Abstract
Streptococcus mutans has been strongly implicated as the principal etiological agent in dental caries. One of the important virulence properties of these organisms is their ability to form biofilms known as dental plaque on tooth surfaces. Since the roles of sucrose and glucosyltransferases in S. mutans biofilm formation have been well documented, we focused our attention on sucrose-independent factors. We have initially identified several mutants that appear to be defective in biofilm formation on abiotic surfaces by an insertional inactivation mutagenesis strategy applied to S. mutans. A total of 27 biofilm-defective mutants were isolated and analyzed in this study. From these mutants, three genes were identified. One of the mutants was defective in the Bacillus subtilis lytR homologue. Another of the biofilm-defective mutants isolated was a yulF homologue, which encodes a hypothetical protein of B. subtilis whose function in biofilm formation is unknown. The vast majority of the mutants were defective in the comB gene required for competence. We therefore have constructed and examined comACDE null mutants. These mutants were also found to be attenuated in biofilm formation. Biofilm formation by several other regulatory gene mutants were also characterized using an in vitro biofilm-forming assay. These results suggest that competence genes as well as the sgp and dgk genes may play important roles in S. mutans biofilm formation.
Biofilms are sessile bacterial communities adherent to a surface, and their formation occurs in response to a variety of environmental cues (32, 34). Biofilm bacteria undergo a developmental program in response to environmental signals that leads to the expression of new phenotypes that distinguish these sessile cells from planktonic cells (10, 32). Of importance with respect to medicine, biofilm cells have been shown to be up to 1,000-fold more tolerant of antibiotics than are planktonic cells and genes (20) and protein expression profiles are altered in planktonic cells relative to those in biofilm-grown cells (10, 32).
Streptococcus mutans has been strongly implicated as the principal etiological agent in human dental caries (18). One of the important virulence properties of these organisms is their ability to form biofilms known as dental plaque on tooth surfaces (24, 48). Dental plaque is one of the best-studied biofilms (12, 22). Dental plaque formation on tooth surfaces involves three distinct steps: (i) formation of the conditioning film or acquired pellicle on the tooth enamel, (ii) subsequent cell-to-surface attachment of the primary colonizers, and (iii) cell-to-cell interactions of late colonizers with one another as well as with the primary colonizers (12).
Biofilm formation is initiated by interactions between planktonic bacteria and a surface in response to appropriate environmental signals (10, 11, 34, 35). In addition to responses to physical and chemical signals, bacteria regulate diverse physiological processes in a cell density-dependent manner, commonly called quorum sensing (3, 11, 19, 36, 43). Molecules called quorum-sensing signals help trigger the regulation of gene expression in biofilms (13, 14). Bacteria constantly secrete low levels of these signals and sense them through the corresponding receptors (30). The receptors do not trigger any behavioral changes until there are enough bacteria to allow the signal concentrations to exceed a critical threshold (15, 37). Once this occurs, bacteria respond by adopting communal behavior, such as forming biofilms.
Isolation and characterization of genes defective in biofilm formation may contribute to understanding how S. mutans responds to environmental signals in the oral cavity, especially in oral biofilms. Previous studies have indicated the role of sucrose and glucosyltransferases (Gtfs) in S. mutans biofilm formation (6, 21, 23, 48). Recent studies have implicated several genes associated with genetic competence (28), the ccpA and brpA (lytR) genes (46), and a putative response regulator as being involved in biofilm formation (4). One purpose of this investigation was therefore to isolate sucrose-independent, biofilm-defective mutants in S. mutans. In addition, signal transduction cascades are responsible for sensing the environment and regulate a variety of cellular processes, including motility, protease production, and biofilm formation (15, 30). Therefore, we have analyzed the role of several signal transduction-associated genes in biofilm formation. The investigations described herein confirm that multiple genes, including the com locus, are associated with biofilm formation.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and plasmids.
All bacterial strains and plasmids used in this study are listed in Table 1. Strains of S. mutans were cultured and maintained in Todd-Hewitt broth (THB; Difco Laboratories, Grand Island, N.Y.) or chemically defined medium (CDM). The CDM contained 2.0 g of l-glutamic acid per liter, 0.2 g of l-cysteine per liter, 0.9 g of l-leucine per liter, 1.0 g of NH4Cl per liter, 2.5 g of K2HPO4 per liter, 2.5 g of KH2PO4 per liter, 4.0 g of NaHCO3 per liter, 1.2 g of MgSO4 · 7H2O per liter, 0.02 g of MnCl2 · 4H2O per liter, 0.02 g of FeSO4 · 7H2O per liter, 0.6 g of sodium pyruvate per liter, 1.0 mg of riboflavin per liter, 0.5 mg of thiamine HCl per liter, 0.1 mg of d-biotin per liter, 1.0 mg of nicotinic acid per liter, 0.1 mg of p-aminobenzoic acid per liter, 0.5 mg of Ca-pantothenate per liter, 1.0 mg of pyridoxal HCl per liter, and 0.1 mg of folic acid per liter, adjusted to pH 7.0 with H3PO4. Transformants of S. mutans were selected following growth on mitis salivarius agar (Difco Laboratories) plates supplemented with erythromycin (10 μg/ml).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| S. mutans strains | ||
| GS-5 | Erys, Kans, serotype c human isolate | SUNYaBa |
| CA1101 | Erys; GS-5::pAYCA1101; ComA− | This study |
| CC1301 | Eryr; GS-5::pAYCC1301; ComC− | This study |
| CD1401 | Eryr; Cmr, GS-5::pAYCD1401; ComD− | This study |
| CE1501 | Eryr; GS-5::pAYCE1501; ComE− | This study |
| GS-5(gtfB)::pSIV2 | Eryr; GS-5::pSIV2 alone | 2 |
| GS-5(gtfB)::pSIV2-SGPAN | Eryr; strain GS-5 expressing antisense sgp RNA from the host chromosome | 2 |
| TnSp-1 | Sptr, Tetr, GS-5 with dgk insertion | Lis and Kuramitsu (SUNYaB) |
| AYRA1 | Eryr; GS-5::pAYRA1; RelA-SpoT− | This study |
| luxS | Eryr, GS-5 with luxS insertion | W. Shi (UCLA)b |
| E. coli DH5α | Cloning host | GIBCO BRL |
| Plasmids | ||
| pResEmMCS10 | Emr; integration vector | 41 |
| pMCL200 | Cmr, cloning vector | 31 |
| pAYCA1101 | Emr, pResEmMCS10 bearing comA upstream and downstream regions | This study |
| pAYCC1301 | Emr, pResEmMCS10 bearing comC upstream and downstream regions | This study |
| pAYCC1302 | Emr, pResEmMCS10 bearing comC upstream regions | This study |
| pAYCD1401 | Cmr, Emr, pMCL200 bearing Emr cassette, comD upstream and downstream regions | This study |
| pAYCE1501 | Emr, pResEmMCS10 bearing comE upstream and downstream regions | This study |
| pSIV2 | Emr; integration vector bearing gtfB 1.083-kb EcoRV fragment | 2 |
| pSIV2-SGPAN | Emr; pSIV2 bearing the sgp gene in antisense orientation downstream of the scrB promoter | 2 |
| pAYRA1 | Emr, pResEmMCS10 bearing relA-spoT 640-bp internal fragment | This study |
SUNYaB, the culture collection in Department of Oral Biology, State University of New York, Buffalo, N.Y.
