Abstract
The influence of grazing by a mixed assemblage of soil protozoa (seven flagellates and one amoeba) on bacterial community structure was studied in soil microcosms amended with a particulate resource (sterile wheat roots) or a soluble resource (a solution of various organic compounds). Sterilized soil was reinoculated with mixed soil bacteria (obtained by filtering and dilution) or with bacteria and protozoa. Denaturing gradient gel electrophoresis (DGGE) of PCR amplifications of 16S rRNA gene fragments, as well as community level physiological profiling (Biolog plates), suggested that the mixed protozoan community had significant effects on the bacterial community structure. Excising and sequencing of bands from the DGGE gels indicated that high-G+C gram-positive bacteria closely related to Arthrobacter spp. were favored by grazing, whereas the excised bands that decreased in intensity were related to gram-negative bacteria. The percentages of intensity found in bands related to high G+C gram positives increased from 4.5 and 12.6% in the ungrazed microcosms amended with roots and nutrient solution, respectively, to 19.3 and 32.9% in the grazed microcosms. Protozoa reduced the average bacterial cell size in microcosms amended with nutrient solution but not in the treatment amended with roots. Hence, size-selective feeding may explain some but not all of the changes in bacterial community structure. Five different protozoan isolates (Acanthamoeba sp., two species of Cercomonas, Thaumatomonas sp., and Spumella sp.) had different effects on the bacterial communities. This suggests that the composition of protozoan communities is important for the effect of protozoan grazing on bacterial communities.
Bacterial communities are central to the functioning of terrestrial ecosystems and consist of a large number of different bacterial types (33). The bacterial populations are heavily grazed by the bacteriophagous microfauna. Grazing decreases bacterial numbers, stimulates mineralization of nutrients (13), and is likely to affect the structures of the bacterial communities. Such effects are poorly studied in soil systems, but a number of reports demonstrate that grazing by protozoa is an important factor in shaping the morphological and taxonomical compositions of bacterioplankton communities in marine and limnic ecosystems (26, 43, 55), as well as in activated sludge (21). Generally, feeding by flagellates and ciliates is size selective (26, 31, 32). Medium-size bacterial cells are most susceptible to predation by flagellates and ciliates, whereas smaller cells and large filamentous forms may be partly resistant to grazing (26, 31). Hence, grazing may lead to a bidirectional shift in the relative distribution of bacterial size classes, with increases in the relative abundance of large filaments and small cells (26). There is also evidence that other factors, such as cell surface properties (38), cell motility (4), and chemical composition (30, 59), may affect the susceptibility of bacteria to grazing. Investigations into the effects of grazing on planktonic communities have focused on changes in bacterial cell size and morphology, in part because these parameters can be determined by direct microscopy. Changes in these characteristics do not necessarily reflect changes in the taxonomic composition of the community, as individual bacterial strains may respond to increased grazing pressure by a change in morphology or cell size (24, 25). However, molecular-fingerprinting techniques, which allow more direct studies of the genetic structures of microbial communities, have now confirmed that grazing affects the taxonomic compositions of planktonic bacterial communities (55).
Many of the interactions between protozoan grazers and their prey are probably similar in aquatic and soil habitats, but there are some important differences. For example, the physical nature of soil means that diffusion, mixing, and movement of organisms is severely constrained compared to aquatic systems. In addition, soil protozoan activity is restricted to water films and water-filled pores, and small pores may protect bacteria from grazing (45, 48, 64). Free-living protozoa fall into three broad categories: filter feeders, raptorial feeders, and diffusion feeders (16). Boenigk and Arndt (2) suggested restriction of the term “raptorial feeding” to forms that actively search for food particles, whereas forms that produce a feeding current and intercept each food particle individually should be referred to as interception feeders. Although filter-feeding ciliates (16) and choanoflagellates (12, 14) are found in soils, the dominant feeding strategies among typical soil protozoa are raptorial feeding and, to some extent, interception feeding. The naked amoebae, and many of the common heterotrophic flagellates, are associated with surfaces and mainly feed on attached bacteria, as demonstrated for Rhyncomonas nasuta and species of the genus Bodo (7, 54). Small chrysophyte flagellates (Spumella spp.) that mainly feed on suspended bacteria by direct interception (2, 54) are also common in soils (14).
Due to the differences between the environments, the results from pelagic systems cannot be directly extrapolated to soil systems. Recently, however, a study using DNA fingerprinting (denaturing gradient gel electrophoresis [DGGE] analysis of PCR-amplified 16S rRNA gene sequences), community level physiological profiling (using Biolog plates), and phospholipid fatty acid analysis demonstrated that protozoan grazing changed the compositions of the microbial communities in soil microcosms (18). Phospholipid fatty acid analysis indicated that the proportion of gram-positive bacteria increased in response to protozoan grazing, but a more specific analysis of grazing effects on the bacterial community was not carried out.
Protozoa are ubiquitous and very abundant in virtually all natural soils (13), but the performance of controlled experiments requires a control treatment with different bacteria but without protozoa. Removal of protozoa from the soil also affects the bacterial community; hence, it is not possible to study the effect of protozoa on an undisturbed bacterial community. In this study, we obtained a soil with a diverse bacterial community, but without protozoa, by reinoculating sterilized soil with two types of bacterial suspensions from which protozoa had been removed by filtering and dilution, respectively.
The aims of this study were (i) to investigate the potential importance of protozoan grazing for bacterial community structure in soil, (ii) to test the hypothesis that protozoan grazing changes the size distribution of bacteria in soil, (iii) to assess whether different protozoa with different feeding ecologies have different effects, and (iv) to obtain more detailed information about how individual types of bacteria respond to grazing. We studied the effects of a mixture of eight protozoan isolates (representing common and abundant morphotypes) in soil microcosms amended with two different carbon substrates (sterilized wheat roots and a nutrient solution). These substrates were chosen to represent two contrasting situations in soil where bacterial and protozoan activity is high, namely, during decomposition of dead plant material and in the rhizosphere zone around growing plant roots. When dead plant material is added to soil, protozoan populations usually increase for 1 to 3 weeks (11, 49, 60) or, in some cases, up to 6 weeks (8). Furthermore, the initial number of protozoa in the inoculum was low, and we therefore sampled after 25 and 52 days to allow full colonization of the soil systems. The taxonomic structure of the bacterial community was assessed by DGGE of PCR-amplified 16S rRNA gene fragments, and the physiological potential of the community was examined with Biolog plates. The bacterial cell size distribution was evaluated by counting and measuring bacterial cells by direct epifluorescence microscopy. To gain information about which bacteria respond to grazing, selected prominent bands from DGGE gels were excised and sequenced. Finally, we compared the effects of five different protozoan isolates representing common and abundant morphotypes; we used one amoeba, three surface-feeding flagellates, and one flagellate that feeds mainly on bacteria in suspension.
MATERIALS AND METHODS
Soil.
A loamy sand (7.7% clay, 9.3% silt, 45.1% fine sand, 34.9% coarse sand, 1.6% organic C, 0.14% N) with a pH (in water) of 6.1 (27) was collected from an integrated arable farming system at the Research Centre Foulum in Jutland, Denmark. The soil was sieved, air dried to a water content of 10% (wt/wt), packed in plastic bags, and sterilized by electron irradiation (two consecutive doses of 15 kGy). Microcosms consisted of 10.0 g of this soil in autoclaved glass Universal bottles.
Isolation of protozoa and preparation of protozoan-free inoculum.
The protozoa were isolated from microtiter plates with dilution cultures (diluted in tryptic soy broth [TSB] to 0.3 g liter−1) prepared from soil samples and incubated at 15°C. The wells were inspected using an inverted microscope several times during a 3-week period, and protozoa were isolated from wells that were free from fungal growth and contained only one protozoan morphotype. Isolates were kept in sterile 50-ml Nunc culture flasks with 10 ml of dilute TSB medium (0.3 g liter−1) and inspected several times during a 4-week period. Eight isolates (one amoeba and seven flagellates) that appeared to be pure cultures were examined by conventional light microscopy and were assigned to the following species or morphotypes: Acanthamoeba sp., Bodo designis, Heteromita globosa, Cercomonas sp. morphotype 1, Cercomonas sp. morphotype 2, Thaumatomonas sp., Spumella sp., and an undescribed species (referred to as type A by Ekelund et al. [14]).
