Abstract
Acinetobacter baumannii is a metabolically versatile pathogen that causes severe infections in compromised patients. However, little is known about the genes and factors involved in its basic physiology and virulence properties. Insertion mutagenesis was used to initiate the identification and characterization of some of these factors and genes in the prototype strain 19606. The utilization of the pLOFKm suicide delivery vector, which harbors a suicide mini-Tn10 derivative, proved to be unsuccessful for this purpose. The EZ::TN 〈R6Kγori/KAN-2〉 Tnp transposome system available from Epicentre was then used in conjunction with electroporation to generate isogenic insertional derivatives of A. baumannii 19606. Replica plating showed that 2% of the colonies that grew after electroporation on agar plates without antibiotics also grew in the presence of 40 μg of kanamycin per ml. DNA hybridization proved that all of the kanamycin-resistant derivatives contained the EZ::TN 〈R6Kγori/KAN-2〉 insertion element, which was mapped to different genomic locations. Replica plating on Simmons citrate agar and microtiter plate-plastic tube assays identified growth- and biofilm-defective derivatives, respectively. The location of the insertion in several of these derivatives was determined by self-ligation of NdeI- or EcoRI-digested genomic DNA and electroporation of Escherichia coli TransforMax EC100D (pir+). Sequence analysis of the recovered plasmids showed that some of the A. baumannii 19606 growth-defective derivatives contain insertions within genes encoding activities required for the generation of energy and cell wall components and for the biosynthesis of amino acids and purines. A gene encoding a protein similar to the GacS sensor kinase was interrupted in four derivatives, while another had an insertion in a gene coding for a hypothetical sensor kinase. A. baumannii 19606 derivatives with defective attachment or biofilm phenotypes had insertions within genes that appear to be part of a chaperone-usher transport system described for other bacteria. DNA hybridization experiments showed that the presence of strain 19606 genes encoding regulatory and attachment or biofilm functions is widespread among other A. baumannii clinical isolates.
Acinetobacter baumannii is being increasingly recognized as an important pathogen that causes severe infections in hospitalized patients (6, 7). In addition, deadly cases of community-acquired pneumonia were reported among compromised patients (3, 9). These infections are difficult to treat due to the expanding antibiotic resistance of the clinical strains (8, 30), which represents one of the most difficult problems confronted by clinicians who deal with the infections caused by this human pathogen.
Literature searches showed that there are numerous reports describing either the different types of infections caused by A. baumannii, the antibiotic resistance profiles of the different clinical strains, the characterization of some of the genetic elements responsible for their antibiotic resistance, or the development and application of typing methods and fingerprinting systems used to identify and trace the sources of clinical isolates of this pathogen. In contrast, only a few reports describing different aspects related to the basic physiology, genetics, and molecular biology of this opportunistic pathogen (2, 12, 13, 19, 20, 41, 58) could be found during these literature searches. The apparent lack of suitable genetic methods such as random mutagenesis with transposable elements, which is a widely used approach to characterize bacteria genetically and functionally, appears to be the reason for the delay of the characterization of this human pathogen. While different basic aspects of the biology and genetics of environmental Acinetobacter strains were analyzed by using transposable elements such as Tn5 (49), Tn10 (14), Tn3171 (55), and mini-Tn10PttKm (35), no work describing the application of this approach to A. baumannii clinical strains could be found. We report here the genetic and molecular analysis of the A. baumannii prototype strain 19606 by electroporation of transposon-transposase complexes. This novel insertional mutagenesis method proved to be efficient for the generation of random mutants affected in metabolic, global regulatory, and attachment and biofilm functions. This convenient approach should facilitate the genetic and functional analysis of this poorly characterized opportunistic human pathogen.
MATERIALS AND METHODS
Bacterial strains, media, and plasmids.
The bacterial strains and plasmids used in this work are listed in Table 1. Bacterial strains were maintained on Luria-Bertani (LB) agar or broth (48) supplemented with the appropriate antibiotics and were incubated overnight at 37°C. Simmons citrate agar (Difco, Detroit, Mich.) was used either without any addition or supplemented with the appropriate amino acids, purines, and meso-α,ɛ-diaminopimelate, all purchased from Sigma (St. Louis, Mo.). These compounds were added to the Simmons citrate agar to final concentrations of 1 mM, 100 μg/ml, and 1 mM, respectively. The inoculated plates were incubated overnight at 37°C. LB broth cultures were incubated at 37°C in a rotary shaker at 200 rpm. The growth curves of the parental and insertion derivatives were determined in triplicate, using fresh samples each time.
TABLE 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Relevant characteristicsa | Source or reference(s) |
|---|---|---|
| Strains | ||
| A. baumannii | ||
| 9235, 8971, 8143, 8114, 8637, 7133, 7138, 8399, 9606, 7931, 9124, 9397 | Clinical isolates representing plasmid profiles A to K and W, respectively | 2, 22 |
| BM4420, BM4421, BM4422, BM4424, BM4427, BM4430, BM4432, BM4436, BM4439 | Clinical isolates representing RAPD types A to F, H, I, and K, respectively | 46 |
| E. coli | ||
| DH5α | Used for recombinant DNA methods | Gibco-BRL (Gaithersburg, Md.) |
| TransforMax EC100D | pir+, used to clone R6K derivatives | Epicentre (Madison, Wis.) |
| CC118, SM10, and S17-1 | λpir, used to maintain and conjugate suicide plasmid pLOFKm | 10, 26 |
| Plasmids | ||
| pLOFKm | Suicide plasmid harboring mini-Tn10Km; Ampr Kmr | 26 |
| pVK100 | Cosmid cloning vector; Kmr Tetr | 33 |
| pRK2073 | Conjugation helper plasmid | 37 |
| pWH1266 | E. coli-Acinetobacter shuttle plasmid; Ampr Tetr | 28 |
| pFVP25 | ColEI, mob+, Ampr; source of the BamHI-PstI fragment harboring gfp | 56 |
| pBCSK+ | Cloning vector; Cmr | Stratagene (La Jolla, Calif.) |
| pMU125 | pWH1266 with gfp under control of the Tet promoter | This work |
Ampr, ampicillin resistant; Kmr, kanamycin resistant; Tetr, tetracycline resistant; RAPD, random amplified polymorphic DNA.
General DNA procedures.