UCLA, University of California at Los Angeles.
DNA manipulations.
DNA isolation, endonuclease restriction, ligation, and transformation of competent Escherichia coli cells were carried out as previously described (40). Transformation of S. mutans was accomplished by procedures routinely carried out in this laboratory (38).
Suicide plasmid-mediated insertional inactivation mutagenesis.
A complete Sau3AI digest of S. mutans GS-5 chromosomal DNA was ligated to BamHI-digested pResEmMCS10 (41). S. mutans GS-5 was randomly mutated by transformation with the S. mutans genomic library. Transformants were selected on mitis salivarius agar plates containing 10 μg of erythromycin/ml.
Biofilm formation assay. (i) Quantitation of biofilm formation.
Biofilm formation was quantified as previously described (35). Flat-bottomed polystyrene microtiter plates (enzyme immunoassay-radioimmunoassay plates, 96-well Easy Wash; Corning Inc., Corning, N.Y.) containing 100 μl of CDM per well were inoculated with S. mutans GS-5 and its mutants (1.7 × 105 CFU per well) from a 24-h growth in THB. After 48 h of incubation at 37°C, 25 μl of 1% (wt/vol) crystal violet (CV) solution was added to each well. After 15 min, wells were rinsed three times with 200 μl of distilled water and air dried. The CV on the abiotic surfaces was solubilized in 95% ethanol, and the optical density at 570 nm was determined. Growth was determined by measuring the turbidities (optical density at 570 nm) of parallel wells following resuspension of the sessile organisms with the planktonic cells.
(ii) Screening for mutants defective in biofilm formation.
The assay used to screen for biofilm-defective mutants is based on the ability of bacteria to form biofilms on a polystyrene surface. The S. mutans GS-5 random mutant library constructed by the insertional inactivation mutagenesis strategy was initially inoculated in 5 ml of CDM-0.5% glucose in six-well polystyrene dishes (Corning Inc.) and was incubated for 48 h anaerobically. Fifty microliters of supernatant fluids containing unattached cells was then transferred into 5 ml of CDM-glucose in another plate, and this procedure was repeated a total of eight times to isolate biofilm-defective mutants. After the final growth stage, individual colonies of the unattached cells were tested for biofilm formation.
(iii) Visualization of the biofilms on abiotic surfaces.
Visualization of bacterial cells attached to the polystyrene surface was performed on six-well polystyrene dishes (Corning Inc.). Briefly, biofilms after 48 h of inoculation were stained with CV and washed several times.
Construction of the com mutants.
The comACDE genes and the relA-spoT gene were identified in the S. mutans UA159 genome database, available from the University of Oklahoma Advanced Center for Genome Technology (OU-ACGT; http://www.genome.ou.edu.smutans.html). The null mutants of the comACDE genes were created by allelic exchange via insertion of an erythromycinresistance (Eryr) determinant into each gene. The plasmids used for disruption of the comACDE genes were prepared as follows: the PCR fragments of the upstream and downstream regions of comA were amplified with pairs of primers, comAUF251(Bam)-comAUR1236(Sal) and comADF3482(Eco)-comADR4315(Bam) (Table 2), respectively, using chromosomal DNA from strain GS-5 as the template. Initially, PCR products of the downstream region of comA (comAD) were cloned into the pResEmMCS10 plasmid (41). Furthermore, PCR products of the upstream region of comA, comAU, were cloned downstream of comAD. The resultant plasmid, pAYCA1101, was digested with BamHI. This resulted in a linear plasmid harboring flanking S. mutans DNA but devoid of the comA gene, which was used to transform S. mutans GS-5. The plasmids for comC and comE null mutants were constructed in a manner similar to that for the comA mutant. Briefly, the PCR fragments outward from the flanking regions of comC or comE were created with pairs of primers: comCUF6917(Bam)-comCUR7575(Hind) for comCU, comCDF7781(Sma)-comCDR8456(Bam) for comCD, comEUF4801(Bam)-comEUR5346(Xba) for comEU, and comEDF5926(Sma)-comEDR6587(Bam) for comED (Table 2). The downstream fragments (comCD and comED) were inserted into pResEmMCS10. Furthermore, the comCU and comEU fragments were inserted downstream of the comCD and comED fragments, respectively. The resultant plasmids, pAYCC1301 and pAYCE1501, were linearized by digestion with BamHI. These linearized plasmids were transformed into S. mutans GS-5. The plasmid for generating the comD null mutant was constructed with DNA fragments upstream and downstream of comD. The PCR primers and constructed fragments were as follows: comDUF4597(Sma)-comDUR6124(Bam) for comDU and comDDF7384(Bam)-comDDR7867(Xba) for comDD. The comDU fragment was initially inserted into pMCL200 (31), and the comDD fragment was inserted downstream of comDU. Finally, the Eryr determinant from pResEmMCS10 was inserted into the BamHI site between the comDU and comDD fragments. The resultant plasmid, pAYCD1401, was linearized and then transformed into S. mutans GS-5.
TABLE 2.
