Abstract
Yersinia pestis is the causative agent of the bubonic, septicemic, and pneumonic plagues (also known as black death) and has been responsible for recurrent devastating pandemics throughout history. To further understand this virulent bacterium and to accelerate an ongoing sequencing project, two whole-genome restriction maps (XhoI and PvuII) of Y. pestis strain KIM were constructed using shotgun optical mapping. This approach constructs ordered restriction maps from randomly sheared individual DNA molecules directly extracted from cells. The two maps served different purposes; the XhoI map facilitated sequence assembly by providing a scaffold for high-resolution alignment, while the PvuII map verified genome sequence assembly. Our results show that such maps facilitated the closure of sequence gaps and, most importantly, provided a purely independent means for sequence validation. Given the recent advancements to the optical mapping system, increased resolution and throughput are enabling such maps to guide sequence assembly at a very early stage of a microbial sequencing project.
There are 11 species in the genus Yersinia, 3 of which are pathogenic for humans (Yersinia pestis, Y. enteroclitica, and Y. pseudotuberculosis). Of the three pathogenic species, Y. pestis, which lives in rodents, transmits this pathogen to humans via fleas, causing bubonic, septicemic, and pneumonic plagues (also known as black death). Y. pestis has been responsible for many recurrent devastating pandemics throughout history, resulting in widespread loss of human life (1, 7, 22). To understand the molecular mechanisms that define the pathogenicity of this bacterial species, sequencing of two strains (KIM and CO-92 biovar Orientalis) of Y. pestis was funded by the National Institute of Allergy and Infectious Diseases and Beowulf Genomics. The two separate sequencing projects were conducted by F. Blattner's laboratory (University of Wisconsin—Madison) and Sanger Center (Hinxton, United Kingdom), using the strategy of whole-genome shotgun sequencing (11). Although this sequencing strategy has racked up an impressive number of completed microbial genomes, it can be further optimized in terms of the cost and effort required during the finishing stages. In this regard, physical maps serve to guide sequence assembly, characterize gaps, and validate the finished sequence. Furthermore, ordered restriction maps are particularly useful when attempting to assemble genomic regions containing repeats, since cleavage patterns can accurately discern such sequence elements.
The costly appellation of “finished,” assumed when dealing with high-quality sequence data, minimally mandates that no genomic region be excluded from the final results (23) and thus requires extensive finishing efforts. The chances that entire regions of a genome might be excluded from “completed” sequence increases in the face of limited budgets and the absence of physical mapping data. Because of these issues, optical maps were used (15) at an early stage of sequence assembly to identify gaps (XhoI) and later to validate the finished sequence (PvuII).
The optical-mapping system has been extensively described elsewhere (5, 6, 9, 13, 15, 16). Briefly, optical mapping is a single-molecule system, which constructs ordered restriction maps from assembles of single DNA molecules mounted on charged glass surfaces. Fluorescence microscopy is used to image DNA molecules after enzymatic cleavage, but, importantly, the restriction fragments maintain their original order. Since the molecules are stained with a fluorochrome, integrated fluorescence intensity measurements are used to accurately determine the mass of each restriction fragment. Maps of entire microbial genomes are then constructed using a map assembler (2, 3), in a process akin to shotgun sequence assembly.
Here we present two optical maps (XhoI and PvuII) of the Y. pestis strain KIM genome that were used as a scaffold for sequence assembly and to validate the finished sequence. Notably, the second (PvuII) optical map is a very high-resolution map with an average fragment size of about 12 kb; it represents the highest resolution attained thus far by optical mapping for an entire genome.
MATERIALS AND METHODS
DNA preparation.
Y. pestis strain KIM genomic DNA gel inserts were prepared from a culture grown in heart infusion broth at 30°C (25) and stored in 0.5 M EDTA (pH 8.0) (17). Prior to use, the DNA gel inserts were washed thoroughly overnight in TE (10 mM Tris, 1 mM EDTA [pH 8.0]) to remove excess EDTA. To release DNA molecules, washed inserts were melted at 72°C for 7 min. A β-agarase solution (100 μl of TE, 1 μl [1 Unit] of β- agarase [New England Biolabs, Beverly, Mass.]), prewarmed to 42°C, was added to the melted inserts, and the mixture was allowed to incubate at 40°C for 2 h. Suitable dilutions were made from this sample with TE to ensure minimal crowding of molecules on optical mapping surfaces. Lambda DASH II bacteriophage DNA (Stratagene, La Jolla, Calif.) was added to the genomic DNA solution (10 pg/μl) as an internal standard for fragment sizing. Such samples were mounted onto an optical mapping surface and examined by fluorescence microscopy to check molecular integrity and concentration.
Surface preparation.
Glass coverslips (22 by 22 mm [Fisher's Finest; Fisher Scientific]) were racked in custom-made Teflon racks, cleaned by boiling in Nano-Strip (sulfuric acid and hydrogen peroxide; Cyantek Corp., Fremont, Calif.) for 50 min (68 to 75°C), and then rinsed extensively with high-purity, dust-free water. After six washes, the surfaces were hydrolyzed in boiling concentrated hydrochloric acid at 98°C for 6 h and rinsed extensively with high-purity water until the wash was neutral. The coverslips were removed from Teflon racks and individually rinsed three times in absolute ethanol; they were then stored in absolute ethanol in polypropylene containers at room temperature.
