Abstract
Cyanobacterial blooms are potential health hazards in water supply reservoirs. This paper reports analyses of a cyanobacterial bloom by use of PCR-based methods for direct detection and identification of strains present and determination of their toxigenicity. Serial samples from Malpas Dam, in the New England region of Australia, were analyzed during a prolonged, mixed cyanobacterial bloom in the summer of 2000 to 2001. Malpas Dam has been shown in the past to have toxic blooms of Microcystis aeruginosa that have caused liver damage in the human population drinking from this water supply reservoir. Cyanobacterial genera were detected at low cell numbers by PCR amplification of the phycocyanin intergenic spacer region between the genes for the β and α subunits. The potential for microcystin production was determined by PCR amplification of a gene in the microcystin biosynthesis pathway. The potential for saxitoxin production was determined by PCR amplification of a region of the 16S rRNA gene of Anabaena circinalis strains. Toxicity of samples was established by mouse bioassay and high-pressure liquid chromatography. We show that bloom components can be identified and monitored for toxigenicity by PCR more effectively than by other methods such as microscopy and mouse bioassay. We also show that toxigenic strains of Anabaena and Microcystis spp. occur at this site and that, over the course of the bloom, the cell types and toxicity changed. This work demonstrates that PCR detection of potential toxicity can enhance the management of a significant public health hazard.
In Australia, most drinking water comes from natural, undeveloped catchment areas and is stored in reservoirs. The water is monitored and managed to ensure good-quality drinking water, free from taste and odor problems and health risks, such as toxins, pathogenic bacteria, and parasites. Bloom-forming cyanobacteria can be a serious problem for water managers, and bloom toxicity needs to be determined early in the bloom development in a drinking water reservoir. Cyanobacteria have been shown to produce a wide range of toxic compounds, including neurotoxins, such as the saxitoxins, and hepatotoxins, such as the microcystins (34). Several incidences of acute effects of hepatotoxins, causing toxic liver injury (13) or death (20), have been reported. Perhaps more seriously, microcystins have been shown to be tumor promoters (14, 19, 29) and pose a serious risk to populations exposed to chronic low-level doses.
Identification of a cyanobacterial genus by microscopic morphology and/or molecular analysis does not indicate the potential for toxin production. Different strains of one species can be morphologically identical but differ in toxigenicity. For example, Microcystis aeruginosa has both toxic and nontoxic strains (23). There have been attempts to refine the identification of strains by using specific gene analysis. Examples include the use of PCR for amplification of the phycocyanin intergenic spacer (PC-IGS) between the β and α subunits of the phycocyanin operon (2, 7, 8, 26), the 16S-23S rRNA internally transcribed spacer region (28, 30), or segments of the ribulose bisphosphate carboxylase genes (11, 32) and the DNA-dependent RNA polymerase (rpoC1) gene (16). The use of PCR for amplification of the phycocyanin intergenic spacer has recently been adapted for direct analysis of environmental samples (2). This technique has the advantage of being specific for cyanobacteria in the presence of other organisms.
Although these molecular techniques have improved the accuracy of strain identification, they have not been able to distinguish toxigenic from nontoxigenic strains. The biosynthetic pathway for microcystin production has now been elucidated (35), and this has enabled the development of specific oligonucleotide primers for genes common to production of all microcystins (36). By using these primers for PCR amplification, toxigenic strains in bloom samples can be distinguished (36).
Besides microcystins, saxitoxins are another important group of toxins produced by Australian cyanobacterial blooms (18). A unique molecular technique for detecting toxigenic strains is not yet available for saxitoxin production because the toxin synthesis pathway has not yet been fully elucidated. New studies show that PCR amplification of regions of the 16S rRNA gene can be useful for distinguishing strains within a genus or species. For example, PCR techniques have allowed separation of strains of common bloom-forming genera, such as Microcystis (27), Anabaena (6), Nodularia (24), and Cylindrospermopsis (33). One of these techniques has segregated Anabaena strains into mostly toxic versus mostly nontoxic strains (6).
In this study we report an analysis of a mixed bloom over the summer of 2000 to 2001 in Malpas Dam, in the New England region of the Northern Tablelands of New South Wales, Australia. This dam has a history of severe cyanobacterial blooms, and a toxic bloom of M. aeruginosa in March 1981 was the basis for an epidemiological study that showed evidence of liver damage in the population of Armidale supplied by water from this dam (13). Records from the local water-monitoring authority over the last decade show that blooms of cyanobacteria on Malpas Dam may be monospecific or mixed, often showing changes in dominance over a period of days, weeks, or months.
We demonstrate the value of using specific PCR methods to analyze bloom samples directly. During the progression of this complex bloom, strains of Anabaena and Microcystis developed and dominated. It was found that PCR-based techniques could identify toxigenic strains of Microcystis and potentially toxic strains of Anabaena circinalis. Early detection of problem organisms can provide a means to improve the management of water quality and treatment to avoid public health risks.
MATERIALS AND METHODS
Study site.
