Abstract
A method of mutagenic and unidirectional reassembly (MURA) that can generate libraries of DNA-shuffled and randomly truncated proteins was developed. The method involved fragmenting the template gene(s) randomly by DNase I and reassembling the small fragments with a unidirectional primer by PCR. The MURA products were treated with T4 DNA polymerase and subsequently with a restriction enzyme whose site was located on the region of the MURA primer. The N-terminal-truncated and DNA-shuffled library of a Serratia sp. phospholipase A1 prepared by this method had an essentially random variation of truncated size and also showed point mutations associated with DNA shuffling. After high-throughput screening on triglyceride-emulsified plates, several mutants exhibiting absolute lipase activity (NPL variants) were obtained. The sequence analysis and the lipase activity assay on the NPL variants revealed that N-terminal truncations at a region beginning with amino acids 61 to 71, together with amino acid substitutions, resulted in the change of substrate specificity from a phospholipase to a lipase. We therefore suggest that the MURA method, which combines incremental truncation with DNA shuffling, can contribute to expanding the searchable sequence space in directed evolution experiments.
The in vitro recombination method, or DNA shuffling, greatly expands the potential applications of the field of protein engineering. Many successfully engineered proteins have proven that various DNA shuffling techniques are effective tools for directed evolution of proteins, such as the improvement of enzyme activity (2) and stability (16) and alteration of the substrate specificity (22). These techniques generate homologous recombination between two or more genes and can also introduce new point mutations during PCR reassembly (17, 24). Point mutations, as well as recombination in DNA shuffling, may provide useful sequence diversity for more rapid adoption of novel properties.
Various methods for incremental truncation or nested deletion have also been developed for the acquisition of novel function in proteins and/or for use as a diagnostic tool for protein structure-function analysis. Nuclease-based methods using controlled digestion of linearized plasmid DNA can generate unidirectional truncations (1, 5, 10). The ITCHY method of creating incremental truncation with exonuclease III was recently reported and is especially well suited for the creation of hybrid enzyme libraries between two nonhomologous sequences (9, 11). In addition, many PCR-based deletion studies have been reported. Unfortunately, most of the PCR-based deletion methods produce only one deletion per experiment and require a series of DNA manipulations (6, 7, 23).
We have developed a method for the creation of DNA-shuffled and truncated enzyme libraries in which the different types of DNA sequences generated by either DNA shuffling or incremental truncation can be simultaneously introduced to a parent gene in a single experiment. The methodology, termed mutagenic and unidirectional reassembly (MURA), involves fragmenting template genes, reassembling their own small-length fragments with a unidirectional primer (MURA primer), and then cloning in the form of blunt- and sticky-ended termini. A unidirectional primer containing an appropriate restriction site was used for a unidirectional reassembly PCR, thus increasing a portion of genetic material truncated in a single desired direction among total reassembled DNA molecules.
Phospholipases A1 (PlaA) (EC 3.1.1.4), such as, for example, PlaA from a Serratia sp. (15), hydrolyze the sn-1 acyl ester bond of 3-sn-phosphoglycerides and show no activity on neutral lipids such as triacylglycerols. In contrast, typical lipases (EC 3.1.1.3) hydrolyze the acyl ester bonds of triacylglycerols to release glycerol and fatty acids and show no or very little activity on phospholipid substrates. However, they have the same catalytic triad residues acting on the same chemical bonds of relatively similar substrates and are believed to share a common structural fold termed the α/β-hydrolase fold (12). Moreover, exceptional lipases with remarkable phospholipase activity, such as the Staphylococcus hyicus lipase and the lidless guinea pig pancreatic lipase, were identified (14, 21) and evolved further to increase their phospholipase/lipase ratios by directed evolution and rational design (18, 19, 20). In this study, the MURA method for creating DNA-shuffled and truncated libraries was used to alter the substrate specificity of PlaA from a phospholipase to a lipase in the reverse direction relative to that of other previous approaches.
