Abstract
Arthrobacter aurescens strain TC1 was isolated without enrichment by plating atrazine-contaminated soil directly onto atrazine-clearing plates. A. aurescens TC1 grew in liquid medium with atrazine as the sole source of nitrogen, carbon, and energy, consuming up to 3,000 mg of atrazine per liter. A. aurescens TC1 is metabolically diverse and grew on a wider range of s-triazine compounds than any bacterium previously characterized. The 23 s-triazine substrates serving as the sole nitrogen source included the herbicides ametryn, atratone, cyanazine, prometryn, and simazine. Moreover, atrazine substrate analogs containing fluorine, mercaptan, and cyano groups in place of the chlorine substituent were also growth substrates. Analogs containing hydrogen, azido, and amino functionalities in place of chlorine were not growth substrates. A. aurescens TC1 also metabolized compounds containing chlorine plus N-ethyl, N-propyl, N-butyl, N-s-butyl, N-isobutyl, or N-t-butyl substituents on the s-triazine ring. Atrazine was metabolized to alkylamines and cyanuric acid, the latter accumulating stoichiometrically. Ethylamine and isopropylamine each served as the source of carbon and nitrogen for growth. PCR experiments identified genes with high sequence identity to atzB and atzC, but not to atzA, from Pseudomonas sp. strain ADP.
s-Triazine rings are common scaffolds for the synthesis of industrial chemicals; they are found in pesticides, plastic resins, dyes, and explosives (20). s-Triazine herbicides are widely used in modern agriculture, where they kill susceptible plants by coordinating to the quinone-binding protein in photosystem II, thereby inhibiting photosynthetic electron transfer (19). Atrazine (2-chloro-4-ethylamino-6-isopropylamino-1,3,5-s-triazine) is one of the most widely used herbicides in the United States for the control of broadleaf weeds in corn, sorghum, and sugarcane (1). Other s-triazine herbicides include ametryn, atratone, cyanazine, prometryn, and simazine.
The fate of s-triazine compounds in the environment depends on the metabolic activities of soil microorganisms (7, 14). Since 1994, a number of laboratories have independently isolated, by enrichment culture, several genera of gram-negative bacteria capable of atrazine dechlorination (4, 16, 22, 31, 33, 35, 36, 38). Subsequent to dechlorination, metabolism of hydroxyatrazine liberates nitrogen to sustain bacterial growth. More recently, gram-positive bacteria from the genera Nocardioides (32) and Arthrobacter (22) have been shown to grow on atrazine, the former using atrazine as the sole source of carbon and nitrogen for growth. Streptomyces strain PS1/5 was also shown to metabolize several s-triazine herbicides in the presence of additional carbon and nitrogen in the growth medium (29).
It is now established that bacteria metabolize melamine and the triazine herbicides such as atrazine via enzyme-catalyzed hydrolytic reactions (6, 11, 12, 35). The enzymatic basis of atrazine mineralization has been most extensively studied in Pseudomonas sp. strain ADP (3, 8, 16, 17, 23). In this strain, displacement of the three substituents on the s-triazine ring is mediated by three enzymatic steps encoded by the genes atzA, atzB, and atzC. The first enzyme, AtzA, catalyzes the hydrolytic dechlorination of atrazine to yield hydroxyatrazine. AtzB subsequently catalyzes hydroxyatrazine deamidation, yielding N-isopropylammelide, and the third enzyme, AtzC, metabolizes N-isopropylammelide stoichiometrically to cyanuric acid and N-isopropylamine. Cyanuric acid is subsequently metabolized to CO2 and NH3 by AtzD, AtzE, and AtzF. All six genes encoding atrazine catabolism in Pseudomonas sp. strain ADP have been localized to plasmid pADP-1, which has been completely sequenced (17). Virtually identical genes and enzymes have been shown to be present in several other gram-negative bacteria (4, 9, 21, 22, 31, 33).
AtzA, AtzB, and AtzC from Pseudomonas sp. strain ADP are members of the amidohydrolase superfamily, a widespread class of enzymes that catalyze numerous hydrolytic reactions in intermediary metabolism (23). There is evidence that these enzymes have recently evolved to handle herbicides (26) and are found in such diverse genera as Ralstonia (21), Agrobacterium (31), Pseudaminobacter (33), Chelatobacter, Aminobacter, Stenotrophomonas (22), and Rhizobium (4). However, it is emerging that not all enzymes that metabolize chlorinated s-triazines via an initial dechlorination reaction are identical. For example, the s-triazine catabolic enzymes in the gram-positive bacterium Nocardioides sp. strain C190 are thought to be different from those found in Pseudomonas sp. strain ADP since atzA, atzB, and atzC gene probes failed to hybridize to Nocardioides DNA (32). The Nocardioides enzyme initiating s-triazine catabolism, designated TrzN, has been purified and shown to have broader substrate specificity than AtzA (32).
In this report, we describe the isolation and characterization of a gram-positive bacterium, Arthrobacter aurescens strain TC1. Strain TC1 was isolated without enrichment by spread plating soil onto an atrazine-containing solid medium. A. aurescens TC1 was shown to metabolize 23 different s-triazine compounds, the broadest catabolism of s-triazine compounds observed for a single strain to date. Moreover, strain TC1 grew on atrazine as the sole source of carbon and nitrogen and transformed 3,000 mg of atrazine per liter in a growing culture. A. aurescens TC1 was shown to contain DNA homologous to the atzB and atzC genes from Pseudomonas sp. strain ADP but not to atzA. Moreover, A. aurescens TC1 was shown to rapidly grow on ethylamine or isopropylamine, metabolic intermediates produced during atrazine metabolism via AtzB- and AtzC-mediated reactions.
MATERIALS AND METHODS
Bacterial isolation and initial characterization.