Oligonucleotide primersa
| Primer or primer pair | Nucleotide sequence | Amplicon | Size (bp) |
|---|---|---|---|
| comAUF251(Xba) | 5′-CCC CCC TCT AGA CAA AAA TCA TAG CAA TAT-3′ | comAU | 986 |
| comAUR1236(Sal) | 5′-CCC CCC GTC GAC ATA AAT AAC TTG TTT CAT-3′ | ||
| comADF3482(Eco) | 5′-CCC CCC GAA TTC ATT ATA ACC TGT TTA ATT-3′ | comAD | 834 |
| comADR4315(Sma) | 5′-CCC CCC CCC GGG TTT GCT ATT TTT CTT AGA-3′ | ||
| comCUF6917(Bam) | 5′-CCC CCC GGA TCC GAT TTA CCA TCA AAT CTT-3′ | comCU | 659 |
| comCUR7575(Hind) | 5′-CAC AAG CTT TGG GAA AAT-3′ | ||
| comCDF7781(Sma) | 5′-CCC CCC CCC GGG TCC TTA GGA CAC ATA ATG-3′ | comCD | 676 |
| comCDR8456(Bam) | 5′-CCC CCC GGA TCC AGC AAA TCT GAA CAA GCA-3′ | ||
| comDUF4597(Sma) | 5′-CCC CCC CCC GGG GTT CAA CTT CTA TCA TTT-3′ | comDU | 1523 |
| comDUR6124(Bam) | 5′-CCC CCC GGA TCC ACC ATT TGA AAG TAT CAT-3′ | ||
| comDDF7384(Bam) | 5′-CCC CCC GGA TCC TCA TTT ATT CAA GCA ACT-3′ | comDD | 483 |
| comDDR7867(Xba) | 5′-CCC CCC TCT AGA AAT GGT AGC TAT TTT GT-3′ | ||
| comEUF4801(Bam) | 5′-CCC CCC GGA TCC TGG CTA TTG TTT TTG TGA-3′ | comEU | 622 |
| comEUR5346(Xba) | 5′-CCC CCC TCT AGA TCA TTA TTT CTC CTT TAA-3′ | ||
| comEDF5926(Sma) | 5′-CCC CCC CCC GGG AGA GAC TTT TTC AGT GCC-3′ | comED | 545 |
| comEDR6587(Bam) | 5′-CCC CCC GGA TCC GGT AGT ATA GAA GCA TAG-3′ | ||
| relA-spoTF3106(Bam) | 5′-CCC CCC GGA TCC TTA CAT GGT GAG ATT TA-3′ | relA-spoTI | 640 |
| relA-spoTR3745(Hind) | 5′-CCC CCC AAG CTT GTT TTG AGC TTA GCT GT-3′ |
Endonuclease recognition sequences are underlined.
The relA-spoT mutant was constructed by insertion-duplication mutagenesis. Briefly, the primer pairs relA-spoTF3106(Bam) and relA-spoTR3745(Hind) were designed to amplify internal regions of relA-spoT. The amplicon, relA-spoTI, was ligated into pResEmMCS10 via BamHI and HindIII sites. The resultant plasmid, designated pAYRA1, was used to transform S. mutans GS-5.
Confirmation of plasmid insertions causing gene disruption was performed either by Southern blotting or by PCR. Southern blotting analysis was performed with digoxigenin (DIG)-labeled PCR products corresponding to the target genes and the Eryr gene as probes, by using the PCR DIG Probe Synthesis Kit and DIG DNA Labeling and Detection Kit (Roche Diagnosis, Indianapolis, Ind.).
CSP synthesis.
The nucleotide sequence of the comC gene was determined by the marker rescue method with the plasmid pAYCC1302 harboring the comCU fragment (Table 2). The competence-stimulating peptide (CSP) precursor amino acid sequence was derived using the obtained sequence. The Gly-Gly cleavage site was deduced, and the resultant mature CSP (SGSLSTFFRLFNRSFTQALGK; 21 amino acids) was synthesized (Sigma-Genosys, Woodlands, Tex.). For complementation of the comC gene by CSP, the synthetic CSP was incubated with bacterial cultures as previously described (1). Briefly, the synthetic peptide was freshly dissolved in distilled water at a concentration of 1 mg/ml. The CSP solution was then added to the cultures at final concentrations ranging from 25 to 2,500 ng/ml.
SEM.
Biofilm formation by S. mutans on polystyrene surfaces was examined by scanning electron microscopy (SEM) to verify the quantitative results observed. Biofilms were aerobically inoculated in the CDM supplemented with 0.5% glucose at 37°C for 48 h on 5- by 5-mm polystyrene tips. Biofilms on the polystyrene tips were washed once with distilled water, fixed by formaldehyde, and incubated at 20°C overnight. Following dehydration through a graded series of ethanol, the polystyrene tips were air dried and sputter coated with gold. Samples were then examined at ×500 to ×7,500 magnification using SEM (JEOL JSM-5400LV; JEOL Techniques Ltd., Tokyo, Japan).
RESULTS
Environmental factors affecting S. mutans biofilm formation.
In order to establish an optimal assay system for S. mutans biofilm formation, biofilm assays were carried out under various conditions. We initially assessed S. mutans GS-5 biofilm formation on relatively hydrophobic and hydrophilic surfaces according to the procedure of O'Toole and Kolter (35). The relatively hydrophobic materials, polyvinyl chloride, polycarbonate, and polypropylene, were utilized as well as borosilicate glass as a hydrophilic surface. Strain GS-5 was allowed to form biofilms on these surfaces in CDM supplemented with 0.5% sucrose. The optimal abiotic material for S. mutans biofilm formation appeared to be polystyrene (data not shown). Biofilm formation on the other surfaces was similar and was equivalent to approximately 25% of that measured on polystyrene surfaces.