Here the feasibility of MURA as a novel method to create DNA-shuffled and truncated enzyme libraries is demonstrated. We used this method to produce DNA-shuffled and N-terminal-truncated variants of PlaA and then performed a screening for lipase activity. In this manner we obtained the shuffled and truncated phospholipase variants that were able to hydrolyze the acyl ester bonds of neutral lipids such as triglycerides, while the wild-type PlaA was not active on the same substrates.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Escherichia coli XL1-Blue {recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 [F′ proAB lacIq ZΔ15 Tn10 (Tetr)]} (Stratagene, San Diego, Calif.) was used as a host strain for DNA manipulation and gene expression. Plasmid pSTV28 (Takara Shuzo, Shiga, Japan) was used to construct a DNA-shuffled and truncated enzyme library. A PlaA gene (plaA) from a Serratia sp. (GenBank accession number U37262) was originated from plasmid pMJ1 (15). Recombinant E. coli cells were grown in Luria-Bertani (LB) medium. When necessary, chloramphenicol (50 μg/ml) was added to the medium.
DNA manipulation and sequencing.
All restriction enzymes, DNA-modifying enzymes, and related reagents used for DNA manipulation were purchased from New England Biolabs Inc., Sigma, Takara, or Boehringer Mannheim. DNA was sequenced by cycle sequencing by using an ABI PRISM BigDye Terminators v3.0 cycle sequencing kit and AmpliTaq DNA polymerase (Perkin-Elmer/Applied Biosystems, Foster City, Calif.).
MURA and library construction.
The substrates for the MURA reaction were 0.96-kb double-stranded DNA PCR products from pMJ1 and were obtained by using a Pfu polymerase (Stratagene) with the primer pair plaf1 (5′-TTGAGTTTTACCTCTGCGATCG-3′, which anneals to the 5′ end of the plaA gene) and plaf2 (5′-TCAGGCATTGGCCATCGCCTC-3′, which anneals to the 3′ end of the plaA gene). About 5 μg of the DNA substrate was digested with 0.3 units of DNase I (Promega, Madison, Wis.) in 50 μl of 40 mM Tris-HCl (pH 8.0), 10 mM MnCl2, and 1 mM CaCl2 for 4 to 5 min at room temperature. After the DNase I digests were purified with a Qiaquick nucleotide removal kit (Qiagen), it was confirmed on a 2.5% Metaphore agarose gel (FMC, Rockland, Minn.) or a 2% agarose gel, the size of which was about 100 bp. About 250 ng of gel-purified 100-bp fragments was reassembled with a unidirectional primer (i.e., MURA primer, 5′-AGCGTCGACTCAGGCATTGGCCATCGCCTCCCATGG-3′), which is annealed to the 3′ end of the plaA gene and is flanked by a SalI restriction site. A 100-μl mixture of MURA reaction contained 20 mM Tris-HCl (pH 8.7), 10 mM KCl, 2 mM MgSO4, 0.1 mM each deoxynucleoside triphosphate (dNTP), 50 to 500 pmol MURA primer, and 2 U of Pfu polymerase. The MURA reaction was performed with an automatic thermal cycler (Ericomp, San Diego, Calif.) for 30 cycles, with each cycle consisting of 94°C for 60 s, 54 to 62°C for 40 s, and 72°C for 60 s.
The MURA products were purified with a Qiaquick PCR purification kit (Qiagen) and then treated with 1 U of T4 DNA polymerase (Takara) in 40 μl of the manufacturer's 1X buffer and 0.17 mM each dNTP for 10 min at 37°C or with 10 U of S1 nuclease (Takara) in 40 μl of the manufacturer's 1X buffer for 10 min at 25°C. After being purified with a Qiaquick PCR purification kit, the blunt-ended MURA products were digested with SalI for the purpose of converting the blunt ends to sticky ends at the 3′ ends of the MURA products. The DNA fragments in the range of 500 to 960 bp were separated by agarose gel electrophoresis and purified by using a Qiaex II kit (Qiagen). The resultant MURA products with 5′ blunt ends and 3′ sticky ends were ligated into SmaI- and SalI-digested pSTV28. After ligation for 16 h at 16°C, E. coli XL1-Blue was transformed and plated on LB or tributyrin agar plates.