Soil samples were obtained from a South Dakota spill site that was exposed to high concentrations of atrazine, up to 29,000 mg per kg of soil, for 18 months (30). Atrazine-degrading bacteria were isolated by directly plating soil onto modified minimal R medium agar plates (13, 27) containing 500 mg of atrazine per liter (R-ATZ) as the sole nitrogen (N) and carbon (C) sources. Agar plates were solidified with 1.5% Noble agar and were incubated at 27°C. A pure culture was obtained by repeated streaking onto R-ATZ plates and was subjected to a Gram stain and MIDI fatty acid methyl ester analysis (Analytical Services Inc, Essex Junction, Vt.).
Chemicals.
Atrazine (>98% purity) and simazine (>98% purity) were kindly provided by Syngenta Crop Protection. The following atrazine analogs had been synthesized previously and were made as described elsewhere (25): fluoroatrazine, cyanoatrazine, azidoatrazine, aminoatrazine, ametryn, and atratone. Other compounds were synthesized as described below.
(i) 2-Isopropylamino-4-ethylamino-1,3,5-triazine (dechloroatrazine).
To a 500-ml flask were added 25 g of atrazine, 1.0 g of 5% palladium on calcium carbonate, 12.5 g of sodium carbonate, and 250 ml of isopropanol. The reaction mixture was stirred under hydrogen at room temperature and atmospheric pressure. After 3 days, an additional 1.0 g of catalyst was added. After an additional 4 days, the collected solid was extracted with 200 ml of hot ethanol. Combined filtrates were evaporated on a rotary evaporator, dissolved in 400 ml of ethyl acetate, and passed through a column (25 by 3 cm) of silica gel and Celite to remove finely divided palladium. The product was recrystallized from ethyl acetate to remove residual atrazine. Repeated crystallizations yielded 13 g of the desired product that was 99% pure as determined by gas chromatography-mass spectrometry (MS).
(ii) 2-Mercapto-4-isopropylamino-6-ethylamino-1,3,5-triazine (mercaptoatrazine).
To a 500-ml flask were added 21.6 g of atrazine, 15.2 g of thiourea, 2.0 ml of H2O, and 250 ml of anhydrous ethanol. The flask was flushed with nitrogen and refluxed overnight (14 h). The reaction mixture was cooled, poured into a solution of 10 g of sodium bicarbonate in 1 liter of water, stirred and shaken until gas evolution ceased, and then filtered. The filtered solid was recrystallized under nitrogen from 1 liter of hot ethanol containing 1 ml of 2-mercaptoethanol to yield 13.9 g, 65% of theoretical yield.
(iii) Di-N,N-alkyldiaminochlorotriazines.
N,N-Dialkylamino-chlorotriazines were produced from N-alkylamino-dichlorotriazine intermediates by a variation of synthetic procedures described previously (25). The N-alkylamino-dichlorotriazine intermediates were formed by reacting cyanuric chloride (2,4,6-trichloro-1,3,5-s-triazine; Aldrich Chemical Company, Milwaukee, Wis.) with the appropriate alkylamine in dichloromethane or anhydrous diethyl ether at 0 to 5°C. The monoalkylamino intermediate was aminated a second time, producing chloro-di-N,N′-alkylamino-triazines.
Products were identified directly after synthesis by MS analysis and, in most cases, following derivatization with 1,1,1,3,3,3-hexamethyldisilazane (Aldrich Chemical Company). Products were of ≥97% purity.
Molecular analyses.
Total genomic DNA was isolated as previously described (2). PCR was performed using primers designed to amplify DNA regions corresponding to the Pseudomonas sp. strain ADP atzA (atrazine chlorohydrolase), atzB (hydroxyatrazine ethylaminohydrolase), and atzC (N-isopropylammelide isopropylamidohydrolase) genes, as described previously (9). The 16S rDNA gene was amplified by PCR using universal 16S rDNA primers (39). PCR products were separated by horizontal electrophoresis on 1% agarose gels. DNA from Pseudomonas sp. strain ADP served as a positive control for each primer pair. PCR products were purified using a QIAquick PCR column purification kit (Qiagen, Valencia, Calif.) and were sequenced at the University of Minnesota Advanced Genetic Sequencing Center. The resulting 16S rDNA sequences were aligned and compared with entries in GenBank using the BLAST algorithm and to the Ribosomal Database using the Ribosomal Database Project server (http://www./RDP/html/index.html). Southern hybridization of total genomic DNA to an atzA gene probe was done with a 0.6-kb ApaI-PstI fragment from pMD4, encoding a conserved internal region of atzA (9). Western blotting was done as previously described (24).
Metabolism of [U-ring-14C]atrazine.
Metabolism of uniformly ring-labeled [14C]atrazine (Sigma Chemical, St. Louis, Mo.) by A. aurescens TC1 was measured in triplicate. Growth was measured in modified R medium with 2,500 mg of unlabeled atrazine per liter supplied as the sole source of C and N for growth. [U-ring-14C]atrazine (17.1 mCi/mmol) was added to yield 8.9 × 10−4 mCi/ml in 40 ml of medium. Cultures were grown at 25°C in 250-ml biometer flasks sealed with Teflon stoppers. The flasks were opened after 24-h intervals, and the amount of 14CO2 evolved and trapped in 2 N NaOH solution was determined by scintillation counting with an HP 1900 TR liquid scintillation analyzer. Prior to resealing and further incubation, the NaOH in each trap was replaced with fresh solution.
Aliquots of stationary-phase cultures from the U-ring-14C growth experiments were filtered though a 0.2-μm filter and analyzed by both high-pressure liquid chromatography (HPLC) protocols described below. Fractions were collected each minute for 30 min, and the radioactivity in each fraction was measured by liquid scintillation counting.
Chromatographic and mass spectral identification of products.
