Abstract
As a result of agricultural activities in regions adjacent to the northern boundary of the Florida Everglades, a nutrient gradient developed that resulted in physicochemical and ecological changes from the original system. Sulfate input from agricultural runoff and groundwater is present in soils of the Northern Everglades, and sulfate-reducing prokaryotes (SRP) may play an important role in biogeochemical processes such as carbon cycling. The goal of this project was to utilize culture-based and non-culture-based approaches to study differences between the composition of assemblages of SRP in eutrophic and pristine areas of the Everglades. Sulfate reduction rates and most-probable-number enumerations revealed SRP populations and activities to be greater in eutrophic zones than in more pristine soils. In eutrophic regions, methanogenesis rates were higher, the addition of acetate stimulated methanogenesis, and SRP able to utilize acetate competed to a limited degree with acetoclastic methanogens. A surprising amount of diversity within clone libraries of PCR-amplified dissimilatory sulfite reductase (DSR) genes was observed, and the majority of DSR sequences were associated with gram-positive spore-forming Desulfotomaculum and uncultured microorganisms. Sequences associated with Desulfotomaculum fall into two categories: in the eutrophic regions, 94.7% of the sequences related to Desulfotomaculum were associated with those able to completely oxidize substrates, and in samples from pristine regions, all Desulfotomaculum-like sequences were related to incomplete oxidizers. This metabolic selection may be linked to the types of substrates that Desulfotomaculum spp. utilize; it may be that complete oxidizers are more versatile and likelier to proliferate in nutrient-rich zones of the Everglades. Desulfotomaculum incomplete oxidizers may outcompete complete oxidizers for substrates such as hydrogen in pristine zones where diverse carbon sources are less available.
Terminal carbon mineralization in low-sulfate freshwater ecosystems is typically controlled by methanogenesis (40); however, sulfate reduction can be important in many freshwater systems and a significant amount of carbon may be mineralized via this pathway (1, 5, 6, 18, 19, 23, 24). In freshwater ecosystems with significant sulfate input, sulfate-reducing prokaryotes (SRP) may play an important role in the carbon cycle and compete with methanogens for electron donors. SRP are a very diverse microbial group and possess several attributes that may be beneficial in these types of environments, including substrate and metabolic versatility, lower Kms than methanogens have for substrates such as acetate and hydrogen, resistance to fluctuating redox potential by sporulation, and a limited ability to survive oxygen exposure (7, 40, 42-45).
The Florida Everglades was historically a low-nutrient ecosystem. Nutrient-enriched runoff from the Everglades Agricultural Area (EAA) resulted in a well-documented phosphorus gradient in Florida Everglades Water Conservation Area 2-A (WCA-2A) (9, 11, 13, 22, 30). These nutrient inputs resulted in changes in vegetation and biogeochemical cycling. P-enriched sites are dominated by cattail (Typha spp.), and areas with lower P content are dominated by sawgrass (Cladium spp.). The vegetation changes resulted in changes in overall productivity and influenced the long-term storage of carbon and phosphorus, which affects water quality and other properties of these ecosystems (30). Several areas impacted by agricultural activities have been shown to have high levels of sulfate, the primary electron acceptor for SRP. Sulfate in the Everglades is derived from natural and anthropogenic activities and may be input through atmospheric deposition and EAA storm water runoff and from groundwater (32). EAA runoff contains high sulfate concentrations due to the use of elemental sulfur as a soil amendment and mineralization of organic S. Two areas in the Everglades with high sulfur concentrations are the EAA and the WCA-2A (3, 4).
Most studies conducted on the biogeochemistry of the Everglades have focused on the process level and not on the microbial level, such that very little is known of the microbial communities responsible for biogeochemical processes or of the impacts of nutrient loading on processes within individual biogeochemical cycles. To our knowledge, the only study that has addressed this issue was conducted by Drake et al. (14), who reported 4 × 1011 SRP per g (dry weight) for soils in P-enriched areas of the WCA-2A and 5 × 107 SRP per g in nonimpacted areas of the WCA-3A. They postulated that the enrichment of SRP in the P-impacted site was due to a high concentration of sulfate coming from EAA runoff.
Most characterization of freshwater SRP populations has been conducted in lakes, rivers, and rice paddies, but to our knowledge, no detailed characterization of SRP assemblages has been conducted in freshwater wetlands. Although SRP play important roles in the carbon, sulfur, and mercury cycles (16), very little information is available regarding SRP or their ecological roles in these wetlands. We are primarily interested in the relationships between the composition of SRP assemblages and eutrophication and in identifying linkages between these assemblages and biogeochemical processes in the Everglades. The overall objective of this study was to investigate differences with regard to the composition and function of SRP assemblages in eutrophic and more pristine regions of the marsh. Specifically, our objectives were to compare sulfate reduction rates, SRP population sizes, and possible differences in the composition of SRP assemblages as defined by diversity in dissimilatory sulfite reductase (DSR) genes. This information will lead to a greater understanding of the impacts of nutrients on pristine wetlands.
MATERIALS AND METHODS
Site characteristics, sampling and biogeochemical characterization.
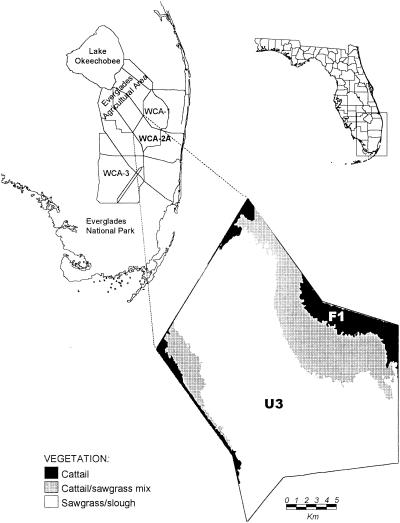
Studies were conducted in samples taken from the WCA-2A (Fig. 1). Samples were collected along the phosphorus gradient, from impacted F1 (cattail-dominated sites) and nonimpacted U3 (sawgrass-dominated sites) zones. The studies were conducted in the 0- to 10-m soil layer, which indicates nutrient impact for longer than 3 years based on peat accretion rates calculated from 137Cs distribution in the soil profile (30, 31). U3 was overlain by a thick (5 to 15 cm) marl and/or periphyton layer. One soil core was collected each on 20 April (spring) 2001, 15 August (summer) 2001, and 7 December (fall) 2001, at F1 and U3 sites of the WCA-2A. Soil cores were sampled by staff of the South Florida Water Management District and were shipped overnight to the Wetland Biogeochemistry Laboratory, Soil and Water Science Department, Gainesville, Fla., where they were sectioned and manually mixed. Subsamples for DNA analysis were kept at −70°C until analysis. Subsamples intended for enumerations and measurement of rates were kept at 4°C until analysis within 2 to 5 days of sampling. Samples were not processed immediately and were kept under anoxic conditions, such that rates and enumerations reported in this study do not represent field conditions. They can be used as good estimators in comparative study between the two sites, however. Total phosphorus, total inorganic phosphorus, ammonium, total Kjeldahl nitrogen, extractable total organic carbon, and microbial biomass carbon were determined according to methods described by White and Reddy (41) and D'Angelo and Reddy (10).