Library analysis.
XL1-Blue cells expressing the DNA-shuffled and truncated PlaA libraries were spread on LB plates containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside, isopropyl-β-d-thiogalactopyranoside, and chloramphenicol. The plasmids were isolated from 90 randomly chosen colonies and digested with EcoRI and SalI. The length variation of truncated genes was determined by agarose gel electrophoresis and the Quantity One software package (Bio-Rad, Hercules, Calif.). DNA from 10 randomly chosen colonies was also sequenced to estimate the location and frequency of mutation.
Screening.
E. coli transformants harboring a library of DNA-shuffled and truncated PlaA variants were spread on LB medium plates containing 1% (wt/vol) tributyrin (Sigma) and chloramphenicol (LB-TB plates). Plates were incubated overnight at 37°C and subsequently placed at 4°C. Lipase activity on the tributyrin substrate in the LB-TB plate was visible as a transparent halo surrounding the colony of interest due to hydrolysis of the turbid tributyrin. Positive colonies, whose activity became visible, were selected to measure enzymatic activity and to determine DNA sequences. Under these conditions, E. coli colonies expressing wild-type PlaA did not produce any detectable halo.
Enzyme activity.
Cell extracts expressing functional variants and wild-type PlaA were prepared as described by Henne et al. (4). Based on the previous methods (3, 16), enzymatic activities were determined in a pH-stat assay with substrate solutions of pure micelles of lecithin as a phospholipase substrate and mixed micelles of tributyrin as a lipase substrate. The enzymatic activities were measured with an autotitrator and an autoburette (Radiometer Copenhagen, Lyon, France) and quantified according to the NaOH consumption that was recorded.
RESULTS AND DISCUSSION
MURA.
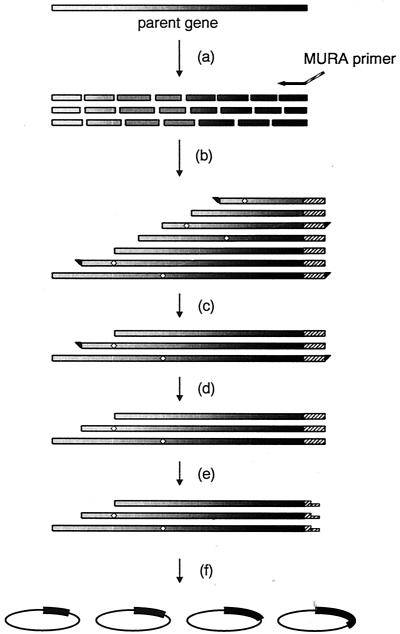
The MURA method described in this study allows the creation of truncated enzyme libraries in which point mutation and homologous recombination of general DNA shuffling are also involved. The MURA process consists of four key steps (Fig. 1). The method starts with random fragmentation of the parental gene obtained by PCR amplification or restriction digestion. The second step involves fragment reassembly in the presence of the unidirectional primer (MURA primer) containing an appropriate restriction site. Next, the MURA products covering the DNA region of interest are isolated by preparative agarose gel electrophoresis, subjected to T4 DNA polymerase or S1 nuclease in order to polish both termini of the fragments, and then digested with a restriction enzyme that has a site in the MURA primer.
FIG. 1.
Schematic overview of the construction of a DNA-shuffled and truncated enzyme library by the MURA method. The sequences of the MURA primer can be chosen on any desired site of the parent gene. This figure represents a procedure using a 3′-complementary MURA primer, and thus N-terminal-truncated and shuffled variants of PlaA are constructed. The steps shown are random fragmentation of the parent gene pool by DNase I (a), unidirectional reassembly with the MURA primer (b), separation of the fragments of interest by preparative agarose gel electrophoresis (c), formation of blunt ends by S1 nuclease or T4 DNA polymerase on both termini (d), sticky-end formation by a restriction enzyme on one terminus (e), and ligation into an expression vector (f). The order of steps c, d, and e can be altered to d, e, and c without affecting the efficiency of library construction. The diamonds on the bars representing genes indicate possible mutations during shuffled and unidirectional reassembly PCR.