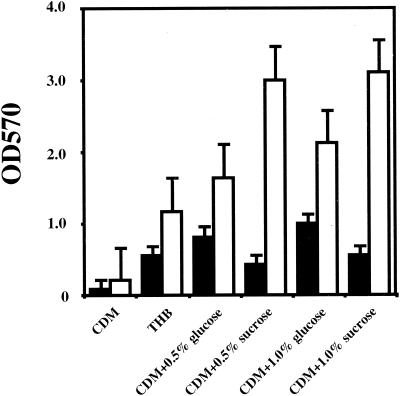
Several media and carbon sources were also examined in the biofilm assay on polystyrene plates (Fig. 1). Negligible growth and biofilm formation were observed in CDM without a carbon source. In THB, there was no significant difference in growth from that found in defined media with glucose or sucrose. However, biofilm formation was apparently optimal in CDM supplemented with sucrose, whereas lower levels of biofilm formation were observed in THB (Fig. 1). Since the role of sucrose in S. mutans biofilm formation has been well documented (6, 21, 48), we focused our attention on sucrose-independent factors that influence this important virulence property of these organisms.
FIG. 1.
Bacterial growth and biofilm formation of S. mutans GS-5 in media with different carbon sources. Bacteria were inoculated in either CDM, CDM supplemented with glucose (0.5 or 1.0%), CDM-sucrose (0.5 or 1.0%), or THB. Growth (black bars) and biofilm formation (white bars) were measured under anaerobic conditions. The data are the averages of three samples, and standard errors are shown.
Isolation of mutants defective in sucrose-independent biofilm formation.
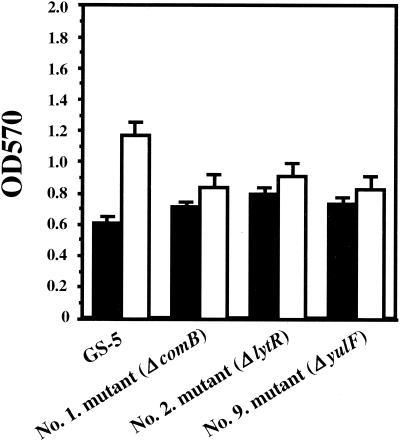
In order to isolate mutants defective in biofilm formation on polystyrene surfaces, we generated a collection of S. mutans GS-5 random mutants using a suicide plasmid-mediated insertional inactivation mutagenesis strategy. A total of 27 (0.23%) biofilm-defective mutants were isolated after visual screening of approximately 12,000 transformants. Southern blot analysis with HindIII-digested chromosomal DNA from each of the mutants with a DIG-labeled Eryr gene from pResEmMCS10 as a probe confirmed the integration of this plasmid (data not shown). Of these mutants, three genetic loci were identified following marker rescue and DNA sequencing. One of the mutants was defective in a homologue of the Bacillus subtilis lytR gene (21% identity) (26). This gene is an attenuator of the expression of both the lytABC and lytR operons, which encode the N-acetylmuramoyl-l-alanine amidase structural gene and modifier. Another class of biofilm-defective mutants was defective in a yulF homologue, which encodes a hypothetical protein of B. subtilis whose function is unknown. Significantly, 25 of the 27 mutants were defective in the comB gene (identified from the University of Oklahoma S. mutans UA159 database). All of these mutants were not markedly attenuated in growth relative to strain GS-5 (Fig. 2).
FIG. 2.
Bacterial growth and biofilm formation of S. mutans GS-5 and biofilm-defective mutants. Assays were performed using CDM-0.5% glucose and polystyrene plates under anaerobic conditions. Growth (black bars) and biofilm formation (white bars) were measured. The data are averages of three samples, and error bars are shown.
The contribution of competence genes to S. mutans biofilm formation.
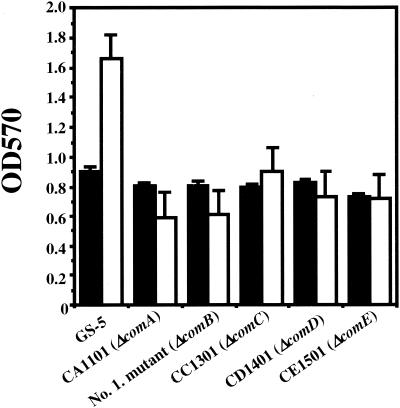
Since one of the mutants altered in biofilm formation was a comB mutant, we hypothesized that the competence regulon affects biofilm formation in S. mutans. To test this hypothesis, we examined the effects of inactivation of the comA, comC, comD, and comE genes on biofilm formation in strain GS-5. Biofilm formation by S. mutans GS-5 and the five mutants defective in each of the com genes was assayed using CDM-0.5% glucose under anaerobic conditions (Fig. 3). These results demonstrated that biofilm formation by each of the mutants was attenuated under these conditions. As demonstrated for the comB mutant, there were no significant differences in the growth rates between the wild-type strain and each of the com mutants (Fig. 3). However, when biofilm formation in the presence of sucrose was measured, there were no significant differences between GS-5 and the com mutants (data not shown).
FIG. 3.
Biofilm formation of S. mutans GS-5 and comA, comB, comC, comD, and comE mutants. Growth (black bars) and biofilm formation (white bars) were measured using CDM-0.5% glucose and polystyrene plates under anaerobic conditions. The data are averages of three samples, and error bars are shown.
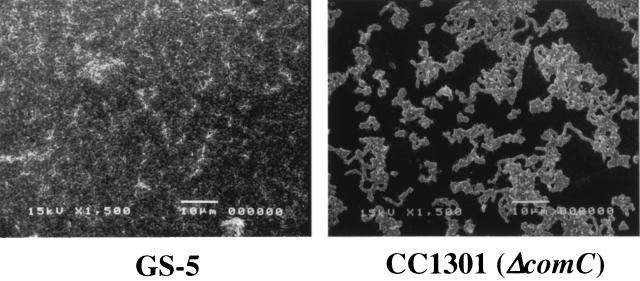
In addition, to further assess the biofilm phenotype of the com mutants, we also employed SEM analysis of the biofilms on polystyrene surfaces. This analysis confirmed that the com mutants were attenuated in biofilm formation on the polystyrene surface compared to GS-5 when relative surface biomasses were measured (data not shown). However, the comA, comB, and comD mutants, as well as GS-5, showed no significant qualitative differences in their biofilm phenotypes on polystyrene surfaces (data not shown). By contrast, the comC mutants formed biofilms that differed from the other strains in morphology. The comC mutants exclusively formed many discrete small microcolonies, and intertwined chains were observed between the microcolonies. In these biofilms, individual cells and chains of cells were not detectable within the microcolonies (Fig. 4).
FIG. 4.