FIG. 1.
Location of sampling sites in WCA-2A. The eutrophic zone, F1, is dominated by cattail (Typha domingensis Pers), and the pristine zone, U3, is dominated by sawgrass (Cladium jamaicense Crantz).
Microbial enumerations and enrichments.
Conventional anaerobic techniques were used throughout the anaerobic microbial work (2, 17). Enumerations were conducted using the most-probable-number (MPN) technique, with three tubes per dilution using basal carbonate yeast extract Trypticase (BCYT) medium (37). Lactate and acetate (20 mM) were added from sterile anoxic stock solutions. FeSO4 (20 mM) was used as the sulfate source and reducing agent. Tubes with black precipitate due to sulfide production were considered positive for MPN calculations. Tubes were incubated at room temperature in the dark for 2 months. Thermophilic enrichments for SRP were established using acetate and lactate as electron donors and were incubated at 60°C.
Sulfate analyses.
Sulfate was measured by ion chromatography with a Dionex Model LC20 equipped with an AS14 column and an ECD50 conductivity detector (Dionex, Sunnyvale, Calif.), with 3.5 mM sodium carbonate-1.0 mM sodium bicarbonate as eluent.
Sulfate reduction measurement.
Sulfate reduction rates were determined using a method described by Ulrich et al. (38) with slight modifications according to the extensive comparison study of radiotracer techniques conducted by Meier et al. (25). Briefly, 1-g soil subsamples were distributed in 120-ml serum bottles containing 1 ml of reduced BCYT medium under a continuous N2 stream. Test tubes (12 by 75 mm) containing 2.5 ml of anoxic 10% zinc acetate were placed as sulfide traps, and the serum bottles were closed with butyl rubber stoppers and aluminum seals (Bellco Glass Inc., Vineland, N.J.). Two microcuries of carrier-free [35S]SO42− (ARC, St. Louis, Mo.) was injected, and the radiochemical was diluted with nonradiolabeled sulfate to render a final sulfate concentration of 100 μM. The flask contents were incubated with shaking at 125 rpm in the dark for 2 h at room temperature. After 2 h, 8 ml of anoxic 6 M HCl and 8 ml of anoxic 1 M CrCl2 in 0.5 M HCl were added via syringe, and the bottles were shaken at 125 rpm for 48 h. Trap contents were mixed with scintillation cocktail (Ecoscint A; National Diagnostics, Atlanta, Ga.), and the radioactivity in the traps and the nonreduced 35SO42− radioactivity remaining in the bottles were measured by liquid scintillation counting (LS 5801; Beckman Coulter, Fullerton, Calif.). Sulfate reduction rates were calculated with the method described by Fossing and Jørgensen (15).
Methanogenesis rates.
One gram of soil was mixed with 2 ml of anoxic BCYT medium under an N2 stream in anaerobic culture tubes that were later closed with butyl rubber stoppers and aluminum seals. The tubes were preincubated for 2 weeks before substrates were added. Sulfate and acetate (20 mM each) were added from anaerobic sterile concentrated stock solutions. Molybdate (an inhibitor of sulfate reduction) was added at a final concentration of 2 mM (27). The tubes were fitted with three-way Luer stopcocks (Cole-Parmer, Vernon Hills, Ill.) for gas sampling and were incubated in the dark at 25°C with shaking at 100 rpm. Methane in the headspace was measured by gas chromatography with a Shimadzu 8A GC equipped with a Carboxen 1000 column (Supelco, Bellefonte, Pa.) and a flame ionization detector operating at 110°C. The carrier gas was N2, and the oven temperature was 160°C. All determinations were carried out in triplicate. Headspace pressure was measured using a digital pressure indicator (DPI 705; Druck, New Fairfield, Conn.).
Nucleic acid extraction.
Nucleic acids were extracted with UltraClean Soil DNA kits (MoBio, Solana Beach, Calif.) according to the manufacturer's instructions. Soil (0.25 g) was used for extraction. Nucleic acid extraction was evaluated on a 1% agarose gel electrophoresed with Tris-acetate-EDTA buffer.
PCR amplification.
PCR was conducted using the primer set designed by Wagner et al. (39), which amplifies a 1.9-kb fragment of the DSR gene; this set consisted of primers DSR1F (5′-ACSCACTGGAAGCACG-3′) and DSR4R (5′-GTGTAGCAGTTACCGCA-3′). The reaction mixture used for PCR amplification contained 7 μl of distilled H2O, 1 μl of each primer (10 pmol/μl), 10 μl of HotStarTaq Master Mix (Qiagen, Valencia, Calif.), and 1 μl of diluted DNA solution. PCR amplification was carried out in a GeneAmp PCR system 2400 (Perkin-Elmer Applied Biosystems, Norwalk, Conn.) with the following conditions: initial enzyme activation and DNA denaturation of 15 min at 95°C, followed by 30 cycles of 30 s at 94°C, 30 s at 58°C, and 90 s of extension at 72°C, and a final extension of 72°C for 7 min. The PCR products were electrophoresed on a 1% agarose gel in Tris-acetate-EDTA buffer to confirm amplification of expected size product.
Cloning of DSR genes and RFLP analysis.