One experimental example of this MURA procedure was demonstrated by the construction of a library for the expression of DNA-shuffled and N-terminal-truncated variants of the PlaA from a Serratia sp. (GenBank accession no. U37262). Figure 2 shows DNase I-digested random fragments of 0.96-kb PCR products and the products of MURA PCR. Purified DNase I digests of about 100 bp were reassembled in the presence of a MURA primer. In order to generate the N-terminal truncation, we used a unidirectional MURA primer annealed to the 3′ end of the plaA gene. In fact, any region of a whole gene can be chosen as sequences of a MURA primer, and in that case any direction of unidirectional reassembly may also be selected.
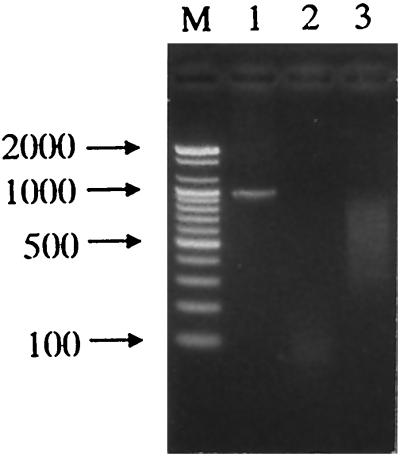
FIG. 2.
Agarose gel analysis of DNA fragments generated during the MURA process. The Serratia sp. phospholipase gene (0.96 kb) was prepared by PCR (lane 1). The template DNA was digested with DNase I and the digests were purified (lane 2) and subjected to the MURA PCR as described in Materials and Methods; the resultant DNA fragments reassembled with a unidirectional primer (lane 3) were purified by using a Qiaquick purification kit. Lane M, 100-bp ladder DNA size marker.
It is believed to be important that the population of reassembled MURA fragments should be obtained without a bias toward a specific size range. Although the MURA primer sequences and the length of parent gene can affect the efficiency in applying the MURA method to other genes, the MURA primer concentration (i.e., the molar ratio of MURA primer and template fragments), annealing temperature, and cycle number can be easily manipulated to obtain a uniform smear. Thus, we tested for the range of 50 to 500 pmol (i.e., about 5- to 50-fold excess of the MURA primer over DNase I-digested fragments) of the MURA primer, and we found that the higher primer concentration yielded a higher efficiency of library construction. Moreover, we obtained a uniform smear without a specific band (i.e., without an uneven distribution of reassembled fragments) on agarose gels when the MURA was performed for 30 cycles at an annealing temperature of 56°C.
After the unidirectionally reassembled DNA fragments were purified, they were blunt-ended on both ends with T4 DNA polymerase or S1 nuclease and then sticky-ended on the 3′ end with SalI. Fragments in the range of about 500 to 960 bp were isolated by preparative agarose gel electrophoresis and cloned into pSTV28. As shown below, the treatment of T4 DNA polymerase and the subsequent restriction digestion guaranteed that the randomly truncated and shuffled DNA fragments having exclusively MURA primer sequences at their 3′ end (i.e., fragments reassembled together with MURA primer) could be correctly ligated into 5′ blunt- and 3′ sticky-ended vector (i.e., SmaI- and SalI-digested pSTV28).
DNA analyses of MURA libraries.
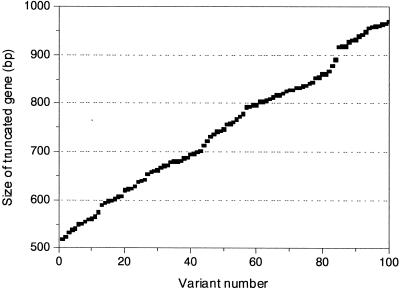
We created N-terminal-truncated and DNA-shuffled PlaA libraries by the MURA method. One hundred randomly chosen colonies from the MURA library were analyzed. When the plasmids from randomly selected members were digested with EcoRI and SalI, we determined that the MURA products were correctly introduced in the form of 5′ blunt- and 3′ sticky-ended fragments, which had been generated by treating T4 DNA polymerase and restriction enzyme SalI. Moreover, the length variation of truncated and shuffled genes was evaluated and found to be essentially random (Fig. 3 and Fig. 4). In other words, the population of truncations covering the DNA region of interest did not have a significant bias against, for example, relatively small fragments or specific size ranges.