To derivatize, 30 cleaned, hydrolyzed surfaces were placed in a flat Teflon block holder in a clean Qorpac container by using forceps and allowed to dry for 5 to 10 min at room temperature. High-purity water (250 ml) was added to a clean polypropylene bottle, to which 62 μl of trimethyl silane (N-trimethylsilylpropyl-N,N,N-trimethylammonium chloride [Gelest Corp., Tullytown, Pa.]) and 3 μl of vinyl silane (vinyltrimethoxysilane [Gelest Corp.]) were added and shaked vigorously for several minutes. The solution was poured into the Qorpac container and incubated at 65°C with gentle shaking (50 rpm) for 17.5 h. The container was then opened in a hood for 1 h to thermally equilibrate. Finally, the silane solution was aspirated off and the surfaces were rinsed three times with high-purity water and once with ethanol and then stored in distilled absolute ethanol. They remain usable for 2 to 3 weeks. Surface properties were assayed by digesting lambda DASH II bacteriophage DNA with 40 units of XhoI diluted in 100 μl of digestion buffer with 0.02% Triton X-100 (Sigma) at 37°C to determine optimal digestion times, which ranged from 40 to 120 min.
DNA mounting, overlay, digestion, and staining.
DNA molecules were mounted on derivatized glass surfaces by capillary action as described by Lim et al. (15). Then a thin layer of acrylamide (3.3%) was applied to the surface, and upon curing, was washed with 400 μl of TE (2×) for 2 min and with 200 μl of polymerization buffer for another 2 min. To set up the digestion, 200 μl of digestion buffer with enzyme (20 μl of NEB [New England Biolabs] buffer 2, 20 μl of bovine serum albumin, 158 μl of high-purity water, 2 μl of NEB-XhoI [20 Units/μl]) was added to the surface, which was incubated in a humidified chamber at 37°C for 40 to 120 min. After digestion, the surface was washed three times by adding 200 μl with TE and aspirated off. The surface was mounted onto a glass slide with 12 μl of 0.2 μM YOYO-1 solution (containing 5 parts of YOYO-1 [1,1′-[1,3-propanediylbis[(dimethyliminio)-3,1-propanediyl]]bis[4-[(3-methyl-2(3H)-benzoxazolylidene)methyl]] tetraiodide; Molecular Probes, Eugene, Oreg.] and 95 parts of β-mercaptoethanol in 20% [vol/vol] TE). The sample was sealed with nail polish and incubated (at room temperature in the dark) for 20 min or overnight for the stain to diffuse before being checked under the fluorescence microscope. For the XhoI map, the DNA mounting was as described as above. The PvuII mapping used a microfluidic device, which is described elsewhere.
Image acquisition and processing.
Samples were imaged by fluorescence microscopy as described previously (15) by using a 63× objective (Zeiss) and high-resolution digital camera (Princeton Instruments). Images were collected for the PvuII map using a fully automated image acquisition system developed by our laboratory. Comounted lambda DASH II (XhoI map) and T7 bacteriophage DNA (PvuII map) molecules were used to estimate the digestion rate and to provide internal fluorescence standards for accurately sizing the DNA fragments (4, 13, 16). The image files were processed to create maps using previously described software (15).
Map assembly.
Individual-molecule restriction maps were overlapped by aligning restriction sites based on fragment sizes by using specially written software “Gentig” (2, 3, 4, 13, 15, 16). Gentig assembles single-molecule restriction maps into a genome-wide contig by using a greedy algorithm with limited backtracking for finding an almost optimal scoring set of map contigs to avoid the high computational complexity that would occur in attempting to find the optimal assembly. Bayesian inference was used to estimate the probability that two distinct single-molecule restriction maps could have been derived from the proposed placement while subject to various data errors such as sizing errors, missing cut sites, and false cut errors. The Bayesian approach requires fine-tuning of these parameters and a known prior statistical distribution of error sources. These parameters, such as standard deviation, digestion rate, false cut, and false match probability, can be reestimated from the data by using a limited number of iterations of Bayesian probability density maximization. After the parameters are correctly estimated from the data, the best offset and alignment between a pair of maps can be computed by an efficient dynamic programming algorithm.
Optical maps versus in silico maps and composite maps.
The XhoI and PvuII optical maps of the Y. pestis KIM genome were initially aligned separately with the in silico XhoI and PvuII maps generated from the finished sequence by using Gentig. The missing fragments and the false cuts or missing cuts were determined based on these alignments. The relative error for each fragment in the optical maps was calculated from the formula [(in silico map fragment size) − (optical map fragment size)]/(in silico map fragment size) and was plotted against in silico map fragment masses to visualize the relationship between fragment size and the relative error.
A composite, in silico, XhoI and PvuII map was generated from the finished sequence by using MapDraw in DNAStar (DNAStar Corp.) by breaking the genomic sequence into 500-kb segements. The optical maps were broken into segments based on the alignments between the single-digest in silico map and the corresponding optical maps. The optical map segments were normalized to the in silico map segment size and were then manually aligned with the in silico double-digested maps generated by using DNAStar software. Then this composite, in silico XhoI and PvuII map was used as a template, and the two separate optical maps were manually aligned to the template.
Sequencing.