Fresh PCR amplicons obtained with primers DSR1F and DSR4R were ligated into the pCRII-TOPO cloning vector and were transformed into chemically competent Escherichia coli TOP10F' cells according to the manufacturer's instructions (Invitrogen, Carlsbad, Calif.). Individual colonies of E. coli were screened by direct PCR amplification, with the DSR primers using the previously described PCR programs. Restriction fragment length polymorphism (RFLP) analyses were conducted using the restriction enzyme HhaI and were analyzed by 2% agarose gel electrophoresis. Clone libraries were analyzed by analytic rarefaction using the software aRarefactWin (version 1.3; S. Holland, Stratigraphy Lab, University of Georgia, Athens [http://www.uga.edu/∼strata/software/]).
Sequencing and phylogenetic analysis.
Selected clones were sequenced at the DNA Sequencing Core Laboratory at the University of Florida using the DSR1F primer. Deduced amino acid sequences of the DSR α-subunit were aligned and analyzed with ClustalX version 1.81 (35). Only unambiguously aligned amino acid positions from the DSR α-subunit gene found in all cloned sequences were used. The final data set consisted of approximately 180 amino acids. Trees based on aligned sequences were constructed using several phylogenetic methods. Protein distances were estimated with PROTDIST with the JTT replacement model, and trees were inferred using FITCH with global rearrangements in PHYLIP software (version 3.6a2; J. L. Felsenstein, Department of Genetics, University of Washington, Seattle). Amino acid alignments were also evaluated with PAUP* version 4.0b8 by using parsimony- and distance matrix-based algorithms with default settings (D. L. Swofford, Sinauer Associates, Sunderland, Mass.). Protein maximum-likelihood trees were obtained with PROML in PHYLIP and with TREE-PUZZLE version 5.0 (33). Bootstrap analysis was performed with 100 resamplings of the amino acid sequences.
Nucleotide sequence accession number.
GenBank accession numbers for partial DSR gene sequences are AY096038 to AY096074.
RESULTS
Biogeochemical characterization.
Pertinent physical and chemical data regarding the two sites studied here are presented in Table 1. Water tables for samples taken in spring were drastically lower than the cumulative April mean for 1996 to 2002; in F1 water levels were −9.1 cm compared to 21.9 cm, and for U3 they were −0.9 cm compared to 21.9 cm. During the wet season, water levels were higher than previously reported means for August 1996 to 2002: 61.3 cm versus a mean of 36.9 cm for F1 and 62.5 cm versus 42.6 for U3. Water levels were lower during December, with 29.3 cm versus a mean of 36.2 cm for F1 and 32.3 cm versus a mean of 41.6 cm for U3. Samples taken from F1, the eutrophic region of the marsh, exhibited significantly higher concentrations of total phosphorus and total inorganic phosphorus than did samples from U3, the pristine region. Nitrogen parameters were similar in both sites. Concentrations of extractable total organic carbon and microbial biomass carbon were higher in F1. These data are in agreement with historical records previously described for this ecosystem (12, 30, 31).
TABLE 1.
Selected chemical characteristics of eutrophic (F1) and pristine (U3) Florida Everglades soilsb
| Sampling time | Field identification | Water table (cm) | Moisture (%) | TP (mg/kg) | TPi (mg/kg) | NH4-N (mg/kg) | TKN (mg/kg) | Extractable TOC (mg/kg) | MBC (mg/kg) |
|---|---|---|---|---|---|---|---|---|---|
| Spring 2001 | F1 | −9.1 | 88.7 (0.4)a | 1,536 (57) | 549 (33) | 52 (28) | 402 (44) | 4,101 (78) | 7,638 (544) |
| U3 | −0.9 | 91.1 (0.3) | 277 (19) | 69 (10) | 94 (7) | 343 (120) | 2,892 (90) | 2,066 (221) | |
| Summer 2001 | F1 | 61.3 | 89.1 (1.0) | 1,089 (205) | 388 (108) | 50 (22) | 309 (29) | 2,798 (278) | 4,043 (1,000) |
| U3 | 62.5 | 90.2 (0.9) | 377 (59) | 82 (15) | 52 (3) | 305 (69) | 1,741 (376) | 2,276 (607) | |
| Fall 2001 | F1 | 29.3 | 90.7 (1.5) | 997 (265) | 368 (155) | 60 (20) | 300 (37) | 2,760 (90) | 3,686 (1,102) |
| U3 | 32.3 | 86.6 (1.7) | 301 (16) | 83 (32) | 44 (6) | 281 (24) | 2,334 (73) | 2,753 (645) |
Values in parentheses represent the standard deviation of triplicate determinations.
Content of different substances is expressed per kilogram (dry weight) of soil. TP, total phosphorus; TPi, total inorganic phosphorus; NH4-N, ammonium; TKN, total Kjeldahl nitrogen; TOC, total organic carbon; and MBC, microbial biomass carbon.
Sulfate concentrations, sulfate reduction rates, and enumeration of sulfate-reducing bacteria.
Results for sulfate concentrations, sulfate reduction rates, and lactate and acetate MPN enumerations for summer samples are summarized in Table 2. Sulfate concentrations were three times higher in samples from F1 than in samples from U3. Sulfate reduction rates observed were significantly higher in F1 than in U3 (P < 0.05). Although the sulfate reduction rate method used in this study does not represent field conditions, this method was used to obtain comparative information of the two type of sites (25). Sulfate reduction rates exhibited high standard deviations typical of those commonly observed with the passive-extraction method (25). SRP enumerations revealed a considerable number of lactate- and acetate-oxidizing SRP in the samples. MPN enumerations were an order of magnitude higher in samples from F1 than in samples from U3. In samples from both sites, MPN estimates for lactate oxidizers were similar to estimates obtained for acetate oxidizers.
TABLE 2.
Sulfate concentrations, sulfate reduction rates, MPN enumerations, and potential methanogenic rates for eutrophic (F1) and pristine (U3) Florida Everglades soilsa
| Soil type | Sulfate concn (mg/g of soil)b | Sulfate reduction rate (nmol · g−1 · day−1)b | Enumerationc (MPN · g−1) for:
|
Potential methanogenic rates (μmol · g−1 · h−1)b,d
|
||||
|---|---|---|---|---|---|---|---|---|
| Lactate | Acetate | Intrinsic soil | Acetate | Acetate + SO42− | Acetate + M0O42− | |||
| F1 (eutrophic) | 0.09 (0.03) | 119 (34) | 9.3 × 106 (2.0 × 106-44 × 106) | 1.5 × 106 (0.3 × 106-6.9 × 106) | 0.015 (0.002) | 0.057 (0.003) | 0.034 (0.008) | 0.050 (0.002) |
| U3 (pristine) | 0.03 (0.01) | 26 (8) | 9.3 × 105 (2.0 × 105-44 × 105) | 4.3 × 105 (1.0 × 105-20 × 105) | 0.0033 (0.0007) | 0.0068 (0.0009) | 0.0065 (0.0005) | 0.0055 (0.0008) |
All values are expressed per gram (wet weight) of soil.