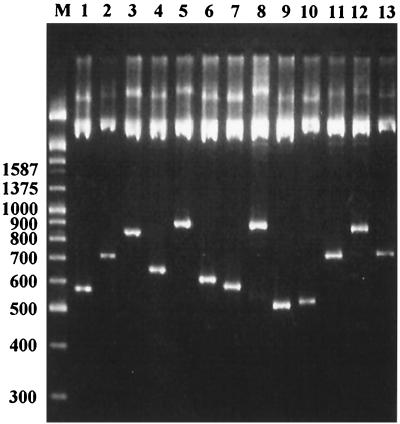
FIG. 3.
An example of restriction enzyme digestion on randomly chosen colonies from the MURA library. E. coli colonies were randomly selected from the library, and their plasmids were isolated. Each sample of plasmid DNA was digested with EcoRI and SalI and then examined by electrophoresis in a 2.5% Metaphore agarose gel (FMC). Lane M, λ/EcoRI and HindIII digests and a 100-bp ladder DNA size marker; lanes 1 to 13, EcoRI and SalI digests of each plasmid.
FIG. 4.
Size distribution of the MURA libraries. The truncated gene sizes of 100 randomly chosen variants in MURA libraries were determined by restriction digestion of each sample. The digest samples were estimated by agarose gel electrophoresis (2.5% Metaphore agarose gel) and the gel analysis software package Quantity One (Bio-Rad). The calculated sizes of the insert genes of randomly selected members were arranged in the order of ascending size.
In addition to the analyses related to the truncation aspect of the MURA library, we determined DNA sequences of 20 randomly selected variants in order to confirm the DNA shuffling aspect of MURA for a single gene. The rate of mutation that was derived from unidirectional reassembly PCR in the MURA procedures, which is a phenomenon similar to that of general DNA shuffling, was 0.11% under the experimental conditions used in this study. Generally, DNA shuffling and the MURA method involve the random fragmentation of target sequences and the reassembly of their own small-length fragments by inevitable mutagenic PCR, except that in the MURA method a unidirectional primer is added to the reassembly PCR. Moreover, it has been noted that the mutagenesis rate associated with DNA shuffling can be adjusted over a wide range, namely, 0.05 to ∼0.7%, by controlling some experimental conditions (24). In fact, when conditions for error-prone PCR, such as a higher concentration of Mg2+, the addition of mutagenic Mn2+, and the use of relatively large amounts of Taq DNA polymerase, were applied to unidirectional PCR reassembly, we obtained a similar pattern of smear on an agarose gel without a significant difference in the level of amplification achieved in PCR reassembly (data not shown). Therefore, the mutagenesis rate in the MURA method is believed to be varied by the application of the same experimental conditions that have already been well established in other previous studies of error-prone PCR and DNA shuffling.
Judging from this type of DNA sequence, the MURA method represents a combination of DNA shuffling and incremental truncation and therefore is believed to add one more method of genetic diversification to the repertoire of methods already available.
Changing the substrate specificity of PlaA by the MURA method.
Following the simultaneous introduction of truncation and shuffling by the MURA method, DNA fragments in the range of 500 to 960 bp were cloned, and E. coli cells that were transformed with this plaA library (2,500 to 3,000 transformants) were plated on an LB-TB plate, on which the wild-type PlaA did not produce a transparent halo indicative of enzymatic hydrolysis of tributyrin. After overnight incubation of the plate at 37°C and a subsequent 6 h at 4°C, the 12 transformants displayed lipase activities. To verify the results from the screening procedure, both lipase and phospholipase activities were measured for cell lysates of these variants (Table 1). To begin with, the lipase activity of the wild-type PlaA was too low to monitor any liberation of fatty acids from triglyceride under the assay conditions used. NPL-0901 showed the highest lipase activity, which was twofold higher than that of NPL-3628, which had the lowest lipase activity among the NPL variants. Surprisingly, the phospholipase activities of all NPL variants were almost equal to that of wild-type PlaA or were somewhat higher. These results indicate that the NPL variants acquired lipase activity without the decrease of their inherent phospholipase activity, thus increasing the lipase/phospholipase activity ratio.