Total DNA was isolated either by ultracentrifugation in CsCl density gradients (38), by a miniscale method adapted from that published previously (4), or with the DNeasy tissue kit from Qiagen (Valencia, Calif.). Plasmid DNA was isolated by ultracentrifugation in CsCl-ethidium bromide density gradients (48) or with a commercial kit (Qiagen). DNA was digested with restriction enzymes as indicated by the supplier (New England Biolabs, Beverly, Mass.) and size fractionated by agarose gel electrophoresis (48). Both strands of cloned DNA were sequenced with BigDye (Applied Biosystems, Foster City, Calif.) and DYEnamic ET (Amersham Pharmacia Biotech, Piscataway, N.J.) chemistries on Applied Biosystems Prism 310 or 3100 instruments and with M13 forward and reverse primers (59) or custom-designed primers. Sequences were examined and assembled with Sequencher 4.1.2 (Gene Codes Corp., Ann Arbor, Mich.). Nucleotide and amino acid sequences were analyzed with DNASTAR (DNASTAR, Inc., Madison, Wis.), BLAST (http://www.ncbi.nlm.nih.gov), and the software available through the ExPASy Molecular Biology Server (http://www.expasy.ch).
Southern blot analyses were conducted by using standard protocols (48) with high-stringency conditions (21). The probe to detect the insertion of transposable elements encoding kanamycin resistance (Kmr) was prepared by PCR amplification of the pUC4K (Amersham Pharmacia Biotech) aph gene, which encodes the aminoglycoside 3′-phosphotransferase that confers this antibiotic resistance, with Pfu DNA polymerase (Stratagene) and primers 133 (5′-GGCGCTGAGGTCTGCCTCGTG-3′) and 134 (5′-GAGCCATATTCAACGGG-3′), which were designed by using the sequence data deposited under GenBank accession number X06404. The primer pairs 1077 (5′-ACATTTACAGGTGGTGTC-3′)-1210 (5′-GCTAGATTTTGCGCAGTG-3′) and 1481 (5′-TACCACGTCAGTTCCACG-3′)-1600 (5′-TTGCGTAGTGGGTTGCTC-3′) were used to amplify internal regions of the gacS-like gene and the hypothetical sensor kinase gene interrupted in the derivative 249, respectively. The primer pairs 1034 (5′-ACACCTACAAACCGTCTG-3′)-1670 (5′-AGGACTTACGCATTGACG-3′) and 1123 (5′-TACTGGTTTGGCCTATCC-3′)-1037 (5′-CGTAAAGCTACTCATGTC-3′) were used for PCR amplification of internal regions of the csuB- and csuE-like genes, respectively. All of these primers were designed by using nucleotide sequences obtained in this work, and the nature of each amplicon was validated by automated DNA sequencing. The amplicons were purified by using the GeneClean II kit (Qbiogene, Carlsbad, Calif.) and labeled with [α-32P]dCTP (15). The radioactive bands were detected with a Storm 860 scanner (Molecular Dynamics, Sunnyvale, Calif.).
Construction of pMU125 and detection of green fluorescent cells.
The pFVP25 BamHI-PstI fragment harboring a promoterless gfp gene was ligated to pBCSK+ and transformed into E. coli DH5α. Plasmid DNA isolated from a green fluorescent colony was digested with BamHI and SalI and cloned in the cognate sites of the shuttle vector pWH1266. An E. coli DH5α colony resistant to 100 μg of ampicillin per ml, sensitive to 20 μg of tetracycline per ml, and expressing green fluorescence due to the fusion of gfp to the pWH1266 Tet promoter was used to isolate the plasmid pMU125. The appropriate structure of the latter construct was confirmed by restriction analysis. The expression of gfp in E. coli DH5α and A. baumannii 19606 cells was examined by epifluorescence microscopy with an Eclipse E400 microscope (Nikon Inc., Melville, N.Y.) equipped with a SPOT digital camera (Diagnostic Instruments Inc., Sterling Heights, Mich.).
Random insertion mutagenesis.
The delivery suicide vector pLOFKm was used as described previously (10, 26, 35) to generate insertion derivatives of A. baumannii 19606. The exconjugants were selected on Simmons citrate agar containing 40 μg of kanamycin per ml. The triparental mating system described before (1) was used as an alternative to mobilize plasmid DNA into A. baumannii 19606. Exconjugants harboring pLOFKm or pVK100 were selected on Simmons citrate agar containing 40 μg of kanamycin per ml or 10 to 20 μg of tetracycline per ml, respectively. The EZ::TN 〈R6Kγori/KAN-2〉 Tnp transposome kit was used as suggested by the manufacturer (Epicentre). The EZ::TN 〈R6Kγori/KAN-2〉 transposon-EZ::TN transposase complexes were introduced by electroporation into A. baumannii 19606 electrocompetent cells. These cells were prepared as described in the Bio-Rad electroporation manual (Bio-Rad, Hercules, Calif.) and electroporated with a 2510 Eppendorf electroporator (Brinkmann Instruments, Westbury, N.Y.) and 2-mm-wide cuvettes. After electroporation at 2.5 kV, the cells were suspended in 1 ml of SOC broth (48) and allowed to recover for 1 h at 37°C with shaking. Dilutions of electroporated cells were plated on LB agar containing 40 μg of kanamycin per ml. The genomic regions harboring the insertion of the EZ::TN 〈R6Kγori/KAN-2〉 transposon were rescued by self-ligation of NdeI- or EcoRI-digested total DNA and electroporation of E. coli TransforMax EC100D (pir+) electrocompetent cells. Transformants were recovered by plating on LB agar containing 40 μg of kanamycin per ml. The nucleotide sequence of the genomic DNA flanking the transposon element was determined by automated DNA sequencing with the primers complementary to this insertion element that were supplied with the mutagenesis kit. Further extension of nucleotide sequences was done by using custom-designed primers and plasmid DNA as a template.
Isolation and characterization of metabolic and adhesion-biofilm mutants.
A. baumannii 19606 derivatives affected in metabolic functions were isolated by replica plating Kmr colonies on Simmons citrate agar plates without the addition of antibiotics. Positive growth was recorded as concomitant detection of abundant bacterial growth on the streaked areas and a change of color from green to blue in the surrounding areas, after incubation at 37°C for 8 h to 12 h. Derivatives affected in their ability to attach to and form biofilms on abiotic surfaces were isolated after 24 h of stagnant incubation at 37°C in polystyrene tubes or microtiter plates as described before (42). Briefly, cells were incubated without shaking in 1 or 0.2 ml of LB broth in sterile polystyrene tubes and 96-well microtiter plates, respectively, at 37°C overnight. The culture supernatants were removed, and the tubes and wells were rinsed with deionized water. Cells attached to the plastic surface were visualized by adding enough 0.1% crystal violet aqueous solution to cover the uppermost level of the original cultures. After incubation at room temperature for 15 min and washing with deionized water, the attached cells were detected visually as a purple band located at or near the LB broth-air interface. Attachment- or biofilm-deficient derivatives were identified by the absence of detectable staining compared with the parental strain.
Nucleotide sequence accession numbers.
The nucleotide sequences of the A. baumannii 19606 cysI, gacS, and trpE genes were deposited in GenBank under accession numbers AF498617, AF497904, and AY094355, respectively.