Standard errors of the mean are shown in parentheses.
MPN 95% confidence limits are shown in parentheses.
For details, see section on methanogenic rates in Materials and Methods.
Methanogenesis versus sulfate reduction.
Potential methanogenesis rates were determined in samples taken in the fall of 2001. Intrinsic initial methanogenesis rates (without added carbon source) were approximately fivefold higher in F1 (0.015 μmol g−1 h−1 measured in 46 h) than in U3 (0.003 μmol g−1 h−1 measured in 22 h) (Table 2). Furthermore, amounts of methane accumulated within 6 days were almost eightfold higher in F1 than in U3 (1.99 μmol versus 0.24 μmol of methane; data not shown). Addition of sulfate or molybdate did not result in significant changes in either the initial methane production rate or in the methane accumulated in 6 days. Addition of acetate resulted in an increase in methane production rates, with approximately four times the amount of methane observed for F1 versus two times for the pristine sites. No significant statistical differences were observed with the combined addition of acetate and sulfate or molybdate (P < 0.05). Due to limited sample availability, these assays were conducted on a small scale (1 g per tube), which may have resulted in high standard deviations due to sample heterogeneity. However, when a P of <0.1 was used, some trends were observed for the eutrophic site F1. Sulfate addition in the presence of acetate resulted in an approximately 60% reduction of initial methane production rates, and the initial rate remained constant with the addition of molybdate.
Phylogenetic analysis of cloned DSR sequences.
An expected ca. 1.9-kb gene product was obtained from all samples, and RFLPs of the clones suggested a high degree of sequence diversity in summer and spring samples. For the spring samples, 27 clones were grouped in 18 RFLP patterns for F1 samples and 15 clones were grouped in 11 distinct RFLP patterns for samples from U3. For F1 samples collected in summer, 20 clones were grouped in 13 RFLP patterns, and for U3 samples, 20 clones were grouped into 12 RFLP patterns. Rarefaction curves approached the plateau typical of a completed clone library (data not shown), and no additional clones were screened. All samples revealed the same degree of diversity as judged by the slope of the rarefaction plots (data not shown). When the clone data were combined, a total of 82 clones with 49 RFLP patterns were observed. No RFLP pattern from F1 was similar to patterns from U3. Only 3 of 28 patterns and 2 of 21 patterns were shared by samples from F1 and U3, respectively, between spring and summer samples.
A total of 46 clones represented by 40 RFLP patterns were partially sequenced, yielding a deduced amino acid sequence of approximately 180 amino acids of the α-subunit of the DSR protein. Some sequences exhibited different RFLP patterns but shared a similar deduced initial 180-amino-acid sequence; therefore, only one representative of each pattern was kept and similar ones were not included in the phylogenetic analysis.
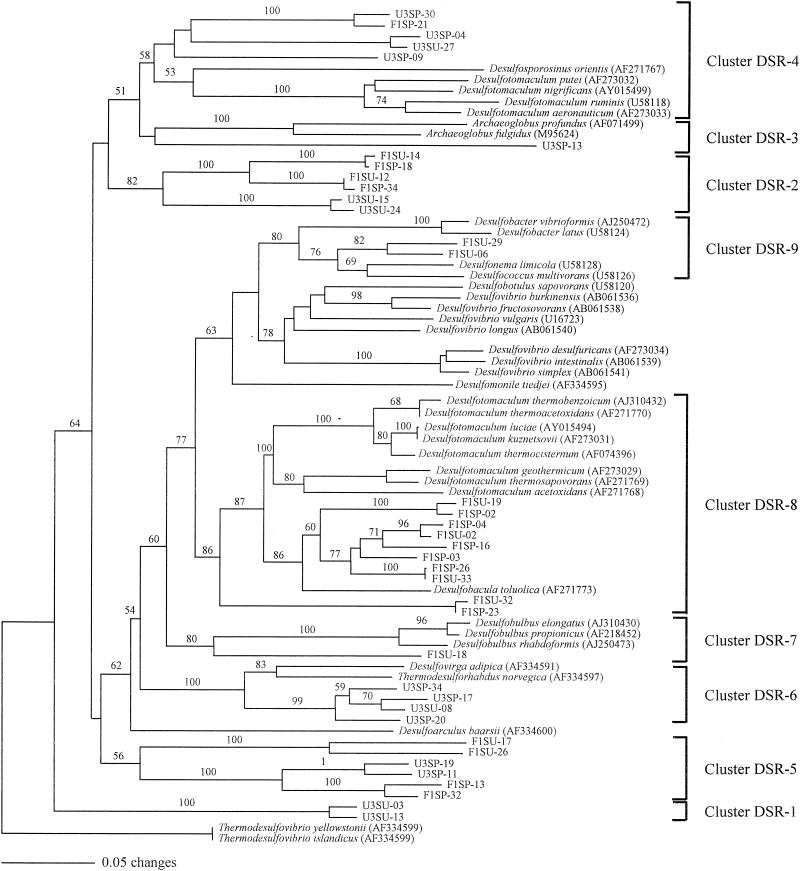
Several phylogenetic approaches were taken to analyze the partial DSR sequences. Most of these approaches yielded similar results, and only minor changes were observed in the placement of some sequences within the major clades (Fig. 2). Bootstrap values for some clades were not high (only bootstrap values over 50 of 100 replicates are presented), but they are not different from previously published bootstrap values for other DSR phylogenetic analyses (8, 20, 21, 26). This is probably due to the fact that partial sequences were used, which would decrease the phylogenetic resolution. Cloned DSR sequences were distributed in a total of nine clades that encompassed gram-negative mesophilic and thermophilic, gram-positive spore-forming, archaeal SRP and two putative clades composed of sequences from uncultured microorganisms. These clades were stable and consistently recovered by distance, parsimony, and maximum-likelihood methods. Thermodesulfovibrio DSR sequences were used as the outgroup in the phylogenetic analysis (21).
FIG. 2.