TABLE 1.
Amino acid sequences and catalytic activities of wild-type PlaA and NPL variants
| Protein | Amino acid positionsa | Relative activity (%)b
|
|
|---|---|---|---|
| Lipase | Phospholipase | ||
| PlaA (wild type) | 1 to 320 | <0.1 | 100.0 |
| NPL-3628 | 61 to 320 | 49.4 | 109.1 |
| NPL-0601 | 62 to 320 | 53.6 | 107.3 |
| NPL-0401 | 63 to 320 | 63.1 | 113.2 |
| NPL-0602 | 63 to 320 (R246C) | 76.8 | 109.3 |
| NPL-0901 | 66 to 320 | 100 | 115.9 |
| NPL-3128 | 67 to 320 | 92.3 | 115.2 |
| NPL-1901 | 68 to 320 | 89.3 | 107.8 |
| NPL-1328 | 70 to 320 | 90.5 | 107.5 |
| NPL-0428 | 71 to 320 | 69.0 | 97.8 |
All numbers are the positions of amino acids from PlaA. Amino acid substitutions are indicated in parentheses.
The E. coli cells expressing the selected variants were harvested when they reached an optical density at 600 nm of 2. Enzymatic activities were measured using at least three independent cell lysates of each variant, and values whose error ranges were within 5% were averaged. The lipase and phospholipase activities were expressed as percentages of the activities of NPL0901 (the variant with the highest lipase activity) and the wild-type enzyme, respectively.
Unlike typical lipases that showed no or little activity towards phospholipid substrates, the lipase from S. hyicus was highly active on phospholipids (13). The conversion of a lipase into a phospholipase by directed evolution and rational design has been also studied for the past several years (8, 18, 19, 20). Originally, two lipases of Staphylococcus aureus and Bacillus thermocatenulatus exhibited a low activity or no activity on phospholipid substrates. These lipases were engineered by directed evolution into a lipase with a high phospholipase activity. Error-prone PCR mutagenesis followed by screening of the resulting mutant libraries on phospholipid plates had yielded several variants with an increased phospholipase/lipase activity ratio (8, 20). Thus, lipolytic enzymes that have hitherto been ascertained to be active both on lipids and on phospholipids were naturally occurring lipases and in vitro-evolved variants of typical lipase, whereas the NPL variants reported here were the first examples that were evolved from a phospholipase toward a lipase. In fact, we are now attempting to further increase the lipase activities of NPL mutants through additional rounds of directed evolution. The detailed results, focusing on the acquisition of a best variant as a compromise between a higher lipase/phospholipase activity ratio and a higher absolute lipase activity, will be described in another report.
Sequence analysis of NPL mutants.
The amino acid sequences of twelve NPL variants with newly obtained lipase activity were determined by DNA sequencing (Table 1). First, each variant was one of the variants with PlaA from which the N-terminal region up to amino acid positions 61 to 71 had been deleted as a result of the MURA process, and 9 different variants were identified among 12 variants sequenced. The number of amino acid substitutions found in the NPL variants with a newly obtained lipase activity was relatively small when compared to those found in the randomly chosen members of the MURA library. Moreover, we assumed that a significant level of structural changes would be required to evolve from the PlaA phospholipase to a lipase, because random point mutations introduced into the whole phospholipase gene by error-prone PCR amplification, in fact, had not yielded any variant with lipase activity (data not shown). Therefore, the drastic structural changes obtained through the truncation aspect of the MURA process were regarded to be more critical, while point mutations were thought to be a matter of secondary importance.