RESULTS AND DISCUSSION
Generation and characterization of A. baumannii 19606 insertion derivatives.
The transposon mutagenesis system based on the delivery vector pLOFKm, which harbors the mini-Tn10Km insertion element (10, 26), was initially used to mutagenize A. baumannii 19606. This decision was based on the fact that pLOFPttKm (26), which is very similar to pLOFKm, was used successfully to obtain insertion derivatives of the Acinetobacter calcoaceticus RAG-1 environmental strain (35). This approach proved to be ineffective for the mutagenesis of the A. baumannii 19606 clinical isolate, since it did not yield Kmr derivatives that harbored the aph gene, although various experimental conditions such as utilization of different donor/recipient ratios, mixing of donor and recipient cells with a sterile loop that was deposited on LB agar, or mixing of different volumes of donor and recipient liquid cultures that were collected on 0.2-μm-pore-size sterile filters that were incubated on LB agar (10, 26, 35), were used. Attempts to generate these derivatives by triparental mating under the conditions described previously (1) also failed. In contrast, A. baumannii 19606 tetracycline-resistant exconjugants could be obtained when the cosmid vector pVK100 was mobilized from an E. coli donor in the presence of the helper plasmid pRK2073 (data not shown). Electroporation of pLOFKm also failed to yield A. baumannii 19606 Kmr derivatives, although this DNA transformation method yielded ampicillin-resistant and green fluorescent cells when this strain was electroporated with pMU125 and plated on LB agar containing ampicillin (data not shown). Taken together, these results demonstrate that plasmid DNA can be transferred to A. baumannii 19606 by conjugation and electroporation and then can be maintained stably without detectable rearrangements even after extensive culture in the absence of selective pressure. Furthermore, this strain can express nonindigenous genes such as those harbored by the pVK100 and pMU125 plasmid vectors as well as the gfp reporter gene from Aequoria victoria. These results also indicate that the failure to obtain A. baumannii 19606 derivatives with pLOFKm could be due to problems associated with this particular insertion mutagenesis system, which has been applied successfully to other gram-negative bacteria.
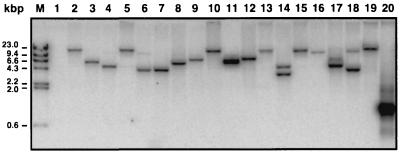
The observation that foreign DNA can be electroporated into A. baumannii 19606 prompted us to use the Epicentre system based on the electroporation of complexes formed between the EZ::TN transposase and the EZ::TN 〈R6Kγori/KAN-2〉 transposon, an approach that has not been applied to any Acinetobacter strain. Replica-plating experiments showed that about 2% of the A. baumannii 19606 colonies recovered on LB agar without antibiotic pressure after electroporation also grew on plates containing 40 μg of kanamycin per ml. DNA hybridization of total DNA isolated from 18 of these A. baumannii 19606 Kmr derivatives with the aph probe showed that all of them (Fig. 1, lanes 2 to 19) harbored this resistance gene, which could not be detected in the genome of the parental strain (lane 1). All of these derivatives appear to contain a single transposon insertion, with the exception of that shown in lane 14, which displays two EcoRI fragments that reacted with the aph probe, suggesting that this derivative harbors two independent transposon insertions. This analysis also shows that about half of the insertions occurred in different EcoRI fragments, while others, such as those shown in lanes 2, 5, 13, 15, and 19; 3 and 11; and 6, 7, and 18, appear to be located in EcoRI fragments of similar size. The nucleotide sequence analysis described below indicates that EZ::TN 〈R6Kγori/KAN-2〉 indeed inserted in different locations of the A. baumannii 19606 genome, although some of them were mapped at different sites within the same gene. This observation explains the detection of EcoRI fragments displaying similar size when tested with the aph probe (Fig. 1).
FIG. 1.
Southern blot analysis of the A. baumannii 19606 parental strain and 18 EZ::TN 〈R6Kγori/KAN-2〉 insertion derivatives. HindIII-digested λ DNA (lane M), EcoRI-digested total DNAs isolated from the parental strain (lane 1) and insertion derivatives (lanes 2 to 19), and the aph amplicon (lane 20) were size fractionated by agarose gel electrophoresis. The DNA fragments were transferred to nitrocellulose and probed with λ DNA and the aph amplicon labeled with [32P]dCTP.
Nucleotide sequence analysis of insertion derivatives.
The random insertion of the EZ::TN 〈R6Kγori/KAN-2〉 transposon in the A. baumannii 19606 strain was assessed further by isolating mutants of several different phenotypes. This was achieved by testing the ability of Kmr derivatives to attach to and form biofilms on plastic surfaces and to grow on Simmons citrate agar. Simmons citrate agar is a standard bacteriological medium that contains only essential inorganic salts, citrate as a carbon source, and a pH indicator, and it therefore should facilitate the isolation of metabolic mutants. Replica plating of 2,800 Kmr derivatives produced 41 colonies that grew well on LB agar but not on Simmons citrate plates. Screening of 3,000 insertion derivatives cultured in plastic tubes and microtiter plates resulted in the identification of mutants slightly affected or completely impaired in their ability to attach to and form biofilms on abiotic surfaces.
Sequence analysis of 21 plasmids rescued from insertion derivatives by self-ligation of EcoRI- or NdeI-digested genomic DNA showed that all of the EZ::TN 〈R6Kγori/KAN-2〉 insertions resulted in a 9-bp duplication of the target sites, with an A+T content that ranged from 33.3 to 77.7% (Table 2). The nucleotide sequences of 17 of these sites were different, while the same nucleotide sequence was determined for the target sites of insertions 11 and 25 and insertions 26 and 28, respectively (Table 2). These results indicate that the EZ::TN 〈R6Kγori/KAN-2〉 transposon inserts most of the time in different regions of the A. baumannii 19606 genome and generates derivatives that display diverse phenotypes. The insertion target sites and the nature of the disrupted genes were characterized further by sequencing the genomic DNA flanking each insertion with primers that anneal near the ends of the transposon as well as custom-made primers that anneal with specific genomic sequences.
TABLE 2.