Neighbor-joining DSR α-subunit tree. The clones are named according to the origin and time of sampling. Scale bar represents 5% change. Numbers at nodes represent percentage of bootstrap resampling based on 100 replicates; only values that are > 50 are presented.
Cluster DSR-1.
Seven clones were related to Thermodesulfovibrio species belonging to the bacterial Nitrospirae phylum. These sequences were present only in the clone library constructed from U3 summer samples. We failed to establish thermophilic enrichments from these samples, suggesting either that these sequences are mesophilic representatives of this clade or that we were unable to cultivate thermophilic strains due to unknown growth requirements.
Cluster DSR-2.
This clade was uniquely composed of our clones and sequences from uncultured microorganisms. The phylogenetic affiliation of these clones remains uncertain. Clade DSR-2 was deeply divergent and was present in all samples except for the spring samples from U3. Similar DSR sequences from GenBank were derived from uncultured sulfate-reducing bacteria from environmental samples, including a marine sediment in Denmark (group II [36]).
Cluster DSR-3.
Three clones clustered with SRP belonging to the Archaea domain. These clones were present only in clone libraries constructed from samples from spring U3 samples.
Cluster DSR-4.
Most clones clustering with the gram-positive Desulfotomaculum incomplete oxidizers were found in clone libraries constructed from U3 (10 clones versus 1 clone from one spring F1 sample). This clade also included uncultured SRP sequences from group III (36) and from a sulfidogenic consortium capable of degrading phenanthrene (28).
Cluster DSR-5.
This clade was composed solely of sequences found in this study and was placed at the bottom of clades encompassing the gram-negative SRP branches of the tree (clusters DSR-6 and DSR-7) and the complete gram-positive oxidizers (cluster DSR-8). These sequences were present in all samples studied.
Cluster DSR-6.
This cluster belongs to one of the δ-proteobacteria SRP clusters and is represented by species of the genera Desulfovirga and Thermodesulforhabdus. This branch was comprised of sequences only found in libraries constructed from U3.
Cluster DSR-7.
This cluster also belongs to the δ-proteobacteria SRP and is represented by species of the genera Desulfobulbus, an incomplete oxidizing genus of the family Desulfobacteraceae. These clones were only found in libraries constructed from F1 samples.
Cluster DSR-8.
This clade is comprised of gram-positive Desulfotomaculum complete oxidizing strains. The 18 clones in this cluster were found exclusively in libraries constructed from F1, with a uniform distribution observed between spring and summer samples.
Cluster DSR-9.
Two clones from libraries constructed from summer F1 clustered in another clade of δ-proteobacteria SRP encompassing the complete oxidizing genus Desulfococcus of the family Desulfobacteraceae.
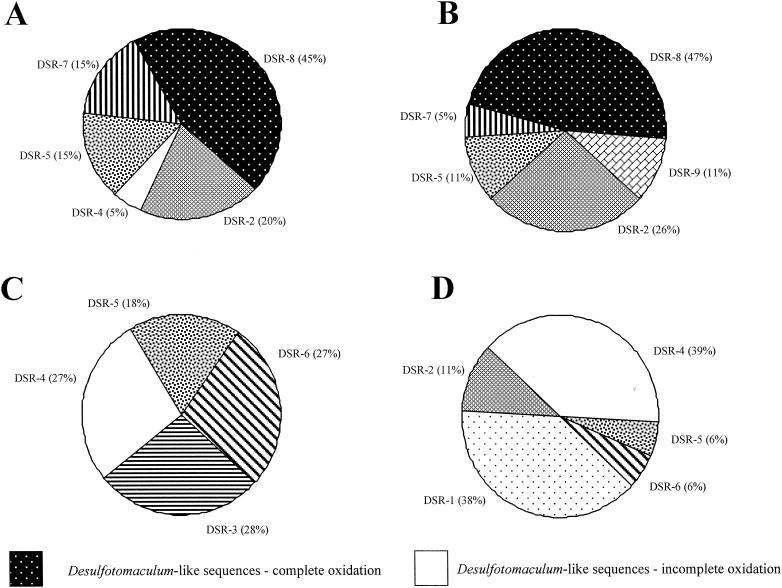
Effect of eutrophication and season on the dynamics of DSR sequences.
F1 samples were dominated by sequences related to cluster DSR-8 of Desulfotomaculum complete oxidizers, followed by sequences belonging to nonculturable SRP of clusters DSR-2 and DSR-5 (Fig. 3A and B). These samples also included sequences related to Desulfobulbus in cluster DSR-7 and minor representation in cluster DSR-4. Seasonal changes between spring and summer resulted in an emergence of SRP related to the Desulfococcus of cluster DSR-9 and disappearance of sequences of cluster DSR-4 but did not result in other major changes of other DSR sequence distributions. SRP dynamics in U3 were less stable (Fig. 3C and D). In spring, no single group was dominant and similar percentages of SRP belonging to clusters DSR-3 (archaeal SRP), DSR-4 (incomplete oxidizing Desulfotomaculum), DSR-5, and DSR-6 (Desulfovirga) were found. In summer, representatives of the cluster DSR-1 emerged and dominated the site with members of the cluster DSR-4. The relative percentages of clones within clusters DSR-5 and DSR-6 decreased. No member of cluster DSR-3 was recovered, and clones representative of cluster DSR-2 emerged.
FIG. 3.
Spatial and seasonal distribution of DSR clones in eutrophic soils for spring (A) and summer (B) and in pristine soils for spring (C) and summer (D).
DISCUSSION
Changes with regard to the effects of nutrient enrichment on biogeochemical cycling in the Everglades have been well documented (9, 11, 13, 22), although detailed characterization at the microbial level is required to understand the effects of eutrophication on processes governing biogeochemical cycles. This is the first microbial study of which we are aware to characterize the diversity and function of SRP in a freshwater wetland such as the Everglades or to characterize potential relationships between phylogenetic groups of SRP and the nutrient status of their environment.
SRP enumerations and sulfate reduction rates.