Scanning electron micrographs comparing biofilm formation of S. mutans GS-5 (left) and CC1301 (right) accumulated on polystyrene tips after 48 h of inoculation. Images were obtained at ×1,500 magnification.
In order to confirm that the phenotype of the comC mutant was indeed the result of the mutation in the comC gene, we performed a complementation analysis. Biofilm formation of the mutant was not increased by the addition of low levels of synthetic CSP (25 to 250 ng/ml). However, 2.5 μg of CSP per ml restored biofilm formation of the comC mutant strain to levels equivalent to those of the wild type (data not shown).
Biofilm formation by other S. mutans regulatory gene mutants.
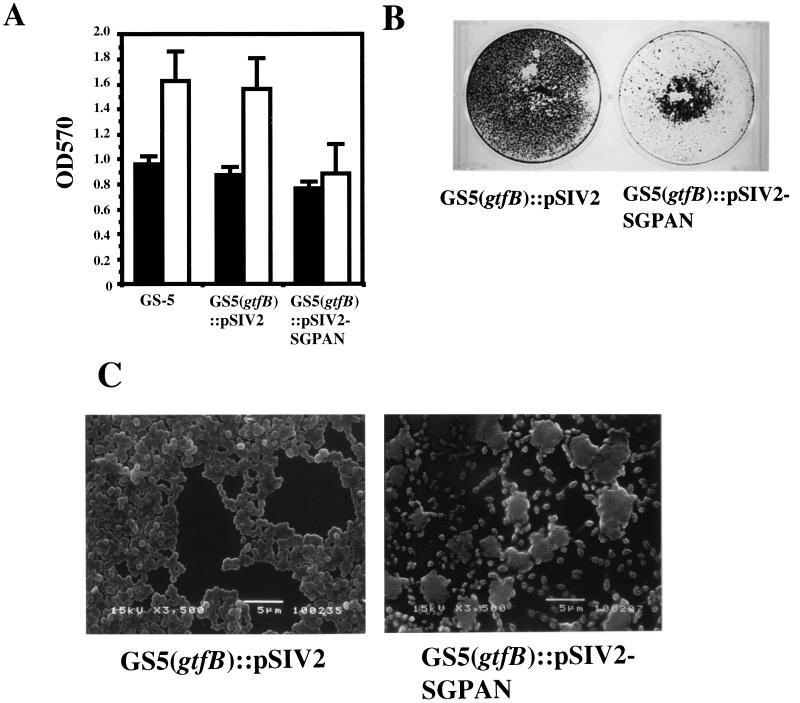
The S. mutans regulatory G protein (SGP) has been recently characterized (47) and was demonstrated to be essential for cell growth. In order to test the effects of SGP on biofilm formation, an S. mutans strain defective in SGP expression (2) was evaluated. This strain expresses sgp antisense RNA that interferes with the normal processing of the sgp gene. Biofilm formation relative to growth by the sgp antisense strain in CDM-0.5% glucose was found to be decreased by approximately 45% from that associated with the wild-type and control strains (harboring the inserted vector but not expressing antisense RNA) (Fig. 5A). This difference was magnified when biofilm formation was compared in polystyrene plates with larger surface areas than the 96-well microtiter plates (Fig. 5B). In addition, the sgp antisense strain exclusively formed many discrete small microcolonies in SEM analysis. Within the microcolonies, individual cells and chains of cells were not detectable. However, pairs, tetrads, and short chains of several cells are visible around the small microcolonies (Fig. 5C).
FIG. 5.
Biofilm formation of sgp antisense strains. (A) Bacterial growth (black bars) and biofilm formation (white bars) of control and sgp antisense strains. Assays were performed using CDM-0.5% glucose and polystyrene plates under anaerobic conditions. The data are averages of three samples, and error bars are shown. (B) Biofilm formation of control strain (left) and sgp antisense strain (right) in CDM supplemented with 0.5% glucose. Biofilm formation was demonstrated using minimal medium supplemented with 0.5% glucose and six-well polystyrene plates under anaerobic conditions. CV-stained biofilm on polystyrene plate wells is shown. (C) Scanning electron micrographs of control strain (left) and sgp antisense strain (right) biofilms accumulated on polystyrene tips after 48 h of inoculation. Images were obtained at ×3,500 magnification.
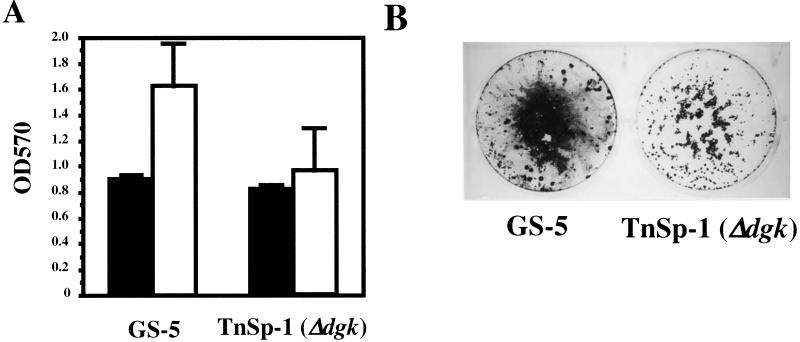
The S. mutans dgk gene codes for undecaprenol kinase activity (M. Lis and H. K. Kuramitsu, submitted for publication), which appears to play an important role in the stress responses of this organism (49). In addition, Dgk is essential for mutacin II production in S. mutans (9). Therefore, we examined the potential role of Dgk as a signal transducer in biofilm formation. Biofilm formation by the dgk null mutant in CDM-0.5% glucose was found to be decreased approximately 40% compared to that by the wild-type strain (Fig. 6A and B). However, the dgk mutants, as well as GS-5, showed no significant qualitative differences in their biofilm phenotypes on polystyrene surfaces following evaluation by SEM analysis (data not shown).
FIG. 6.
Biofilm formation by the dgk mutant. (A) Bacterial growth (black bars) and biofilm formation (white bars) of S. mutans GS-5 and its dgk mutant TnSp-1. Assays were performed as described in the text. The data are averages of three samples, and error bars are shown. (B) Biofilm formation of S. mutans GS-5 (left) and TnSp-1 dgk mutant (right) in CDM-0.5% glucose in six-well polystyrene plates under anaerobic conditions. CV-stained biofilm on polystyrene plate wells is shown.