Characterization of transposome insertion derivatives of A. baumannii 19606
| Insertion | Target site | Gene, function disrupteda |
|---|---|---|
| 1 | GCCCTAAAA | dapA, dihydrodipicolinate synthase |
| 2 | GATCATAGT | gacS/lemA, histidine kinase sensor |
| 4 | GTAGTGACT | csuB, chaperone-usher secretion system |
| 5 | CTACAATCA | gacS/lemA, histidine kinase sensor |
| 9 | CGCGGATAC | trpD, anthranilate phosphoribosyltransferase |
| 10 | GTTTATTCA | argF, ornithine carbamoyltransferase |
| 11 | GAACATGAT | gacS/lemA, histidine kinase sensor |
| 12 | GGGCCATAC | argG, argininosuccinate synthase |
| 13 | ATAGAATGG | aceElaceA, pyruvate dehydrogenase E1 |
| 15 | AATGGAAAC | proA, γ-glutamyl phosphate reductase |
| 16 | AATATACGT | cysI/nirA/sir, sulfite reductase |
| 19 | CTACGATGC | trpE, anthranilate synthase |
| 23 | CTATTCACA | trpE, anthranilate synthase |
| 25 | GAACATGAT | gacS/lemA, histidine kinase sensor |
| 26 | CATGTGAAT | cysI/nirA/sir, sulfite reductase |
| 28 | CATGTGAAT | cysI/nirA/sir, sulfite reductase |
| 30 | GCATTGGGT | hisA/his4, phosphoribosylformimino-5-aminoimidazole carboxamide ribotide isomerase |
| 31 | CTGCAAACC | hisH, glutamine amidotransferase |
| 44 | GTTTTACGT | cysI/nirA/sir, sulfite reductase |
| 144 | GTCACAAAC | csuE, chaperone-usher secretion system |
| 249 | ATGGTAAAA | Hypothetical sensor histidine kinase |
Potential genes and functions were predicted by BLASTp and BLASTx searches.
Amino acid biosynthesis-defective derivatives.
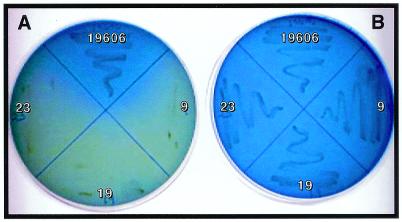
A. baumannii 19606 derivative 9 harbors the insertion of the transposon element within the trpD gene homolog described for A. calcoaceticus (31), which is part of the trpGDC gene cluster required for the tryptophan biosynthesis. The product of the trpD is the anthranilate phosphoribosyltransferase that is involved in the second step of the biosynthesis of this amino acid (45). Insertions 19 and 23 interrupted the A. baumannii 19606 trpE homolog, which has been described for A. calcoaceticus (23) and encodes anthranilate synthase component I. This enzymatic activity is involved in the first step of tryptophan biosynthesis from chorismate. These two insertions were mapped in two different regions of the trpE homolog, and their combined nucleotide sequences produced the entire sequence of this 1,317-nucleotide (nt) open reading frame (ORF), which is predicted to encode a 48.3-kDa protein highly similar (score, 701; e value, 0.0) to that reported for the A. calcoaceticus homolog (23). The metabolic deficiency of these three insertion derivatives was confirmed further by the restoration of their growth in Simmons citrate agar only when supplemented with tryptophan (Fig. 2B). However, these three insertion derivatives showed growth curves identical to that of the 19606 parental strain when cultured in LB broth (data not shown).
FIG. 2.
Tryptophan complementation assay of trp mutants. The 19606 parental strain and the trpD (derivative 9) and trpE (derivatives 19 and 23) mutants were streaked on Simmons citrate agar (A) and Simmons citrate agar supplemented with 1 mM tryptophan (B).
The EZ::TN 〈R6Kγori/KAN-2〉 transposon interrupted genes encoding products that are highly similar to those encoded by the Pseudomonas syringae pv. phaseolicola argF (24) and the Haemophilus influenzae Rd argG (16) genes in derivatives 10 and 12, respectively. These two genes encode ornithine carbamoyltransferase and argininosuccinate synthase, respectively, which are involved in the biosynthesis of arginine (18). The arginine biosynthesis defect of these two derivatives was further confirmed by the observation that the addition of arginine to Simmons citrate agar plates restored their ability to grow in this chemically defined medium, while no growth defects were observed when they were tested with LB broth (data not shown).
Insertion 15 was mapped within the A. baumannii 19606 proA-like gene, which encodes a protein highly related to the γ-glutamyl phosphate reductase described for Pseudomonas aeruginosa PAO1 (51). This enzymatic activity is required to complete the second step of the biosynthesis of proline from glutamate (36). Accordingly, this mutant grew as the parental strain did when cultured in LB broth, although it could be cultured on Simmons citrate agar only after the addition of proline (data not shown). Insertions 16, 26 (same as 28), and 44 were mapped within a gene that showed very high similarity to the A. calcoaceticus RAG-1 cysI gene (GenBank accession number AAK96890). The combined nucleotide sequences of these insertion derivatives resulted in the identification of the A. baumannii 19606 cysI homolog, which comprises a 1,644-nt ORF encoding a 547-amino-acid protein with a predicted molecular size of 62.1 kDa. This predicted protein is highly similar (score, 1030; e value, 0.0) to those reported for A. calcoaceticus RAG-1 (GenBank accession number AAK96890) and other bacteria such as P. aeruginosa PAO1 (51) and the plant pathogen Ralstonia solanacearum (47). The sulfite reductase activity of this protein is essential for the biosynthesis of cysteine in other bacteria (34), an A. baumannii 19606 requirement that was confirmed by the ability of these four derivatives to grow on Simmons citrate agar only after supplementation with cysteine, without detectable growth defects in LB broth (data not shown).
Derivative 1 harbors an insertion within the A. baumannii 19606 dapA-like gene, whose deduced translation product showed the highest similarity with the P. aeruginosa dihydrodipicolinate synthase (51). This mutation should result in a more complex phenotype, because inactivation of dapA impairs lysine biosynthesis as well as the production of murein monomers, which are required for the formation of the bacterial cell wall (43). This hypothesis was supported by the observation that this derivative produced colonies with different sizes, which in general were smaller than the parental strain when plated on LB agar (data not shown). The growth rate of this derivative was slightly reduced but not significantly different from that displayed by the parental strain when cultured in LB broth (data not shown). Furthermore, the growth of this derivative on Simmons citrate agar was similar to that of the parental strain only after the simultaneous addition of lysine and meso-α,ɛ-diaminopimelate, which restored the amino acid and cell wall precursor deficiencies of this mutant.
The transposome insertions 30 and 31 were mapped within genomic regions encoding proteins with the highest similarity to those coded for by the Rhodobacter sphaeroides hisA gene (GenBank accession number X87256) and the E. coli O157:H7 hisH gene (44). These genes encode the phosphoribosylformimino-5-aminoimidazole carboxamide ribotide isomerase and glutamine amidotransferase, respectively. These enzymatic activities are involved in the fourth and fifth steps, respectively, of the histidine biosynthesis from 5-phosphorybosyl-α-pyrophosphate (57). In addition, this metabolic pathway generates the purine biosynthetic precursor 5-aminoimidazole-4-carboxamide ribonucleotide, and therefore these two derivatives must be affected in histidine as well as purine biosynthesis. While their growth was similar to that of the parental strain when tested in LB broth, their ability to grow on Simmons citrate agar was restored only after the simultaneous addition of histidine, adenine, and guanine (data not shown), a response that is compatible with the locations of these insertions in the genome of this bacterium.