HPLC analyses were performed by using a Hewlett-Packard HP1100 liquid chromatograph system equipped with a photodiode array detector and interfaced to an HP 79994A Chemstation. A. aurescens strain TC1 was grown for 48 h in minimal R medium containing 2,000 mg of atrazine per liter. The supernatant was filtered through a 0.2-μm filter, and one of two protocols was used to resolve atrazine and its metabolites. Protocol A resolved atrazine and hydroxyatrazine with an analytical C18 reverse-phase NovaPak HPLC column and an acetonitrile gradient in water, as described previously (8). Protocol B resolved N-isopropylammelide, biuret, and cyanuric acid by using an Inertsil 5μ (150 by 4.6 mm) phenyl column (Alltech Associates, Deerfield, Ill.) with an isocratic mobile phase containing 5 mM sodium octane sulfate in 0.05% H3PO4 at pH 2.8. Atrazine-free uninoculated media, authentic atrazine, hydroxyatrazine, N-isopropylammelide, cyanuric acid, and biuret standards were run for each protocol. The HPLC retention times for authentic standards analyzed by protocol A were as follows: N-isopropylammelide, 4.8 min; hydroxyatrazine, 15.3 min; and authentic atrazine, 18.5 min. The retention times from HPLC analysis of authentic standards analyzed by protocol B were 2.6 min for cyanuric acid and 10.2 min for N-isopropylammelide; biuret exhibited phase segregation and showed three characteristic peaks at 2.6, 3.6, and 4.0 min.
A Finnigan MAT Spectra System HPLC mass spectrometer was used to positively identify metabolites produced by A. aurescens strain TC1 growing on atrazine. Cultures of A. aurescens growing in modified R medium with atrazine as the sole source of N and C were sampled 42, 90, 162, and 200 h after inoculation, and analyzed by using protocol A as described above. The mass spectrometer was run in electrospray protonating mode. Authentic standards were run for atrazine (M + 1: 216.5), hydroxyatrazine (M + 1: 198.5), N-isopropylammelide (M + 1: 171.5), and cyanuric acid (M + 1: 166.5).
Gas chromatography for routine analysis of atrazine was conducted as previously described (30).
Metabolic studies.
Growth curves were obtained for strain TC1 by using liquid minimal R medium containing 500 to 3,000 mg of atrazine per liter as the sole source of C and N at 30°C. When atrazine was added sequentially during growth, it was added as a finely divided powder directly to the culture and made a fine suspension. Because atrazine is soluble only to 33 mg/liter in aqueous media, growth data were obtained either by measuring absorbance of the culture at 600 nm (A600) with a Beckman DU-64 spectrophotometer when the atrazine was fully utilized, e.g., at stationary phase, or by filtering an aliquot of the culture medium through 1 cm of glass wool to remove suspended crystalline atrazine. This was shown to remove particulate atrazine and render turbidometric growth measurements accurate. For growth rate determinations, data for the initial growth in 500 mg of atrazine per liter were fitted with a population logistic equation (34) with an r2 value of 0.90. The parameters from this fit were then applied to a second curve obtained after supplemental atrazine (500 mg/liter) was added to the cultures.
The use of atrazine as a sole C and N source for growth was determined by measuring the dry weight of cells obtained from 250 ml of culture grown at 30°C for 7 days in liquid minimal R medium containing 3,000 mg of atrazine per liter (n = 4). Cells were dried in an oven at 60°C until a constant weight was obtained. Additional growth studies were performed to determine if A. aurescens strain TC1 could grow on the N-alkyl substituents of atrazine (N-isopropylamine or N-ethylamine) as sources of C, N, and energy. Growth curves were obtained at 30°C in modified liquid minimal R medium containing N-isopropylamine or N-ethylamine as the sole source of C and N or augmented with an additional N source. Cell growth was determined with a Klett-Summerson electrophotometric colorimeter using a green 54 filter. Klett flasks were sealed with gas-tight seals to prevent evaporation of N-isopropylamine and N-ethylamine. The four media used were (i) 7.5 mM N-isopropylamine as the C source with 15 mM NH4NO3 as a supplementary N source, (ii) 7.5 mM N-isopropylamine as the sole C and N source, (iii) 7.5 mM N-ethylamine as the C source with 15 mM NH4NO3 as a supplementary N source, and (iv) 7.5 mM N-ethylamine as the sole C and N source.
Growth of A. aurescens TC1 on a variety of s-triazine growth substrates was tested in R medium containing 10 mM glucose and 200 mg of the specified s-triazine compound per liter as the sole source of N. The cultures were incubated at 30°C until a maximum A600 was observed. Data are presented as the observed absorbances. Negative control cultures, containing no nitrogen source, showed a maximum absorbance of 0.17. With s-triazines as the nitrogen source, absorbance values of less than 0.2 were interpreted as indicating that growth was not supported.
RESULTS
Bacterial isolation and identification.
Initially, 41 bacterial colonies that showed clearing zones around the colonies on agar plates that were supersaturated with atrazine, ametryn, or prometryn were picked. One isolate showed apparent, very rapid removal of atrazine from the medium and was selected for further study. The atrazine-degrading bacterium was a gram-positive rod that appeared to be coccus-like when in stationary phase. Fatty acid methyl ester analysis showed that this strain had similarity indices of 0.686 with Arthrobacter histidinolovorans, 0.665 with Curtobacterium citreum, and 0.593 with Micrococcus varians. To more precisely determine the taxonomic identity of the organism, a gene region of 1,279 bp encoding 16S rRNA was amplified by PCR and sequenced. This was found to comprise 92% of a gene region for A. aurescens (GenBank accession number AF467106) to which it showed 99.8% sequence identity (15). The second strongest match was with another A. aurescens strain (GenBank accession number AF388032). Consequently, this bacterium was designated A. aurescens strain TC1.
Range of s-triazine compounds used.