All sequencing operations were performed at the Blattner laboratory (8, 21). Briefly, a whole-genome shotgun library of Y. pestis was prepared by random shearing of genomic DNA. Fragments between 1 and 2.5 kb were fractionated and cloned into the M-13 Janus vector for sequencing. A pBluescript plasmid library with an insert size of 5 to 6 kb was also built for this project. Some of the clones from each library were dual-end sequenced and used as linking clones.
RESULTS
Acquisition of optical map data.
We used whole-genome shotgun optical mapping (13, 16) to construct the restriction maps presented here, and this mapping scheme is similar in many respects to whole genome shotgun sequencing (11, 18, 24). However, shotgun optical mapping assembles maps derived from single DNA molecules into gap-free maps, which span entire genomes. Moreover, since shotgun optical mapping directly constructs maps from genomic DNA, the system obviates the need for clones, PCR, or separation techniques. Like shotgun sequencing, a number of single-molecule maps (versus “sequence reads”) are used to cover any given locus to deal with errors and to establish continuity across a genome.
Our cell lysis procedures (see Materials and Methods) naturally break DNA molecules into large, random fragments, and such molecules were mounted onto optical mapping surfaces for digestion by restriction endonucleases. Images of digested DNA molecules, ranging in size from 265 to 2,600 kb were acquired at the microscope by using Gencol (see Materials and Methods), a software package designed to control image acquisition. Maps derived from single molecules were assembled into consensus maps covering the entire genome by using the Gentig map assembler (2, 3, 4, 13).
A total of 251 digested DNA molecules were imaged and processed to construct the XhoI map. Of this total, 208 went into the final map contig, thus showing a favorable contig rate of 83%. This high rate of contig formation was due to the availability of large (918.9-kb [average molecular size]) well-digested molecules (87.15% of the available cleavage sites were cut). In other words, the mapped molecules contained large, well-defined patterns, which facilitated the alignment process. The total mass of these molecules was 230.66 Mb, which represents approximately 50× coverage of the Y. pestis genome.
Figure 1 shows the finished XhoI map contig containing 208 molecules, with an average restriction fragment size of 20.4 kb. The map was circularized without gaps, and a typical restriction fragment was computed from the average of about 30 molecules. The precision of the mapping process (sizing error per single molecule) was estimated from the standard deviation calculated from the ensemble of restriction fragments used to determine each map position. The average standard deviation of the fragment size about the mean was 3.11 kb, based on the finished map contig or consensus map. Finally, the genome size of this strain of Y. pestis was calculated to be 4.57 Mb, based on summing all restriction fragments comprising the XhoI consensus map.
FIG. 1.
Whole-genome XhoI map contig of Y. pestis KIM displayed by the DNAStar software package. The outermost color circle is the consensus map generated by Gentig and is built from the underlying maps represented as arcs. These maps were constructed from individual DNA molecules cleaved with XhoI. Congruent restriction fragments shown in the consensus map are denoted by a common color; the color-ordering scheme is random to provide contrast.
A total of 283 molecules were collected and processed to construct the PvuII map. As with the XhoI map, a high percentage of the molecules were assembled into the final consensus map (225 molecules, or 79.5%). The summed mass of these collected molecules was 208.79 Mb, which represents about 45× coverage. The average molecule size was 737.79 kb and the average restriction fragment size was 15.27 kb using the pool of collected molecule maps. The finished PvuII consensus map (constructed with Gentig) was composed of 225 molecules (not shown). This map was also circularized with no gaps, and a typical restriction fragment was computed from the average of 25 molecules. The digestion rate was calculated at 77.46%, and the average standard deviation about the mean fragment size was 1.56 kb based on the final circularized consensus map. The genome size of this strain of Y. pestis was calculated to be 4.68 Mb, again after summing all of the PvuII fragments contained within the consensus map.
This high degree of genome coverage provided strong confidence in determining restriction sites, ensured accurate fragment sizing, and enabled accurate size estimates for the circularized genome map. The accuracy of the optical map versus the genome sequence is compared below. Since Gentig (see Materials and Methods) statistically evaluates a series of hypothetical maps based on scoring them against the molecule data set, with priors based on experimental error models, the correctness of a final map can be evaluated. Accordingly, the false-positive probability of the maps reported here was 0.00004 and 0.01257 for the circularization of the XhoI and PvuII optical maps, respectively. Given that the PvuII map had higher resolution, its higher false-positive probability probably reflects errors associated with the optical sizing of small restriction fragments (19) and the increase of the random circularization probability for high resolution map. However, for both maps, the false-positive probabilities for the circularization were below 0.05, which has been found to be an acceptable value for reliable map circularization. The restriction patterns generated by both XhoI and PvuII were apparently random; hence, no specific restriction patterns or structural features were discerned.
Uses of optical maps in sequence assembly.
The XhoI map was used to guide and validate the genome sequence assembly when a number of expansive (on average 243-kb) sequence contigs were generated by the sequence assembly program (10). Here, the XhoI map aided in the clarification of repetitive regions that are commonly prone to sequence misassembly. Due to clone size limitations, paired sequence reads are often unable to properly span such regions with flanks that contain unique sequence, which can be used to ensure correct assembly.