Malpas Dam (30°16′S, 151°75′E) lies on the Gara River in the New England region of the Northern Tablelands of New South Wales, Australia. The dam is at an elevation of 1,178 m, and it has a catchment area of about 20,000 ha and a capacity of 13,000 Ml. This dam is the water supply reservoir for the city of Armidale, a town with approximately 22,000 inhabitants. Annual average rainfall in the catchment area is 890 mm. The rainfall pattern is variable, but rainfall peaks in the summer months. Average summer temperatures range from 10 to 23°C, and the winter range is between −1 and 10°C. Malpas Dam is classified as eutrophic on the basis of phosphorus, nitrogen, and chlorophyll levels (5). The plentiful nutrients are a product of agricultural land use and management and the basic geology of the catchment area. The soil landscapes of the Malpas catchment area include basalt with a phosphorus content 2 to 3 times higher than the worldwide mean (39). An analysis of the dam water in 1995 showed that levels of soluble reactive phosphorus ranged from 87 to 95 μg liter−1 (9), exceeding the level of 50 μg liter−1 considered conducive to cyanobacterial blooms (37). In 2001, levels of soluble reactive phosphorus ranged from 6 to 11 μg liter−1 (P. Wilson, unpublished data), but total phosphorus levels in sediments remained high (measured at 1.6 mg g−1 [dry weight], compared to 1.2 mg g−1 [dry weight] in 1995 [9]). Improved catchment management is probably responsible for reduced nutrient inflow into the dam and thus for the lower level of soluble reactive phosphorus. The pH of Malpas Dam water averages 8.8. High pHs (pH 7 to 9) favor proliferation of cyanobacteria over other phytoplankton (37).
Reference cultures.
The specific cyanobacterial strains used as references are listed in Table 1.
TABLE 1.
Reference cultures, bloom samples and database entries used in this study
| Code | Straina (toxicity)b | Origin | Reference or (mo.day.yr) |
|---|---|---|---|
| Reference cultures | |||
| AWQC118C | A. circinalis (S-P) | Australia | 6 |
| AWQC131C | A. circinalis (S-P) | Australia | 6 |
| AWQC306A | A. circinalis (NT) | Australia | 6 |
| AWQC271C | A. circinalis (NT) | Australia | 6 |
| NIES 80 | Anabaena solitaria (ND) | Japan | 38 |
| PCC 7806 | M. aeruginosa (M-P) | The Netherlands | 31 |
| Bloom samplesc | |||
| T-96 (clone) | Microcystis sp. | Malpas Dam | 2.12.96 |
| MD-34 | M. flos-aquae | Malpas Dam | 1.24.00 |
| BD-1 | Microcystis sp. | Burrendong Dam | 7.7.98 |
| CD-1 | Microcystis sp. | Chaffey Dam | 4.2.96 |
| CaD-1 | Microcystis sp. | Cania Dam | 5.23.00 |
| WW-1 | Microcystis sp. | Wagga Wagga | 1.14.99 |
| Database entries | |||
| AF195159 | M. aeruginosa (−) | United States | 36 |
| AF195161 | M. flos-aquae (−) | United States | 36 |
| AF195162 | M. flos-aquae (−) | United States | 36 |
| AF195167 | M. aeruginosa (+) | South Africa | 36 |
| AF195174 | M. aeruginosa (+) | United States | 36 |
| AF195175 | M. aeruginosa (+) | Canada | 36 |
| AF195176 | M. aeruginosa (+) | Scotland | 36 |
| AF195177 (PCC 7806) | M. aeruginosa (+) | The Netherlands | 36 |
Species designation based on morphology.
Toxicity: S-P, saxitoxin-producing strain; NT, non-toxin-producing strain; M-P, microcystin-producing strain; ND, not determined; −, absence of the microcystin synthetase NMT domain; +, presence of the microcystin synthetase NMT domain.
Australian bloom samples other than the 2000-to-2001 Malpas Dam samples.
Environmental water samples.
The local water authority, Armidale Dumaresq Council (ADC), takes monitoring samples at four sites in the dam, on the surface and at 1-m depth intervals at each site, on a thrice-weekly basis during the summer. For this study, only surface samples at the site adjacent to the drinking water off-take tower have been examined and analyzed. Once a week, for the duration of this study, two samples were taken at this site. One sample was used for analysis in our laboratory, and the other sample (the regular monitoring sample) was used for cell counts and mouse bioassay by the water authority. Within 24 h of collection, a 1-ml aliquot of each sample was examined microscopically in a Sedgwick-Rafter chamber at a magnification of ×400 under phase contrast. The number of cells of each bloom species was estimated, and a ratio of the component species was derived. Features, such as the presence of akinetes, were also noted. Species identification was based on morphological characteristics as described in the Australian reference text of Baker and Fabbro (3). A subset of the weekly samples (Table 2) was subjected to detailed analysis.
TABLE 2.