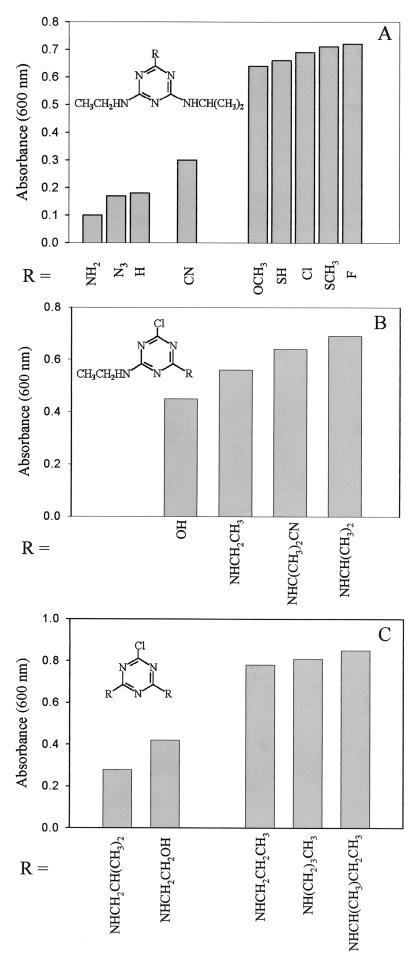
The range of s-triazine growth substrates metabolized by A. aurescens TC1 was determined in media containing limiting concentrations of the s-triazine compound supplied as the sole N source and glucose as the C source. In the first set of experiments, the chlorine substituent of atrazine was replaced with other substituents; some of these substrates are commercially available herbicides, and others were synthesized for testing the specificity of s-triazine-metabolizing enzymes. Figure 1A shows that substrates which clearly supported growth contained fluorine, chlorine, mercaptan, methylmercaptan, cyano, or O-methyl substituents. However, substrates containing hydrogen, amino, and azido substituents in place of chlorine did not support significant growth.
FIG. 1.
Specificity of s-triazine utilization by A. aurescens strain TC1. Substrates were added at a concentration of 200 mg per liter and used as the sole source of nitrogen. (A) Growth of strain TC1 on substrates where the chlorine substituent of atrazine was replaced with fluoro, methylmercapto, mercapto, O-methyl, cyano, hydrogen, azido, and amino functional groups; (B) growth of strain TC1 where the N-isopropyl group of atrazine was replaced with OH, NHCH2CH3, and NHC(CH3)2CN functional groups; (C) growth of strain TC1 on substrate variants containing symmetrical N-alkyl substituents.
Replacement of the N-isopropyl group of atrazine with OH, NHCH2CH3, and NHC(CH3)2CN groups yielded significant growth with all three alternative substrates tested (Fig. 1B). Atrazine [R = NHCH(CH3)2, on the far right] is shown as a positive control. The compound containing the cyanoisopropyl group [R = NHC(CH3)2CN] is the herbicide cyanazine.
A wider range of substrate variants containing symmetrical N-alkyl substituents were synthesized and tested as substrates for strain TC1 (Fig. 1C). All of the compounds tested were substrates, including those with s-butyl, isobutyl, and t-butyl groups. Other s-triazine compounds shown to support growth as the sole nitrogen source were ammeline, ammelide, chlorodiaminotriazine, and fluorodiaminotriazine (data not shown).
Growth on atrazine.
A. aurescens strain TC1 was grown in liquid minimal R medium containing 3,000 mg of atrazine per liter as the sole C and N source. After 7 days of growth at 30°C, the culture reached an A600 of 2.2, which corresponded to 2 × 1010 cells per ml by direct counting. The final cell dry weight was 566 mg per liter of culture. All 3,000 mg of atrazine per liter was consumed. This corresponds to 0.833 g of available C/liter and 0.389 g of available N/liter, assuming that each atrazine molecule has 5 C atoms and 2 N atoms that are assimilated to support growth (based on metabolic studies shown below). The ring carbon atoms of atrazine are at the same oxidation state as CO2 and therefore do not provide an oxidizable energy source. The molar growth yield was calculated to be 18.1 g of cells per mol of C. This value compares favorably to published values of 12 g of cells/mol of C (6) and 16 g of cells/mol of C (38) for the utilization of s-triazine compounds.
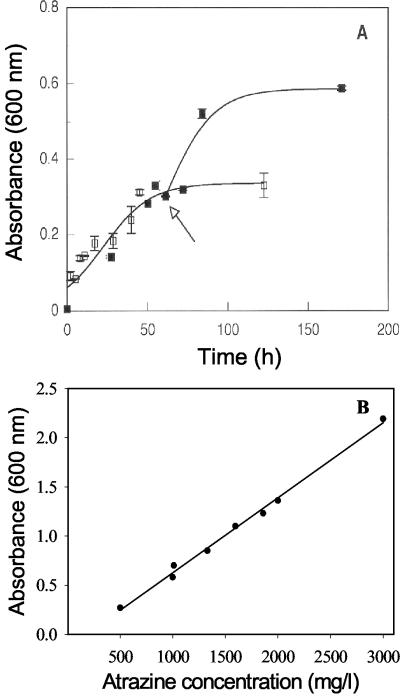
A growth curve for strain TC1, cultured at 30°C in minimal R medium with a successive addition of atrazine as the sole nitrogen and carbon source is shown in Fig. 2A. The initial culture received 500 mg of atrazine per liter and reached an A600 of 0.3 before growth subsided. At this time, additional atrazine (500 mg/liter) was added to one set of replicate cultures; growth resumed and reached a final A600 of 0.6. Control cultures receiving no additional atrazine showed no increase in absorbance. These data indicated that strain TC1 had a doubling time of 22 h when grown on atrazine as the sole source of C and N.
FIG. 2.
Influence of different concentrations of atrazine on growth of A. aurescens TC1. Strain TC1 was grown in minimal medium using atrazine as a sole source of carbon, nitrogen, and energy. (A) Culture grown with 500 mg of atrazine per liter (□) and culture initially grown on 500 mg of atrazine per liter and receiving an additional 500 mg of atrazine per liter at the time indicated by the arrow (▪); r2 = 0.90. (B) Linear correlation of growth of A. aurescens TC1 on several concentrations of atrazine; r2 = 0.99.
Figure 2B shows the stationary-phase A600 values for strain TC1 obtained with different concentrations of atrazine as the sole source of N and C for growth and energy. Cell density at stationary phase increased linearly as a function of increasing atrazine concentration in the medium (r2 = 0.99), up to the 3,000 mg/liter tested. In all cases, the final atrazine concentration in the culture medium at stationary phase decreased to below the detection limits of gas chromatography (1 ng of atrazine per μl of analyte).
Growth on alkylamines.