Insertion derivatives affected in central metabolic and global regulatory functions.
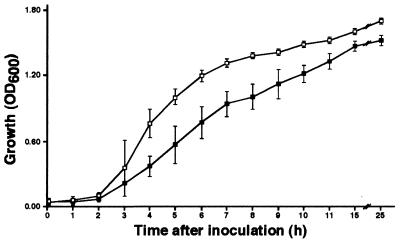
Insertion derivative 13 did not grow as fast and well as the parental strain when cultured on LB agar (data not shown) or broth medium (Fig. 3). This growth behavior, which was reproducible in replicate experiments, suggested the disruption of a central metabolic function, a hypothesis that was confirmed when the insertion harbored by this derivative was mapped within the aceE-like gene that encodes a predicted protein highly similar to the P. aeruginosa E1 component of the pyruvate dehydrogenase complex (51). Therefore, disruption of this essential metabolic function, which participates in the conversion of pyruvate into acetyl coenzyme A and CO2 (17), should affect bacterial growth even when the cells are cultured in rich media, as was observed with this A. baumannii 19606 derivative.
FIG. 3.
Growth curves of the parental strain and the aceE insertion derivative. Overnight cultures of the parental strain 19606 (open squares) and the insertion derivative 13 (closed squares) were diluted 100-fold in LB broth and incubated at 37°C in a rotary shaker. Samples were drawn at the times shown, and bacterial growth was determined by optical density at 600 nm (OD600). Error bars represent standard deviations of the mean from one representative experiment.
EZ::TN 〈R6Kγori/KAN-2〉 insertions 2, 5, 11, and 25 were mapped within a genomic region of A. baumannii 19606 encoding a protein that initially showed the highest similarity to sensor kinases described for different Pseudomonas strains. The combined nucleotide sequences of these four insertions produced the entire sequence of a 2,808-nt ORF predicted to encode a 107.1-kDa protein that showed the highest similarity (score, 410; e value, −113) to the Pseudomonas tolaasii RtpA (39) and the P. fluorescens GacS sensor kinase (GenBank accession number AAG13658) (score, 401; e value, −110). This protein is also known as the LemA regulatory protein in the plant pathogen Pseudomonas syringae pv. syringae (27), which is the sensor element of a two-component regulatory system that includes the GacA transcriptional response regulator. This system plays a role in the virulence of this plant pathogen, being required for lesion formation, swarming, production of proteases, and N-acyl-l-homoserine lactone compounds involved in quorum sensing (27, 32). Some of the other bacterial systems similar to GacA-GacS (LemA) include the BarA sensor protein, which could activate OmpR by phosphorylation (40); the RcsB-RcsC system, which controls the biosynthesis of capsule in E. coli (50); the RpfC protein, which positively controls the synthesis of extracellular enzymes and polysaccharides in Xanthomonas campestris pv. campestris (52); the E. coli aerobic respiration control sensor protein ArcB (29); and the Vibrio harveyi LuxQ sensor protein (5). Whether the disruption of the A. baumannii 19606 gacS-like gene results in phenotype changes similar to those described for other bacteria remains to be determined, since this is, to the best of our knowledge, the first work that reports the presence of this two-component regulatory system in Acinetobacter.
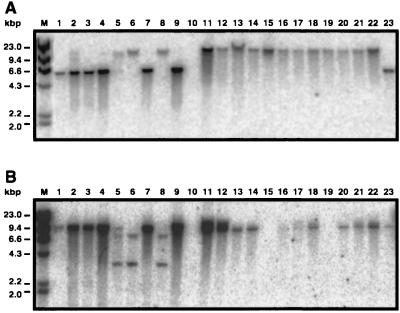
DNA hybridization showed that with the exception of strain BM4439 (Fig. 4A, lane 10), all of the A. baumannii clinical isolates tested positive when probed with gacS. This gene was located in a 6.3-kbp EcoRI fragment in strain 19606 (lanes 1 and 23) and the European isolates BM4420 to BM4422, BM4439, and BM4436 (lanes 2 to 4, 7, and 9, respectively), while the probe reacted with EcoRI fragments ranging from 20 to 23 kbp in the European strains BM4424, BM4427, and BM4432 (lanes 5, 6, and 8, respectively) and all of the Oregon isolates (lanes 11 to 22).
FIG. 4.
Detection of sensor kinase genes in A. baumannii clinical strains. HindIII-digested λ DNA (lane M) and EcoRI-digested total DNAs isolated from the strains 19606 (lanes 1 and 23), BM4420 (lane 2), BM4421 (lane 3), BM4422 (lane 4), BM4424 (lane 5), BM4427 (lane 6), BM4430 (lane 7), BM4432 (lane 8), BM4436 (lane 9), and BM4439 (lane 10) represent the European isolates. Strains 9235 (lane 11), 8971 (lane 12), 8143 (lane 13), 8114 (lane 14), 8637 (lane 15), 7133 (lane 16), 7138 (lane 17), 8399 (lane 18), 9606 (lane 19), 7931 (lane 20), 9124 (lane 21), and 9397 (lane 22) represent the Oregon isolates. DNA blots were probed with radiolabeled λ DNA and either a fragment of the gacS-like gene (A) or a fragment of the hypothetical sensor histidine kinase gene disrupted in the insertion derivative 249 (B).
Derivative 249 is a mutant that was initially identified by its reduced ability to attach to and form biofilm on microtiter plates. However, nucleotide sequence analysis showed that EZ::TN 〈R6Kγori/KAN-2〉 interrupted an ORF encoding a protein highly similar (score, 323; e value, −138) to a hypothetical sensor histidine kinase found in Vibrio cholerae (25) and other gram-negative bacteria such as P. aeruginosa PAO1 (51) and Aeromonas hydrophila (GenBank accession number AF388670). The latter was annotated as the proline sensor PrlS that appears to be involved in the regulation of bacterial motility. It is noteworthy that insertion derivative 249 grew well when cultured on Simmons citrate agar, while derivatives 2, 5, 11, 25, in which the transposon interrupted the gacS-like gene, were not able to grow in this culture medium. Furthermore, no significant similarity was detected between these two predicted genes when they were compared by using BLAST. These results indicate that the GacS-like sensor kinase and the hypothetical sensor kinase are components of two unrelated regulatory systems that appear to sense different environmental signals.
A Southern blot hybridization analysis showed that, with the exception of strains 8637, 7133, 7138, and 9606 (Fig. 4B, lanes 15 to 17 and 19), the Oregon strains harbor sequences related to this hypothetical sensor kinase gene, which were located within a 9.4-kbp EcoRI fragment. A restriction fragment of similar size was also detected in the prototype strain 19606 (lanes 1 and 23). EcoRI fragments of similar size were also detected in the European isolates BM4420, BM4421, BM4422, BM4430, and BM4436 (lanes 2 to 4, 7, and 9), while strains BM4424, BM4427, and BM4432 (lanes 5, 6, and 8) displayed a different hybridization pattern. No signal was detected in the DNA sample of the European strain BM4439 (lane 10).
