Abstract
Reverse transcription-PCR assays have been established for a quick, sensitive, and specific diagnosis of acute bee paralysis virus (ABPV), a common virus of the honeybee (Apis mellifera), directly from clinical samples. A 3,071-nucleotide fragment of the ABPV genome, which includes the entire capsid polyprotein gene, was amplified from Austrian, German, Polish, and Hungarian ABPV samples and sequenced, and the sequences were compared. The alignment of a smaller fragment with ABPV sequences from the United States and the United Kingdom revealed nucleotide identity rates between 89 and 96%, respectively. Phylogenetic trees which display the molecular relationship between the viruses of different geographic origin were constructed.
Acute bee paralysis virus (ABPV) was first described as a cause of inapparent infections of the honeybee (Apis mellifera) (3). The presence of the virus has been reported from several countries worldwide (12, 15, 22, 31, 35). ABPV is considered to be a common infective agent of bees which is frequently detected in apparently healthy colonies. However, it has been presumed that this virus plays a role in cases of sudden collapse of honeybee colonies infested with the parasitic mite Varroa destructor (former name, Varroa jacobsoni) (6, 31). ABPV was suggested to be a primary cause of bee mortality in such colonies in Germany (5), Yugoslavia (27), France (15), and the United States of America (21). The worldwide spread of V. destructor in honeybee colonies has a significant influence on virus infection of bees. On the one hand, the Varroa mite is a possible vector for the virus (5, 8); on the other hand, the mite weakens the bees and activates the viral infection, leading to clinical symptoms and severe losses in apiaries (5, 10, 32). Some scientists, however, doubt the essential roles of both the mites (1, 22) and the viruses (23, 24) in the so-called bee parasitic mite syndrome (joint infection of viruses, Acarapis woodi, and V. destructor) (33). Interestingly, in the United Kingdom, not ABPV but rather slow paralysis virus was identified as an agent responsible for the rapid decline and death of many colonies infested with the Varroa mite (4).
ABPV has a single-stranded, positive-sense, polyadenylated RNA genome comprising 9,491 nucleotides (nt). The complete nucleotide sequence was determined recently (18). The genome contains two open reading frames (ORFs). ORF1 encodes the nonstructural proteins (RNA-dependent RNA polymerase, helicase, and protease), while ORF2 encodes the three major structural proteins (35, 33, and 24 kDa) and a minor protein (9.4 kDa) transcribed together in a capsid polyprotein (18). ABPV has not yet been assigned to a certain virus genus or family; it is generally referred to as a picorna-like positive-strand RNA virus. Its genomic RNA is considerably longer than that of classical picornaviruses (approximately 9,500 versus 7,500 nt), and it also differs in its genome organization from picornaviruses. It was therefore suggested that ABPV be classified together with some other picorna-like viruses infecting insects in a novel taxonomic group called cricket paralysis-like viruses (18). ABPV shares some antigenic properties (2) and sequence similarities (73 and 79% at the nucleotide and amino acid levels, respectively [13]) with the Kashmir bee virus, another picorna-like virus infecting honeybees.
The importance of viruses in diseases of bees has been investigated in several studies. To date, 18 different honeybee viruses have been described (1, 19). Most of them cause inapparent infections. Such infections are sometimes exacerbated and activated by subservient environmental factors. Besides mite infestation and bacterial infections, pollution and the comprehensive use of chemicals and insecticides in agricultural technology trigger environmental stress in bees (11, 16, 26). In certain cases, even the acaricides used against Varroa mites have to be blamed for the suppression of the bee's immune system (9). In addition, the notable decrease of natural pollinator species—also due to environmental pollution—emphasizes the significance of honeybees in the pollination of plants, both in the natural ecosystem and in agricultural pollination (29); thus, the importance of healthy bees is far beyond honey production (34).