A mixed protozoan-free community of soil bacteria was obtained by homogenizing 60 g of soil with 200 ml of amoeba saline (40) for 2 min in a kitchen blender and separating bacteria from soil particles by flotation in Percoll (Pharmacia Fine Chemicals, Uppsala, Sweden) according to the method of Priemé et al. (46). The resulting suspension was filtered through Nuclepore filters (Whatman International Ltd., Maidstone, United Kingdom) with successively smaller pore diameters (5, 3, 2, and 1 μm) in order to eliminate protozoa. The filtered suspensions were kept in culture flasks until the start of the experiment and were inspected for protozoan contamination prior to inoculation. Some flasks contained small flagellates and were discarded. The filtered suspensions were supplemented with bacteria obtained from protozoan-free wells in the microtiter plates used for isolation of protozoa to minimize the number of bacterial types present in the protozoan cultures but absent in the control soils. To reduce this number further, protozoan cultures in microtiter plates were diluted again and bacteria and protozoa were isolated as described above. This procedure was repeated twice.
Experimental design.
The first experiment was set up with a 2-by-2 factorial design with two different carbon substrates, each with and without a mixture of eight isolates of soil protozoa. Half the microcosms received an initial amendment of 0.1 g of sterilized ground wheat roots (N content, 6.9 mg g−1; C/N ratio, ca. 60) mixed into the soil. The other half was amended with 50 μl of a nutrient solution (12.3 g of fructose liter−1, 12.3 g of glucose liter−1, 23.4 g of sucrose liter−1, 4.05 g of succinic acid liter−1, 3.95 g of malic acid liter−1, 3.0 g of arginine liter−1, 2.05 g of cysteine liter−1, 6.25 g of malt extract liter−1, 3.75 g of TSB liter−1) at intervals of 3 to 5 days during the first 6 weeks of the experiment. The simple sugars, organic acids, and amino acids in the nutrient solution are typical of those found in root exudates (19). A total of 0.88 mg of N and 18.6 mg of C was added during the experiment, corresponding to a loading rate of 39 μg of C g of soil−1 day−1, reflecting projected levels of carbon exudation in the rhizosphere of living plant roots (19). All microcosms were inoculated with 1.95 ml of a protozoan-free inoculum of mixed soil bacteria. Half the microcosms were additionally inoculated with 50 μl of a mixture of soil protozoa, while control microcosms received 50 μl of the bacterial inoculum. After incubation at 15°C for 25 and 52 days, three replicate microcosms from each of the four treatments were destructively sampled and analyzed as described below.
The second experiment was designed to compare the effects of five individual protozoan isolates (Acanthamoeba sp., Cercomonas sp. morphotype 1, Cercomonas sp. morphotype 2, Thaumatomonas sp., and Spumella sp.) with the effect of the mixed community used in the first experiment. The microcosms were established as described above for those amended with nutrient solution, with sampling after 52 days.
Extraction and purification of DNA from soil samples.
DNA was extracted from 0.5-g soil samples using phenol and disruption by a Hybaid Ribolyser Cell Disrupter as described by Webster et al. (61). After centrifugation at 10,000 × g for 5 min, the supernatant was removed and mixed with an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1). The suspension was centrifuged, and the supernatant was removed and mixed with an equal volume of chloroform-isoamyl alcohol (24:1). After centrifugation, the DNA present in the aqueous supernatant was purified by gel electrophoresis through a 1% (wt/vol) low-melting-point agarose gel. A band containing DNA was excised, and the agarose was digested with agarase (Roche Diagnostics Ltd., Lewes, United Kingdom) using the manufacturer's protocol.
PCR amplification.
PCR amplification of bacterial DNA was performed using primers p3 and p2 (39), which amplify a 194-bp fragment of the 16S rRNA gene, including the variable V3 region, and incorporating a 40-bp GC clamp at the 5′ end of p3. PCR amplification was carried out in two stages, with bovine serum albumin omitted in the second step, to reduce background during silver staining. In the first stage, amplification was performed in 25-μl reaction mixtures containing ∼10 ng of soil DNA, 0.5 U of Taq polymerase (Promega UK Ltd., Southampton, United Kingdom), each deoxynucleoside triphosphate at a concentration of 50 μM, each primer at a concentration of 0.08 μM, 1.5 mM MgCl2, appropriate dilutions of the manufacturer's buffer, and 0.5 μl of a bovine serum albumin solution (10 mg ml−1; Roche Diagnostics Ltd.). The cycling parameters were 95°C for 5 min, followed by 10 cycles of 94°C for 30 s, 55°C for 30 s and 72°C for 30 s. In the second stage, 1 μl of the reaction mixture from the first stage was used as a template, and amplification was performed in 50-μl reaction mixtures as described by McCaig et al. (37) with the exception that only 25 cycles were applied. The amplification products were visualized on ethidium bromide-stained agarose gels.
DGGE.
DGGE analysis was carried out as described by McCaig et al. (37) using a 40 to 60% vertical denaturing gradient. Approximately 50 ng of each PCR product was loaded, and the gels were electrophoresed for 16 h at 75 V and 60°C. The gels were either stained in ethidium bromide (10 μg ml−1) for 15 min (for excising bands) or silver stained and fixed (37) for image analysis using Phoretix 1D gel analysis software version 4.00 (Phoretix International, Newcastle upon Tyne, United Kingdom) as described previously (37). To correct for variations in DNA loading between lanes, the band intensity in each lane was normalized to that of the lane with the lowest DNA load (37). The data for the relative intensities of the individual bands were analyzed by correspondence analysis (CANOCO for Windows version 4.0) on square-root-transformed values and with downweighting of rare species. Ordination diagrams were generated with scaling focusing on intersample distances. Data from the first experiment were analyzed using detrended correspondence analysis to remove an arch effect (29).
Excising of bands, cloning, and sequencing.
Selected bands were excised from the gels, stained with ethidium bromide, and placed in sterilized Eppendorf vials with 10 μl of sterilized distilled water. The gel slices were crushed, and the vials were left at 5°C overnight to allow diffusion of DNA from the gel slices. One microliter of the eluted DNA was used as a template in a PCR as described above. The resulting PCR product was rerun on a gel alongside the original sample to confirm the band position. Bands were excised for a second time, eluted, and reamplified as before. Where the second DGGE resulted in further resolution of the original band into two bands, both bands were excised. The amplified products were then sequenced with p1 and p2 (39), and the sequences were compared to the EMBL database using FastA searches.
Biolog.
Profiling of the physiological potential of the microbial community using Biolog (Biolog, Hayward, Calif.) was done at the sampling after 52 days for all four treatments of experiment 1 and for the treatment with Acanthamoeba and Cercomonas sp. morphotype 1 of experiment 2. Soil samples (2.5 g) were shaken with 20 ml of sterile amoeba saline for 15 min on a Stuart flask shaker. The resulting soil suspension was further diluted in sterile amoeba saline to give an absorbance of 0.4 at a wavelength of 595 nm, and 150 μl was inoculated into each well of a Biolog GN1 plate. The plates were incubated at 15°C, and the absorbance of each well at 595 nm was measured with an automatic plate reader after 3, 4, and 5 days. Wells with an optical density >1.4-fold greater than the control well were considered positive. The time course profiles of the Biolog data were analyzed from the area under the color development profile (23). The 95 areas (i.e., one for each substrate) were analyzed using principal-component analysis with GENSTAT version 5 release 3.2 (41).
Enumeration of bacteria and protozoa and measurement of inorganic nitrogen.
Soil samples (2.5 g) were shaken with 30 ml of sterile amoeba saline for 15 min on a Stuart flask shaker. Appropriate dilutions of this suspension were spread in triplicate on agar plates (0.3 g of TSB and 15 g of agar liter of amoeba saline−1). The plates were incubated at 14°C, and bacterial colonies were enumerated after 2, 4, 7, 9, 13, 17, 22, and 31 days. Protozoa were enumerated with a modified version of the most-probable-number method (10, 47). Threefold dilution series were prepared in microtiter plates (8 by 12 wells; Costar 3598; Biotech Line ag., Slangerup, Denmark) containing TSB (0.3 g · liter−1). The plates were incubated at 14°C, and individual wells were inspected for the presence or absence of protozoa using an inverted microscope (×200 magnification) after 1 and 4 weeks. A 4-ml sample of the soil suspension was fixed with formaldehyde (final concentration, 4%), stained with acridine orange (final concentration, 0.1 mg ml−1), and filtered through 0.2-μm-pore-size polycarbonate membrane filters (Nuclepore). The bacteria on the filters were enumerated by fluorescence microscopy at ×1,250 magnification (49) and scored into size classes using a Porton graticule (36). This graticule contains 11 globes increasing in size with a √2 progression so that, at the given magnification, the size group limits are 0.19, 0.27, 0.38, 0.55, 0.77, 1.1, 1.5, 2.2, 3.1, 4.4, and 6.2 μm. Inorganic nitrogen was extracted and measured as described by Webster et al. (61).