Recent studies have reported that the stringent response activates the quorum-sensing systems of Pseudomonas aeruginosa independently of cell density (44). We therefore investigated the effects of the inactivation of the relA-spoT gene on biofilm formation in strain GS-5. However, biofilm formation by the relA-spoT mutant was equivalent to that of the wild type in the CDM-0.5% glucose (data not shown). Besides the com quorum-sensing system, S. mutans also contains a luxS gene (W. Shi, personal communication), which mediates density-dependent signaling in a variety of bacteria (3, 8). We therefore analyzed biofilm formation by the luxS mutant and observed that this mutant was not attenuated compared to the wild-type strain in CDM-glucose medium (data not shown).
DISCUSSION
The colonization of tooth surfaces by S. mutans appears to result from two distinct processes: initial sucrose-independent attachment and enhancement of attachment by sucrose-dependent mechanisms involving Gtfs (23). The role of sucrose and Gtfs in S. mutans biofilm formation has been well documented (6, 21, 24, 48). In addition to Gtfs, several other genes associated with biofilm formation have been reported in recent investigations (4, 28, 46). Therefore, in order to examine the genetic basis for sucrose-independent biofilm formation, we have isolated and characterized sucrose-independent, biofilm-defective mutants by an insertional inactivation mutagenesis strategy. The mutants identified by this strategy can be divided into three groups: (i) those involved in competence, (ii) insertions into genes involved in amidase activity, and (iii) those with insertions in a gene with an unknown function.
Biofilm formation was assayed by measuring the ability of S. mutans to attach to the abiotic surfaces. In the process of the bacterial biofilm formation, bacterial cells migrate over the solid substrate, sense the surface, and subsequently produce exopolysaccharide that allows bacteria to accumulate in multiple layers of the biofilm (11). The biofilm assay that we have employed recapitulates this process with monoculture bacteria. Biofilm formation by S. mutans was significant on polystyrene surfaces; however, other bacteria such as Pseudomonas fluorescens WCS365 form weak biofilms on such surfaces (35). This result is consistent with the biofilm formation by Streptococcus gordonii Challis (29). Carbon source availability is one of the important environmental signals that play a role in biofilm development (8). S. mutans GS-5 biofilm formation was enhanced in minimal medium relative to that in nutritionally rich environments. This result is also consistent with previous results for S. gordonii Challis biofilm formation (29). These results further suggest that sessile growth may represent a survival strategy in a nutritionally limited environment (10, 11).
One of the 27 biofilm-defective mutants contains a plasmid integrated in the open reading frame which is the homologue of the B. subtilis lytR gene (http://genolist.pasteur.fr/SubtiList/). LytR is a 35-kDa protein that acts as an attenuator of the expression of both lytABC and lytR operons (26). The lytABC operon encodes a lipoprotein (LytA), a modifier of the amidase (LytB), and an N-acetylmuramoyl-l-alanine amidase (amidase, LytC/CwlB), one of the two major vegetative-phase autolysins (25, 26, 42). However, no apparent lytABC homologue was found in the S. mutans UA159 database. Previous studies reported that the Staphylococcus aureus lytS and lytR genes, whose products are members of the two-component regulator family of proteins, are involved in the control of peptidoglycan hydrolase activity (5, 16). These genes are distinct from the B. subtilis lytR gene. However, several genes involved in biofilm-defective mutants of S. gordonii Challis are associated with peptidoglycan biosynthesis (29). The S. mutans lytSR homologue was identified in the UA159 database (46). This suggests that peptidoglycan formation might also specifically affect biofilm formation. Furthermore, one of the mutants is the homologue of B. subtilis yulF, which is a hypothetical gene (http://genolist.pasteur.fr/SubtiList/). It is not clear yet how these genes are involved in biofilm formation. Further analysis of these genes might provide additional information on S. mutans biofilm formation.
Twenty-five of the 27 biofilm-defective mutants isolated in the screening process contained a plasmid integration within the comB gene. The comB gene encodes the accessory protein for the ComA ABC transporter, which is required for secretion of the CSP. Competence for genetic transformation is regulated by a CSP-mediated quorum-sensing system in a large group of closely related streptococci (27). Recent studies have also suggested that cell-to-cell signaling is also required for the differentiation of individual cells of P. aeruginosa into biofilms (13). In addition, screening for S. gordonii Challis biofilm-defective mutants by Tn916 transposon mutagenesis revealed that one of the genes associated with biofilm formation is comD, the cognate receptor for CSP (29). Our results demonstrate that the com genes of S. mutans are involved in biofilm formation and are consistent with that report.
Null mutants of the comACDE genes were individually constructed, and each mutant was also attenuated in sucrose-independent biofilm formation. A recent study has also reported that the comC, comD, and comE knockout mutants of S. mutans NG8 exhibited decreased transformation efficiencies that were approximately 100-fold lower than that of the parent strain (27). Furthermore, it was observed that higher frequencies of transformation occurred in actively growing biofilms than in planktonic cells (27). However, sucrose-dependent biofilm formation of each of the com mutants was very similar to that of strain GS-5. Therefore, the presence of sucrose and subsequent glucan synthesis likely compensate for the com-dependent requirements for biofilm formation. Since plaque formation, in part, may depend on initial sucrose-independent attachment followed by glucan formation, the com-dependent biofilm properties could play a significant role in initial attachment in vivo.
In order to compare the biofilm structures of the com mutants, we performed SEM analysis of strain GS-5 and the S. mutans biofilms on polystyrene surfaces. Biofilm formation is a complex developmental process involving attachment and immobilization on a surface, cell-to-cell interactions, microcolony formation, formation of a confluent biofilm, and development of a characteristic three-dimensional biofilm structure (33, 45). In order to monitor the differences in three-dimensional biofilm structure, we observed the SEM images after 48 h of inoculation. By direct observation of these strains, we observed that the biofilm morphology of the comC mutant was quite distinct from that of other strains. However, it is not clear why some of the other com mutants displayed biofilm morphologies distinct from that of the comC mutant. The S. mutans comC, comD, and comE genes, respectively, encode a CSP precursor, its histidine kinase sensor, and an intracellular response regulator (27). The mutation of comC that blocked the generation of the signal molecules hindered the normal differentiation of S. mutans biofilms. As an explanation for this phenotype, Li et al. suggested the existence of a second receptor that also responds to CSP (28). Complementation of biofilm formation by the comC mutant with CSP also provides further evidence for the role of this gene in this process. In the present study, we initially tested 2.5 ng of CSP/ml for complementation analysis according to the results of Alloing et al. (1). However, the amount of biofilm and biofilm morphology were normalized relative to strain GS-5 only by concentrations of CSP greater than 2.5 μg/ml. This amount is 1,000 times higher than that required for the complementation of the Streptococcus pneumoniae comC mutant. The reason of this discrepancy is still unclear and may result from the differences in the physiological responses of the two organisms to quorum sensing.