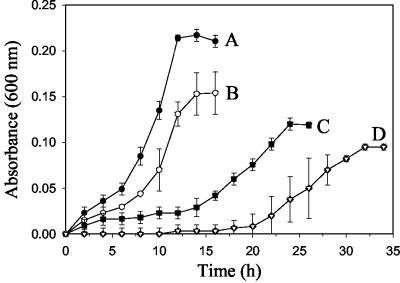
A. aurescens strain TC1 was tested for its ability to use the N-alkyl substituents of atrazine as N and C sources for growth. Results in Fig. 3 show that strain TC1 used N-isopropylamine or N-ethylamine as the sole C and N source for growth, with doubling times of approximately 2 and 5 h, respectively. Doubling times were slightly less with the addition of nitrate. Overall, growth on the alkylamines was substantially faster than with atrazine, suggesting that utilization of the alkylamines was not rate determining during growth on atrazine as the sole C and N source.
FIG. 3.
Utilization of N-alkyl amines as a source of C and N by A. aurescens TC1. Cells were grown in minimal medium containing 7.5 mM N-isopropylamine plus 15 mM NH4NO3 (A), 7.5 mM N-isopropylamine (B), 7.5 mM ethylamine plus 15 mM NH4NO3 (C), and 7.5 mM ethylamine (D).
Metabolism of atrazine.
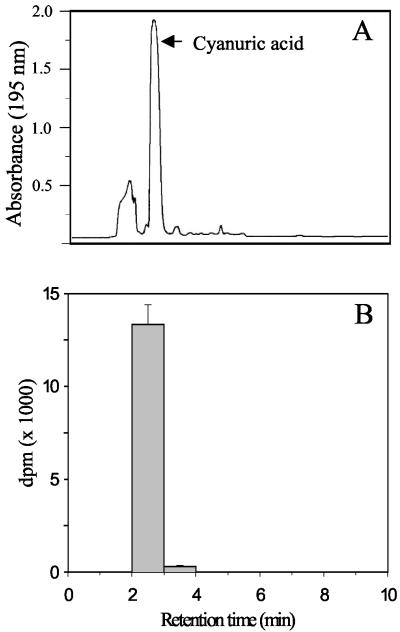
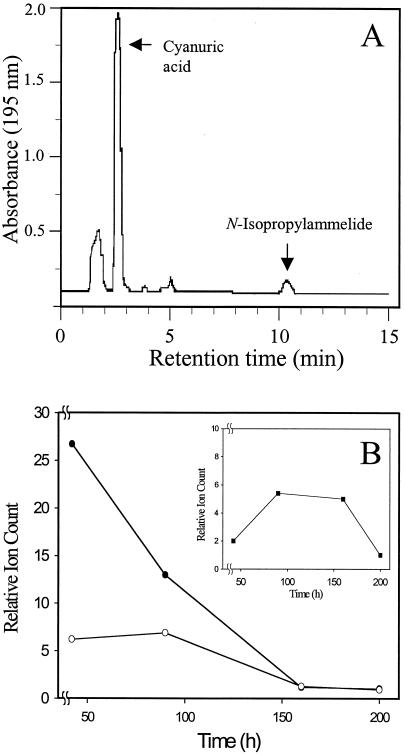
A. aurescens strain TC1 did not produce discernible [14C]CO2 from [U-ring-14C]atrazine. When growth was allowed to proceed for an extended period, a single major metabolite was detected by HPLC. Figure 4 shows both the UV absorbance and radioactivity as a function of HPLC retention time. Absorbance peaks are evident for material that was eluted in the void volume (1.9 min) and cyanuric acid (2.6 min). The latter was consistent with the elution peak of an authentic cyanuric acid standard. No additional absorbance peaks or radioactivity were observed out to a 30-min elution time. The fraction that was coeluted with cyanuric acid contained 13,350 dpm, accounting for 95% of the theoretical yield. Thus, atrazine was transformed to cyanuric acid with a 1:1 stoichiometry. Standard N-isopropylammelide was eluted at 10.4 min.
FIG. 4.
Atrazine metabolite(s) detected in stationary-phase cultures of A. aurescens TC1. UV absorbance and radioactivity as a function of HPLC retention time are shown for culture supernatants of stationary-phase A. aurescens TC1 grown in minimal medium containing cold and ring-labeled [14C]atrazine. (A) HPLC elution profile at 195 nm; (B) corresponding radioactivity.
To look for metabolic precursors of cyanuric acid, culture supernatants taken from late-exponential-growth-phase cultures of A. aurescens strain TC1 were analyzed by HPLC protocol B, which is capable of resolving cyanuric acid, biuret, and N-isopropylammelide. Figure 5A shows a peak with an HPLC retention time identical to that of standard N-isopropylammelide. In this same experiment, a major metabolite with a retention time identical to that of cyanuric acid was also observed (Fig. 5A).
FIG. 5.
Metabolites produced by A. aurescens strain TC1 during growth on atrazine. (A) HPLC elution profile of metabolites from culture supernatant at late exponential growth phase. Atrazine and hydroxyatrazine were eluted with the medium front. (B) MS of atrazine and metabolic intermediates produced by strain TC1 in culture filtrates sampled during exponential through stationary growth phase. The relative ion count for each compound is presented as a ratio of the ion count at each time point divided by the minimum ion count observed over the time course. •, atrazine; ○, hydroxyatrazine; ▪ (inset), N-isopropylammelide.
HPLC-MS (Fig. 5B) was used to more positively identify putative metabolites and to determine if they are true pathway intermediates, that is, if their production will increase and then fall as expected for intermediates. Culture supernatants were sampled from exponential through stationary growth phase. A metabolite with an m/z (M + 1) of 198 was identified as hydroxyatrazine, and another metabolite with an m/z (M + 1) of 171 was identified as N-isopropylammelide. No N-ethylammelide, the major metabolite of Nocardioides sp. strain C190 (31), was detected. The concentration of atrazine was observed to decrease throughout the experiment. Hydroxyatrazine was maximally present at 42 and 90 h, and N-isopropylammelide was observed at significant levels from 90 to 160 h (Fig. 5B, inset). Both metabolites decreased to negligible levels when stationary phase was reached (190 h). These observations are consistent with the observation that atrazine had been stoichiometrically transformed to cyanuric acid when the culture was sampled late in stationary phase.