Previously, it has been shown that each S. aureus and B. thermocatenulatus variant with a higher phospholipase activity contained two to eight mutations at the amino acid level (8, 20). Moreover, the amino acid exchanges of the B. thermocatenulatus variants with increased phospholipase activity were predominantly in regions of the N-terminal part corresponding to putative structural elements for efficient interaction with the phospholipid substrate and/or interface (8). In fact, the substrates for phospholipase, like glycerophospholipid, are believed to require a more strict recognition for the enzyme-substrate binding (e.g., a structural element for the interaction with the phosphate head group) compared to triglyceride. Although the estimate obtained without precise crystallographic information is somewhat imperfect, the newly obtained lipase activity of N-terminal-truncated and DNA-shuffled variants (NPL variants) is therefore considered to arise from the deletion of the N-terminal region responsible for the recognition of phospholipid substrates, which is achieved without a significant loss of catalytic activity acting on the ester bond. In other words, PlaA could discriminate the substrate for phospholipase (i.e., glycerophospholipid) from that for lipase (i.e., triglyceride), while the NPL variants seemed to be unable to discriminate between them because the important structural motif no longer existed as a result of truncation.
Conclusions.
We developed a MURA method that can create a library of DNA-shuffled and truncated proteins and then used this to alter substrate specificity from a phospholipase to a lipase. As shown in the insert size distribution and the DNA sequences of all of the randomly selected members, the different types of DNA sequence generated by either DNA shuffling or general incremental truncation could be simultaneously obtained by employing the MURA method. Furthermore, judging from the fact that the NPL variants newly obtained lipase activity through the shuffled truncation of PlaA by the MURA method, the combination of DNA shuffling and incremental truncation could allow workers to access a more diverse sequence space for directed evolution.
Meanwhile, of various methods established for directed evolution, the MURA method is a very flexible and easy-to-implement alternative technique for generating mutant libraries. Just as DNA family shuffling can recombine a family of homologous sequences, for instance, so the MURA method can recombine several homologous sequences along with a concurrent truncation. Furthermore, by applying the MURA method to the existent methods for using recombination in vitro, it should be also possible to create hybrid enzyme libraries with multiple crossovers from multiple parents irrespective of sequence identity.
Acknowledgments
We thank Keehyuk Kim for his helpful discussion.
B. Chung and J. K. Song contributed equally to this study.
J. K. Song was supported by a research fellowship from the Brain Korea 21 (BK21) program.
REFERENCES
- 1.Breault, D. T., M. L. Stover, M. Li, A. C. Lichtler, and D. W. Rowe. 1995. Novel use for BAL 31 nuclease-generated nested deletions: sequencing from the inside out. BioTechniques 18:614-617. [PubMed] [Google Scholar]
- 2.Crameri, A., E. A. Whitehorn, E. Tate, and W. P. Stemmer. 1996. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat. Biotechnol. 14:315-319. [DOI] [PubMed] [Google Scholar]
- 3.Eggert, T., G. Pencreac'h, I. Douchet, R. Verger, and K. E. Jaeger. 2000. A novel extracellular esterase from Bacillus subtilis and its conversion to a monoacylglycerol hydrolase. Eur. J. Biochem. 267:6459-6469. [DOI] [PubMed] [Google Scholar]
- 4.Henne, A., R. Daniel, R. A. Schmitz, and G. Gottschalk.1999. Construction of environmental DNA libraries in Escherichia coli and screening for the presence of genes conferring utilization of 4-hydroxybutyrate. Appl. Environ. Microbiol. 65:3901-3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henriquez, V., and M. L. Gennaro. 1990. A simple strategy to generate small deletions using Bal31 nuclease. Nucleic Acids Res. 18:6735-6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Imai, Y., Y. Matsushima, T. Sugimura, and M. Terada. 1991. A simple and rapid method for generating a deletion by PCR. Nucleic Acids Res. 19:2785.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karreman, C. 1998. Fusion PCR, a one-step variant of the “megaprimer” method of mutagenesis. BioTechniques 24:736, 740, 742. [DOI] [PubMed] [Google Scholar]