Isolation of attachment or biofilm mutants.
Insertion derivative 144 was isolated by its inability to attach to and form biofilm in LB broth cultures when incubated stagnantly in polystyrene tubes (Fig. 5A) and microtiter plates (data not shown). Nucleotide sequence analysis showed that the EZ::TN 〈R6Kγori/KAN-2〉 transposon interrupted an ORF encoding a polypeptide highly similar to the Vibrio parahaemolyticus CsuE protein (GenBank accession number AAK37524.1), which is encoded within the apparently polycistronic locus csuABCDE (GenBank accession number AF339087). Figure 5B shows that with the exception of strains 8114 and 8637 (lanes 14 and 15), all of the Oregon isolates (lanes 11 to 22) harbor sequences related to the csuE-like gene. While these sequences were located in 20- to 23-kbp EcoRI fragments in nine of these strains (lanes 11 to 13 and 16 to 21), that present in strain 9397 (lane 22) was detected in a 9.4-kbp EcoRI fragment. Only three, strains BM4420, BM4427, and BM4439 (lanes 2, 6, and 10, respectively), of the nine European isolates (lanes 2 to 10) showed the presence of csuE-like sequences in their genomes.
FIG. 5.
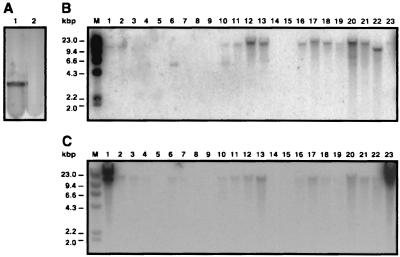
Detection of bacterial attachment and biofilm formation and of secretion genes. (A) Attachment to and biofilm formation by the parental strain 19606 (lane 1) and the insertion mutant 144 (lane 2) were tested by incubation in polystyrene tubes and crystal violet staining. (B and C) Detection of csuE-like genes (A) and csuB-like genes (B) in A. baumannii clinical isolates. Lanes are as described for Fig. 4.
Insertion derivative 4 was isolated by its slightly reduced expression of the attachment-biofilm phenotype compared with that of the parental strain (data not shown). Sequence analysis showed that the EZ::TN 〈R6Kγori/KAN-2〉 transposon interrupted a gene highly similar to csuB, which is part of the V. parahaemolyticus csuABCDE locus (GenBank accession number AF339087) that is related to other bacterial chaperone-usher secretion systems. DNA hybridization showed that the 10 Oregon isolates that tested positive with the csuE probe (Fig. 5B, lanes 11 to 13 and 16 to 22) also tested positive with the csuB probe (same lanes of Fig. 5C). The European strains BM4420, BM4421, BM4422, BM4427, BM4430, and BM4439 (lanes 2 to 4, 6, 7, and 10, respectively) also produced detectable signals with the latter probe, although strains BM4421, BM4422, and BM4430 (compare lanes 3, 4, and 7 of Fig. 5B and C) did not show the presence of csuE-like determinants in their genomes. Interestingly, the EcoRI fragments hybridizing with the csuB and csuE probes displayed similar sizes (ca. 15 kbp) in all strains that tested positive with both probes. This observation indicates that these two genes are located within the same chromosomal region, suggesting a potential organization similar to that described for the V. parahaemolyticus csuABCDE locus. The detection of two A. baumannii 19606 EcoRI fragments with the csuB probe (Fig. 5C, lanes 1 and 23), with one of them matching the size of the fragments detected in other clinical isolates, is another interesting result of these hybridization experiments. The presence of a second csuB copy in the 19606 genome may help explain the observation that the adhesion and biofilm phenotypes of insertion 4 were reduced slightly but not abolished, as was the case with the disruption of csuE. The latter appears to be present as a single-copy gene in all positive strains (Fig. 5B), and therefore its interruption must result in a more drastic effect as was observed with insertion derivative 144.
Concluding remarks.
The A. baumannii 19606 prototype strain proved to be amenable to the electroporation of the EZ::TN 〈R6Kγori/KAN-2〉 transposon complexed with the EZ::TN transposase, an approach that resulted in the generation of random insertion derivatives, some of which were affected in primary metabolic functions. This insertion mutagenesis approach also led to the identification of a gacS-like gene and a hypothetical gene that encode sensor kinases that are part of global regulatory systems present in other, unrelated bacteria. The widespread presence of these genes in different A. baumannii clinical isolates suggests that these genetic traits play an important regulatory function or functions that could determine the ability of this pathogen to use different nutrient sources and respond to changes in environmental signals. The potential regulatory function of the A. baumannii 19606 GacS-like sensor kinase is supported further by the observation that isogenic derivatives with transposon insertions within the gene encoding this protein were not able to use citrate as a sole carbon source. A similar influence of the gacS-gacA system on the utilization of carbon sources was found recently in P. fluorescens (11), indicating that this two-component global regulatory system influences primary as well as secondary metabolism in bacteria. Insertions in the csuB- and csuE-like genes, which affected to different degrees the expression of attachment and biofilm functions, indicate that A. baumannii contains a csuABCDE locus. The role of this predicted locus is supported further by the fact that the csuC and csuD predicted translation products are highly related to chaperone and usher assembly proteins required for the secretion of folded proteins across bacterial outer membranes (53). This type of protein secretion system, which has been described for other bacteria (54), is involved in the assembly of a variety of surface structures that play a role in the interactions between bacterial cells and biotic and abiotic surfaces.
Acknowledgments
Miami University research funds and Public Health grants AI44776-01A1 and DE13657-02 supported this work.
We thank M.-C. Ploy (Laboratoire de Bactériologie-Virologie-Hygiène, CHU Dupuytren, Limoges, France) for providing A. baumannii strains BM4420, BM4421, BM4422, BM4424, BM4427, BM4430, BM4432, BM4436, and BM4439. We also thank V. de Lorenzo (Centro Nacional de Biotecnología, Madrid, Spain) for providing E. coli strains CC118 λpir, SM10 λpir, and S17-1 λpir and the plasmid pLOFKm, and we thank W. Hillen (Friedich-Alexander Universität Erlangen, Nürnberg, Germany) and R. H. Valdivia (Stanford University School of Medicine, Palo Alto, Calif.) for providing plasmids pWH1266 and pFVP25, respectively. We are grateful to C. Wood, coordinator of the Miami University Center of Bioinformatics and Functional Genomics, for his support and assistance with automated DNA sequencing and nucleotide sequence analysis.