Calculation of diversity indices.
DGGE banding data were used to estimate diversity indices. The Shannon diversity index (H′) was calculated from the equation H′= −Σpi · ln pi, where pi is the proportion of the total intensity of the ith band (35). Evenness (E) was calculated as E = H′/Hmax = H′/ln S (35), where S is the number of bands. The evenness varies between 0 and 1 and gives a measure of the ratio of the observed diversity (H′) to the maximum diversity (Hmax = ln S). Simpson's index of dominance (D = Σpi2) estimates the probability that any two individuals drawn from a community belong to different species. It is often expressed as 1/D so that an increase in the index reflects an increase in diversity (35). The Berger-Parker index (d) measures dominance as the proportion of the most abundant species, hence, d = Nmax/N, where Nmax represents the intensity of the most intense band.
An index based on the morphologies of the bacterial colonies appearing on the agar plates was calculated at the final counting. This index, the sequential-sampling index, does not require an absolute identification of colony morphology types. Colonies were inspected in a sequence, and decisions were made as to whether each new colony was different from the last one inspected. The index is calculated as the number of times an observed individual is different from the previous one observed divided by the total number of individuals (63). It has a range between 0 and 1, with a value of 1 representing the situation where each new individual observed is different from the last one observed.
Statistical analyses were performed using SigmaStat for Windows (version 2.03 package).
Nucleotide sequence accession numbers.
The sequences in this study were submitted to the EMBL database and assigned accession numbers AJ488008 through AJ488032.
RESULTS
Growth of protozoa.
The initial number of protozoa was low (<100 g of soil−1; data not shown) for all inoculated treatments. After 25 days, the numbers had increased to high levels in the treatments with protozoa whereas no protozoa were detected in control treatments (Fig. 1a and 2a). All eight of the inoculated protozoan types were observed in all microtiter plates prepared from the protozoan treatments of experiment 1 on both sampling dates, indicating that all of the morphotypes grew in the microcosms. While the most-probable-number method allows estimation of total protozoan numbers, it is not suitable for precise enumeration of individual species in a community (13, 14, 47). However, there was a clear trend at both samplings, with the small flagellates (Cercomonas spp., Bodo, Heteromita, Spumella, and type A) occurring in higher dilutions than the amoeba and Thaumatomonas, indicating that the small flagellates numerically dominated the protozoan community.
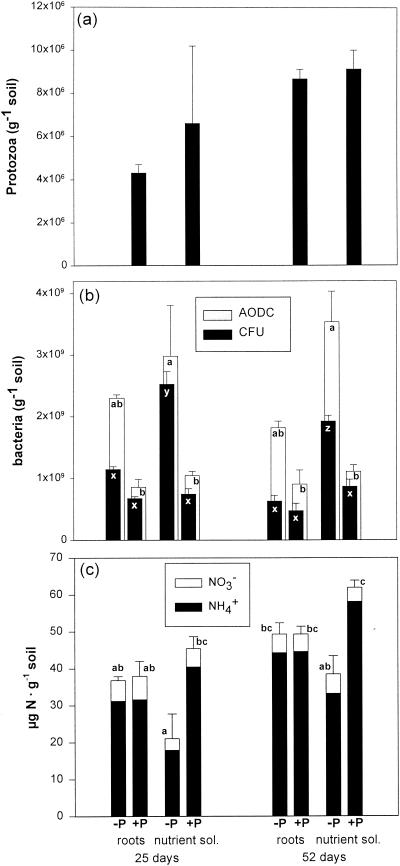
FIG. 1.
Numbers of protozoa (a) and bacteria (b) and amount of inorganic nitrogen (c) in microcosms amended either with sterilized wheat roots or with a nutrient solution (sol.) and with (+P) and without (−P) a mixed protozoan community. The number of bacteria was estimated both by direct epifluorescence microscopy (AODC) and by plate counts (CFU). The error bars represent 1 standard error. Values marked with different letters are significantly different (P < 0.05; Tukey). For the bacterial counts, the letters a and b refer only to AODC values and the letters x, y, and z refer only to CFU values. For inorganic nitrogen (panel c), the error bars and letters (a, b, and c) refer to values for total inorganic nitrogen (NH4+ + NO3−).
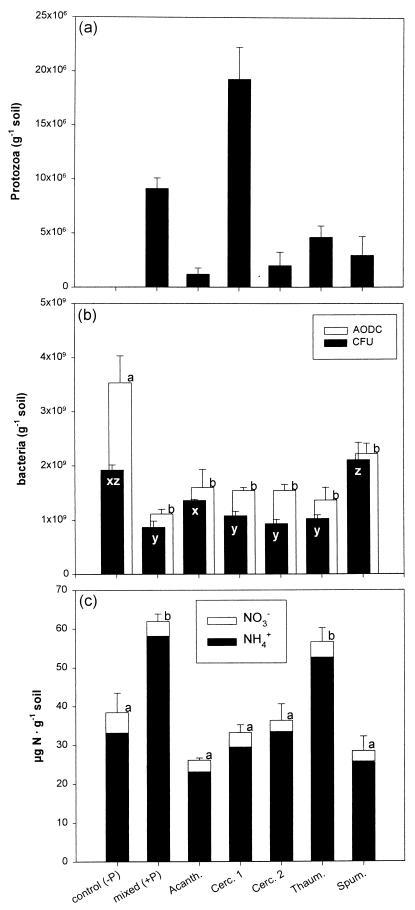
FIG. 2.
Numbers of protozoa (a) and bacteria (b) and amount of inorganic nitrogen (c) in microcosms without protozoa (control), with a mixed community of eight isolates of soil protozoa (mixed), or with only one protozoan isolate (Acanth., Acanthamoeba; Cerc. 1 and 2, Cercomonas sp. morphotypes 1 and 2; Thaum., Thaumatomonas sp.; Spum., Spumella sp.). The data refer to the sampling after 52 days. For further details, see the legend to Fig. 1.
Bacterial abundance, bacterial cell size, and mineralization of nitrogen.
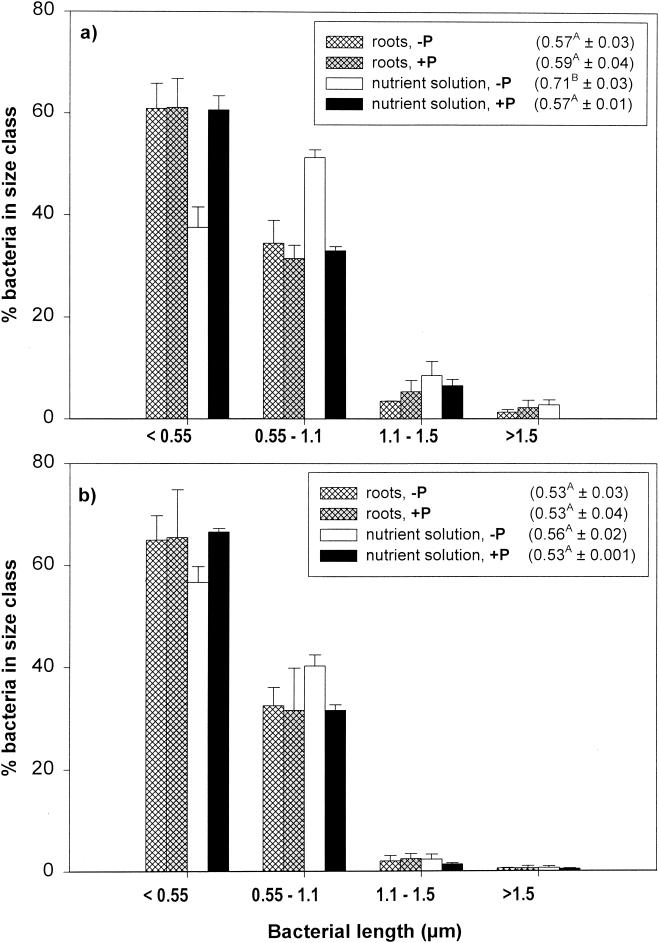
Immediately after inoculation, the numbers of viable bacteria (CFU) were low in all treatments (1.2 × 107 ± 0.2 × 107 [standard error] g of soil−1), but it increased >60-fold within 25 days. The presence of protozoa reduced the number of CFU significantly in the treatment amended with a nutrient solution, and total bacterial counts (acridine orange direct counts [AODC]) were reduced significantly for both substrate types (Fig. 1b). Each of the five single protozoan isolates reduced total bacterial numbers and CFU, with the exception of Spumella sp. (Fig. 2b). In both experiments, protozoa increased the ratio of viable to total direct counts (Fig. 1b and 2b), but this difference was not statistically significant (P = 0.400). The mixed protozoan community increased the amount of inorganic nitrogen in microcosms amended with nutrient solution but not in those amended with wheat roots (Fig. 1c). Only one of the protozoan isolates (Thaumatomonas sp.) had a significant effect on the amount of extractable nitrogen (Fig. 2c). Bacteria shorter than 1.1 μm dominated in the total bacterial counts (Fig. 3). Grazing significantly reduced the number of bacteria in all four bacterial size classes. In microcosms amended with root material, protozoa did not significantly affect the average bacterial cell size or the frequency distribution of bacteria (Fig. 3). In the amendments with nutrient solution, however, there was a significant decrease in the proportion of cells between 0.55 and 1.1 μm long (Fig. 3). In experiment 2, grazing had no significant effect on bacterial cell size (data not shown).