Reverse transcription-PCR (RT-PCR) assays have been developed for the detection of certain honeybee viruses such as sacbrood virus (SBV) (20), the Kashmir bee virus (13, 25), black queen cell virus (BQCV) (7), and ABPV (7, 13). RT-PCR proved to be a quick, specific, sensitive, and reliable technique for the diagnosis of honeybee virus infections. A further advantage of RT-PCR is that genetic comparison and classification of different virus strains can be rapidly carried out by sequencing of the appropriate PCR products (20). This approach may even allow the prediction of virus taxonomies, as already successfully applied by Evans and Hung (14).
The aims of this study were to establish RT-PCR assays for the sensitive direct detection of ABPV in clinical samples, to reveal and compare the nucleotide sequences of the ABPV capsid polyprotein gene region of different European isolates, and to assess the genetic relationship between ABPV strains of various geographic origins.
Samples.
The ABPV isolates originated from infected honeybees collected in four different European countries. The samples from Austria (one sample) and Germany (three samples) were collected following outbreaks of acute bee paralysis; the Hungarian and Polish samples were taken from colonies showing clinical symptoms (Hungary, six samples; Poland, one sample) but also from obviously healthy colonies from apiaries participating in an ABPV survey (seven and three samples, respectively). The specimens were collected within a 5-year period (1996 to 2000). Virus identification was carried out by the agar gel immunodiffusion test (6) and electron microscopy. The samples were also tested for other picorna-like honeybee viruses such as SBV and BQCV. Each sample contained 50 to 60 dead honeybees, which were stored at −80°C until investigated.
Isolation of RNA.
The bees were homogenized either in liquid nitrogen or in sterile mortars by using sterile phosphate-buffered saline. The homogenates were centrifuged at 20,000 × g for 10 min, and the supernatant was used for RNA extraction. In some cases, the virus-containing samples were propagated by inoculation of virus-free pupae and purified by cesium chloride gradient ultracentrifugation followed by dialysis against phosphate-buffered saline. RNA was extracted from a 140-μl virus suspension using a QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions.
Primer design.
Six different primer pairs were selected based on the complete ABPV sequence deposited at the GenBank database (accession no. AF150629) with the help of a primer designer program (Scientific and Educational Software, version 3.0). The oligonucleotide primers were designed in order to amplify overlapping PCR products comprising the entire structural protein gene region of ABPV. The primer sequences, orientations, and locations as well as the expected sizes of the amplicons are shown in Table 1. Nucleotide positions refer to the ABPV sequence with the GenBank accession no. AF150629. The oligonucleotides were synthesized by GibcoBRL Life Technologies, Ltd. (Paisley, Scotland, United Kingdom).
TABLE 1.
Oligonucleotide primer pairs selected for ABPV RT-PCR
| Primer namea | Primer sequence (5′ to 3′) | Nucleotide positionsb | Length of the amplified product (bp) |
|---|---|---|---|
| APV 17 f | TAT CAG AAG GCC ACT GGA GA | 6242-6261 | 722 |
| APV 18 r | TCC ACT CGG TCA TCA TAA GG | 6995-7014 | |
| APV 19 f | TCT TGG ACA TTG CCT TCA GT | 6848-6867 | 778 |
| APV 20 r | ATA CCA TTC GCC ACC TTG TT | 7607-7626 | |
| APV 21 f | TGC AGT TCC AGA AGT TAA GA | 7447-7466 | 686 |
| APV 22 r | ATA GTR GCT CGC CAA TAT GA | 8114-8133 | |
| APV 23 nf | GTG CTA TCT TGG AAT ACT AC | 7928-7947 | 618 |
| APV 24 nr | AAG GYT TAG GTT CTA CTA CT | 8527-8546 | |
| APV 25 f | GGA ACA TGG AAG CAT TAT TG | 8694-8713 | 687 |
| APV 26 r | AAT GTC TTC TCG AAC CAT AG | 9362-9381 | |
| APV 27 f | ATT GGC GAG CYA CTA TGT GC | 8118-8137 | 858 |
| APV 28 r | CGC GGT AYT AAG AAG CTA CG | 8957-8976 |
RT-PCR.