MPN enumerations showed that SRP were present in considerable numbers and active in both zones of the marsh. F1 contained slightly higher sulfate concentrations (0.09 mg/g of soil, ca. 1 mM) than most typical freshwater systems (ca. 0.1 to 0.3 mM). These relatively high sulfate concentrations correlated with high numbers of lactate- and acetate-utilizing SRP and with high sulfate reduction rates. In U3, the sulfate concentration was lower (0.03 mg/g of soil, ca. 0.3 mM) than those found in F1 and in the upper range of a typical freshwater ecosystem. As expected, lower sulfate reduction rates and SRP numbers were also observed in this site. Similar trends were observed in a previous study conducted in eutrophic areas of WCA-2A and pristine sites of WCA-3A (14). Our enumerations are lower than those reported by Drake et al. (14), but they used a combination of lactate and hydrogen as electron donors and we used lactate or acetate only individually. Lactate was selected as a general electron donor for SRP, since lactate can be used as an electron donor by most species, with the exception of Desulfobacter and some Desulfobacterium species. Many SRP capable of complete oxidation of electron donors utilize acetate, including strains of the genus Desulfobacter and some Desulfotomaculum strains. In our samples, the numbers of lactate- and acetate-utilizing SRP were similar and DSR sequence analysis did not reveal sequences related to Desulfobacter or Desulfobacterium. It can be assumed, therefore, that most of the cultivable SRP in this enumeration medium were complete oxidizers. This finding contradicts the analysis of DSR sequences from the pristine zones, where a great number of sequences are associated with Desulfotomaculum incomplete oxidizers.
Clones related to Desulfotomaculum incomplete oxidizers may represent SRP that could not be grown under the conditions of this study. Other clones present in U3 are either related to unculturable SRP or to poorly characterized SRP clades that are presently represented only by a few cultivable SRP (Fig. 2) (i.e., Archaeoglobus, Desulfovirga, and Thermodesulfovibrio species), making it difficult to infer much from about their physiology.
Methanogenesis versus sulfate reduction.
Methanogenesis rates and accumulated methane in 6-day incubations were higher in F1 than in U3, which is in agreement with previously published reports (14, 31). Methanogenesis in U3 did not respond to addition of acetate, suggesting that acetoclastic methanogenesis may not be a major process in these regions of the marsh. This finding is in agreement with Drake et al. (14), who reported numbers of acetoclastic methanogens that were 6 orders of magnitude greater in eutrophic site F1 than in a more pristine site in WCA-3A. In U3, no major changes were observed upon addition of acetate and molybdate or sulfate, suggesting that acetate-utilizing SRP may not be important in this region of the marsh. On the contrary, acetate-utilizing SRP may play a role in eutrophic zones of the marsh, as suggested by the observations that addition of sulfate to F1 microcosms inhibited methanogenesis to some extent. The observed lack of response to acetate in the presence of sulfate suggests that SRP present in the pristine zones are using electron donors other than acetate or that they may be using another type of metabolism such as fermentation. Incomplete oxidizing Desulfotomaculum strains can use electron donors such as hydrogen, formate, and ethanol (43). Moreover, incomplete oxidizers would likely outcompete complete oxidizers for substrates used by both groups, such as intermediates of the anaerobic degradation of organic matter such as hydrogen and/or lactate (42).
Phylogenetic analysis of DSR sequences.
DSR sequence analysis revealed a great deal of diversity, and nine phylogenetic clusters can be identified in these libraries. Samples from F1 exhibited a stable distribution of DSR sequences between spring and summer sampling times. F1 libraries were dominated by sequences related to uncultured SRP and Desulfotomaculum complete oxidizers. Relatively few phylotypes belonging to Desulfobulbus-, Desulfococcus-, and Desulfotomaculum-like incomplete oxidizer sequences were cloned. In U3, the distribution was less stable between spring and summer. Desulfotomaculum comprised an important proportion of the SRP population, and Desulfotomaculum incomplete oxidizers dominated the distribution in the clone library. A similar dominance of DSR Desulfotomaculum-like sequences was reported in groundwater at a uranium mill tailing site (8), which may suggest a greater role for gram-positive SRP in the sulfur and carbon cycle than previously ascribed to this group. The Everglades ecosystem is characterized by seasonal water table fluctuations, and the ability of Desulfotomaculum to sporulate as a survival mechanism may explain the relatively high number of phylotypes belonging to this group. The selection of Desulfotomaculum due to alternating oxic and anoxic conditions has been observed in rice paddies (34, 43, 46, 47).
Other groups present in the eutrophic zones with a known phylogenetic association were related to Desulfobulbus and Desulfococcus, members of the family Desulfobacteraceae. Desulfobulbus is an incomplete oxidizer able to use propionate and to ferment lactate or ethanol in sulfate-free media and is found in anaerobic freshwater mud. Desulfococcus species are complete oxidizers that can ferment lactate and pyruvate and are found in anaerobic mud and marine habitats (44, 45).
The DSR gene has been extensively used as a genetic marker in microbial diversity studies (8, 20, 26, 28, 36). It has been proposed that the DSR gene was subject to multiple lateral gene transfer events that may confuse the use of this genetic marker in phylogenetic studies (21). One such group in question is Desulfotomaculum, a genus that is not monophyletic but rather is divided between those Desulfotomaculum organisms able to perform complete oxidation and those limited to incomplete oxidation. In our study, this distinction facilitated correlation between two different types of metabolism with two regions of the marsh exposed to different levels of nutrient impact. This metabolic differentiation may be linked to the type of substrates that different Desulfotomaculum spp. utilize. Complete oxidizers are generally more versatile and may utilize a broader range of substrates than do incomplete oxidizers (43). In our findings, Desulfotomaculum complete oxidizers dominate in eutrophic regions of the marsh and Desulfotomaculum incomplete oxidizers dominate in pristine regions. A general concept in ecology is that specialists outcompete generalists for specific nutrients (29). Therefore, it may be that the more specialist Desulfotomaculum incomplete oxidizers outcompete generalist complete oxidizers for other substrates such as hydrogen and lactate that can be metabolized by both groups of Desulfotomaculum (42). This would explain the greater abundance of incomplete oxidizers in clone libraries constructed from pristine zones of the marsh (U3), which may have a narrower range of electron donors than would be found in the eutrophic region (F1).