The Janus sequencing approach (see Materials and Methods) enabled the dual-ended sequencing of clones. However, when this resource was exhausted during the assembly process, 19 contigs were generated, which ranged in size from 39.83 to 671.06 kb. At this stage, Gentig was used to align the in silico XhoI sequence contig maps with the XhoI optical map. These alignments provided contig order, orientation, and the identification of gaps in terms of position and breadth (data not shown). Of the 19 in silico sequence contig maps, 8 were aligned with the XhoI optical map by using Gentig while 7 required manual alignment. This was necessary since the rigorous alignments that Gentig affords require more than seven restriction fragments to be present in the optical map. The remaining four contigs presented in silico sequence contig maps that initially did not agree with the optical map. These were later identified to contain misassemblies due to the repetitive sequences. This validation process lead to reassembly of the contigs to enable the alignment of the in silico sequence contig maps with the optical map.
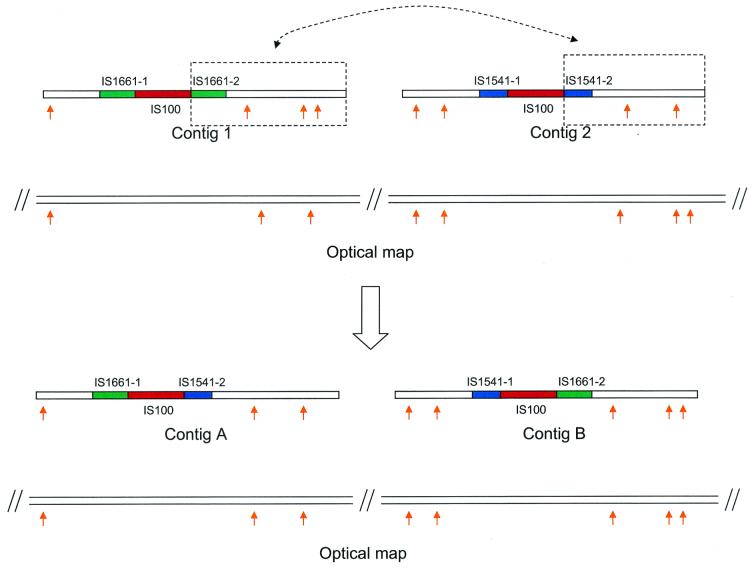
As shown in Fig. 2, contigs A and B were originally assembled with each contig having an IS element (IS1661 and IS1541, respectively) disrupted by the insertion of a copy of IS100. The in silico sequence contig maps of these two contigs were not consistent with the optical map. Apparently a homologous recombination event between the two copies of IS100 generated two hybridized IS structures (10), resulting in two new contigs whose in silico sequence contig maps perfectly matched the optical map. The new assemblies were later confirmed by four pairs of sequences obtained from pBluescript plasmid clones. Initially, these missassemblies were not apparent since only ∼2× of the plasmid clones were dual-end sequenced.
FIG. 2.
Example of sequence validation and correction via optical mapping. Orange arrows indicate XhoI restriction sites. Partial IS elements IS1661-1 and IS1661-2 together make an intact IS1661 element, and partial IS elements IS1541-1 and IS1541-2 together make an intact IS1541 element. Contigs 1 and 2 were originally assembled with each bearing an IS element (IS1661 and IS1541, respectively) disrupted by the insertion of a copy of IS100. The in silico sequence contig maps were not consistent with the optical map. Allowing for a homologous recombination event between the two copies of IS100, however, generated two hybridized IS structures, which was confirmed by new contigs (A and B) whose in silico sequence contig maps perfectly matched the optical map.
Once the misassemblies were corrected, all of the in silico sequence contig maps were aligned with the optical map, leading to the identification of the contig order, orientation, and gaps. The gaps were then closed by techniques such as genomic PCR and primer walking based on the alignments of the in silico sequence contig maps with the optical map. As a final step in the sequence validation process, an in silico PvuII map was constructed from the genome sequence, which included XhoI optical map information, and compared with the PvuII optical map. This comparison provided a purely independent means of sequence validation, which showed minor differences and is described below.
Assessment of optical mapping errors.