- 8.Kauffmann, I., and C. Schmidt-Dannert. 2001. Conversion of Bacillus thermocatenulatus lipase into an efficient phospholipase with increased activity towards long-chain fatty acyl substrates by directed evolution and rational design. Protein Eng. 14:919-928. [DOI] [PubMed] [Google Scholar]
- 9.Lutz, S., M. Ostermeier, and S. J. Benkovic. 2001. Rapid generation of incremental truncation libraries for protein engineering using alpha-phosphothioate nucleotides. Nucleic Acids Res. 29:E16.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murphy, G. 1993. Generation of a nested set of deletions using exonuclease III. Methods Mol. Biol. 23:51-59. [DOI] [PubMed] [Google Scholar]
- 11.Ostermeier, M., A. E. Nixon, and S. J. Benkovic.1999. Incremental truncation as a strategy in the engineering of novel biocatalysts. Bioorg. Med. Chem. 7:2139-2144. [DOI] [PubMed] [Google Scholar]
- 12.Schrag, J. D., and M. Cygler. 1997. Lipases and alpha/beta hydrolase fold. Methods Enzymol. 284:85-107. [DOI] [PubMed] [Google Scholar]
- 13.Simons, J. W., H. Adams, R. C. Cox, N. Dekker, F. Gotz, A. J. Slotboom, and H. M. Verheij.1996. The lipase from Staphylococcus aureus. Expression in Escherichia coli, large-scale purification and comparison of substrate specificity to Staphylococcus hyicus lipase. Eur. J. Biochem. 242:760-769. [DOI] [PubMed] [Google Scholar]
- 14.Simons, J. W., F. Gotz, M. R. Egmond, and H. M. Verheij. 1998. Biochemical properties of staphylococcal (phospho)lipases. Chem. Phys. Lipids 93:27-37. [DOI] [PubMed] [Google Scholar]
- 15.Song, J. K., M. K. Kim, and J. S. Rhee.1999. Cloning and expression of the gene encoding phospholipase A1 from Serratia sp. MK1 in Escherichia coli. J. Biotechnol. 72:103-114. [DOI] [PubMed] [Google Scholar]
- 16.Song, J. K., and J. S. Rhee. 2000. Simultaneous enhancement of thermostability and catalytic activity of phospholipase A1 by evolutionary molecular engineering. Appl. Environ. Microbiol. 66:890-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stemmer, W. P. 1994. DNA shuffling by random fragmentation and reassembly: in vitro recombination for molecular evolution. Proc. Natl. Acad. Sci. USA 91:10747-10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Kampen, M. D., J. W. Simons, N. Dekker, M. R. Egmond, and H. M. Verheij.1998. The phospholipase activity of Staphylococcus hyicus lipase strongly depends on a single Ser to Val mutation. Chem. Phys. Lipids 93:39-45. [DOI] [PubMed] [Google Scholar]
- 19.van Kampen, M. D., H. M. Verheij, and M. R. Egmond. 1999. Modifying the substrate specificity of staphylococcal lipases. Biochemistry 38:9524-9532. [DOI] [PubMed] [Google Scholar]
- 20.van Kampen, M. D., and M. R. Egmond.2000. Directed evolution: from a staphylococcal lipase to a phospholipase. Eur. J. Lipid Sci. Technol. 102:717-726. [Google Scholar]
- 21.Withers-Martinez, C., F. Carriere, R. Verger, D. Bourgeois, and C. Cambillau.1996. A pancreatic lipase with a phospholipase A1 activity: crystal structure of a chimeric pancreatic lipase-related protein 2 from guinea pig. Structure 4:1363-1374. [DOI] [PubMed] [Google Scholar]
- 22.Yano, T., S. Oue, and H. Kagamiyama. 1998. Directed evolution of an aspartate aminotransferase with new substrate specificities. Proc. Natl. Acad. Sci. USA 95:5511-5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yohda, M., N. Kato, and I. Endo. 1995. Solid-phase nested deletion: a new subcloning-less method for generating nested deletions. DNA Res. 2:175-181. [DOI] [PubMed] [Google Scholar]
- 24.Zhao, H., and F. H. Arnold. 1997. Optimization of DNA shuffling for high fidelity recombination. Nucleic Acids Res. 25:1307-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]