REFERENCES
- 1.Actis, L. A., S. Potter, and J. H. Crosa. 1985. Iron-regulated outer membrane protein OM2 of Vibrio anguillarum is encoded by virulence plasmid pJM1. J. Bacteriol. 161:736-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Actis, L. A., M. E. Tolmasky, L. M. Crosa, and J. H. Crosa. 1993. Effect of iron-limiting conditions on growth of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol. 31:2812-2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anstey, N. M., B. J. Currie, and K. M. Withnall. 1991. Community-acquired Acinetobacter pneumonia in the northern territory of Australia. Clin. Infect. Dis. 14:83-91. [DOI] [PubMed] [Google Scholar]
- 4.Barcak, J. G., M. S. Chandler, R. J. Redfield, and J. F. Tomb. 1991. Genetic systems in Haemophilus influenzae. Methods Enzymol. 204:321-432. [DOI] [PubMed] [Google Scholar]
- 5.Bassler, B. L., M. Wright, and M. R. Silverman. 1994. Multiple signaling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol. Microbiol. 13:273-286. [DOI] [PubMed] [Google Scholar]
- 6.Bergogne-Berenzin, E., M. L. Joly-Guillou, and K. J. Towner. 1996. History and importance of Acinetobacter spp., role in infection, treatment and cost implications, p. 2-12. In E. Bergogne-Berenzin, M. L. Joly-Guillou, and K. J. Towner (ed.), Acinetobacter: microbiology, epidemiology, infections, management. CRC Press, Inc., Boca Raton, Fla.
- 7.Bergogne-Berenzin, E., and K. J. Towner. 1996. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin. Microbiol. Rev. 9:148-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlquist, J. F., M. Conti, and J. P. Burke. 1982. Progressive resistance in a single strain of Acinetobacter calcoaceticus recovered during a nosocomial outbreak. Am. J. Infect. Control 10:43-48. [DOI] [PubMed] [Google Scholar]
- 9.Chang, W. N., C. Lu, C. R. Huang, and Y. C. Chuang. 2000. Community-acquired Acinetobacter meningitis in adults. Infection 28:395-397. [DOI] [PubMed] [Google Scholar]
- 10.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235:386-405. [DOI] [PubMed] [Google Scholar]
- 11.Duffy, B. K., and G. Defago. 2000. Controlling instability in gacS-gacA regulatory genes during inoculant production of Pseudomonas fluorescens biocontrol strains. Appl. Environ. Microbiol. 66:3142-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Echenique, J. R., H. Arienti, M. E. Tolmasky, R. Read, J. Staneloni, J. H. Crosa, and L. A. Actis. 1992. Characterization of a high-affinity iron transport system in Acinetobacter baumannii. J. Bacteriol. 174:7670-7679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Echenique, J. R., C. W. Dorsey, L. C. Patrito, A. Petroni, M. E. Tolmasky, and L. A. Actis. 2001. Acinetobacter baumannii has two genes encoding glutathione-dependent formaldehyde dehydrogenase: evidence for differential regulation in response to iron. Microbiology 147:2805-2815. [DOI] [PubMed] [Google Scholar]
- 14.Ely, B. 1985. Vectors for transposon mutagenesis of non-enteric bacteria. Mol. Gen. Genet. 200:302-304. [DOI] [PubMed] [Google Scholar]
- 15.Feinberg, A. P., and B. Vogelstein. 1983. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132:6-13. [DOI] [PubMed] [Google Scholar]
- 16.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J.-F. Tomb, B. A. Dougherty, J. M. Merrick, K. McKenney, G. Sutton, W. FitzHugh, C. Fields, J. D. Gocayne, J. Scott, R. Shirley, L.-I. Liu, A. Glodek, J. M. Kelley, J. F. Weidman, C. A. Phillips, T. Spriggs, E. Hedblom, M. D. Cotton, T. R. Utterback, M. C. Hanna, T. Nguyen, D. M. Saudek, R. C. Brandon, L. D. Fine, J. L. Fritchman, J. L. Fuhrmann, N. S. M. Geoghagen, C. L. Gnehm, L. A. McDonald, K. V. Small, C. M. Fraser, H. O. Smith, and J. C. Venter. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 17.Fraenkel, D. G. 1996. Glycolysis, p. 189-198. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 18.Glansdorff, N. 1996. Biosynthesis of arginine and polyamines, p. 408-433. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 19.Goel, V. K., and A. Kapil. 2001. Monoclonal antibodies against the iron regulated outer membrane proteins of Acinetobacter baumannii are bactericidal. BMC Microbiol. 1:16-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goel, V. K., A. Kapil, B. Das, and D. N. Rao. 1998. Influence of iron on growth and extracellular products of Acinetobacter baumannii. Jpn. J. Med. Sci. Biol. 51:25-33. [DOI] [PubMed] [Google Scholar]
- 21.Graber, K., L. M. Smoot, and L. A. Actis. 1998. Expression of iron binding proteins and hemin binding activity in the dental pathogen Actinobacillus actinomycetemcomitans. FEMS Microbiol. Lett. 163:135-142. [DOI] [PubMed] [Google Scholar]
- 22.Hartstein, A. I., A. L. Rashad, J. M. Liebler, L. A. Actis, J. Freeman, J. W. Rourke, Jr., T. B. Stibolt, M. E. Tolmasky, G. R. Ellis, and J. H. Crosa. 1988. Multiple intensive care unit outbreak of Acinetobacter calcoaceticus subspecies anitratus respiratory infection and colonization associated with contaminated, reusable ventilators and resuscitation bags. Am. J. Med. 85:624-631. [DOI] [PubMed] [Google Scholar]
- 23.Haspel, G., M. Hunger, R. Schmucker, and W. Hillen. 1990. Identification and nucleotide sequence of the Acinetobacter calcoaceticus encoded trpE gene. Mol. Gen. Genet. 220:475-477. [DOI] [PubMed] [Google Scholar]
- 24.Hatziloukas, E., and N. J. Panopoulos. 1992. Origin, structure, and regulation of argK, encoding the phaseolotoxin-resistant ornithine carbamoyltransferase in Pseudomonas syringae pv. Phaseolicola, and functional expression of argK in transgenic tobacco. J. Bacteriol. 174:5895-5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. A. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herrero, M., V. De Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hrabak, E. M., and D. K. Willis. 1992. The lemA gene required for pathogenicity of Pseudomonas syringae pv. Syringae on bean is a member of a family of two-component regulators. J. Bacteriol. 174:3011-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hunger, M., R. Schumucker, V. Kishan, and W. Hillen. 1990. Analysis and nucleotide sequence of an origin of DNA replication in Acinetobacter calcoaceticus and its use for Escherichia coli shuttle plasmids. Gene 87:45-51. [DOI] [PubMed] [Google Scholar]