Composition of selected bloom samples by microscopy
| Code | Sampling date (mo.day.yr) | A. circinalis/ M. aeruginosa/ M. flos-aquae ratioa | 103 cell counts ml−1b
|
|
|---|---|---|---|---|
| Anabaena | Microcystis | |||
| MD-78 | 11.28.00 | 0:0:0 | 0 | 0 |
| MD-80 | 12.8.00 | 100:0:0 | 9.8 | 0 |
| MD-84 | 12.22.00 | 100:0:0 | 630 | 0 |
| MD-87 | 1.5.01 | 99:0:1 | 11 | 0 |
| MD-88 | 1.12.01 | 1:0:99 | 33 | 0 |
| MD-90 | 1.25.01 | 4:1:95 | 120 | 1,000 |
| MD-93 | 2.15.01 | 5:0:95 | 2,500 | 10 |
| MD-96 | 2.26.01 | 5:85:10 | 380 | 65 |
| MD-103 | 3.12.01 | 1:90:9 | 21 | 36 |
| MD-108 | 3.26.01 | 1:99:0 | 0.4 | 23 |
| MD-109 | 3.30.01 | 0:100:0 | 0 | 13 |
Obtained by microscopic examination of samples taken for molecular analysis in our laboratory (as described in Materials and Methods).
Samples taken separately, at the same time and place, by the Armidale Dumaresq Council. These counts are from council records.
Sample preparation for PCR.
The cyanobacterial cells in bloom samples were concentrated by centrifugation, washed, and subjected to a freeze-thaw treatment for PCR template preparation (2). All PCRs described in this study were carried out after this treatment, by using approximately 1,000 cells per reaction. This method is simple and quick and has been proven effective with fresh bloom material, when most cells are intact.
DNA amplification, analysis, and sequencing for identification.
PCR amplification of the PC-IGS was performed as previously described (2, 26).
Samples of DNA from PCR amplification were purified using a QIAquick PCR Purification kit (Qiagen) and sequenced using the ABI Prism BigDye Terminator v3.0 Ready Reaction Cycle Sequencing kit (Perkin-Elmer Applied Biosystems) and an ABI Prism 3700 DNA Analyzer (Perkin-Elmer Applied Biosystems).
Sequence analysis was performed with the programs available through the Australian National Genomic Information Service (ANGIS). “Pileup” was used for multiple sequence alignments by use of a simplification of the progressive alignment method (15). PC-IGS nucleotide sequences were compared to entries deposited in GenBank by using “BlastN” (1).
Assessing toxigenicity by PCR.
PCR amplification of the N-methyltransferase (NMT) region in the microcystin polyketide-peptide biosynthetic pathway in cyanobacteria was performed as described previously (36). Thermal cycling was carried out as published except that the annealing temperature was modified to 55°C to refine the method for conditions in our laboratory. This was confirmed with microcystin-producing cell lines.
Analysis of strains of A. circinalis was carried out to determine their toxic grouping by PCR-based detection of a specific region of the 16S rRNA (6). The PCR conditions for this method were modified to use an annealing temperature of 65°C. This was confirmed with Anabaena cultures known to produce saxitoxin.
Electrophoresis of the PCR products was carried out in 1% agarose gels in TAE buffer (0.04 M Tris-acetate-0.001 M EDTA [pH 8.0]), and the DNA was stained with ethidium bromide and visualized and photographed under UV illumination.
Toxicity testing.
A mouse bioassay was employed for rapid, broad-spectrum toxin detection (12). Baker and Humpage (4) have reported the quantity of cells of the common toxigenic bloom-forming cyanobacteria Anabaena, Microcystis, Nodularia, and Cylindrospermopsis that gave a toxic response when extracted and injected intraperitoneally into a 16- to 23-g mouse. Based on their results, our water samples were concentrated by centrifugation to achieve approximately 108 cells ml−1 and then the extract from 1 ml was injected and assayed according to the work of Falconer (12). When mice died within 15 min with neurotoxic symptoms, the sample was recorded as “neurotoxic.” When mice died within 24 h and showed enlarged, blood-filled livers, the sample was recorded as “lethal hepatotoxic.” If the mice did not die within 24 h, the sample was recorded as “nontoxic,” or as “nonlethal hepatotoxic” if the mice had mottled, enlarged livers (>20% heavier than the livers of control mice).
Toxins were identified and quantified by high-pressure liquid chromatography (HPLC) analysis. Samples (50 ml) of bloom material were analyzed for microcystins by using sonication, concentration on a C18 cartridge, and subsequent HPLC based on the method of Lawton et al. (22). Saxitoxins were identified and quantified by using a peroxide oxidation method of sample preparation, based on the work of Lawrence, Menard, and Cleroux (21). Samples (50 ml) of bloom material were sonicated, filtered, and then subjected to the peroxide oxidation, followed by HPLC.
Nucleotide sequence accession numbers.
The PC-IGS nucleotide sequences described in this study have been deposited in GenBank under accession numbers AY117039 to AY117047; A. circinalis AWQC118C has been assigned accession number AF426004; and the accession numbers of Microcystis PC-IGS sequences used in comparisons to Australian Microcystis bloom samples are listed in Table 1.