FIG. 3.
Frequency distribution of bacterial size classes in experiment 1 after 25 (a) and 52 days (b). The mean bacterial cell length (± standard error) for each treatment is given in parentheses. Values followed by different letters are significantly different (P < 0.05; Tukey). +P and −P, with and without a mixed protozoan community.
DGGE.
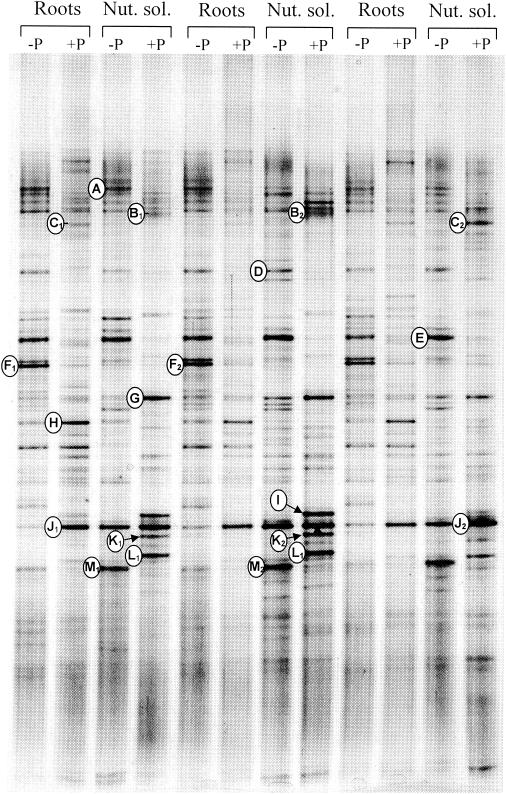
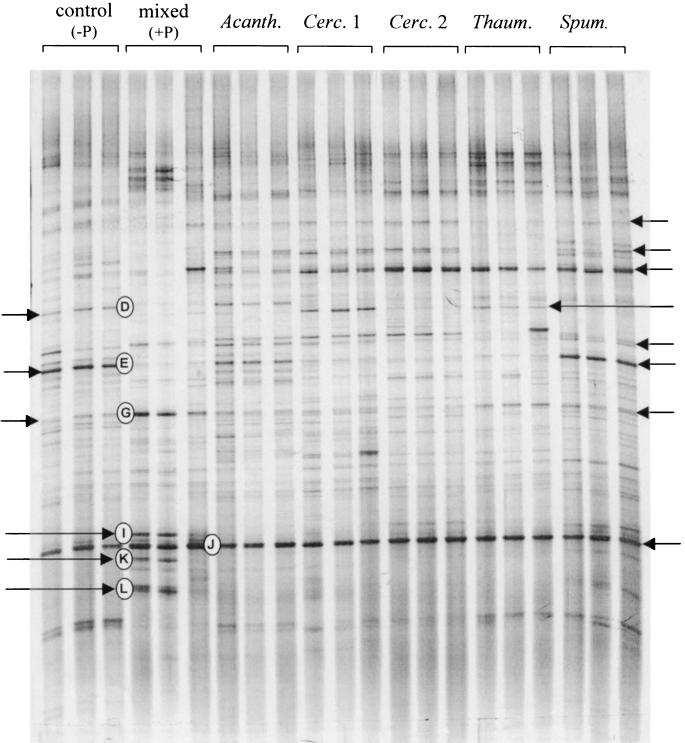
In experiment 1, the three replicates from each treatment showed good reproducibility, and differences among the four treatments were clearly discernible by visual comparison of lanes at samplings after 25 days (data not shown) and after 52 days (Fig. 4). A plot of the detrended correspondence analysis likewise showed that the analysis separated the four treatments along the first axis (data not shown). Coordinates on the first axis (mean ± standard error) for ungrazed and grazed microcosms were 0.05 ± 0.03 and 0.80 ± 0.04, respectively, for the root treatment and 0.57 ± 0.09 and 1.80 ± 0.18, respectively, for the nutrient solution treatment. The presence of protozoa decreased the intensities of some bands (e.g., bands A, D, E, F, and M) and increased the intensities of others (e.g., bands H, J, K, and L) (Fig. 4 and Table 1). Likewise, some bands (e.g., F and H) were more prominent in microcosms amended with wheat root material, while others (e.g., G, L, and M) showed higher intensities in the nutrient solution treatments. Sequence analysis of the excised bands showed that most of the bands that increased in intensity contained DNA closely related to high-G+C gram-positive bacteria (bands G, I, J, K, and L [Table 1]), in particular Arthrobacter. Bands that decreased in intensity during grazing showed affinity to gram-negative proteobacteria belonging to the β, γ, and δ subdivisions or the Cytophaga-Flavobacterium group. However, bands C and H, which were related to gram-negative bacteria of the genera Pseudomonas and Achromobacter, tended to increase in intensity during grazing. The sequenced bands accounted for 33 to 43% of the total intensity in the lanes. The percentage of intensity found in bands related to high-G+C gram-positive bacteria was significantly higher in the nutrient solution treatment than in the root treatments and increased significantly in response to grazing (Table 2), indicating that these bacteria had a selective advantage in the presence of protozoa.
FIG. 4.
DGGE analysis of 16S rRNA gene fragments amplified from DNA extracted from three replicate microcosms of the four treatments of experiment 1 using eubacterial primers. Sterile soil microcosms were amended with either sterile wheat roots (Roots) or repeated additions of a liquid nutrient solution (Nut. sol.) containing various carbohydrates, organic acids, and amino acids (see the text) and inoculated with a mixture of soil bacteria (−P) or with bacteria and a mixture of eight soil protozoan isolates (+P). Bands marked with letters to the left of the lane correspond to bands that were excised from an ethidium bromide-stained gel with the same amplified DNA.
TABLE 1.