RT and amplifications were performed with a continuous RT-PCR method by employing a OneStep RT-PCR kit (Qiagen) according to the manufacturer's recommendations. RT-PCR, gel electrophoresis, and DNA extraction as well as nucleotide sequencing were carried out essentially as described by Grabensteiner et al. (20).
Phylogenetic analysis.
Phylogenetic analysis was performed with the Phylogeny Inference Program package (PHYLIP), version 3.57c. Bootstrap resampling analyses of 100 replicates were performed with the SEQBOOT program to prove the stability of the trees. Distance matrices were generated by the DNADIST/Neighbor-Joining and Fitch programs, using a translation/transversion ratio of 2.0.
Analysis of ABPV samples by RT-PCR.
Six different RT-PCR assays employing primer pairs designed for the amplification of overlapping fragments which cover the entire capsid polyprotein gene region of ABPV (Table 1) have been developed. To ensure that the RT-PCR assays detected nucleic acid from diverse ABPV strains, the reactions were performed with 11 samples, which originated from different European countries; besides 1 Austrian sample and 3 German samples, 3 samples from Poland and 4 samples from Hungary were tested. All samples proved positive in all RT-PCR assays. The amplicons could be observed as clear and distinct bands of the expected molecular sizes, even in the case of one German sample which contained a mixture of ABPV and BQCV. Amplification products never occurred in the negative controls. For the amplification of the ABPV genomic region from nt 8044 to 8512, the initially designed forward (APV 23 f) and reverse (APV 24 r) primers gave only weak signals in RT-PCR and failed at sequencing. Therefore, a new primer pair (APV 23 nf and APV 24 nr) covering the same region (nt 7928 to 8546) was designed. The new oligonucleotides operated properly; thus, they were applied hereafter.
Sequence analysis and comparison.
In total, 66 RT-PCR amplification products were sequenced in both directions. The sequences were identified as ABPV sequences by a BLAST search (National Center for Biotechnology Information, National Institutes of Health, Bethesda, Md.). The sequences derived from the six overlapping PCR products of all analyzed ABPV samples were compiled and aligned, using the published complete ABPV sequence (18) as a reference. In total, sequence information for a 3,071-nt fragment, which covers 32.4% of the ABPV genome, including the entire capsid protein gene region, was available from every investigated isolate. The isolates were 94 to 95% identical to the reference strain (data not shown; all complete nucleotide and amino acid sequence alignments are available upon request). When we analyzed the sequences of the strains, two German isolates proved to be fully identical; thus, only one of them was used in the further studies. Deduced amino acid sequences were generated for the complete capsid polyprotein (nt 6509 to 9253), resulting in 914-amino-acid-long polypeptide sequences. The amino acid sequences were aligned and compared to the reference strain. They exhibited an identity rate of 97 to 98%. Phylogenetic trees were constructed based on the nucleotide sequences of the various isolates, and the stability of the trees was tested by bootstrap analysis of 100 replicates. The optimizing tree reconstruction method (Fitch) proposed the phylogenetic tree presented in Fig. 1.
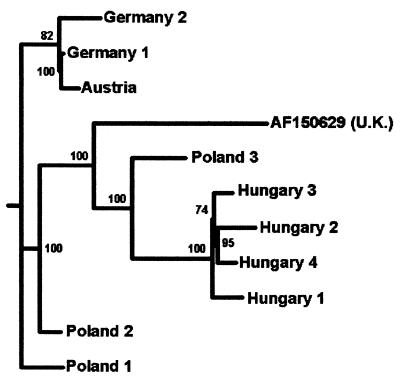
FIG. 1.
Phylogenetic tree illustrating the genetic relationship between European ABPV samples based on a 3,071-nt fragment of the ABPV genome which includes the entire capsid polyprotein gene region (nucleotide positions 6283 to 9353 according to numbering for reference strain AF150629 from the United Kingdom [U.K.]).