RESULTS
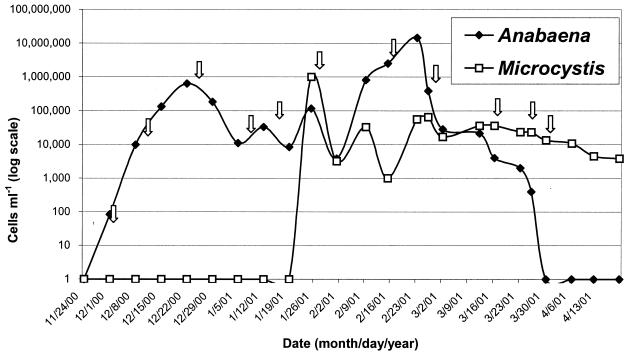
During the weekly monitoring of Malpas Dam, samples were selected for molecular analysis, based on the occurrence of particular cell types as they bloomed over the course of the summer. Samples can be identified by the dates in Fig. 1 and Table 2 and are indicated by arrows in Fig. 1.
FIG. 1.
Cyanobacterial cell counts for site 1 surface samples, summer 2000 to 2001. Arrows indicate sampling dates shown in Table 2. Data are summarized from ADC records.
Detection of cyanobacteria in selected samples.
The samples listed in Table 2 were subjected to microscopic examination, and the cyanobacterial species observed were recorded, together with their relative frequencies. Table 2 also shows cell counts determined by the ADC on separate samples taken at the same time and place. For samples MD-88, MD-93, and MD-96, there are obvious discrepancies between the ratios observed in our laboratory samples and the cell counts for the samples taken by the ADC. This has occurred where Microcystis flos-aquae was present in the samples. In Malpas Dam this species is very buoyant, floating right on the surface of the water. When replicate surface samples are taken, some samples may contain many M. flos-aquae colonies and other samples may contain few of this species. We have found no mention of this point in the literature. This was an unexpected sampling problem, revealed by the discrepancies shown in Table 2.
The samples in Table 2 were subjected to PCR amplification of the PC-IGS, and products specific for cyanobacteria were obtained for all samples except MD-78, which had no observable cyanobacterial cells present. The results agreed qualitatively with those of the microscopic examination.
Identification of bloom species by using the phycocyanin region.
Samples MD-80, MD-88, and MD-108 were taken at points during the bloom when only one cyanobacterial type was dominant. These samples were subjected to PC-IGS amplification, and the products were used directly in sequencing reactions. The subsequent chromatograms showed single base peaks throughout, indicating that DNA from a single cyanobacterial type dominated. Alignment of these sequences with reference culture sequences allowed taxonomic identification to the genus level, based upon both the length and the sequence of the PC-IGS (2) (Fig. 2). Figure 2 shows that sample MD-80 was identical to the A. circinalis reference culture AWQC118C, in agreement with microscopy results. The sequences of samples MD-88 and MD-108, identified as Microcystis species by microscopy, were almost identical to the sequences of the reference culture M. aeruginosa PCC 7806 (AF195177) and an M. flos-aquae database entry (AF195162), respectively.
FIG. 2.
Identification of bloom samples by PC-IGS sequences. Key bloom samples MD-80, MD-88, and MD-108 were compared with a reference culture and two database entries (see Tables 1 and 2 for more information). Sample MD-80 was confirmed as A. circinalis, and samples MD-88 and MD-108 are Microcystis species.
Alignment of the PC-IGS sequences of Malpas Dam Microcystis samples with those of overseas strains and Australian Microcystis bloom samples showed no grouping according to species or toxicity, as was found also by Tillett et al. (36). Thus, this method could not be used to differentiate species of Microcystis in the bloom.
Microcystin-producing potential in bloom samples.
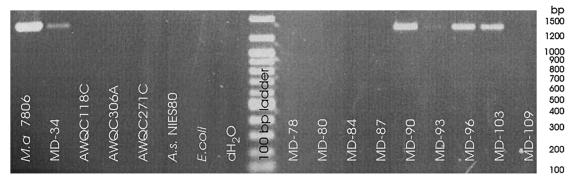
We employed the PCR technique for amplifying the NMT domain of the gene for microcystin synthetase A (mcyA) in bloom samples to identify which samples contained potentially microcystin producing strains (36). The results in Fig. 3 showed that samples MD-90 and MD-93 (dominated by M. flos-aquae), and samples MD-96 and MD-103 (dominated by M aeruginosa), each formed a PCR product identical in size to that formed from the toxin-producing reference culture. Thus, at least some of the Microcystis strains in each of these samples were capable of producing microcystin. Bloom samples MD-80, MD-84, and MD-87 (which were dominated by A. circinalis) and sample MD-88 (dominated by M. flos-aquae) (data not shown) did not produce PCR products. This suggested that the A. circinalis strains in this bloom were not microcystin producing. The lack of product from sample MD-88 suggested that there was more than one strain of M. flos-aquae within this bloom, some toxigenic (MD-90 and MD-93) and others not producing toxin (MD-88). There was no discernible PCR product from sample MD-109, which contained M aeruginosa and had high numbers of the diatom Aulocoseira.
FIG. 3.
Detection of the NMT domain of the microcystin synthetase gene, mcyA, in Malpas Dam bloom samples (Table 2) and control samples (Table 1). Of the control samples, only M. aeruginosa PCC 7806 (M.a. 7806) is a microcystin producer.
Testing of A. circinalis in bloom samples by using 16S rRNA analysis.