Molecular analysis of s-triazine metabolic genes.
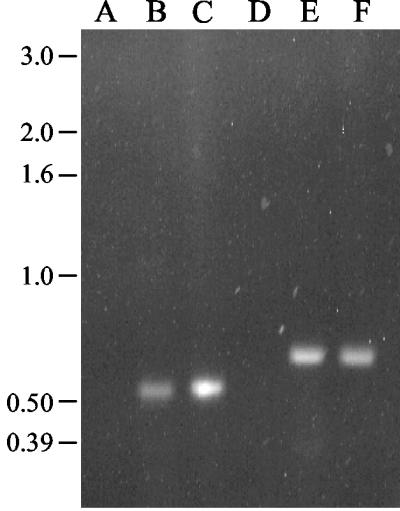
To determine if A. aurescens strain TC1 contains atrazine degradation genes similar to those found in Pseudomonas strain ADP and other gram-positive bacteria, we performed PCR analyses with primers specific for internal regions of the atzA, atzB, and atzC genes. Results in Fig. 6 show that A. aurescens strain TC1 has PCR products of the same size as the atzB and atzC genes of Pseudomonas sp. strain ADP. Moreover, sequence analysis of the PCR product obtained with atzB and atzC primers showed that A. aurescens TC1 contained genes with 100% sequence identity to the corresponding genes from Pseudomonas sp. strain ADP. Primers effective in amplifying atzA DNA from other atrazine-metabolizing bacteria (9) did not amplify DNA from A. aurescens strain TC1. Southern hybridization experiments done with an atzA probe under conditions of low stringency failed to demonstrate that strain TC1 has an atzA homolog. Moreover, cell-free protein extracts from TC1 were tested for the presence of an immunologically cross-reacting AtzA protein by Western blotting. No AtzA protein was detected under conditions that readily detected AtzA in Pseudomonas sp. strain ADP.
FIG. 6.
PCR amplification of A. aurescens TC1 total genomic DNA with primers for the atzB and atzC genes from Pseudomonas sp. strain ADP. Lanes: A, negative control (atzB primers and buffer); B, atzB primers with genomic DNA from A. aurescens TC1; C, atzB primers with genomic DNA from Pseudomonas sp. strain ADP; D, negative control (atzC primers and buffer); E, atzC primers with genomic DNA from A. aurescens TC1; F, atzC primers and genomic DNA from Pseudomonas sp. strain ADP.
DISCUSSION
A. aurescens TC1 was shown here to have the broadest specificity for the catabolism of s-triazine compounds of any atrazine-degrading bacterium characterized to date. Strain TC1 grew at the expense of 23 distinct s-triazine compounds as the sole N source. A. aurescens TC1 grew with atrazine as the sole C and N source and consumed 3,000 mg of atrazine per liter in liquid culture during its growth phase. Studies with other strains have shown consumption of 0.1 to 96 mg of atrazine per liter (16, 21, 31, 36, 38).
A. aurescens TC1 can apparently grow on atrazine efficiently because of its ability to use both alkylamine side chain substituents. A previously described Nocardioides sp. metabolized atrazine with the accumulation of N-ethylammelide (32), indicating that only isopropylamine was released to support growth. With A. aurescens TC1, growth on ethylamine or isopropylamine was substantially faster than growth on atrazine, indicating that removal of one or more substituents from the s-triazine ring is likely rate determining for growth. It cannot currently be determined which of the enzymatic steps catalyzed by the initial enzyme or the AtzB and AtzC homologs are rate determining. Topp and coworkers (33) showed that the rates for metabolism of simazine, atrazine, and terbuthylazine decreased in linear proportion to the size of the variable side chain. These and other experiments with purified AtzC (28) support the idea that AtzC determines the rate of growth on each of the different s-triazine herbicides with a variable N-alkyl side chain. Confirmation of this will additionally require purification of AtzB and comparative steady-state kinetic studies with AtzC.
A. aurescens TC1 also metabolized a large number of s-triazine substrates containing other alkylamine side chains, including N-propyl, -butyl, -s-butyl, -isobutyl, -t-butyl, and -hydroxyethyl. These alkylamines have all been shown to be liberated from N-alkylammelides by purified AtzC from Pseudomonas sp. strain ADP (28). PCR experiments conducted here showed that, in A. aurescens TC1, the atzB and atzC genes are identical to the homologous genes in Pseudomonas sp. strain ADP over the region amplified by PCR. Most bacteria that have been described to catabolize alkylamines are high-G+C gram-positive bacteria such as Arthrobacter or Mycobacterium strains (5, 37). Facile metabolism of alkylamines might underlie the more efficient metabolism of s-triazines by A. aurescens TC1 compared to gram-negative bacteria, which use the ring N atoms of s-triazines.
In a previous study, three Arthrobacter crystallopoietes strains were isolated from soil by enrichment culture on atrazine as the sole N source and citrate as the C source (22). These strains were shown to metabolize atrazine to cyanuric acid. Moreover, the A. crystallopoietes strains contained genes with high sequence identity to atzB and atzC from Pseudomonas sp. strain ADP, similar to the observations here with A. aurescens TC1. With A. crystallopoietes, total DNA was shown to contain a gene that hybridizes weakly with an atzA gene probe. In this study, an atzA gene probe failed to hybridize to A. aurescens TC1 genomic DNA under less stringent hybridization conditions, and PCR amplification experiments failed to amplify an atzA homolog. Additionally, Western blotting experiments failed to detect an AtzA protein in strain TC1. The nature of the enzyme initiating the metabolism of the diverse s-triazine substrates tested here remains undefined.