Similarities of sequences obtained from excised DGGE bands (see Fig. 4) to sequences in the EMBL database, Gram status and phylogenetic position of the OTU, and response of the band intensity to presence of protozoa in microcosms amended with wheat root material or nutrient solution
| Band | Sequencea | Accession no. | Nearest relativeb | Accession no.c | Similarity (%) | Phylogenetic group | Gram status | Response to grazingd
|
|
|---|---|---|---|---|---|---|---|---|---|
| Roots | Nut. sol. | ||||||||
| A | 1 | AJ488008 | Uncultured bacterium clone P3 (1) | AF414577 | 98.7 | CFB groupe | G− | − | − |
| 2 | AJ488009 | Uncultured bacterium clone P3 (1) | AF414577 | 98.1 | |||||
| B1 | 1 | AJ488010 | Flavobacterium (1) | AF375835 | 98.7 | CFB group | |||
| 2 | AJ488011 | Uncultured Bacteriodaceae bacterium (1) | AJ318151 | 98.7 | G− | ND | ND | ||
| B2 | AJ488012 | Flavobacterium (1); unidentified (1) | AY039834 | 100 | CFB group | ||||
| C1 | AJ488013 | Pseudomonas (7) | AF105387 | 98.7 | γ Proteobacteria | G− | (+) | ND | |
| C2 | AJ488014 | Pseudomonas (7) | AF105387 | 100 | γ Proteobacteria | ||||
| D | AJ488015 | Bdellovibrio (1) | AF084853 | 98.8 | δ Proteobacteria | G− | − | − | |
| E | AJ488016 | Buttiauxella (3); Enterobacter (4); Hafnia (1); Obesumbacterium (1) | AJ233406 | 100 | γ Proteobacteria | G− | − | − | |
| F1 | AJ488017 | Bdellovibrio bacteriovorous (15); Bdellovibrio (6) | AF084850 | 100 | δ Proteobacteria | G− | − | − | |
| F2 | AJ488018 | B. bacteriovorous (15); Bdellovibrio (6) | AF084850 | 100 | δ Proteobacteria | ||||
| G | AJ488019 | Leifsonia poae (1); Agrococcus jenensis (1); unidentified (1); uncultured (1) | AF116342 | 96.7 | High G + C gram positive | G+ | ND | + | |
| H | AJ488020 | Achromobacter xylosoxidans (1); unidentified (1); uncultured (1) | AF225979 | 98.0 | β Proteobacteria | G− | + | ND | |
| I | AJ488021 | Arthrobacter (27); Methylomicrobium (1); uncultured (4); unidentified (2) | X83408 | 100 | High G + C gram positive | G+ | ND | + | |
| J1 | AJ488022 | Arthrobacter rhombi (1) | Y15884 | 99.3 | High G + C gram positive | G+ | + | no | |
| J2 | AJ488023 | Arthrobacter (13); Brachybacterium (8); Brevibacterium (1); unidentified (1) | M23411 | 100 | High G + C gram positive | ||||
| K1 | AJ488024 | Arthrobacter (13); Brachybacterium (8); Brevibacterium (1); unidentified (1) | M23411 | 100 | High G + C gram positive | G+ | ND | + | |
| K2 | 1 | AJ488025 | Arthrobacter (13); Brachybacterium (8); Brevibacterium (1); unidentified (1) | M23411 | 100 | High G + C gram positive | |||
| 2 | AJ488026 | Arthrobacter psychrolactophilus (1) | AF134181 | 96.7 | |||||
| L1 | 1 | AJ488027 | Arthrobacter (13); Brachybacterium (2); Brevibacterium (1); unidentified (1) | M23411 | 98.6 | High G + C gram positive | G+ | ND | + |
| 2 | AJ488028 | A. psychrolactophilus (1) | AF134181 | 95.9 | |||||
| L2 | 1 | AJ488029 | Arthrobacter (13); Brachybacterium (8); Brevibacterium (1); unidentified (1) | M23411 | 100 | High G + C gram positive | |||
| 2 | AJ488030 | A. psychrolactophilus (1) | AF134181 | 96.6 | |||||
| M1 | AJ488031 | Alcaligenes (1) | X92415 | 97.5 | β Proteobacteria | G− | ND | − | |
| M2 | AJ488032 | Alcaligenes (1) | X92415 | 96.2 | β Proteobacteria | ||||
From some bands, two separate bands were obtained after rerunning the excised band. In these cases, sequences from both bands are included.
Number of hits to a species or genus with the indicated similarity is given in parentheses.
Accession number of the nearest relative. When more than one sequence had the same similarity, only the accession number of the first sequence is given.
Effect of the presence of protozoa on the intensity of the band in microcosms amended with the two types of substrate. The effect was evaluated by two-way analysis of variance on the percent intensity of the bands. +, significant positive effect of grazing (P < 0.01); −, significant negative effect of grazing; (+), discernible but nonsignificant positive effect; no, no effect of grazing; ND, not detectable due to very low or variable band intensity. Nut. sol., nutrient solution.
CFB, Cytophaga-Flavobacterium.
TABLE 2.
Relative intensities of sequenced bandsa
| Band | Relative intensity (% of total intensity in lane)
|
|||
|---|---|---|---|---|
| Roots
|
Nutrient solution
|
|||
| No protozoa (−P) | Mixed protozoa (+P) | No protozoa (−P) | Mixed protozoa (+P) | |
| High G + C gram positive | 4.5 ± 0.41A | 19.3 ± 0.96B | 12.6 ± 1.2C | 32.9 ± 1.7D |
| Gram negative | 25.7 ± 2.3A | 9.4 ± 0.71B | 26.6 ± 1.2A | 11.1 ± 2.9B |
| Sum of sequenced bands | 30.3 ± 1.9AB | 28.6 ± 1.5A | 39.2 ± 1.2BC | 44.0 ± 3.0C |
Related to high G + C gram-positive bacteria and to gram-negative bacteria and the sum of all the sequenced bands (means ± standard errors). Values in a row are significantly different (P < 0.05; Tukey) if they are followed by different letters.
Each of the individual protozoan isolates caused clearly discernible differences in the banding pattern compared to the control (Fig. 5). Bands were not excised from the DGGE gels from this experiment, but some of the bands could be tentatively identified by comparison with the banding patterns obtained in the experiment with mixed protozoa. Increases in intensity of bands related to high-G+C gram-positive bacteria in the presence of the individual protozoa were less than those following inoculation with the mixed community (bands G, I, K, and L).
FIG. 5.
DGGE analysis of 16S rRNA gene sequences amplified from DNA extracted from three replicate microcosms amended with nutrient solution and inoculated with bacteria only (control), bacteria and a mixture of eight soil protozoan isolates (mixed), or bacteria and one of five individual protozoan isolates (Acanth., Acanthamoeba sp.; Cerc. 1 and 2, Cercomonas sp. morphotypes 1 and 2; Thaum., Thaumatomonas sp.; Spum., Spumella sp.). The bands marked with letters are presumed to be similar to the excised bands marked in Fig. 4. The bands marked with arrows are bands that respond differently in the different treatments.
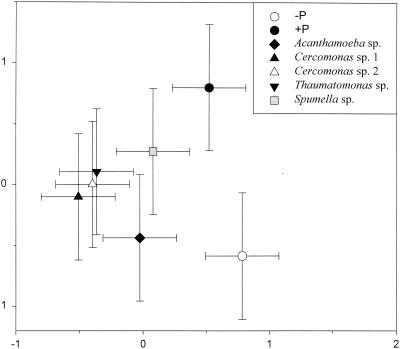
Correspondence analysis of the overall banding patterns (Fig. 6) separated treatments with protozoa from the ungrazed treatment, and the mixed community was separated from all treatments with a single protozoan. Treatments with Acanthamoeba and Spumella were separated from the other treatments, whereas the two Cercomonas isolates and Thaumatomonas were not clearly separated from each other.
FIG. 6.
Correspondence analysis plot of the DGGE banding patterns obtained from soil microcosms inoculated with bacteria alone (−P), with mixed protozoa (+P), or with one of the five individual protozoan isolates (see the legend to Fig. 5). The error bars represent the least significant difference (Tukey); hence, the coordinates are significantly different (P < 0.05) when the error bars do not overlap the mean from another treatment.
Biolog.
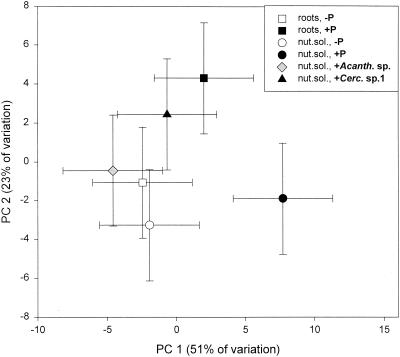
The presence of protozoa correlated with utilization of α-lactose, while the utilization of the other substrates varied according to protozoa and environment (Table 3). Principal-component analysis based on rates of substrate utilization could not distinguish the microbial communities in the two treatments without protozoa (Fig. 7). However, the presence of a mixed protozoan inoculum resulted in a significantly different pattern, and likewise, the patterns of utilization in the treatments with mixed protozoa were significantly different for the two substrate types. The patterns of utilization were also significantly different in the nutrient treatments amended with mixed protozoa, Acanthamoeba, and Cercomonas.
TABLE 3.
Substrates from Biolog GN plates utilized differentially by the four treatments of experiment 1 and the treatments with Acanthamoeba and Cercomonas sp. morphotype 1 of experiment 2
| Substrate | Utilizationa
|
|||||
|---|---|---|---|---|---|---|
| Roots
|
Nutrient solution
|
|||||
| No protozoa | Mixed protozoa | No protozoa | Mixed protozoa | Acanthamoeba | Cercomonas sp. morphotype 1 | |
| Dextrin | + | + | + | + | − | + |
| α-Lactose | + | − | + | − | − | − |
| Hydroxy l-proline | + | + | + | − | + | + |
| l-Leucine | + | + | + | − | + | + |
| Uridine | + | − | + | − | + | − |
| Putrescine | + | + | + | − | + | + |
| Glycerol | + | + | + | − | + | − |
+, utilized; −, not utilized.