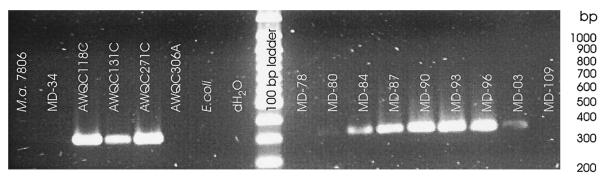
In Australia, within the genus Anabaena, strains of A. circinalis have been identified as producers of a neurotoxin (4), which has subsequently been shown to be saxitoxin (18). To indicate which bloom samples may contain toxic A. circinalis, we employed a set of PCR primers that were designed to anneal to specific sequences within the 16S rRNA gene. One of these primers was shown to be specific for a group of A. circinalis strains which are mostly toxic (6). The results are shown in Fig. 4. A PCR product was formed from all bloom samples except MD-78 and MD-109, which were not shown by microscopy to contain Anabaena (see Table 2). On the basis of this analysis, all of the samples that gave a PCR product contained potentially toxic A. circinalis.
FIG. 4.
Detection of potentially saxitoxin producing A. circinalis strains in Malpas Dam bloom samples by amplification of a region of the 16S rRNA gene. For descriptions of strain and bloom samples, see Tables 1 and 2. Of the four reference strains of A. circinalis, AWQC118C and AWQC131C are known saxitoxin producers while AWQC271C and AWQC306A are not known to produce saxitoxin, although AWQC271C gives a positive result in this test (6).
Toxicity analysis of bloom samples.
The bloom samples listed in Table 2 were analyzed for toxins by mouse bioassay and HPLC (see Table 3 for results).
TABLE 3.
Summary of toxin assays and PCR results for selected Malpas Dam bloom samplesa
| Sample | A. circinalis/M. aeruginosa/ M. flos-aquae ratio | Mouse bioassay resultb | Concn (μg/liter−1) of the indicated toxin by HPLCc
|
PCR test resultd
|
||
|---|---|---|---|---|---|---|
| Microcystins | Saxitoxin | Anabaena | Microcystins | |||
| MD-34 | 0:0:100 | n-l H | ND | NT | ND | + |
| MD-78 | 0:0:0 | n-t | ND | ND | ND | ND |
| MD-80 | 100:0:0 | n-t | ND | ND | + | ND |
| MD-84 | 100:0:0 | n-t | ND | 33 | + | ND |
| MD-87 | 99:0:1 | n-t | ND | 5 | + | ND |
| MD-88 | 1:0:99 | n-t | ND | 7 | NT | ND |
| MD-90 | 4:1:95 | n-l H | 32 | 40 | + | + |
| MD-93 | 5:0:95 | N | 1 | 135 | + | + |
| MD-96 | 5:85:10 | n-l H | 8 | 6 | + | + |
| MD-103 | 1:90:9 | H | 80 | 4 | + | + |
| MD-108 | 1:99:0 | NT | ND | ND | NT | ND |
| MD-109 | 0:100:0 | n-l H | ND | ND | ND | ND |
See also Table 2.
n-t, nontoxic; N, neurotoxic; H, hepatotoxic; n-l H, nonlethal hepatotoxic; NT, not tested.
The limit of detection was 10 μg liter−1 because of interfering peaks. ND, peak(s) or band not detected.
+, band detected.
Mouse bioassays showed that, from sample MD-90 onward, some degree of hepatotoxicity was apparent. This corresponded with the presence of Microcystis strains in the samples (see Table 2). Hepatotoxicity was not established for sample MD-93 because the mice died rapidly with neurotoxic symptoms before symptoms of hepatotoxicity were apparent.
Measurement of saxitoxin by HPLC (Table 3) showed the presence of the toxins in the same samples that gave positive results in the PCR analysis (Fig. 4), and the sample with the highest toxin concentration was the sample that was neurotoxic by mouse bioassay. Similarly, detection of microcystin by use of HPLC was consistent with positive results in the PCR analysis (Fig. 3), and the highest microcystin concentration corresponded with the strongly hepatotoxic mouse bioassay result.
DISCUSSION
This study has shown that Malpas Dam, during the summer of 2000 to 2001, contained toxigenic strains of cyanobacteria from mid-December to mid-March. During this time cell counts continuously exceeded 10,000 cells ml−1. Changing levels of different species and toxins were observed over the course of the bloom. The untreated water of Malpas Dam remains a public health risk during such bloom periods.
Determination of the toxigenicity of the cyanobacteria present provides a warning of possible toxicity development and allows early intervention to avoid health problems. We have demonstrated that PCR-based tests, combined with microscopy, quickly and simply monitored the succession of cyanobacterial types in this bloom and indicated their toxigenicities. Previous work has shown that the NMT PCR assay reliably indicates microcystin toxigenicity (36). We have adapted this technique for use with bloom samples, and we have found that it provides a positive result with 1,000 toxigenic cells and that environmental cell densities as low as 10 cells ml−1 can provide sufficient cells for this assay. The NMT PCR assay signaled the presence of toxigenic Microcystis strains as soon as Microcystis was evident in samples by microscopy (Fig. 1; Table 3) and demonstrated that toxigenic cells continued to be present throughout the remainder of the bloom. By comparison, the mouse bioassay (the method which has routinely been employed to test Malpas Dam samples) showed lethal hepatotoxicity only in sample MD-103, which was shown by HPLC to contain the highest level of microcystins (see Table 3). The mouse bioassay is not effective in indicating the presence of microcystins when neurotoxins are also present because the neurotoxins rapidly kill the mice before microcystins cause pathological effects (12). In mixed blooms such as the one in this study, this bioassay could fail to detect the presence of microcystin producers. The negative result in the NMT PCR assay of sample MD-109, which contained Microcystis (Table 2), was consistent with the negative result by HPLC and contrasted with the assessment of the possible presence of a low level of hepatotoxins in the mouse bioassay (Table 3).