- 29.Iuchi, S., Z. Matsuda, T. Fujiwara, and E. C. C. Lin. 1990. The arcB gene of Escherichia coli encodes a sensor-regulator protein for anaerobic repression of the arc modulon. Mol. Microbiol. 4:715-727. [DOI] [PubMed] [Google Scholar]
- 30.Joly-Guillou, M. L., E. Bergogne-Berenzin, and J. F. Vieu. 1990. Epidemiology of Acinetobacter and resistance to antibiotics in hospitals. A 5-year evaluation. Presse Med. 19:357-361. [PubMed] [Google Scholar]
- 31.Kaplan, J. B., P. Goncharoff, A. M. Seibold, and B. P. Nichols. 1984. Nucleotide sequence of the Acinetobacter calcoaceticus trpGDC gene cluster. Mol. Biol. Evol. 1:456-472. [DOI] [PubMed] [Google Scholar]
- 32.Kinscherf, T. G., and D. K. Willis. 1999. Swarming by Pseudomonas syringae B728a requires gacS (lemA) and gacA but not the acyl-homoserine lactone biosynthetic gene ahlI. J. Bacteriol. 181:4133-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knauf, V. C., and E. W. Nester. 1982. Wide host range cloning vectors: a cosmid clone bank of an Agrobacterium Ti plasmid. Plasmid 8:45-54. [DOI] [PubMed] [Google Scholar]
- 34.Kredich, N. M. 1996. Biosynthesis of cysteine, p. 514-527. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 35.Leahy, J. G., J. M. Jones-Meehan, E. L. Pullias, and R. R. Colwell. 1993. Transposon mutagenesis in Acinetobacter calcoaceticus RAG-1. J. Bacteriol. 175:1838-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leisinger, T. 1996. Biosynthesis of proline, p. 434-441. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 37.Leong, S., G. S. Ditta, and D. R. Helinski. 1982. Heme biosynthesis in Rhizobium: identification of a cloned gene coding for an amino levulininc acid synthetase from Rhizobium meliloti. J. Biol. Chem. 257:8724-8730. [PubMed] [Google Scholar]
- 38.Meade, H. M., S. R. Long, S. E. Ruvkum, S. E. Brown, and F. M. Ausubel. 1982. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J. Bacteriol. 149:114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murata, H., T. Tsukamoto, and A. Shirata. 1998. rtpA, a gene encoding a bacterial two-component sensor kinase, determines pathogenic traits of Pseudomonas tolaasii, the causal agent of brown blotch disease of a cultivated mushroom, Pleurotus ostreatus. Mycoscience 39:261-272. [Google Scholar]
- 40.Nagasawa, S., S. Tokishita, H. Aiba, and T. Mizuno. 1992. A novel sensor-regulator protein that belongs to the homologous family of signal-transduction proteins involved in adaptive responses in Escherichia coli. Mol. Microbiol. 6:799-807. [DOI] [PubMed] [Google Scholar]
- 41.Obana, Y. 1986. Pathogenic significance of Acinetobacter calcoaceticus: analysis of experimental infection in mice. Microbiol. Immunol. 30:645-657. [DOI] [PubMed] [Google Scholar]
- 42.O'Toole, G. A., L. A. Pratt, P. I. Watnick, D. K. Newman, V. B. Weaver, and R. Kolter. 1999. Genetic approaches to study of biofilms. Methods Enzymol. 310:91-109. [DOI] [PubMed] [Google Scholar]
- 43.Patte, J.-C. 1996. Biosynthesis of threonine and lysine, p. 528-541. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 44.Perna, N. T., G. I. Plunkett, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. Dimalanta, K. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 45.Pittard, J., and B. J. Wallace. 1966. Distribution and function of genes concerned with aromatic biosynthesis in Escherichia coli. J. Bacteriol. 91:1494-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ploy, M. C., F. Denis, P. Courvalin, and T. Lambert. 2000. Molecular characterization of integrons in Acinetobacter baumannii: description of a hybrid class 2 integron. Antimicrob. Agents Chemother. 44:2684-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salanoubat, M., S. Genin, F. Artiguenave, J. Gouzy, S. Mangenot, M. Arlat, A. Billault, P. Brottier, J. C. Camus, L. Cattolico, M. Chandler, N. Choisne, C. Claudel-Renard, S. Cunnac, N. Demange, C. Gaspin, M. Lavie, A. Moisan, C. Robert, W. Saurin, T. Schiex, P. Siguier, P. Thebault, M. Whalen, P. Wincker, M. Levy, J. Weissenbach, and C. A. Boucher. 2002. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415:497-502. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 49.Singer, J. T., and W. R. Finnerty. 1984. Insertional specificity of transposon Tn5 in Acinetobacter sp. J. Bacteriol. 157:607-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stout, V., and S. Gottesman. 1990. RcsB and RcsC: a two-component regulator of capsule synthesis in Escherichia coli. J. Bacteriol. 172:659-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. L. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrook-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. M. Lim, K. A. Smith, D. H. Spencer, G. K. S. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 52.Tang, J.-L., Y.-N. Liu, C. E. Barber, J. M. Dow, J. C. Wootton, and M. J. Daniels. 1991. Genetic and molecular analysis of a cluster of rpf genes involved in positive regulation of synthesis of extracellular enzymes and polysaccharide in Xanthomonas campestris pathovar campestris. Mol. Gen. Genet. 226:409-417. [DOI] [PubMed] [Google Scholar]
- 53.Thanassi, D. G. 2002. Ushers and secretins: channels for the secretion of folded proteins across the bacterial outer membrane. J. Mol. Microbiol. Biotechnol. 4:11-20. [PubMed] [Google Scholar]
- 54.Thanassi, D. G., E. T. Saulino, and S. J. Hultgren. 1998. The chaperone/usher pathway: a major terminal branch of the general secretory pathway. Curr. Opin. Microbiol. 1:223-231. [DOI] [PubMed] [Google Scholar]
- 55.Towner, K. J. 1983. Transposon-directed mutagenesis and chromosome mobilization in Acinetobacter calcoaceticus EBF65/65. Genet. Res. 41:97-102. [DOI] [PubMed] [Google Scholar]
- 56.Valdivia, R. H., and S. Falkow. 1996. Bacterial genetics by flow cytometry: rapid isolation of Salmonella typhimurium acid-inducible promoters by differential fluorescence induction. Mol. Microbiol. 22:367-378. [DOI] [PubMed] [Google Scholar]
- 57.Winkler, M. E. 1996. Biosynthesis of histidine, p. 485-505. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 58.Yamamoto, S., N. Okujo, and Y. Sakakibara. 1994. Isolation and structure elucidation of acinetobactin, a novel siderophore from Acinetobacter baumannii. Arch. Microbiol. 162:249-252. [DOI] [PubMed] [Google Scholar]
- 59.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-109. [DOI] [PubMed] [Google Scholar]