In conclusion, cultivation-based techniques revealed greater SRP activity and higher SRP numbers in eutrophic sites than in pristine sites of the marsh, likely as a result of nutrient enrichment that resulted in a microbial enrichment. In F1, sulfate additions inhibited acetoclastic methanogenesis somewhat, suggesting possible substrate competition between SRP and methanogens for acetate. This competition was not observed in U3. Diversity within DSR sequences was found in both zones of the marshes. Significantly, Desulfotomaculum-like sequences from eutrophic regions were related to those Desulfotomaculum organisms able to carry out complete oxidation and in pristine regions they were related to those unable to carry out complete oxidation. Therefore, molecular and conventional cultivation techniques revealed that nutrient loading resulted in a selection of different SRP populations. Molecular techniques revealed a selection in the type of Desulfotomaculum populations present in eutrophic versus pristine regions of the marsh, suggesting that Desulfotomaculum complete oxidizers are better adapted to eutrophic conditions than to pristine sites, where a greater number of Desulfotomaculum incomplete oxidizing strains are present.
Acknowledgments
This study was supported by a grant from the National Science Foundation.
We are grateful to Joe Prenger and Yu Wang, Wetland Biogeochemistry Laboratory, Soil and Water Science Department, for sampling coordination and providing the geochemical data. We also thank Sue Newman, South Florida Water Management District, for assistance with field sampling.
Footnotes
Florida Agricultural Experimental Station Journal Series No. R-09041.
REFERENCES
- 1.Bak, F., and N. Pfennig. 1991. Microbial sulfate reduction in littoral sediment of Lake Constance. FEMS Microbiol. Ecol. 85:31-42. [Google Scholar]
- 2.Balch, W. E., G. E. Fox, L. J. Magrum, C. R. Woese, and R. S. Wolfe. 1979. Methanogens: reevaluation of a unique biological group. Microbiol. Rev. 43:260-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bates, A. L., W. H. Orem, J. W. Harvey, and E. C. Spiker. 2002. Tracing sources of sulfur in the Florida Everglades. J. Environ. Qual. 31:287-299. [DOI] [PubMed] [Google Scholar]
- 4.Bates, A. L., E. C. Spiker, and C. W. Holmes. 1998. Speciation and isotopic composition of sedimentary sulfur in the Everglades, Florida, USA. Chem. Geol. 146:155-170. [Google Scholar]
- 5.Cappenberg, T. E. 1974. Interrelations between sulfate-reducing and methane-producing bacteria in bottom deposits of a freshwater lake. I. Field observations. Antonie Leeuwenhoek 40:285-295. [DOI] [PubMed] [Google Scholar]
- 6.Cappenberg, T. E. 1974. Interrelations between sulfate-reducing and methane-producing bacteria in bottom deposits of a freshwater lake. II. Inhibition experiments. Antonie Leeuwenhoek 40:297-306. [DOI] [PubMed] [Google Scholar]
- 7.Castro, H. F., N. H. Williams, and A. Ogram. 2000. Phylogeny of sulfate-reducing bacteria. FEMS Microbiol. Ecol. 31:1-9. [DOI] [PubMed] [Google Scholar]
- 8.Chang, Y.-J., A. D. Peacock, P. E. Long, J. R. Stephen, J. P. McKinley, S. J. Macnaughton, A. K. M. A. Hussain, A. M. Saxton, and D. C. White. 2001. Diversity and characterization of sulfate-reducing bacteria in groundwater at a uranium mill tailings site. Appl. Environ. Microbiol. 67:3149-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Craft, C. B., and C. J. Richardson. 1993. Peat accretion and phosphorus accumulation along a eutrophication gradient in the northern Everglades. Biogeochemistry 22:133-156. [Google Scholar]
- 10.D'Angelo, E. M., and K. R. Reddy. 1999. Regulators of heterotrophic microbial potentials in wetland soils. Soil Biol. Biochem. 31:815-830. [Google Scholar]
- 11.DeBusk, W. F., S. Newman, and K. R. Reddy. 2001. Spatio-temporal patterns of soil phosphorus enrichment in Everglades Water Conservation Area 2A. J. Environ. Qual. 30:1438-1446. [DOI] [PubMed] [Google Scholar]
- 12.DeBusk, W. F., and K. R. Reddy. 1998. Turnover of detrital organic carbon in a nutrient-impacted Everglades marsh. Soil Sci. Soc. Am. J. 62:1460-1468. [Google Scholar]
- 13.DeBusk, W. F., K. R. Reddy, M. S. Koch, and Y. Wang. 1994. Spatial distribution of soil nutrients in a northern Everglades marsh: Water Conservation Area 2A. Soil Sci. Soc. Am. J. 58:543-552. [Google Scholar]
- 14.Drake, H. L., N. G. Aumen, C. Kuhner, C. Wagner, A. Grießhammer, and M. Schmittroth. 1996. Anaerobic microflora of Everglades sediments: effects of nutrients on population profiles and activities. Appl. Environ. Microbiol. 62:486-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fossing, H., and B. B. Jørgensen. 1989. Measurement of bacterial sulfate reduction in sediments: evaluation of a single-step chromium reduction method. Biogeochemistry 8:205-222. [Google Scholar]
- 16.Gilmour, C. C., G. S. Riedel, M. C. Ederington, J. T. Bell, J. M. Benoit, G. A. Gill, and M. C. Stordal. 1998. Methylmercury concentrations and production rates across a trophic gradient in the northern Everglades. Biogeochemistry 40:327-345. [Google Scholar]
- 17.Hungate, R. E. 1969. A roll tube method for cultivation of strict anaerobes, p. 117-132. In J. R. Norris and D. W. Ribbons (ed.), Methods in microbiology. Academic Press, New York, N.Y.