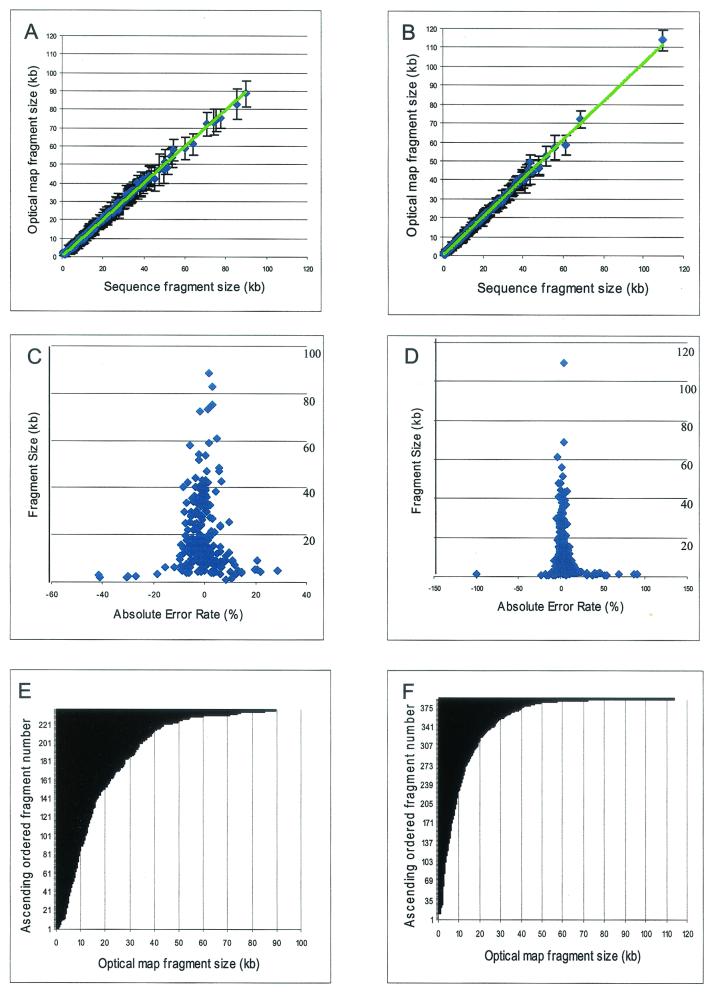
To assess the errors and accuracy of the XhoI and PvuII maps, comparisons were made between optical mapping and sequence data (Fig. 3). The relative sizing error was determined by the alignment of optical maps with in silico maps constructed from the available sequence data (Fig. 3A to D). The error bars indicate the standard deviation about the means of the restriction fragment sizes used to calculate the consensus map fragments shown in Fig. 1 and 4. Generally, a high degree of correspondence was found between optical map fragment sizes and the in silico map fragment sizes, and the trend line is almost identical to the diagonal line. Figures 3C and D are scatter plots showing how the absolute relative fragment sizing error (optical map versus in silico map) varies with restriction fragment size. Overall, relative sizing errors were as expected, found to be inversely proportional to the fragment size (19). For the XhoI map, the average relative sizing error [100 × (|optical map fragment size − corresponding in silico map fragment size|)/corresponding in silico map fragment size] was 4.88% for fragments smaller than 5 kb and 6.62% for fragments larger than 5 kb. A similar assessment of the PvuII map showed an average relative sizing error of 12.22% for fragments smaller than 5 kb and 3.04% for fragments larger than 5 kb.
FIG. 3.
Comparisons of XhoI and PvuII optical map to sequence data. (A and B) Plots of optical map fragment sizes versus the in silico map fragment sizes from finished sequence for XhoI (A) and PvuII (B). The error bars represent the standard deviation of optical map fragment size on the means. (C and D) Plots of the absolute error rates of optical fragment size versus sequence for XhoI (C) and PvuII (D). (E and F) Cumulative histograms of optical map fragments, by size, for XhoI (E) and PvuII (F).
FIG. 4.
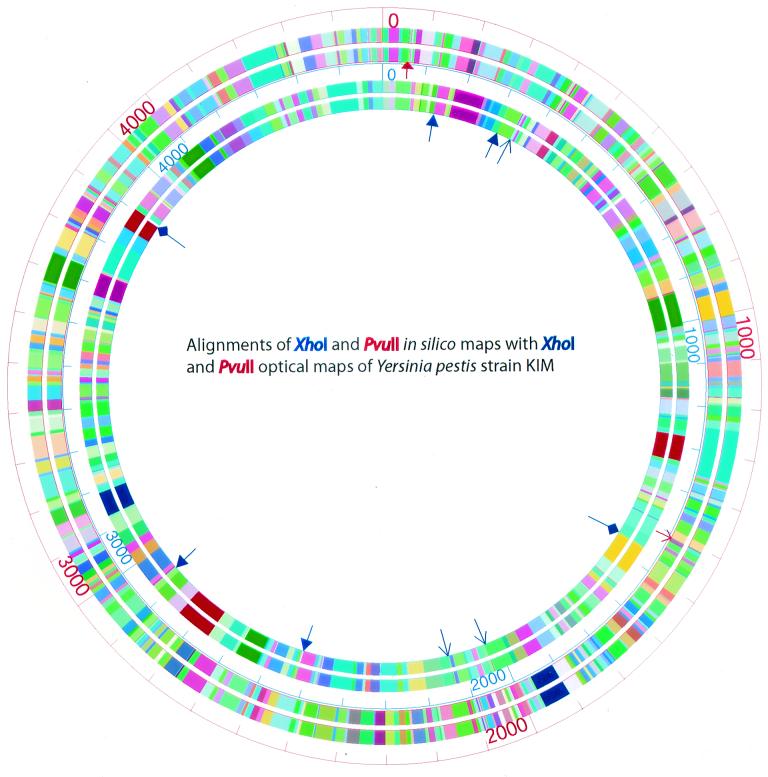
Alignments of the XhoI and PvuII consensus optical maps with the corresponding in silico maps. The outer two circles are the alignment of the in silico PvuII map (the outermost circle) with the PvuII optical map, while the inner two circles show the alignment of the in silico XhoI map (the outer one) with the XhoI optical map. Arrows with solid triangular heads denote missing fragments (missing fragments less than 1 kb are not shown); slender arrows denote missing cuts; arrows with diamonds denote false cuts. Blue and red arrows refer to the XhoI and PvuII maps, respectively.