Oral biofilms are especially subject to a number of environmental fluctuations, such as nutrient availability, aerobic-to-anaerobic transitions, and pH changes (7, 10). Therefore, studying biofilms in the context of environmental stress is essential to reveal the role of signal transduction systems in biofilm formation. From this point of view, we also examined the role of the regulatory G protein SGP in biofilm formation. Previous studies in this laboratory have indicated that SGP is required for viability and plays a role in the environmental stress response of S. mutans (2). To our knowledge, the present results represent the first report of a possible role for a G protein in biofilm formation. Previous studies indicated that sgp antisense RNA expression in S. mutans resulted in hypersensitivity to environmental stress conditions (44°C, pH 5.5, and high osmolarity) (2). In the present study, biofilm formation by the SGP-defective strain in CDM-0.5% glucose relative to that by control strains was somewhat decreased, compared to the growth rates of these strains. The role of the SGP in biofilm formation is presently under investigation in this laboratory.
Another gene in S. mutans GS-5 dgk, encoding undecaprenol kinase activity, was also shown to play a role in responding to environmental stress (49). Therefore, we examined the role of Dgk as a possible signal transducer in biofilm formation. Inactivation of the dgk gene also differentially attenuated biofilm formation. Chen et al. recently reported that Dgk also plays a role in mutacin II production in S. mutans (9). In addition, a correlation between environmental stress responses and bacteriocin production was reported for S. mutans JH1005 (17). These observations suggest that common mechanisms modulate several different stress-regulated responses in S. mutans, including biofilm development and bacteriocin production.
In the view of the present results, we speculated that the stringent response and luxS-mediated quorum sensing also might be involved in biofilm formation. However, both the relA-spoT and luxS global regulatory genes do not appear to play a role in sucrose-independent biofilm formation. Mutants of each of these genes formed biofilms, as did wild-type GS-5 in the present assay system.
Several genes involved in biofilm formation have been identified in a variety of organisms (4, 28, 29, 34, 39, 46). In the present study, we have identified several genes associated with biofilm formation. However, little is known regarding the molecular mechanisms necessary to transduce environmental signals that trigger biofilm development. The present study has identified several regulatory genes, dgk, sgp, and the com genes, which may play significant roles in communicating these signals in S. mutans. How these environmental signals are sensed and transduced by biofilm-forming bacteria and what are the molecular mechanisms utilized to initiate the development of a biofilm in response to these cues are crucial questions that still remain unanswered.
Acknowledgments
This investigation was supported in part by NIH grant DE03258.
Footnotes
This article is dedicated to the memory of Toshihiko Kaga.
REFERENCES
- 1.Alloing, G., B. Martin, C. Granadel, and J. P. Claverys. 1998. Development of competence in Streptococcus pneumonaie: pheromone autoinduction and control of quorum sensing by the oligopeptide permease. Mol. Microbiol. 29:75-83. [DOI] [PubMed] [Google Scholar]
- 2.Baev, D., R. England, and H. K. Kuramitsu. 1999. Stress-induced membrane association of the Streptococcus mutans GTP-binding protein, an essential G protein, and investigation of its physiological role by utilizing an antisense RNA strategy. Infect. Immun. 67:4510-4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bassler, B. L. 1999. How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr. Opin. Microbiol. 2:582-587. [DOI] [PubMed] [Google Scholar]
- 4.Bhagwat, S. P., J. Nary, and R. A. Burne. 2001. Effects of mutating putative two-component systems on biofilm formation by Streptococcus mutans UA159. FEMS Microbiol. Lett. 205:225-230. [DOI] [PubMed] [Google Scholar]
- 5.Brunskill, E. W., and K. W. Bayles. 1996. Identification of LytSR-regulated genes from Staphylococcus aureus. J. Bacteriol. 178:5810-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burne, R. A., Y. Y. Chen, and J. E. Penders. 1997. Analysis of gene expression in Streptococcus mutans in biofilms in vitro. Adv. Dent. Res. 11:100-109. [DOI] [PubMed] [Google Scholar]
- 7.Carlsson, J. 1997. Bacterial metabolism in dental biofilms. Adv. Dent. Res. 11:75-80. [DOI] [PubMed] [Google Scholar]
- 8.Carlsson, J. 2000. Growth and nutrition as ecological factors, p. 67-130. In H. K. Kuramitsu and R. P. Ellen (ed.), Oral bacterial ecology: the molecular basis. Horizon Scientific Press, Wymondham, United Kingdom.