HPLC analysis of these bloom samples clearly demonstrated that at least some of the A. circinalis cells present throughout the bloom produced saxitoxins and were thus neurotoxic (see Tables 2 and 3). By comparison, the mouse bioassay showed neurotoxicity only in the sample (MD-93) which contained the very highest concentration of saxitoxin. The 16S rRNA-based PCR assay consistently agreed with the HPLC data, with the exception of sample MD-80. This PCR test for toxic type A. circinalis is not a definitive toxigenicity test, as it may give occasional false-positive or false-negative results (6). In the absence of a PCR test to detect the presence of a gene in the saxitoxin synthesis pathway, the 16S rRNA-based test is the best available method for rapid screening of environmental samples. Based on the results in this report, the 16S rRNA-based method was valuable in providing an early signal of potentially saxitoxin producing Anabaena. Like the NMT PCR assay for microcystins, the 16S rRNA-based assay for toxic Anabaena types is effective at a level of 10 cells ml−1 and can indicate the possible neurotoxicity of the bloom well before the cell counts reach the local high-alert level of 20,000 cells ml−1. The local water authority responsible for Malpas Dam has an action alert at a cell density of 2,000 ml−1, as recommended by the Australian Drinking Water Guidelines (25), and a high-alert level of 20,000 cells ml−1, where blooms may contain sufficient toxin to be of concern for human health (17).
In the work presented here, the PCR assays, applied directly to environmental samples, were as sensitive as HPLC in providing a useful indicator of toxicity (see Table 3). Although HPLC provides a direct measure of toxins present, it does require a large capital investment and considerable sample preparation. The PCR-based assays employed here detect toxigenic cells rather than toxins and require little sample preparation and modest capital costs. When regular monitoring of a problem water supply reservoir is required, we have shown that both HPLC and PCR-based assays are more sensitive than mouse bioassays. The only advantage of bioassays lies in broad-spectrum toxin detection, when the type of toxin present is unknown.
In the analysis of the Malpas Dam bloom, we employed two methods of identifying the cyanobacteria present: microscopy and the PC-IGS analysis (see Table 2 and Fig. 2). Microscopy is rapid and sensitive. However, even with skilled and experienced operators, identification is sometimes uncertain. The PC-IGS analysis takes considerable time and effort but provides certainty in the identification of genera by using both the length and the sequence of the PC-IGS region (2). This method provides a valuable tool for confirming the identity of cell types in this and other blooms.
This bloom showed a complex pattern of cyanobacterial species succession. Figure 1 shows that the bloom began with the development of large numbers of A. circinalis, which peaked in mid-December and peaked again at even higher numbers in late February. A Microcystis population suddenly appeared in mid-January and was identified as M. flos-aquae by microscopic examination. M. flos-aquae numbers declined and were replaced by M. aeruginosa (as identified by microscopy) in late February. The cells identified as M. flos-aquae and M. aeruginosa by microscopy were found to exhibit slightly different PC-IGS sequences (Fig. 2), which confirms that at least two different Microcystis types were present in this bloom. Unfortunately, the length and sequence of the PC-IGS are unable to identify Microcystis species (2). Bloom sample MD-90 corresponds to the peak identified as M. flos-aquae, was toxigenic as detected by NMT PCR, and contained microcystins as measured by HPLC (see Table 3). A sample identified as M. flos-aquae in a Malpas Dam bloom in 1999 to 2000, MD-34, gave a positive result in the NMT PCR assay, but microcystins were not detected by HPLC. Sample MD-34 differed from sample MD-88 (identified as M. flos-aquae by microscopic examination) in the sequence of the PC-IGS, indicating that these samples may contain different strains of M. flos-aquae. This is the first study to indicate toxigenicity in blooms of M. flos-aquae, although this has been suggested in a Swedish study (10). Tillett et al. (36) tested the toxigenicities of many M. flos-aquae strains by using the NMT PCR assay and the protein phosphatase inhibition assay, and all strains examined were nontoxic.
Acknowledgments
J.A.B. acknowledges the support of an Australian Postgraduate Scholarship. She also gratefully acknowledges the ADC both for financial support and for help with collection of samples and access to Council records.
We thank Mandy Choice (ADC) for performing the mouse bioassays and Geoff Eaglesham and Brad Davis (Queensland Health Scientific Services) for HPLC analysis.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, J. A., B. A. Neilan, B. Entsch, and D. B. McKay. 2001. Identification of cyanobacteria and their toxigenicity in environmental samples by rapid molecular analysis. Environ. Toxicol. 16:472-482. [PubMed] [Google Scholar]
- 3.Baker, P. D., and L. D. Fabbro. 1999. A guide to the identification of common blue-green algae (Cyanoprokaryotes) in Australian freshwaters. Cooperative Research Centre for Freshwater Ecology, Albury, Australia.