Figure 3E and F show the distribution of XhoI and PvuII restriction fragments contained within the finished consensus maps. These data show an exponential distribution, as expected, with no apparent discontinuities. Further examination of these distributions reveals a major difference in the frequency of fragments smaller than 2 kb and between 2 and 5 kb, which may account for the increased errors associated with small fragments produced by PvuII. For the PvuII map, 86 of 444 fragments were smaller than 2 kb and 84 were between 2 and 5 kb, while for the XhoI map, 30 of 267 fragments were smaller than 2 kb and 29 were between 2 and 5 kb. The error rates for fragments smaller than than 2 kb were 19.76% for the PvuII map and 28.11% for the XhoI map. The error rates for fragments between 2 and 5 kb were 6.86% for the PvuII map and 10.21% for the XhoI map.
Comparing optical maps to the sequence.
Although the previous analysis portrays overall errors, it is important to assess such errors in the context of consensus map locations versus sequence. The alignment of the XhoI optical map with the corresponding in silico map (Fig. 4) showed that there were only 2 false cuts (present in the optical map but not in the in silico map) and 4 missing cuts out of a total of 267 XhoI cuts, assuming that the in silico maps were correct. Missing fragments were likewise compared, and here we determined that 24 of 267 fragments were missing in the XhoI optical map. Normally, optical maps do not report restriction fragments below 500 bp and they report attenuated recording of fragments smaller than 1 kb (19). Further analysis showed that 20 fragments were smaller than 1 kb.
The alignment of the PvuII optical map with the in silico PvuII map (Fig. 4) showed no false cuts. However, the optical map omitted 13 cleavage sites in relation to the in silico map, and 22 of 444 fragments (2 were between 1 and 2 kb, and the rest were smaller than 0.5 kb) were also omitted.
Composite optical maps.
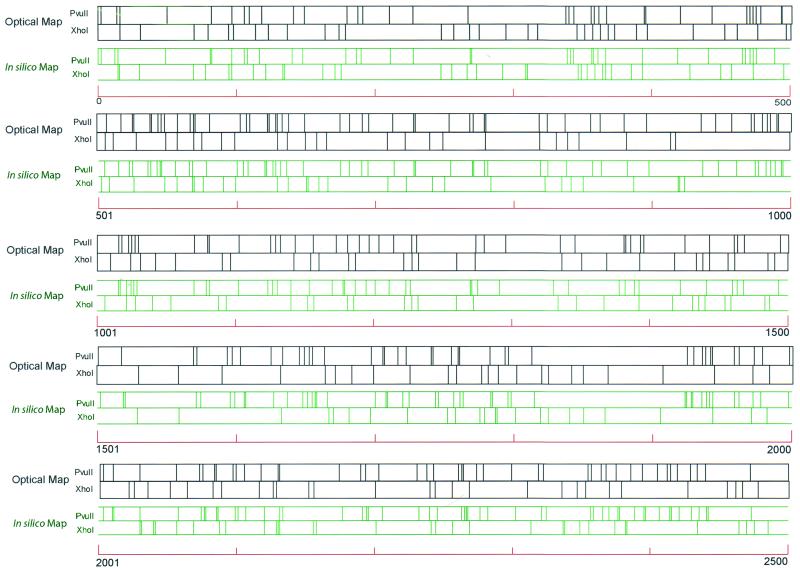
Figure 5 shows the optical composite map and the in silico composite map of Y. pestis strain KIM with both XhoI and PvuII restriction sites marked on the maps. These maps were aligned with an in silico composite map, which facilitates comparison (see Materials and Methods). Due to sizing errors, the occurrence of some cleavage site reversals between the contiguous restriction sites (XhoI and PvuII) is unavoidable. In other words, for a given map locus, does a XhoI site follow or precede a neighboring PvuII site? Accordingly, we identified the following instances of restriction sites reversals (number of reversals/total number of contiguous sites per 500 kb of alignment): 6/37, 5/37, 9/37, 2/24, 7/33, 9/33, 4/35, 11/38, 10/36, and 0/4. Collectively, this analysis showed a reversal rate of about 20%.
FIG. 5.
XhoI and PvuII composite optical-map alignment with the in silico XhoI and PvuII map. The composite optical map was generated by normalizing the single-enzyme maps to the same size. Both the in silico and optical maps were broken into 500-kb segments and then leftmost aligned (in a pairwise fashion), using the sequence as the guiding template. The green and blue lines represent the in silico and optical maps, respectively. Scale = 100 kb.
DISCUSSION
Whole-genome XhoI and PvuII optical maps of Y. pestis strain KIM were constructed to validate sequence assemblies and simplify the gap closure aspects of a parallel-sequencing project. The XhoI map was used solely to guide sequence assembly through the validation of nascent sequence contigs. This process worked in several ways: (i) nascent sequence contigs were aligned with the map to assess sequence assembly errors, (ii) validated sequence contigs were placed and oriented on the map, and (iii) gaps were characterized between mapped sequence contigs. Since the finished sequence contained information gleaned from the XhoI optical map, the PvuII map was reserved and used as a purely independent means of sequence validation.
The finished sequence of Y. pestis strain KIM5 is 4.60 Mb (10), very close to our estimate of 4.57 Mb based on the XhoI optical restriction map, this shows that there was a mere 0.7% difference or map error. This genome sizing error is generally superior to that associated with other whole-genome physical maps constructed using pulsed-field gel electrophoresis (12, 13, 19). Previously, the genome size of Y. pestis strain KIM was estimated by two-dimensional pulsed-field gel electrophoresis (SpeI) (17) to be 4.21 Mb, which indicates a sizing error of about 8.5% compared to sequence data. An accurate and independent estimation of genome size is critical when embarking on or concluding a microbial sequencing project, since goals and end points can be precisely known. We found that the in silico PvuII map constructed from the finished sequence of Y. pestis strain KIM was almost congruent with its PvuII optical restriction map counterpart. The differences between them stemmed mainly from the absence of, or errors associated with, small restriction fragments (smaller than 2 kb).