- 9.Chen, P., J. Novak, F. Qi, and P. W. Caufield. 1998. Diacylglycerol kinase is involved in regulation of expression of the lantibiotic mutacin II of Streptococcus mutans. J. Bacteriol. 180:167-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 11.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 12.Davey, M. E., and G. A. O'Toole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 14.de Kievit, T. R., R. Gillis, S. Marx, C. Brown, and B. H. Iglewski. 2001. Quorum-sensing genes in Pseudomonas aeruginosa biofilms: their role and expression patterns. Appl. Environ. Microbiol. 67:1865-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Kievit, T. R., and B. H. Iglewski. 2000. Bacterial quorum sensing in pathogenic relationships. Infect. Immun. 68:4839-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujimoto, D. F., E. W. Brunskill, and K. W. Bayles. 2000. Analysis of genetic elements controlling Staphylococcus aureus lrgAB expression: potential role of DNA topology in SarA regulation. J. Bacteriol. 182:4822-4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gutierrez, J. A., P. J. Crowley, D. P. Brown, J. D. Hillman, P. Youngman, and A. S. Bleiweis. 1996. Insertional mutagenesis and recovery of interrupted genes of Streptococcus mutans by using transposon Tn917: preliminary characterization of mutants displaying acid sensitivity and nutritional requirements. J. Bacteriol. 178:4166-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamada, S., and H. D. Slade. 1980. Biology, immunology, and cariogenicity of Streptococcus mutans. Microbiol. Rev. 44:331-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hassett, D. J., J. F. Ma, J. G. Elkins, T. R. McDermott, U. A. Ochsner, S. E. West, C. T. Huang, J. Fredericks, S. Burnett, P. S. Stewart, G. McFeters, L. Passador, and B. H. Iglewski. 1999. Quorum sensing in Pseudomonas aeruginosa controls expression of catalase and superoxide dismutase genes and mediates biofilm susceptibility to hydrogen peroxide. Mol. Microbiol. 34:1082-1093. [DOI] [PubMed] [Google Scholar]
- 20.Hoyle, B. D., and J. W. Costerton. 1991. Bacterial resistance to antibiotics: the role of biofilms. Prog. Drug Res. 37:91-105. [DOI] [PubMed] [Google Scholar]
- 21.Hudson, M. C., and R. Curtiss III. 1990. Regulation of expression of Streptococcus mutans genes important to virulence. Infect. Immun. 58:464-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolenbrander, P. E. 2000. Oral microbial communities: biofilms, interactions, and genetic systems. Annu. Rev. Microbiol. 54:413-437. [DOI] [PubMed] [Google Scholar]
- 23.Kuramitsu, H. K. 2000. Streptococcus mutans: molecular genetic analysis, p. 280-286. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. American Society for Microbiology, Washington, D.C.
- 24.Kuramitsu, H. K. 1993. Virulence factors of mutans streptococci: role of molecular genetics. Crit. Rev. Oral Biol. Med. 4:159-176. [DOI] [PubMed] [Google Scholar]
- 25.Kuroda, A., and J. Sekiguchi. 1991. Molecular cloning and sequencing of a major Bacillus subtilis autolysin gene. J. Bacteriol. 173:7304-7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lazarevic, V., P. Margot, B. Soldo, and D. Karamata. 1992. Sequencing and analysis of the Bacillus subtilis lytRABC divergon: a regulatory unit encompassing the structural genes of the N-acetylmuramoyl-l-alanine amidase and its modifier. J. Gen. Microbiol. 138:1949-1961. [DOI] [PubMed] [Google Scholar]
- 27.Li, Y. H., P. C. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2001. Natural genetic transformation of Streptococcus mutans growing in biofilms. J. Bacteriol. 183:897-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, Y. H., N. Tang, M. B. Aspiras, P. C. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2002. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J. Bacteriol. 184:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loo, C. Y., D. A. Corliss, and N. Ganeshkumar. 2000. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J. Bacteriol. 182:1374-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165-199. [DOI] [PubMed] [Google Scholar]
- 31.Nakano, Y., Y. Yoshida, Y. Yamashita, and T. Koga. 1995. Construction of a series of pACYC-derived plasmid vectors. Gene 162:157-158. [DOI] [PubMed] [Google Scholar]
- 32.O'Toole, G. A., K. A. Gibbs, P. W. Hager, P. V. Phibbs, Jr., and R. Kolter. 2000. The global carbon metabolism regulator Crc is a component of a signal transduction pathway required for biofilm development by Pseudomonas aeruginosa. J. Bacteriol. 182:425-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Toole, G. A., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 34.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 35.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 36.Otto, M., R. Sussmuth, C. Vuong, G. Jung, and F. Gotz. 1999. Inhibition of virulence factor expression in Staphylococcus aureus by the Staphylococcus epidermidis agr pheromone and derivatives. FEBS Lett. 450:257-262. [DOI] [PubMed] [Google Scholar]
- 37.Parsek, M. R., and E. P. Greenberg. 2000. Acyl-homoserine lactone quorum sensing in gram-negative bacteria: a signaling mechanism involved in associations with higher organisms. Proc. Natl. Acad. Sci. USA 97:8789-8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perry, D., L. M. Wondrack, and H. K. Kuramitsu. 1983. Genetic transformation of putative cariogenic properties in Streptococcus mutans. Infect. Immun. 41:722-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rashid, M. H., K. Rumbaugh, L. Passador, D. G. Davies, A. N. Hamood, B. H. Iglewski, and A. Kornberg. 2000. Polyphosphate kinase is essential for biofilm development, quorum sensing, and virulence of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 97:9636-9641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Shiroza, T., and H. K. Kuramitsu. 1993. Construction of a model secretion system for oral streptococci. Infect. Immun. 61:3745-3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith, T. J., S. A. Blackman, and S. J. Foster. 2000. Autolysins of Bacillus subtilis: multiple enzymes with multiple functions. Microbiology 146:249-262. [DOI] [PubMed] [Google Scholar]
- 43.Surette, M. G., M. B. Miller, and B. L. Bassler. 1999. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA 96:1639-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Delden, C., R. Comte, and A. M. Bally. 2001. Stringent response activates quorum sensing and modulates cell density-dependent gene expression in Pseudomonas aeruginosa. J. Bacteriol. 183:5376-5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watnick, P., and R. Kolter. 2000. Biofilm, city of microbes. J. Bacteriol. 182:2675-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wen, Z. T., and R. A. Burne. 2002. Functional genomics approach to identifying genes required for biofilm development by Streptococcus mutans. Appl. Environ. Microbiol. 68:1196-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu, J., M.-I. Cho, and H. K. Kuramitsu. 1995. Expression, purification, and characterization of a novel G protein, SGP, from Streptococcus mutans. Infect. Immun. 63:2516-2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamashita, Y., W. H. Bowen, R. A. Burne, and H. K. Kuramitsu. 1993. Role of the Streptococcus mutans gtf genes in caries induction in the specific-pathogen-free rat model. Infect. Immun. 61:3811-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamashita, Y., T. Takehara, and H. K. Kuramitsu. 1993. Molecular characterization of a Streptococcus mutans mutant altered in environmental stress responses. J. Bacteriol. 175:6220-6228. [DOI] [PMC free article] [PubMed] [Google Scholar]