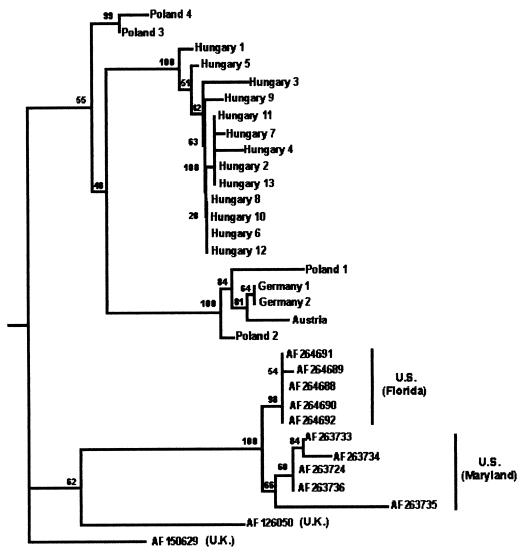
Since additional (shorter) ABPV sequences have been deposited in the GenBank database by other research groups (one sequence from the United Kingdom [strain Rothamsted, GenBank accession no. AF126050, deposited by Ghosh et al.; 17] and 10 sequences from the United States [sequences from samples originating from the state of Maryland with the accession no. AF263724 and AF263733 to AF263736 and sequences from samples from Florida with accession no. AF264688 to AF264692]; all U.S. sequences were deposited by Hung et al. [13]), we amplified the same region from an additional nine ABPV samples from Hungary and one from Poland by employing the newly designed primer pair APV 23 nf and APV 24 nr and sequenced them in order to compose a shorter but more comprehensive comparison of the different isolates. In total, 32 ABPV nucleotide sequences were aligned using a 401-nt stretch from positions 8121 to 8521. The sequences exhibited between 89 and 96% identity with the reference strain. Deduced amino acid sequences from these 32 ABPV strains were also compiled and aligned. The 133-amino-acid-long polypeptide sequences showed 90 to 96% identity with the reference strain. Phylogenetic trees were constructed, and bootstrap analysis was performed. A representative tree generated by the Neighbor-Joining method is shown in Fig. 2.
FIG. 2.
Phylogenetic tree illustrating the genetic relationship of ABPV isolates from different European countries and from the United States, based on a 401-nt stretch within the capsid protein gene region (nucleotide positions 8121 to 8521 of the ABPV reference strain AF150629 from the United Kingdom [U.K.]).
The scientific interest in honeybee viruses is increasing. The majority of bee viruses have been formerly characterized by classical virological methods (1, 30), and their presence has been reported worldwide; our knowledge of the molecular properties and genome organizations of some of these viruses is the result of recent studies (16, 17, 28). There is still very limited information available on the taxonomic classification of honeybee viruses and on the existence of serotypes or variants within one species, as well as on the role of the viruses in bee diseases. Molecular comparison of various isolates is an exact and reliable method to study the relationships between and within different species, since it directly detects the changes of the genetic information in the course of evolution and employs statistical evaluation of the data. We should even think about using molecular phylogenetics to clarify the relationships of bee viruses as a whole (14).
This study presents a phylogenetic analysis of the structural protein gene region of ABPV strains. Sequences of 10 central European ABPV isolates were aligned and compared to the sequence of the reference strain, which originated from the United Kingdom. We focused our investigations on the structural protein gene, because this genomic region usually shows a higher degree of divergence than nonstructural genomic regions. The analyzed isolates exhibited 94 to 95% sequence identity with the United Kingdom reference strain. The nucleotide changes were distributed fairly evenly over the entire region that we have sequenced, although some stretches showed a slightly higher sequence divergence. The probable relationship between the viruses is illustrated by a phylogenetic tree (Fig. 1). The strains form at least three groups. The Austrian and German isolates cluster together in one branch, the Hungarian strains belong to a second distinct cluster, while the Polish ABPV samples exhibit a higher diversity. The reference strain from the United Kingdom shows a relatively lower-level genetic relationship to the central European ABPV strains.