Advancements in DNA mounting, image collection, processing, and map assembly software have resulted in a new optical mapping system capable of high-resolution analysis. These advancements are evident in the map data presented here. For example, the XhoI map of Y. pestis strain KIM was generated by the old system before modifications. The average fragment sizing error was 3.11 kb, and more than half of the fragments smaller than 2 kb based on the in silico map were missing. However, the higher-resolution PvuII map (average fragment size, 12.06 kb) was constructed using the modified system, with the results showing twice the precision (1.56 kb, [Fig. 3B]). The recent success in the construction of a BamHI optical map of Shigella flexneri with an average fragment size of 10.72 kb based on the finished optical map (unpublished results) and an average fragment size standard deviation of 1.48 kb provides another example that the resolution and precision of optical mapping have been greatly improved. It seems contradictory that the sizing accuracy of optical mapping has been reduced in the modified system because the average relative sizing error (optical map versus in silico map) is smaller for the XhoI map (5.14%) than for the PvuII map (6.00%). The reason for this result is that both the old and the modified optical mapping systems have reduced accuracy when sizing small fragments. In the XhoI optical map, less than 1/10 of the fragments are smaller than 5 kb, while in PvuII map, about 1/3 of the fragments are smaller than 5 kb, as shown in Fig. 3E and F. We therefore see a slight increase in the relative sizing error in the PvuII optical map.
With increasing resolution of the optical mapping system, maps could be used for comprehensive genotyping. Since most genes are highly conserved among different strains of the same bacterial species or even closely related species (20, 21), this provides the basis for optical maps to identify large syntenic regions and instances of chromosomal rearrangements across closely related microbial genomes. Such rearrangements may be discerned as large insertions, deletions, or translocations. Given the sequence of a prototypic or reference strain, this would enable molecular studies without the need to sequence each microbe used in the comparison. For example, the flanking regions around a translocation event identified by a map versus sequence analysis could be sequenced by the pinpoint generation of appropriate amplicons. We are actively making progress toward this goal.
A composite map constructed from the linear addition of independently derived restriction enzyme maps is more informative than a single-enzyme map having the same average restriction fragment size (14). The construction of such maps is generally not problematic for clones or for bacterial genomes, where macro- and microrestriction maps are commonly combined. However, expansive, high-resolution optical maps of entire genomes present new technical challenges in the alignment of congruent fragments from different restriction enzyme maps. What is sought is the proper registration of restriction fragments relative to each other (i.e., does the XhoI fragment come before or after the PvuII fragment?). Our previous efforts to align restriction sites of different enzymes have produced some unavoidable errors, especially for small fragments (smaller than 5 kb) (12, 15). In this study, the composite optical map of Y. pestis KIM with XhoI and PvuII was constructed based on the alignments of the two separate optical maps with the in silico map made from the finished sequence. We conclude from this analysis that a new algorithm should be developed to systematically align multiple optical maps with sequence data, which takes into account both map and sequence errors.
In summary, the high-resolution maps we have presented were constructed using a newly advanced optical mapping system, and we have shown how such maps can facilitate large-scale sequencing efforts. However, we plan to use these same advancements to lay the groundwork for large-scale comparative genome studies that would evade analysis by microarray-based approaches.
Acknowledgments
This work was supported by grant AI44387 from NIH/NIAID to F.B., subcontract to D.C.S.
REFERENCES
- 1.Achtman, M., K. Zurth, G. Morelli, G. Torrea, A. Guiyoule, and E. Carniel. 1999. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 96:14043-14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anantharaman, T. S., B. Mishra, and D. C. Schwartz. 1999. Genomics via optical mapping. III. Contiging genomic DNA and variations. The Seventh International Conference on Intelligent Systems for Molecular Biology, vol. 7, p. 18-27. [PubMed]
- 3.Anantharaman, T. S., B. Mishra, and D. C. Schwartz. 1998. Genomics via optical mapping. III. Contiging genomic DNA and variations. Courant Technical Report 760. Courant Institute, New York University, New York, NY.