- 18.Ingvorsen, K., and T. D. Brock. 1982. Electron flow via sulfate reduction and methanogenesis in the anaerobic hypolimnion of Lake Mendota. Limnol. Oceanogr. 27:559-564. [Google Scholar]
- 19.Ingvorsen, K., J. G. Zeikus, and T. D. Brock. 1981. Dynamics of bacterial sulfate reduction in a eutrophic lake. Appl. Environ. Microbiol. 42:1029-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joulian, C., N. B. Ramsing, and K. Ingvorsen. 2001. Congruent phylogenies of most common small-subunit rRNA and dissimilatory sulfite reductase gene sequences retrieved from estuarine sediments. Appl. Environ. Microbiol. 67:3314-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klein, M., M. Friedrich, A. J. Roger, P. Hugenholtz, S. Fishbain, H. Abicht, L. L. Blackall, D. A. Stahl, and M. Wagner. 2001. Multiple lateral transfers of dissimilatory sulfite reductase genes between major lineages of sulfate-reducing prokaryotes. J. Bacteriol. 183:6028-6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koch, M. S., and K. R. Reddy. 1992. Distribution of soil and plant nutrients along a trophic gradient in the Florida Everglades. Soil Sci. Soc. Am. J. 56:1492-1499. [Google Scholar]
- 23.Kuivila, K. M., J. W. Murray, A. H. Devol, and P. C. Novelli. 1989. Methane production, sulfate reduction and competition for substrates in the sediments of Lake Washington. Geochim. Cosmochim. Acta 53:409-416. [Google Scholar]
- 24.Lovley, D. R., and M. J. Klug. 1983. Sulfate reducers can outcompete methanogens at freshwater sulfate concentrations. Appl. Environ. Microbiol. 45:187-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meier, J., A. Voigt, and H. D. Babenzien. 2000. A comparison of 35S-SO42− radiotracer techniques to determine sulphate reduction rates in laminated sediments. J. Microbiol. Methods 41:9-18. [DOI] [PubMed] [Google Scholar]
- 26.Minz, D., J. L. Flax, S. J. Green, G. Muyzer, Y. Cohen, M. Wagner, B. E. Rittmann, and D. A. Stahl. 1999. Diversity of sulfate-reducing bacteria in oxic and anoxic regions of a microbial mat characterized by comparative analysis of dissimilatory sulfite reductase genes. Appl. Environ. Microbiol. 65:4666-4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oremland, R. S., and D. G. Capone. 1988. Use of specific inhibitors in biogeochemistry and microbial ecology. Adv. Microb. Ecol. 10:285-383. [Google Scholar]
- 28.Perez-Jimenez, J. R., L. Y. Young, and L. J. Kerkhof. 2001. Molecular characterization of sulfate-reducing bacteria in anaerobic hydrocarbon-degrading consortia and pure cultures using the dissimilatory sulfite reductase (dsr AB) genes. FEMS Microbiol. Ecol. 35:145-150. [DOI] [PubMed] [Google Scholar]
- 29.Pianka, E. R. 1974. Evolutionary ecology. Harper & Row, Publishers, Inc., New York, N.Y.
- 30.Reddy, K. R., R. D. DeLaune, W. F. DeBusk, and M. S. Koch. 1993. Long-term nutrient accumulation rates in the Everglades. Soil Sci. Soc. Am. J. 57:1147-1155. [Google Scholar]
- 31.Reddy, K. R., J. R. White, A. Wright, and T. Chua. 1999. Influence of phosphorus loading on microbial processes in soil and water column of wetlands, p. 249-273. In K. R. Reddy, G. A. O'Connor, and C. L. Schelske (ed.), Phosphorus biogeochemistry in subtropical ecosystems. Lewis Publishers, New York, N.Y.
- 32.South Florida Water Management District. 2001. Executive summary. Everglades consolidated report. 2001. South Florida Water Management District, West Palm Beach, Fla.
- 33.Strimmer, K., and A. von Haeseler. 1996. Quartet puzzling: a quartet maximum likelihood method for reconstructing tree topologies. Mol. Biol. Evol. 13:964-969. [Google Scholar]
- 34.Stubner, S., and K. Meuser. 2000. Detection of Desulfotomaculum in an Italian rice paddy soil by 16S ribosomal nucleic acid analyses. FEMS Microbiol. Ecol. 34:73-80. [DOI] [PubMed] [Google Scholar]
- 35.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomsen, T. R., K. Finster, and N. B. Ramsing. 2001. Biogeochemical and molecular signatures of anaerobic methane oxidation in a marine sediment. Appl. Environ. Microbiol. 67:1646-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Touzel, J. P., and G. Albagnac. 1983. Isolation and characterization of Methanococcus mazei strain MC3. FEMS Microbiol. Lett. 16:241-245. [Google Scholar]
- 38.Ulrich, G. A., L. R. Krumholz, and J. M. Suflita. 1997. A rapid and simple method for estimating sulfate reduction activity and quantifying inorganic sulfides. Appl. Environ. Microbiol. 63:1627-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wagner, M., A. J. Roger, J. L. Flax, G. A. Brusseau, and D. A. Stahl. 1998. Phylogeny of dissimilatory sulfite reductases supports an early origin of sulfate respiration. J. Bacteriol. 180:2975-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ward, D. M., and M. R. Winfrey. 1985. Interactions between methanogenic and sulfate-reducing bacteria in sediments, p. 141-179. In H. W. Jannasch and P. J. Williams (ed.), Advances in aquatic microbiology. Academic Press, London, United Kingdom.
- 41.White, J. R., and K. R. Reddy. 1999. Influence of nitrate and phosphorus loading on denitrifying enzyme activity in Everglades wetland soils. Soil Sci. Soc. Am. J. 63:1945-1954. [Google Scholar]
- 42.Widdel, F. 1988. Microbiology and ecology of sulfate- and sulfur-reducing bacteria, p. 469-585. In A. J. B. Zehnder (ed.), Biology of anaerobic microorganisms. John Wiley & Sons, New York, N.Y.
- 43.Widdel, F. 1992. The genus Desulfotomaculum, p. 1792-1799. In A. Balows, H. G. Truper, M. Dworkin, W. Harder, and K. H. Schleifer (ed.), The prokaryotes. Springer-Verlag, New York, N.Y.
- 44.Widdel, F., and F. Bak. 1992. Gram-negative mesophilic sulfate-reducing bacteria, p. 3352-3378. In A. Balows, H. G. Truper, M. Dworkin, W. Harder, and K. H. Schleifer (ed.), The prokaryotes. Springer-Verlag, New York, N.Y.
- 45.Widdel, F., and N. Pfennig. 1984. Dissimilatory sulfate- or sulfur-reducing bacteria, p. 663-679. In N. R. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology. Williams & Wilkins, Baltimore, Md.
- 46.Wind, T., and R. Conrad. 1995. Sulfur compounds, potential turnover of sulfate and thiosulfate, and numbers of sulfate-reducing bacteria in planted and unplanted paddy soil. FEMS Microbiol. Ecol. 18:257-266. [Google Scholar]
- 47.Wind, T., S. Stubner, and R. Conrad. 1999. Sulfate-reducing bacteria in rice field soil and on rice roots. Syst. Appl. Microbiol. 22:269-279. [DOI] [PubMed] [Google Scholar]