- 4.Baker, P. D., and A. R. Humpage. 1994. Toxicity associated with commonly occurring cyanobacteria in surface waters of the Murray-Darling Basin, Australia. Aust. J. Mar. Freshwater Res. 45:773-786. [Google Scholar]
- 5.Banens, R. J. 1989. A comparative limnological study of New England reservoirs, with particular reference to water quality. Ph.D. thesis. University of New England, Armidale, New South Wales, Australia.
- 6.Beltran, E. C., and B. A. Neilan. 2000. Geographical segregation of the neurotoxin-producing cyanobacterium Anabaena circinalis. Appl. Environ. Microbiol. 66:4468-4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolch, C. J. S., S. I. Blackburn, B. A. Neilan, and P. M. Grewe. 1996. Genetic characterization of strains of cyanobacteria using PCR-RFLP of the cpcBA intergenic spacer and flanking regions. J. Phycol. 32:445-451. [Google Scholar]
- 8.Bolch, C. J. S., P. T. Orr, G. J. Jones, and S. I. Blackburn. 1999. Genetic, morphological, and toxicological variation among globally distributed strains of Nodularia (cyanobacteria). J. Phycol. 35:339-355. [Google Scholar]
- 9.Boulton, A., R. Faulkner, and P. Southcott. 1996. Survey of the potential compartments of phosphorus in Malpas Dam and its major tributaries. University of New England, Armidale, New South Wales, Australia.
- 10.Cronberg, G., H. Annadotter, and L. A. Lawton. 1999. The occurrence of toxic blue-green algae in Lake Ringsjon, southern Sweden, despite nutrient reduction and fish biomanipulation. Hydrobiologia 404:123-129. [Google Scholar]
- 11.Delwiche, C. F., and J. D. Palmer. 1996. Rampant horizontal transfer and duplication of rubisco genes in eubacteria and plastids. Mol. Biol. Evol. 13:873-882. [DOI] [PubMed] [Google Scholar]
- 12.Falconer, I. R. 1993. Measurement of toxins from blue-green algae in water and foodstuffs, p. 165-175. In I. R. Falconer (ed.), Algal toxins in seafood and drinking water. Academic Press Ltd., London, United Kingdom.
- 13.Falconer, I. R., A. M. Beresford, and M. T. C. Runnegar. 1983. Evidence of liver damage by toxin from a bloom of the blue-green alga Microcystis aeruginosa. Med. J. Aust. 1:511-514. [DOI] [PubMed] [Google Scholar]
- 14.Falconer, I. R., and A. R. Humpage. 1996. Tumour promotion by cyanobacterial toxins. Phycologia 35:74-79. [Google Scholar]
- 15.Feng, D. F., and R. F. Doolittle. 1987. Progressive sequence alignment as a prerequisite to correct phylogenetic trees. J. Mol. Evol. 25:351-360. [DOI] [PubMed] [Google Scholar]
- 16.Fergusson, K. M., and C. P. Saint. 2000. Molecular phylogeny of Anabaena circinalis and its identification in environmental samples by PCR. Appl. Environ. Microbiol. 66:4145-4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fitzgerald, D. J., D. A. Cunliffe, and M. D. Burch. 1999. Development of health alerts for cyanobacteria and related toxins in drinking water in South Australia. Environ. Toxicol. 14:203-209. [Google Scholar]
- 18.Humpage, A. R., J. Rositano, A. H. Bretag, J. Brown, P. D. Baker, B. C. Nicholson, and D. A. Steffensen. 1994. Paralytic shellfish poisons from Australian cyanobacterial blooms. Aust. J. Mar. Freshwater Res. 45:761-771. [Google Scholar]
- 19.Humpage, A. R., S. J. Hardy, E. J. Moore, S. M. Froscio, and I. R. Falconer. 2000. Microcystins (cyanobacterial toxins) in drinking water enhance the growth of aberrant crypt foci in the mouse colon. J. Toxicol. Environ. Health A 61:155-165. [DOI] [PubMed] [Google Scholar]
- 20.Jochimsen, E. M., W. W. Carmichael, J. S. An, D. M. Cardo, S. T. Cookson, C. E. M. Holmes, M. B. D. Antunes, D. A. DeMelo Filho, T. M. Lyra, V. S. T. Barreto, S. M. F. O. Azevedo, and W. R. Jarvis. 1998. Liver failure and death after exposure to microcystins at a hemodialysis center in Brazil. N. Engl. J. Med. 338:873-878. [DOI] [PubMed] [Google Scholar]
- 21.Lawrence, J. F., C. Menard, and C. Cleroux. 1995. Evaluation of prechromatographic oxidation for liquid chromatographic determination of paralytic shellfish poisons in shellfish. J. Assoc. Off. Anal. Chem. 78:514-520. [PubMed] [Google Scholar]
- 22.Lawton, L. A., K. A. Beattie, S. P. Hawser, D. L. Campbell, and G. A. Codd. 1994. Evaluation of assay methods for the determination of cyanobacterial hepatotoxicity, p. 111-116. In G. A. Codd, T. M. Jeffries, C. W. Keevil, and E. Potter (ed.), Detection methods for cyanobacterial toxins. The Royal Society of Chemistry, Cambridge, United Kingdom.