A triazine hydrolase acting on atrazine, but distinct from AtzA, was isolated from a Nocardioides sp. which metabolized atrazine stoichiometrically to N-ethylammelide (18, 32). DNA from this strain did not hybridize to atzA, atzB, or atzC gene probes, indicating that its s-triazine catabolic genes were distinct from those of Pseudomonas sp. strain ADP and the atzB and atzC genes found in the Arthrobacter strains. The Nocardioides sp. was found to hydrolyze eight other s-triazine herbicides, including those with S-methyl and O-methyl substituents as tested here. Atrazine analogs with fluorine, hydrogen, cyano, azido, amino, and mercaptan substituents were not tested with the Nocardioides sp., so it is not possible to directly compare the specificities of the Nocardioides enzyme with that from A. aurescens TC1.
To our knowledge, A. aurescens TC1 is the first atrazine-degrading bacterium directly isolated from contaminated soil without enrichment. Active enrichment procedures have previously been shown to bias diversity studies and artificially select for fast-growing catabolic genotypes (10). Our results suggest that A. aurescens strain TC1 represents one of the numerically dominant and catabolically significant atrazine degraders in this contaminated soil. The soil was contaminated from a spill of 1,000 lb of atrazine and contained 29,000 mg of atrazine per kg of soil (30). While atrazine in this spill site soil was being biodegraded by microorganisms indigenous to the soil, atrazine biodegradation was inhibited by additions of C or N. This is consistent with the idea that organisms such as A. aurescens TC1, using atrazine as the sole C and N source, were important for indigenous metabolism at this site.
The observation that A. aurescens TC1 consumed 3,000 mg of atrazine per liter in liquid culture further demonstrated the potential of this organism to metabolize substantial quantities of s-triazine compounds in the environment. Moreover, the metabolic studies conducted here indicate that different s-triazine metabolism genes can be incorporated into different metabolic backgrounds to generate efficient metabolic pathways. While atrazine genes in Pseudomonas sp. strain ADP and other gram-negative bacteria are localized on a large plasmid, the localization of the genes in A. aurescens TC1 and other gram-positive organisms is currently unknown. Ongoing studies should reveal more about the evolution and distribution of the s-triazine catabolism genes in this and other gram-positive bacteria.
Acknowledgments
This work was supported, in part, by a grant from Syngenta Crop Protection.
We thank Carol Somody and Janis McFarland for providing s-triazine compounds and for helpful suggestions for improving the manuscript. We thank Hilary Major for assistance with the growth experiments. We thank Tom Krick and the Mass Spectrometry Consortium for the Life Sciences Center for help with MS analyses, Jennifer Seffernick for help with the HPLC protocols, and Adam Judd for helpful discussions.
REFERENCES
- 1.Aspelin, A. L., and A. H. Grube. 1999. Pesticides industry sales and usage 1996 and 1997 market estimates. Document 733-R-99-001. Office of Prevention, Pesticides and Toxic Substances, U.S. Environmental Protection Agency, Washington, D.C.
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, and D. Moore. 1999. Short protocols in molecular biology: a compendium of methods from Current Protocols in Molecular Biology, 4th ed. John Wiley & Sons, New York, N.Y.
- 3.Boundy-Mills, K. L., M. L. deSouza, R. T. Mandelbaum, L. P. Wackett, and M. J. Sadowsky. 1997. The atzB gene of Pseudomonas sp. strain ADP encodes hydroxyatrazine ethylaminohydrolase, the second step of a novel atrazine degradation pathway. Appl. Environ. Microbiol. 63:916-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouquard, C., J. Ouazzani, J. C. Prome, Y. Michel-Briand, and P. Plesiat. 1997. Dechlorination of atrazine by a Rhizobium sp. isolate. Appl. Environ. Microbiol. 63:862-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cerniglia, C. E., and J. J. Perry. 1975. Metabolism of N-propylamine, isopropylamine and 1,3-propane diamine by Mycobacterium convolutum. J. Bacteriol. 124:285-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cook, A. M. 1987. Biodegradation of s-triazine xenobiotics. FEMS Microbiol. Rev. 46:93-99. [Google Scholar]
- 7.Cook, A. M., and R. Hutter. 1981. s-Triazines as nitrogen sources for bacteria. J. Agric. Food Chem. 29:1135-1143. [Google Scholar]
- 8.de Souza, M. L., M. J. Sadowsky, and L. P. Wackett. 1996. Atrazine chlorohydrolase from Pseudomonas sp. strain ADP: gene sequence, enzyme purification and protein characterization. J. Bacteriol. 178:4894-4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Souza, M. L., J. Seffernick, B. Martinez, M. J. Sadowsky, and L. P. Wackett. 1998. The atrazine catabolism genes atzABC are widespread and highly conserved. Appl. Environ. Microbiol. 64:178-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunbar, J., S. White, and L. Forney. 1997. Genetic diversity through the looking glass: effect of enrichment bias. Appl. Environ. Microbiol. 63:1326-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eaton, R. W., and J. S. Karns. 1991. Cloning and comparison of the DNA encoding ammelide aminohydrolase and cyanuric acid amidohydrolase from three s-triazine-degrading bacterial strains. J. Bacteriol. 173:1363-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eaton, R. W., and J. S. Karns. 1991. Cloning and analysis of s-triazine catabolic genes from Pseudomonas sp. strain NRRLB-12227. J. Bacteriol. 173:1215-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eaton, R. W., and D. W. Ribbons. 1982. Metabolism of dibutylphthalate and phthalate by Micrococcus sp. strain 12B. J. Bacteriol. 151:48-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erickson, E. L., and K. H. Lee. 1989. Degradation of atrazine and related s-triazines. Crit. Rev. Environ. Control 19:1-13. [Google Scholar]
- 15.Koch, C., P. Schumann, and E. Stackebrandt. 1995. Reclassification of Micrococcus agilis (Ali-Cohen 1889) to the genus Arthrobacter as Arthrobacter agilis comb. nov. and emendation of the genus Arthrobacter. Int. J. Syst. Bacteriol. 45:837-839. [DOI] [PubMed] [Google Scholar]
- 16.Mandelbaum, R. T., D. L. Allan, and L. P. Wackett. 1995. Isolation and characterization of a Pseudomonas sp. that mineralizes the s-triazine herbicide atrazine. Appl. Environ. Microbiol. 61:1451-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez, B., J. Tomkins, L. P. Wackett, R. Wing, and M. J. Sadowsky. 2001. Complete nucleotide sequence and organization of the atrazine catabolic plasmid pADP-1 from Pseudomonas sp. strain ADP. J. Bacteriol. 183:5684-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mulbry, W. W., H. Zhu, S. M. Nour, and E. Topp. 2002. The triazine hydrolase gene trzN from Nocardioides sp. strain C190: cloning and construction of gene-specific primers. FEMS Microbiol. Lett. 206:75-79. [DOI] [PubMed] [Google Scholar]
- 19.Mullet, J. E., and C. J. Arntzen. 1981. Identification of a 32-34-kilodalton polypeptide as a herbicide receptor protein in photosystem II. Biochim. Biophys. Acta 635:236-248. [DOI] [PubMed] [Google Scholar]
- 20.Quirke, J. M. E. 1984. 1,3,5-Triazines, p. 459-529. In A. R. Katrizky and C. W. Rees (ed.), Comprehensive heterocyclic chemistry. Pergamon Press, New York, N.Y.