FIG. 7.
Principal-component (PC) plot of the community level physiological profile of soil microcosms amended with sterile roots and with (+P) or without (−P) the presence of a mixed protozoan community and soil microcosms amended with a nutrient solution (nut. sol.) without protozoa, with mixed protozoa, or with either Acanthamoeba (+Acanth. sp.) or Cercomonas sp. morphotype 1 (+Cerc. sp.1). The error bars represent the least significant difference (see the legend to Fig. 6).
Diversity indices.
Diversity indices calculated using band intensities of the DGGE profiles indicated that the genetic diversity of the bacterial communities was lower for the nutrient solution treatment than for the root treatment (Table 4). The sequential-sampling index likewise indicated a lower diversity of colony morphology types in nutrient solution treatments. Grazing also affected diversity negatively, most noticeably in the nutrient solution treatment.
TABLE 4.
Diversity indices based on DGGE data, colony morphology, and Biolog data for the four treatments of experiment 1
| Analysis | Diversity indexa | Value
|
ANOVAb
|
|||||
|---|---|---|---|---|---|---|---|---|
| Roots
|
Nutrient solution
|
|||||||
| No protozoa | Mixed protozoa | No protozoa | Mixed protozoa | Subst. | Prot. | Int. | ||
| DGGE | No. of bands (S) | 46.7 | 45.3 | 43.7 | 40.3 | NS | NS | NS |
| Shannon (H) | 1.51 | 1.49 | 1.42 | 1.36 | *** | * | NS | |
| Evenness (E) | 0.91 | 0.90 | 0.87 | 0.85 | *** | NS | NS | |
| Simpson (1/D) | 25.8 | 21.4 | 18.0 | 15.1 | *** | ** | NS | |
| Berger-Parker (1/d) | 14.6 | 7.5 | 8.1 | 6.0 | * | * | NS | |
| CFU | Sequential-sampling index | 0.83 | 0.77 | 0.70 | 0.64 | *** | * | NS |
| Biolog | No. of positive wells | 69.7 | 68.3 | 68.0 | 55.3 | ** | *** | * |
For formulas for calculation of indices, see the text.
Significance levels (two-way analysis of variance) for effects of substrate (Subst.), presence of protozoa (Prot.), and interaction between these factors (Int.) (***, P < 0.001; **, P < 0.01; *, P < 0.05; NS, not significant).
DISCUSSION
The bacterial community structure in our experiments was determined both by the species compositions in our inocula and the environmental conditions during the incubation. Organic soil carbon from killed microbial biomass and added nutrients probably favored bacteria adapted to high substrate availability. Nevertheless, the complex banding patterns obtained in the DGGE profiles suggest that a diverse bacterial community was successfully established in the soil. The finding of sequences affiliated with several major phylogenetic groups in the excised bands further supports this.
For analysis of the DGGE profiles, it was assumed that each band represented 1 bacterial operational taxonomic unit (OTU) and that the intensity of the band reflected the relative abundance of this OTU in the community. However, factors such as differences in DNA extraction efficiency, primer affinity (52), amplification efficiencies of different sequences (44), and differences in the numbers of ribosomal DNA copies per cell (15) may introduce bias. Consequently, DGGE profiles do not necessarily reflect the structures of the actual bacterial communities. Furthermore, as DGGE separates sequences only on the basis of migration distance, each band may represent different sequences which comigrate to the same position in the gel. However, mixed sequences were found only rarely in excised bands and were closely related, belonging to the same phylogenetic group, while consistency in treatment and analysis of samples will reduce differential biases. The clear difference in the banding patterns of DGGE profiles for the two different substrates and between grazed and ungrazed systems indicates that the bacterial community structure was significantly affected both by bottom-up effects (substrate quality) and top-down effects (predation). Similarly, the Biolog data indicate that grazing affected the physiological profiles of the communities for both substrate types. The assay suggested a decreased physiological potential of the grazed communities, particularly in the nutrient solution treatment, where the number of positive wells was significantly lower in the presence of protozoa. Similarly, the diversity indices based on the DGGE banding pattern and on colony morphology both suggested that grazing decreased bacterial diversity (Table 4). The decreased diversity is mainly due to a decrease in evenness, presumably caused by the dominance of a few high-G+C gram-positive bacteria related to Arthrobacter. The larger proportion of high-G+C gram-positive bacteria in the presence of protozoa is consistent with previous reports (18).
Several mechanisms may contribute to the effect that grazing has on the bacterial community structure. Generally, size-selective predation has an important influence on the composition of planktonic bacterial communities (26, 31, 42, 43, 55), and we found evidence for preferential feeding on cells larger than 0.5 μm in the nutrient solution amendments. This is in accordance with the previous finding that small cells may escape predation (26). For example, a community of small flagellates dominated by Spumella guttula showed a higher rate of uptake of bacteria between 0.8 and 1.5 μm in length than of smaller or larger bacteria (34). Cells larger than 0.5 μm probably have higher growth rates than smaller cells (53), and hence, the importance of size selectivity could be underestimated by simply comparing the distributions of cell sizes. However, grazing did not affect bacterial cell size following amendment with roots, and the abundances of all bacterial size classes decreased significantly in the presence of grazers in all treatments of both experiments. Furthermore, the effect of grazing on bacterial size distribution was most evident at the first sampling and rather low at the last sampling, when the effect of grazing on DGGE and community level physiological profiles was estimated. Therefore, preferential feeding on the larger cells is probably not the most important factor for the observed changes in the taxonomic compositions of the bacterial communities. Similarly, we found no evidence for increased abundance of long grazing-resistant filamentous bacteria, as observed in plankton communities grazed by flagellates (25). Gram-positive bacteria may be less suitable protozoan food than gram-negative bacteria (1, 62), and some, e.g., the rod-shaped high-G+C gram-positive bacterium Mycobacterium chlorophenolicum, appear to be completely unaffected by grazing by indigenous soil protozoa (50). The lower edibility of gram-positive bacteria may be related to a lower rate of digestion of the gram-positive cell wall (17, 28), which may enable survival during passage through the protozoan cell (28). The flagellates, Ochromonas and Spumella, egested indigestible particles 2 to 3 min after ingestion (4, 5). Rapid excretion of unsuitable bacteria provides a very plausible explanation for active food selectivity among protozoa (5). Hence, the increased proportion of high-G+C gram-positive bacteria affiliated with Arthrobacter could be due to a low protozoan preference for these cells. Many gram-negative bacteria are good food sources for protozoa, and nonpigmented Enterobacteriaceae produce high protozoan growth yields (47, 62). If protozoa show a preference for these cells, this may explain the decrease in band E in grazed microcosms, as this band showed close affiliation with species of the Enterobacteriaceae. The intensities of two bands (D and F) related to Bdellovibrio spp., bacterial predators of gram-negative bacteria, decreased in the presence of protozoa, possibly through reductions in the abundance of their host bacteria. The direct predation pressure on the free-living stages of Bdellovibrio is probably low, since they are highly motile (6).
Although Gram status may relate to the edibility of bacteria, it is by no means an absolute factor, as many gram-positive bacteria are edible (58, 62) and many Gram-negative bacteria are completely unsuitable as food for protozoa. For example, representatives of both groups produce substances that are toxic to protozoa (9, 20), and several other factors that affect the grazing resistance of bacteria (e.g., size, cell morphology, and motility) are not related to Gram status. Furthermore, the bacterial community structure may be affected by grazing even if protozoan feeding is not selective. For example, grazing may favor bacteria with high growth rates because they will be able to replace cells lost to predation (22, 57). Grazing will also affect bacterial community structure indirectly as bacterial numbers decrease and nutrients are mineralized; hence, the competition for substrates and limiting nutrients is reduced. In this study, grazing reduced bacterial numbers for all treatments, and inorganic nitrogen increased in the nutrient solution treatment in the presence of the mixed protozoan community and Thaumatomonas. Nitrogen, however, was probably never a strong limiting factor for microbial growth, since nitrate was always present. Nitrifying bacteria are sensitive and very slow colonizers, so when sterilized soils are reinoculated, the nitrifying potential of the soil is reestablished only very slowly (51). Protozoan grazing may also affect the interaction between bacteria and other organisms in the soil. For example, protozoa were found to stimulate viral activity in freshwater systems (56). The mechanism behind this interaction is presently unknown but could be related to reduced bacterial diversity in grazing-enhanced treatments or to a change in the properties of individual bacterial cells caused by higher bacterial growth rates (56). It is possible that virus-induced mortality also changed in response to the presence of grazers in our systems, but we have no direct evidence to support this.