- 23.Meissner, K., E. Dittmann, and T. Borner. 1996. Toxic and non-toxic strains of the cyanobacterium Microcystis aeruginosa contain sequences homologous to peptide synthetase genes. FEMS Microbiol. Lett. 135:295-303. [DOI] [PubMed] [Google Scholar]
- 24.Moffitt, M. C., S. I. Blackburn, and B. A. Neilan. 2001. rRNA sequences reflect the ecophysiology and define the toxic cyanobacteria of the genus Nodularia. Int. J. Syst. E vol. Microbiol. 51:505-512. [DOI] [PubMed] [Google Scholar]
- 25.National Health and Medical Research Council/Agriculture and Resource Management Council of Australia and New Zealand. 1996. Australian drinking water guidelines. National Health and Medical Research Council, and Agriculture and Resource Management Council of Australia and New Zealand, Canberra, Australia.
- 26.Neilan, B. A., D. Jacobs, and A. E. Goodman. 1995. Genetic diversity and phylogeny of toxic cyanobacteria determined by DNA polymorphisms within the phycocyanin locus. Appl. Environ. Microbiol. 61:3875-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neilan, B. A., D. Jacobs, T. Deldot, L. L. Blackall, P. R. Hawkins, P. T. Cox, and A. E. Goodman. 1997. rRNA sequences and evolutionary relationships among toxic and nontoxic cyanobacteria of the genus Microcystis. Int. J. Syst. Bacteriol. 47:693-697. [DOI] [PubMed] [Google Scholar]
- 28.Neilan, B. A., J. L. Stuart, A. E. Goodman, P. T. Cox, and P. R. Hawkins. 1997. Specific amplification and restriction polymorphisms of the cyanobacterial rRNA operon spacer region. Syst. Appl. Microbiol. 20:612-621. [Google Scholar]
- 29.Nishiwaki-Matsushima, R., T. Ohta, S. Nishiwaki, M. Suganuma, K. Kohyama, T. Ishikawa, W. W. Carmichael, and H. Fujiki. 1992. Liver tumour promotion by the cyanobacterial cyclic peptide toxin microcystin-LR. J. Cancer Res. Clin. Oncol. 118:420-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otsuka, S., S. Suda, R. H. Li, M. Watanabe, H. Oyaizu, S. Matsumoto, and M. M. Watanabe. 1999. Phylogenetic relationships between toxic and non-toxic strains of the genus Microcystis based on 16S to 23S internal transcribed spacer sequence. FEMS Microbiol. Lett. 172:15-21. [DOI] [PubMed] [Google Scholar]
- 31.Rippka, R., and M. Herdman. 1992. Pasteur Culture Collection (PCC) of cyanobacterial strains in axenic culture, vol. 1. Catalogue of strains. Institut Pasteur, Paris, France.
- 32.Rudi, K., O. M. Skulberg, and K. S. Jakobsen. 1998. Evolution of cyanobacteria by exchange of genetic material among phyletically related strains. J. Bacteriol. 180:3453-3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saker, M. L., and B. A. Neilan. 2001. Varied diazotrophies, morphologies, and toxicities of genetically similar isolates of Cylindrospermopsis raciborskii (Nostocales, Cyanophyceae) from northern Australia. Appl. Environ. Microbiol. 67:1839-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sivonen, K. 1996. Cyanobacterial toxins and toxin production. Phycologia 35:12-24. [Google Scholar]
- 35.Tillett, D., E. Dittmann, M. Erhard, H. von Dohren, T. Borner, and B. A. Neilan. 2000. Structural organization of microcystin biosynthesis in Microcystis aeruginosa PCC7806: an integrated peptide-polyketide synthetase system. Chem. Biol. 7:753-764. [DOI] [PubMed] [Google Scholar]
- 36.Tillett, D., D. L. Parker, and B. A. Neilan. 2001. Detection of toxigenicity by a probe for the microcystin synthetase A gene (mcyA) of the cyanobacterial genus Microcystis: comparison of toxicities with 16S rRNA and phycocyanin operon (phycocyanin intergenic spacer) phylogenies. Appl. Environ. Microbiol. 67:2810-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wasson, R., R. Banens, P. Davies, W. Maher, S. Robinson, R. Volker, D. Tait, and S. Watson-Brown. 1996. Inland waters, p. 7.1-7.55. In R. Taylor (ed.), Australia: state of the environment 1996. Commonwealth Scientific and Industrial Research Organisation, Collingwood, Australia.
- 38.Watanabe, M. M., and M. Hiroki. 1997. NIES—collection list of strains. Microalgae and protozoa. National Institute for Environmental Studies, Tsukuba, Japan.
- 39.Wilkinson, J. F. G. 1986. Classification of average chemical compositions of common basalts and andersites. J. Petrol. 27:31-62. [Google Scholar]