In the comparison of the sequences of the partial capsid protein gene region (nt 8121 to 8521), we included partial sequences of the above-mentioned ABPV strains as well as the sequences of one additional Polish strain and nine additional Hungarian strains. We also included 10 sequences of ABPV strains from the United States (13) as well as one sequence from the United Kingdom which had been deposited in the GenBank database. In this 401-nt stretch, the strains exhibited between 89 and 96% identity with the United Kingdom reference strain. The phylogenetic tree comprising a total of 32 isolates suggests the separation of at least two distinct genetic lineages of ABPV (Fig. 2). One major branch is formed by the U.S. (and United Kingdom) strains, while the other contains the continental-European isolates. Within the U.S.-United Kingdom genotype, there is a clear segregation of United Kingdom isolates from U.S. isolates, while the U.S. strains are closely related and subcluster in two groups, one consisting of the samples originating from Florida and the other containing the Maryland ABPVs. The continental-European genotype is subdivided into at least three distinct major subgroups. The Austrian and German isolates cluster together, showing a low degree of divergence similar to that in the previous tree, but two Polish strains (Poland 1 and 2) also cluster close to them. The Hungarian strains form a distinct, homogeneous branch, while two closely related Polish isolates (Poland 3 and 4) form a unique branch within the continental-European genotype.
The deduced amino acid sequences of the ABPV strains in both analyses showed a quite high degree of identity (97 to 98 and 90 to 96%, respectively); within the genotypes, the degree of amino acid variability was even lower. Nonsense mutations were not observed, and a 4-nt deletion detected in one Hungarian strain was downstream of the coding region; thus, it did not affect the transcribed polyprotein.
Although it turned out that the ABPV capsid polyprotein gene seems to be a relatively conserved genomic region, the results of this study revealed that the observed sequence diversity is a convenient indicator for the identification and classification of ABPV isolates of different geographic origins. The molecular comparison gave insight into the variability of the ABPV isolates collected from various geographic regions and exhibited genomic relationships between the strains. Similar genetic divergence was observed in the case of SBV (20), thus supporting the concept of geographical segregation of bee virus strains. Since ABPV is a worldwide-distributed honeybee virus, the genomic analysis of further isolates from other continents may supplement our knowledge of the diversity of this virus. The RT-PCR assays described in this paper proved to be sensitive and reliable methods for the detection and classification of ABPVs from various countries; we can assume that these RT-PCR assays will be capable of identifying at least all European ABPV isolates but will also likely identify viruses from other continents. The assays should provide an appropriate tool for further investigations.
Nucleotide sequence accession numbers.
The ABPV sequences described in this study were submitted to the GenBank database under accession no. AY053366 to AY053385.
Acknowledgments
This study was supported by the Hungarian National Grants OTKA (grants T030335, C077, and M027651), and by a grant from the Austrian Federal Office and Research Centre for Agriculture (BFL), no. 921/99. Tamás Bakonyi's stay at the Clinical Virology Group, Institute of Virology, University of Veterinary Medicine, Vienna, Austria, was financed by a CEEPUS grant from the European Union, Network H19, stipends for Austria.
REFERENCES
- 1.Allen, M., and B. Ball. 1996. The incidence and world distribution of the honey bee viruses. Bee World 77:141-162. [Google Scholar]
- 2.Allen, M. F., and B. V. Ball. 1995. Characterisation and serological relationships of strains of Kashmir bee virus. Ann. Appl. Biol. 126:471-484. [Google Scholar]
- 3.Bailey, L., A. J. Gibbs, and R. D. Woods. 1963. Two viruses from adult honey bees (Apis mellifera Linnaeus). Virology 21:390-395. [DOI] [PubMed] [Google Scholar]
- 4.Ball, B. V. 1997. Varroa and viruses, p. 11-15. In P. Munn and R. Jones (ed.), Varroa! Fight the mite. International Bee Research Association, Cardiff, Wales, United Kingdom.