- 21.Radosevich, M., S. J. Traina, H. Yue-Li, and O. H. Tuovinen. 1995. Degradation and mineralization of atrazine by a soil bacterial isolate. Appl. Environ. Microbiol. 61:297-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rousseaux, S., A. Hartmann, and G. Soulas. 2001. Isolation and characterisation of new Gram-negative and Gram-positive atrazine degrading bacteria from different French soils. FEMS Microbiol. Ecol. 36:211-222. [DOI] [PubMed] [Google Scholar]
- 23.Sadowsky, M. J., Z. Tong, M. de Souza, and L. P. Wackett. 1998. AtzC is a new member of the amidohydrolase protein superfamily and is homologous to other atrazine-metabolizing enzymes. J. Bacteriol. 180:152-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seffernick, J. L., M. L. de Souza, M. J. Sadowsky, and L. P. Wackett. 2001. Melamine deaminase and atrazine chlorohydrolase: 98 percent identical but functionally different. J. Bacteriol. 183:2405-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seffernick, J. L., G. Johnson, M. J. Sadowsky, and L. P. Wackett. 2001. Substrate specificity of atrazine chlorohydrolase and atrazine-catabolizing bacteria. Appl. Environ. Microbiol. 66:4247-4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seffernick, J. L., and L. P. Wackett. 2001. Rapid evolution of bacterial catabolic enzymes: a case study with atrazine chlorohydrolase. Biochemistry 40:12747-12753. [DOI] [PubMed] [Google Scholar]
- 27.Selifonova, O., R. Burlage, and T. Barkay. 1993. Bioluminescent sensors for detection of bioavailable Hg(II) in the environment. Appl. Environ. Microbiol. 59:3083-3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shapir, N., J. P. Osborne, G. Johnson, M. J. Sadowsky, and L. P. Wackett. 2002. Purification, substrate range and metal center of AtzC: the N-isopropylammelide hydrolase involved in bacterial atrazine metabolism. J. Bacteriol. 184:5376-5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shelton, D. R., S. Khader, J. S. Karns, and B. M. Pogel. 1996. Metabolism of twelve herbicides by Streptomyces. Biodegradation 7:129-136. [DOI] [PubMed] [Google Scholar]
- 30.Strong, L. C., H. McTavish, M. J. Sadowsky, and L. P. Wackett. 2000. Field-scale remediation of atrazine-contaminated soil using recombinant Escherichia coli expressing atrazine chlorohydrolase. Environ. Microbiol. 2:91-98. [DOI] [PubMed] [Google Scholar]
- 31.Struthers, J. K., K. Jayachandran, and T. B. Moorman. 1998. Biodegradation of atrazine by Agrobacterium radiobacter J14a and use of this strain in bioremediation of contaminated soil. Appl. Environ. Microbiol. 64:3368-3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Topp, E., W. M. Mulbry, H. Zhu, S. M. Nour, and D. Cuppels. 2000. Characterization of s-triazine herbicide metabolism by a Nocardioides sp. isolated from agricultural soils. Appl. Environ. Microbiol. 66:3134-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Topp, E., H. Zhu, S. M. Nour, S. Houot, M. Lewis, and D. Cuppels. 2000. Characterization of an atrazine-degrading Pseudaminobacter sp. isolated from Canadian and French agricultural soils. Appl. Environ. Microbiol. 66:2773-2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vandermeer, J. 1981. Elementary mathematical ecology. Wiley & Sons, New York, N.Y.
- 35.Wackett, L. P., M. J. Sadowsky, B. Martinez, and N. Shapir. 2002. Biodegradation of atrazine and related triazine compounds: from enzymes to field studies. Appl. Microbiol. Biotechnol. 58:39-45. [DOI] [PubMed] [Google Scholar]
- 36.Wenk, M., T. Baumgartner, J. Dobovsek, T. Fuchs, J. Kucsera, J. Zopfi, and G. Stucki. 1998. Rapid atrazine mineralization in soil slurry and moist soil by inoculation of an atrazine-degrading Pseudomonas sp. strain. Appl. Environ. Microbiol. 49:624-630. [DOI] [PubMed] [Google Scholar]
- 37.Wilce, M. C., D. M. Dooley, H. C. Freeman, J. M. Guss, H. Matsunami, W. S. McIntire, C. E. Ruggiero, K. Tanizawa, and H. Yamaguchi. 1997. Crystal structures of the copper-containing amine oxidase from Arthrobacter globiformis in the holo and apo forms: implications for the biogenesis of topaquinone. Biochemistry 36:16116-16133. [DOI] [PubMed] [Google Scholar]
- 38.Yanze-Kontchou, C., and N. Gschwind. 1994. Mineralization of the herbicide atrazine as a carbon source by a Pseudomonas strain. Appl. Environ. Microbiol. 60:4297-4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng, D., E. W. Alm, D. A. Stahl, and L. Raskin. 1996. Characterization of universal small-subunit rRNA hybridization probes for quantitative molecular microbial ecology studies. Appl. Environ. Microbiol. 62:4504-4513. [DOI] [PMC free article] [PubMed] [Google Scholar]