The protozoa chosen for these experiments are common and abundant in soil (13, 40), and their combined effects may represent that of a natural soil protozoan community. Nevertheless, the observation that the five single protozoan isolates affected the bacterial community in different ways suggests that the actual composition of the protozoan communities is important for the impact of grazing on the microbial communities. Interestingly the correspondence analysis of the DGGE profiles clearly separated the bacterial communities treated with amoebae from those treated with the suspension-feeding Spumella, whereas the effects of the three surface-feeding flagellates were similar. This suggests a correlation between the overall feeding ecology of the flagellates and their effect on the bacterial community. However, even among the three surface-feeding species, differences in banding patterns could be observed by direct visual inspection of the DGGE profiles, in agreement with the results of Boenigk and Arndt (3) and Boenigk et al. (5), who found considerable species-specific differences in the feeding behaviors of different flagellates.
The short generation times of bacteria and protozoa mean that changes in population sizes may occur rapidly, and sampling on only two occasions would not have detected all fluctuations in bacterial community structure. In aquatic systems, significant changes in bacterial community structure have been observed after a few days with increased grazing pressure (32). However, the physical constraints on mixing and movement in soil probably mean that oscillations in the overall dynamics of protozoan and bacterial populations are reduced compared to aquatic systems. Furthermore, effects of grazing were found on both samplings, and even if fluctuations in community structure occurred, the results clearly indicate that protozoan grazing may have an important impact on the bacterial community structure in soil. The observation that high-G+C gram-positive bacteria related to Arthrobacter appeared to be favored by grazing was interesting, given their high relative abundance in soil. Protozoan grazing probably affects bacterial communities through a wide array of interacting mechanisms, such as selective feeding, differences in the susceptibilities of bacteria to predation, and indirect effects on the conditions for growth of bacterial populations (e.g., nutrient and substrate availability). The finding that even rather similar flagellate species had different effects on the community structure indicates that species-specific food preferences may be important. In future studies, it will be of great interest to gain deeper insight into the bacterial characteristics that affect their responses to grazers with different feeding ecologies. This would provide a better basis for understanding how microfaunal grazing on microbial communities affects the functioning of those communities.
Acknowledgments
Regin Rønn was funded by the Danish Natural Science Research Council.
We thank Gordon Webster for analysis of inorganic nitrogen and Flemming Ekelund for help with identification of flagellates and for comments on the manuscript.
REFERENCES
- 1.Anscombe, F. J., and B. N. Singh. 1948. Limitation of bacteria by micro-predators in soil. Nature 161:140-141. [DOI] [PubMed] [Google Scholar]
- 2.Boenigk, J., and H. Arndt. 2000. Comparative studies of the feeding behavior of two heterotrophic nanoflagellates: the filter-feeding choanoflagellate Monosiga ovata and the raptorial-feeding kinetoplastid Rhyncomonas nasuta. Aquat. Microb. Ecol. 22:243-249. [Google Scholar]
- 3.Boenigk, J., and H. Arndt. 2000. Particle handling during interception feeding by four species of heterotrophic nanoflagellates. J. Eukaryot. Microbiol. 47:350-358. [DOI] [PubMed] [Google Scholar]
- 4.Boenigk, J., C. Matz, K. Jürgens, and H. Arndt. 2001. Confusing selective feeding with differential digestion in bacterivorous nanoflagellates. J. Eukaryot. Microbiol. 48:425-432. [DOI] [PubMed] [Google Scholar]
- 5.Boenigk, J., C. Matz, K. Jürgens, and H. Arndt. 2001. The influence of preculture conditions and food quality on the ingestion and digestion process of three species of heterotrophic nanoflagellates. Microb. Ecol. 42:168-176. [DOI] [PubMed] [Google Scholar]
- 6.Brock, T. D., M. T. Madigan, J. M. Martinko, and J. Parker. 1994. Biology of microorganisms, 7th ed. Prentice-Hall International, London, United Kingdom.
- 7.Caron, D. A. 1987. Grazing of attached bacteria by heterotrophic microflagellates. Microb. Ecol. 13:203-218. [DOI] [PubMed] [Google Scholar]
- 8.Christensen, S., B. Griffiths, F. Ekelund, and R. Rønn. 1992. Huge increase in bacterivores on freshly killed barley roots. FEMS Microbiol. Ecol. 86:303-310. [Google Scholar]
- 9.Cordovilla, P., E. Valdiva, A. Gonzalea-Segura, A. Galvez, M. Martinez-Bueno, and M. Maqueda. 1993. Antagonistic action of the bacterium Bacillus licheniformis M-4 toward the amoeba Naegleria fowleri. J. Eukaryot. Microbiol. 40:323-328. [DOI] [PubMed] [Google Scholar]
- 10.Darbyshire, J. F., R. E. Wheatley, M. P. Greaves, and R. H. E. Inkson. 1974. A rapid micromethod for estimating bacterial and protozoan populations in soil. Rev. Écol. Biol. Sol 11:465-475. [Google Scholar]
- 11.Ekelund, F., H. B. Frederiksen, and R. Rønn. 2002. Population dynamics of active and total ciliate populations in arable soil amended with wheat. Appl. Environ. Microbiol. 68:1096-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ekelund, F., and D. J. Patterson. 1997. Some heterotrophic flagellates from a cultivated garden soil in Australia. Arch. Protistenkd. 148:461-478. [Google Scholar]
- 13.Ekelund, F., and R. Rønn. 1994. Notes on protozoa in agricultural soil, with emphasis on heterotrophic flagellates and naked amoebae and their ecology. FEMS Microbiol. Rev. 15:321-363. [DOI] [PubMed] [Google Scholar]
- 14.Ekelund, F., R. Rønn, and B. S. Griffiths. 2001. Quantitative estimation of flagellate community structure and diversity in soil samples. Protist 152:301-314. [DOI] [PubMed] [Google Scholar]
- 15.Farrelly, V., F. A. Rainey, and E. Stackebrandt. 1995. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl. Environ. Microbiol. 61:2798-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fenchel, T. 1987. Ecology of protozoa. The biology of free-living phagotrophic protists. Science Tech Publishers, Madison, Wis.
- 17.Gonzáles, J. M., J. Iriberri, L. Egea, and I. Barcina. 1990. Differential rates of digestion of bacteria by freshwater and marine phagotrophic protozoa. Appl. Environ. Microbiol. 56:1851-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griffiths, B. S., M. Bonkowski, G. Dobson, and S. Caul. 1999. Changes in soil microbial community structure in the presence of microbial-feeding nematodes and protozoa. Pedobiologia 43:297-304. [Google Scholar]
- 19.Griffiths, B. S., K. Ritz, N. Ebblewhite, and G. Dobson. 1999. Soil microbial community structure: effects of substrate loading rates. Soil Biol. Biochem. 31:145-153. [Google Scholar]
- 20.Groscop, J. A., and M. M. Brent. 1964. The effects of selected strains of pigmented microorganisms on small free-living amoebae. Can. J. Microbiol. 10:579-584. [DOI] [PubMed] [Google Scholar]
- 21.Güde, H. 1979. Grazing by protozoa as selection factor for activated sludge bacteria. Microb. Ecol. 5:225-237. [DOI] [PubMed] [Google Scholar]
- 22.Gurijala, K. R., and M. Alexander. 1987. Effect of growth rate and hydrophobicity on bacteria surviving protozoan grazing. Appl. Environ. Microbiol. 56:1631-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hackett, C. A., and B. S. Griffiths. 1997. Statistical analysis of the time-course of Biolog substrate utilisation. J. Microbiol. Methods 30:63-69. [Google Scholar]
- 24.Hahn, M. W., and M. G. Höfle. 1998. Grazing pressure by a bacterivorous flagellate reverses the relative abundance of Comomonas acidovorans PX54 and Vibrio strain CB5 in chemostat cocultures. Appl. Environ. Microbiol. 64:1910-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hahn, M. W., E. R. B. Moore, and M. G. Höfle. 1999. Bacterial filament formation, a defense mechanism against flagellate grazing, is growth rate controlled in bacteria of different phyla. Appl. Environ. Microbiol. 65:25-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hahn, M. W., and M. G. Höfle. 2001. Grazing of protozoa and its effect on populations of aquatic bacteria. FEMS Microbiol. Ecol. 35:113-121. [DOI] [PubMed] [Google Scholar]
- 27.Heidmann, T. 1989. Startkarakterisering af arealer til systemforskning. II. Resultater fra arealet ved Foulum. Tidsskr. Planteavls Specialserie S2007:1-186.