- 5.Ball, B. V., and M. F. Allen. 1988. The prevalence of pathogens in the honey bee (Apis mellifera) colonies infested with the parasitic mite Varroa jacobsoni. Ann. Appl. Biol. 113:237-244. [Google Scholar]
- 6.Békési, L., B. V. Ball, M. Dobos-Kovács, T. Bakonyi, and M. Rusvai. 1999. Occurrence of acute paralysis virus of the honeybee (Apis mellifera) in a Hungarian apiary infested with the parasitic mite Varroa jacobsoni. Acta Vet. Hung. 47:319-324. [DOI] [PubMed] [Google Scholar]
- 7.Benjeddou, M., N. Leat, M. Allsopp, and S. Davison. 2001. Detection of acute bee paralysis virus and black queen cell virus from honeybees by reverse transcriptase PCR. Appl. Environ. Microbiol. 67:2384-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowen-Walker, P. L., S. J. Martin, and A. Gunn. 1999. The transmission of deformed wing virus between honeybees (Apis mellifera L.) by the ectoparasitic mite Varroa jacobsoni Oud. J. Invertebr. Pathol. 73:101-106. [DOI] [PubMed] [Google Scholar]
- 9.Brødsgaard, C. J., and H. Hansen. 1996. Virus and varroa—a deadly combination. Gron Viden Landburg 162:4. [Google Scholar]
- 10.Brødsgaard, C. J., W. Ritter, H. Hansen, and H. F. Brødsgaard. 2000. Interactions among Varroa jacobsoni mites, acute paralysis virus, and Paenibacillus larvae larvae and their influence on mortality of larval honeybees in vitro. Apidologie 31:543-554. [Google Scholar]
- 11.Bromenshenk, J. J., J. L. Gudatis, S. R. Carlson, M. J. Thomas, and A. M. Simmons. 1991. Population dynamics of honey bee nucleus colonies exposed to industrial pollutants. Apidologie 22:359-370. [Google Scholar]
- 12.Carpana, E., M. A. Vecchi, A. Lavazza, S. Bassi, M. Dottori, and W. Ritter. 1991. Prevalence of acute paralysis virus (APV) and other viral infections in honeybees in Italy, p. 155-165. In Proceedings of the International Symposium on Recent Research on Bee Pathology, Ghent, Belgium.
- 13.Evans, J. D. 2001. Genetic evidence for coinfection of honey bees by acute bee paralysis and Kashmir bee viruses. J. Invertebr. Pathol. 78:189-193. [DOI] [PubMed] [Google Scholar]
- 14.Evans, J. D., and A. C. Hung. 2000. Molecular phylogenetics and the classification of honey bee viruses. Arch. Virol. 145:2015-2026. [DOI] [PubMed] [Google Scholar]
- 15.Faucon, J. P., C. Vitu, P. Russo, and M. Vignoni. 1992. Diagnosis of acute paralysis: application to the epidemiology of honey bee viral diseases in France in 1990. Apidologie 23:139-146. [Google Scholar]
- 16.Fleche, C., M. C. Clement, S. Zeggane, and J. P. Faucon. 1997. Contamination of bee products and risk for human health: the situation in France. Rev. Sci. Tech. O. I. E. 16:609-619. [PubMed] [Google Scholar]
- 17.Ghosh, R. C., B. V. Ball, M. M. Willcocks, and M. J. Carter. 1999. The nucleotide sequence of sacbrood virus of the honey bee: an insect picorna-like virus. J. Gen. Virol. 80:1514-1549. [DOI] [PubMed] [Google Scholar]
- 18.Govan, V. A., N. Leat, M. Allsop, and S. Davison. 2000. Analysis of the complete genome sequence of acute bee paralysis virus shows that it belongs to the novel group of insect-infecting RNA viruses. Virology 277:457-463. [DOI] [PubMed] [Google Scholar]
