides for 18S rDNA. Mycoses 39:25-30. [DOI] [PubMed] [Google Scholar]
- 16.Kirkpatrick, S., C. D. Gelatt, and M. Vecchi. 1983. Optimization by simulated annealing. Science 220:671-680. [DOI] [PubMed] [Google Scholar]
- 17.Kowalchuk, G. A., S. Gerards, and J. W. Woldendorp. 1997. Detection and characterization of fungal infections of Ammophila arenaria (Marram grass) roots by denaturing gradient gel electrophoresis of specifically amplified 18S rDNA. Appl. Environ. Microbiol 63:3858-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Makimura, K., S. Y. Murayama, and H. Yamaguchi. 1994. Detection of a wide range of medically important fungi by the polymerase chain reaction. J. Med. Microbiol. 40:358-364. [DOI] [PubMed] [Google Scholar]
- 19.May, L. A., B. Smiley, and M. G. Schmidt. 2001. Comparative denaturing gel electrophoresis analysis of fungal communities associated with whole plant corn silage. Can. J. Microbiol. 47:829-841. [DOI] [PubMed] [Google Scholar]
- 20.Pace, N. R. 1997. A molecular view of microbial diversity and the biosphere. Science 276:734-740. [DOI] [PubMed] [Google Scholar]
- 21.Simon, L., M. Lalonde, and T. D. Bruns. 1992. Specific amplification of 18S fungal ribosomal genes from vesicular-arbuscular endomycorrhizal fungi colonizing roots. Appl. Environ. Microbiol. 58:291-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smit, E., P. Leeflang, B. Glandorf, J. D. van Elsas, and K. Wernars. 1999. Analysis of fungal diversity in the wheat rhizosphere by sequencing of cloned PCR-amplified genes encoding 18S rRNA and temperature gradient gel electrophoresis. Appl. Environ. Microbiol. 65:2614-2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tuomela, M., M. Vikman, A. Hatakka, and M. Itavaara. 2000. Biodegradation of lignin in a compost environment: a review. Bioresour. Technol. 72:169-183. [Google Scholar]
- 24.Vainio, E. J., and J. Hantula. 2000. Direct analysis of wood-inhabiting fungi using denaturing gradient gel electrophoresis of amplified ribosomal DNA. Mycol. Res. 104:927-936. [Google Scholar]
- 25.Valinsky, L., G. Della Vedova, A. J. Scupham, S. Alvey, A. Figueroa, B. Yin, R. J. Hartin, M. Chrobak, D. E. Crowley, T. Jiang, and J. Borneman. 2002. Analysis of bacterial community composition by oligonucleotide fingerprinting of rRNA genes. Appl. Environ. Microbiol. 68:3243-3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vandenkoornhuyse, P., S. L. Baldauf, C. Leyval, J. Straczek, and J. P. W. Young. 2002. Extensive fungal diversity in plant roots. Science 295:20.. [DOI] [PubMed] [Google Scholar]
- 27.Ward, D. M., M. M. Bateson, R. Weller, and A. L. Ruff-Roberts. 1992. Ribosomal RNA analysis of microorganisms as they occur in nature. Adv. Microb. Ecol. 12:219-286. [Google Scholar]
- 28.Westphal, A., and J. O. Becker. 1999. Biological suppression and natural population decline of Heterodera schachtii in a California field. Phytopathology 89:434-440. [DOI] [PubMed] [Google Scholar]