- 28.Iriberri, J., I. Azúa, A. Labirua-Iturburu, I. Artolozaga, and I. Barcina. 1994. Differential elimination of enteric bacteria by protists in a freshwater system. J. Appl. Bacteriol. 77:476-483. [DOI] [PubMed] [Google Scholar]
- 29.Jongman, R. H. G., C. J. F. ter Braak, and O. F. R. van Tongeren. 1987. Data analysis in community and landscape ecology. Centre for Agricultural Publishing and Documentation (Pudoc), Wageningen, The Netherlands.
- 30.Jürgens, K., and W. R. DeMott. 1995. Behavioral flexibility in prey selection by bacterivorous nanoflagellates. Limnol. Oceanogr. 40:1503-1507. [Google Scholar]
- 31.Jürgens, K., and H. Güde. 1994. The potential importance of grazing-resistant bacteria in planktonic systems. Mar. Ecol. Prog. Ser. 112:169-188. [Google Scholar]
- 32.Jürgens, K., J. Pernthaler, S. Schalla, and R. Amann. 1999. Morphological and compositional changes in a planktonic bacterial community in response to enhanced protozoan grazing. Appl. Environ. Microbiol. 65:1241-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kennedy, A. C., and V. L. Gewin. 1997. Soil microbial diversity: present and future considerations. Soil Science 162:607-617. [Google Scholar]
- 34.Kinner, N. E., R. W. Harvey. K. Blakeslee, G. Novarino, and L. D. Meeker. 1998. Size-selective predation on groundwater bacteria by nanoflagellates in an organic-contaminated aquifer. Appl. Environ. Microbiol. 64:618-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Magurran, A. E. 1988. Ecological diversity and its measurement. Chapman & Hall, London, United Kingdom.
- 36.May, K. R. 1965. A new graticule for particle counting and sizing. J. Scientific Instrum. 42:500-501. [Google Scholar]
- 37.McCaig, A. E., L. A. Glover, and J. I. Prosser. 2001. Numerical analysis of grassland bacterial community structure under different land management regimens by using 16S ribosomal DNA sequence data and denaturing gradient gel electrophoresis banding patterns. Appl. Environ. Microbiol. 67:4554-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Monger, B. C., M. R. Landry, and S. L. Brown. 1999. Feeding selection of heterotrophic marine nanoflagellates based on the surface hydrophobicity of their picoplankton prey. Limnol. Oceanogr. 44:1917-1927. [Google Scholar]
- 39.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Page, F. C. 1988. A new key to freshwater and soil gymnamoebae. Freshwater Biological Association, Ambleside, United Kingdom.
- 41.Payne, R. W., P. W. Lane, P. G. N. Digny, S. A. Harding, P. K. Leech, G. W. Morgan, A. D. Todd, R. Thompson, G. T. Wilson, S. J. Welham, and R. P. White. 1993. Genstat 5 release 3 reference manual. Oxford University Press, Oxford, United Kingdom.
- 42.Pernthaler, J., B. Sattler, K. Šimek, A. Schwarzenbacher, and R. Psenner. 1996. Top-down effects on the size-biomass distribution of a freshwater bacterioplankton community. Aquat. Microb. Ecol. 10:255-263. [Google Scholar]
- 43.Pernthaler, J., T. Posch, K. Šimek, J. Vrba, A. Pernthaler, F. O. Glöckner, U. Nübel, R. Psenner, and R. Amann. 2001. Predator-specific enrichment of actinobacteria from a cosmopolitan freshwater clade in mixed continuous culture. Appl. Environ. Microbiol. 67:2145-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Polz, M. F., and C. M. Cavanaugh. 1998. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 64:3724-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Postma, J., and J. A. van Veen. 1990. Habitable pore space and survival of Rhizobium leguminosarum biovar trifolii introduced into soil. Microb. Ecol. 19:149-161. [DOI] [PubMed] [Google Scholar]
- 46.Priemé, A., J. I. B. Sitaula, Å. K. Klemedtsson, and L. R. Bakken. 1996. Extraction of methane-oxidizing bacteria from soil particles. FEMS Microbiol. Ecol. 21:59-68. [Google Scholar]
- 47.Rønn, R., F. Ekelund, and S. Christensen. 1995. Optimizing soil extract and broth media for MPN-enumeration of naked amoebae and heterotrophic flagellates in soil. Pedobiologia 39:10-19. [Google Scholar]
- 48.Rønn, R., I. K. Thomsen, and B. Jensen. 1995. Naked amoebae, flagellates, and nematodes in soils of different texture. Eur. J. Soil Biol. 31:135-141. [Google Scholar]
- 49.Rønn, R., B. S. Griffiths, F. Ekelund, and S. Christensen. 1996. Spatial distribution and successional pattern of microbial activity and micro-faunal populations on decomposing barley roots. J. Appl. Ecol. 33:662-672. [Google Scholar]
- 50.Rønn, R., J. Grunert, and F. Ekelund. 2001. Protozoan response to addition of the bacteria Mycobacterium chlorophenolicum and Pseudomonas chlororaphis to soil microcosms. Biol. Fertil. Soils 33:126-131. [Google Scholar]
- 51.Rønn, R. M., B. S. Griffiths, and I. M. Young. 2001. Protozoa, nematodes and N-mineralization across a prescribed soil textural gradient. Pedobiologia 45:481-495. [Google Scholar]
- 52.Schmalenberger, A., F. Schweiger, and C. C. Tebbe. 2001. Effect of primers hybridising to different evolutionarily conserved regions of the small-subunit rRNA gene in PCR-based microbial community analyses and genetic profiling. Appl. Environ. Microbiol. 67:3557-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sherr, B. F., E. B. Sherr, and J. McDaniel. 1992. Effect of protistan grazing on the frequency of dviding cells in bacterioplankton assemblages. Appl. Environ. Microbiol. 58:2381-2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sibbald, M. J., and L. J. Albright. 1988. Aggregated and free bacteria as food sources for heterotrophic microflagellates. Appl. Environ. Microbiol. 54:613-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Šimek, K., J. Vrba, J. Pernthaler, T. Posch, P. Hartman, J. Nedoma, and R. Psenner. 1997. Morphological and compositional shifts in an experimental bacterial community influenced by protists with contrasting feeding modes. Appl. Environ. Microbiol. 63:587-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Šimek, K., J. Pernthaler, M. G. Weinbauer, K. Hornák, J. R. Dolan, J. Nedoma, M. Mašín, and R. Amann. 2001. Changes in bacterial community composition and dynamics and viral mortality rates associated with enhanced flagellate grazing in a mesoeutrophic reservoir. Appl. Environ. Microbiol. 67:2723-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sinclair, J. L., and M. Alexander. 1989. Effect of protozoan predation on relative abundance of fast- and slow-growing bacteria. Can. J. Microbiol. 35:578-582. [Google Scholar]
- 58.Singh, B. N. 1941. Selectivity in bacterial food by soil amoebae in pure mixed culture and in sterilized soil. Ann. Appl. Biol. 28:52-64. [Google Scholar]
- 59.Verity, P. G. 1991. Feeding in planktonic protozoans: evidence for non-random acquisition of prey. J. Protozool. 38:69-76. [Google Scholar]
- 60.Vestergaard, P., R. Rønn, and S. Christensen. 2001. Reduced particle size of plant material does not stimulate decomposition, but affects the microbivorous microfauna. Soil Biol. Biochem. 33:1805-1810. [Google Scholar]
- 61.Webster, G., T. M. Embley, and J. I. Prosser. 2002. Grassland management regimens reduce small-scale heterogeneity and species diversity of β-proteobacterial ammonia oxidizer populations. Appl. Environ. Microbiol. 68:20-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weekers, P. H. H., P. L. E. Bodelier, J. P. H. Wijen, and G. D. Vogels. 1993. Effects of grazing by the free-living soil amoebae Acanthamoeba castellanii, Acanthamoeba polyphaga, and Hartmannella vermiformis on various bacteria. Appl. Environ. Microbiol. 59:2317-2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wratten, S. D., and G. L. A. Fry. 1980. Field and laboratory exercises in ecology. Edward Arnold, London, United Kingdom.
- 64.Wright, D., K. Killham, L. A., Glover, and J. I. Prosser. 1993. The effects of location on protozoan grazing of a genetically modified bacterial inoculum in soil. Geoderma 56:633-640. [Google Scholar]