- 19.Grabensteiner, E., and N. Nowotny. 2001. Virusinfektionen bei der Honigbiene (Apis mellifera). Wien. Tieraerztl. Monschr. 88:79-87. [Google Scholar]
- 20.Grabensteiner, E., W. Ritter, M. J. Carter, S. Davison, H. Pechhacker, J. Kolodziejek, O. Boecking, I. Derakhshifar, R. Moosbeckhofer, E. Licek, and N. Nowotny. 2001. Sacbrood virus of the honeybee (Apis mellifera): rapid identification and phylogenetic analysis using reverse transcription-PCR. Clin. Diagn. Lab. Immunol. 8:93-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hung, A. C. F., B. V. Ball, J. R. Adams, H. Shimanuki, and D. A. Knox. 1996. A scientific note on the detection of American strains of acute paralysis virus and Kashmir bee virus in dead bees in one US honey-bee (Apis mellifera) colony. Apidologie 27:55-56. [Google Scholar]
- 22.Hung, A. C. F., J. R. Adams, and H. Shimanuki. 1995. Bee parasitic mite syndrome (II): the role of varroa mite and viruses. Am. Bee J. 135:702-704. [Google Scholar]
- 23.Hung, A. C. F., and H. Shimanuki. 1999. A scientific note on the detection of Kashmir bee virus in individual honey bees and Varroa jacobsoni mites. Apidologie 30:353-354. [Google Scholar]
- 24.Hung, A. C. F., H. Shimanuki, and D. A. Knox. 1996. The role of viruses in bee parasitic mite syndrome. Am. Bee J. 136:731-732. [Google Scholar]
- 25.Hung, A. C. F., H. Shimanuki, and D. A. Knox. 1996. Inapparent infection of acute paralysis virus and Kashmir bee virus in the U.S. honey bees. Am. Bee J. 136:874-876. [Google Scholar]
- 26.Kevan, P. G. 1999. Pollinators as bioindicators of the environment: species, activity and diversity. Agric. Ecosyst. Environ. 74:373-393. [Google Scholar]
- 27.Kulnicevic, J., B. V. Ball, and V. Mladjan. 1990. Viruses in honey-bee colonies infested with Varroa jacobsoni: first findings in Yugoslavia. Acta Vet. 40:37-42. [Google Scholar]
- 28.Leat, N., B. Ball, V. Govan, and S. Davison. 2000. Analysis of the complete genome sequence of black queen-cell virus, a picorna-like virus of honey bees. J. Gen. Virol. 81:2111-2119. [DOI] [PubMed] [Google Scholar]
- 29.Morse, R. A., and N. W. Calderone. 2000. The value of honey bee pollination in the United States. Bee Cult. 128:1-15. [Google Scholar]
- 30.Newman, J. F. E., F. Brown, L. Bailey, and A. J. Gibbs. 1973. Some physico-chemical properties of two honey-bee picornaviruses. J. Gen. Virol. 19:405-409. [Google Scholar]
- 31.Nordstrom, S., I. Fries, A. Aarhus, H. Hansen, and S. Korpela. 1999. Virus infections in Nordic honey bee colonies with no, low or severe Varroa jacobsoni infestation. Apidologie 30:457-466. [Google Scholar]
- 32.Ritter, W., E. Lecrecq, and W. Koch. 1984. Observations on bee and Varroa mite populations in infested honey bee colonies. Apidologie 15:389-399. [Google Scholar]
- 33.Shimanuki, H., N. W. Calderone, and D. A. Knox. 1994. Parasitic mite syndrome: the symptoms. Am. Bee J. 134:827-828. [Google Scholar]
- 34.Spira, T. P. 2001. Plant-pollinator interactions: a threatened mutualism with implications for the ecology and management of rare plants. Nat. Areas J. 21:78-88. [Google Scholar]
- 35.Topolska, G., B. Ball, and M. Allen. 1995. Identification of viruses in bees from two Warsaw apiaries. Med. Weter. 51:145-147. [Google Scholar]