- 4.Anantharaman, T. S., B. Mishra, and D. C. Schwartz. 1997. Genomics via optical mapping. 2. Ordered restriction maps. J. Comput. Biol. 4:91-118. [DOI] [PubMed] [Google Scholar]
- 5.Aston, C., C. Hiort, and D. C. Schwartz. 1999. Optical mapping: an approach for fine mapping. Methods Enzymol. 303:55-73. [DOI] [PubMed] [Google Scholar]
- 6.Aston, C., B. Mishra, and D. C. Schwartz. 1999. Optical mapping and its potential for large-scale sequencing projects. Trends Biotechnol. 17:297-302. [DOI] [PubMed] [Google Scholar]
- 7.Bearden, S. W., J. D. Fetherston, and R. D. Perry. 1997. Genetic organization of the yersiniabactin biosynthetic region and construction of avirulent mutants in Yersinia pestis. Infect. Immun. 65:1659-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1462. [DOI] [PubMed] [Google Scholar]
- 9.Cai, W., J. Jing, B. Irvin, L. Ohler, E. Rose, H. Shizuya, U. Kim, M. Simon, T. Anantharaman, B. Mishra, and D. C. Schwartz. 1998. High-resolution restriction maps of bacterial artificial chromosomes constructed by optical mapping. Proc. Natl. Acad. Sci. USA 95:3390-3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng, W., V. Burland, G. Plunkett III, A. Boutin, G. F. Mayhew, P. Liss, N. T. Perna, D. J. Rose, B. Mau, D. C. Schwartz, S. Zhou, J. D. Fetherston, L. E. Lindler, R. R. Brubaker, G. V. Plano, S. C. Straley, K. A. McDonough, M. L. Nilles, J. S. Matson, F. R. Blattner, and R. D. Perry. 2002. Genome sequence of Yersinia pestis KIM. J. Bacteriol. 184:4601-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, J. M. McKenney, K. Sutton, G. FitzHugh, W. Fields, C. Gocayne, J. D. Scott, J. Shirley, R. Liu, L.-I. Glodek, A. Kelley, J. M. Weidman, J. F. Phillips, C. A. Spriggs, T. Hedblom, E. Cotton, M. D. Utterback, T. R. Hanna, M. C. Nguyen, D. T. Saudek, D. M. Brandon, R. C. Fine, L. D. Fritchman, J. L. Fuhrmann, J. L. Geoghagen, N. S. M. Gnehm, C. L. McDonald, L. A. Small, K. V. Fraser, C. M. Smith, and J. C. Venter. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 12.Jing, J., Z. Lai, C. Aston, J. Lin, D. J. Carucci, M. J. Gardner, B. Mishra, T. Anantharaman, H. Tettelin, L. M. Cummings, S. L. Hoffman, J. C. Venter, and D. C. Schwartz. 1999. Optical mapping of Plasmodium falciparum chromosome 2. Genome Res. 9:175-181. [PMC free article] [PubMed] [Google Scholar]
- 13.Lai, Z., J. Jing, C. Aston, V. Clarke, J. Apodaca, E. T. Dimalanta, D. J. Carucci, M. J. Gardner, B. Mishra, T. S. Anatharaman, S. Paxia, S. L. Hoffman, J. C. Venter, E. J. Huff, and D. C. Schwartz. 1999. A shotgun optical map of the entire Plasmodium falciparum genome. Nat. Gene. 23:309-313. [DOI] [PubMed] [Google Scholar]
- 14.Lander, E. S., and M. S. Waterman. 1988. Genomic mapping by fingerprinting random clones: a mathematical analysis. Genomics 2:231-239. [DOI] [PubMed] [Google Scholar]
- 15.Lim, A., E. T. Dimalanta, K. D. Potamousis, G. Yen, J. Apodoca, C. Tao, J. Lin, R. Qi, J. Skiadas, A. Ramanathan, N. T. Perna, G. Plunkett III, V. Burland, B. Mau, J. Hackett, F. R. Blattner, T. S. Anantharaman, B. Mishra, and D. C. Schwartz. 2001. Shotgun optical maps of the whole Escherichia coli O157:H7 genome. Genome Res. 11:1584-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin, J., R. Qi, C. Aston, J. Jing, T. S. Anatharaman, B. Mishra, O. White, M. J. Daly, K. W. Minton, J. C. Venter, and D. C. Schwartz. 1999. Whole genome shotgun optical mapping of Deinococcus radiodurans. Science 285:1558-1562. [DOI] [PubMed] [Google Scholar]
- 17.Lucier, T. S., and R. R. Brubaker. 1992. Determination of genome size, macrorestriction pattern polymorphism, and nonpigmentation-specific deletion in Yersinia pestis by pulsed-field gel electrophoresis. J. Bacteriol. 174:2078-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marra, M. A., T. A. Kucaba, N. L. Dietrich, E. D. Green, B. Brownstein, R. K. Wilson, K. M. McDonald, L. W. Hillier, J. D. McPherson, and R. H. Waterston, 1997. High throughput fingerprint analysis of large-insert clones. Genome Res. 7:1072-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meng, X., K. Benson, K. Chada, J. E. Huff, and D. C. Schwartz. 1995. Optical mapping of lambda bacteriophage clones using restriction endonucleases. Nat. Genet. 9:432-438. [DOI] [PubMed] [Google Scholar]
- 20.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. G. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, et al. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 21.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. Dimalanta, K. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2000. Genome sequence of entrohemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 22.Perry, R. D., and J. D. Fetherston. 1997. Yersinia pestis—etiologic agent of plague. Clin. Microbiol. Rev. 10:35-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sensen, C. W. 1999. Sequencing microbial genomes, p. 1-9. In R. L. Charlebois (ed.), Organization of the prokaryotic genome. ASM Press, Washington, D.C.
- 24.Soderlund, C., I. Longden, and R. Mott. 1997. FPC: a system for building contigs from restriction fingerprinted clones. Comput. Appl. Biosci. 13:523-535. [DOI] [PubMed] [Google Scholar]
- 25.Staggs, T. M., and R. D. Perry. 1991. Identification and cloning of a fur regulatory gene in Yersinia pestis. J. Bacteriol. 173:417-425. [DOI] [PMC free article] [PubMed] [Google Scholar]