Abstract
A 2,665-bp cryptic plasmid, pTXL1, isolated from Leuconostoc mesenteroides subsp. mesenteroides Y110 was identified. This plasmid harbors a replicon localized on a 1,300-bp fragment. Two observations suggested that pTXL1 does not belong to rolling-circle replication (RCR)-type plasmids and most likely replicates via a theta mechanism. These hypotheses are supported by the observation that no detectable single-stranded intermediate was found for the replicon and that, unlike in RCR-type plasmids, the pTXL1 replicon sequence lacks an open reading frame encoding a replicase. The small-sized pTXL1 plasmid is stable and, according to its origin, can be considered in the “generally recognized as safe” category. Its ability to replicate in several lactic acid bacteria was exploited to develop a vector producing mesentericin Y105, a class II anti-Listeria bacteriocin. With this new vector, a recombinant industrial Leuconostoc cremoris strain able to produce mesentericin Y105 was constructed.
Plasmids from lactic acid bacteria (LAB) have become a focus of numerous studies, thus leading to the development of families of cloning vectors. Most vectors for LAB are rolling-circle replication (RCR) plasmids, which replicate by using single-stranded intermediates similar to those from other gram-positive bacteria (2, 15, 22). While these plasmids have been indispensable in designing new gene cloning systems in LAB, problems linked to their stability are well documented (14, 21). Cloning vectors that replicate without the use of RCR display several advantages, as exemplified by those derived from the enterococcal pAMβ1 plasmid (9). The replication of pAMβ1 and other LAB plasmids, such as pWV02 (20) and pCI305 (17), by the theta mechanism was reviewed by Jannière et al. (19).
Leuconostoc spp. are a diverse group of heterofermentative LAB of considerable industrial importance. Many Leuconostoc species harbor one or more native plasmids of various sizes, but to date, only a few reports have dealt with RCR plasmids in species of the Leuconostoc genus (5, 7, 10, 11, 30) and none concerns identification of a non-RCR plasmid. The aim of the present study was to analyze the mode of replication of the Leuconostoc plasmid pTXL1 (6) in order to develop families of vectors designed for specific industrial applications. Plasmid pTXL1 was rather small and stable during attempts to cure its host strain, Leuconostoc mesenteroides subsp. mesenteroides Y110, with novobiocin treatment. Moreover, it seemed compatible with various vectors derived either from the pWV01 replicon or from the pAMβ1 replicon (data not shown), and we consequently hypothesized that pTXL1 might be a good candidate for elaborating new food-grade vectors for Leuconostoc.
Bacterial strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristic(s) | Source or referencea |
|---|---|---|
| Strains | ||
| Escherichia coli DH5α | recA endA1 gyrA96 thi-1 hsdR17 supE44 Δlac U169(φ80 ΔlacZΔM15) deoR F− λ− | Gibco-BRL |
| Leuconostoc mesenteroides subsp. mesenteroides | ||
| Y105 | Wild-type strain | 18 |
| Y110 | Wild-type strain | CNCM I-1936a |
| FR52 | Wild-type strain | 24 |
| Leuconostoc mesenteroides subsp. dextranicum DSM20484 | Wild-type strain | DSMZb |
| Leuconostoc cremoris LC | Wild-type industrial strain | Rhodia Food |
| Lactococcus lactis | ||
| IL1403 | Lab strain | 8 |
| MG1363 | Lab strain | 13 |
| RD230 | Wild-type industrial strain | Rhodia Food |
| Lactobacillus sake 23K | Plasmid cured | 3 |
| Listeria ivanovii BUG 497 | Lab strain | Pasteur Institute |
| Pediococcus acidilactici P120 | Wild-type industrial strain | Rhodia Food |
| Plasmids | ||
| pBSSKII+ | pBluescript SK II+ cloning vector, 2.96 kb, AprlacZ | Stratagene |
| pGhost9:ISS1 | pWV01 derivative replicon | 23 |
| pFR18 | 1.8-kb cryptic plasmid from L. mesenteroides FR52 | 5 |
| pFBYC018 | pBSSKII+::1.8-kb HindIII pFR18, 4.8 kb, Apr Ems | 5 |
| pFBYC018E | pFBYC018::1.1-kb BamHI pGhost9:ISS1 (Emr), 5.9 kb, Apr Emr | 5 |
| pFBYC18E | pFBYC018E with 2.5-kb PvuII pBSSKII+ deleted, 3.5 kb, Emr | 5 |
| pTXL1 | 2.66-kb cryptic plasmid from L. mesenteroides Y110 | 5 |
| pFBYC050 | pBSSKII+::2.66-kb SalI pTXL1, 5.62 kb, Apr Ems | This study |
| pFBYC051 | pBSSKII+::2.66-kb SpeI pTXL1, 5.62 kb, Apr Ems | This study |
| pFBYC052 | pBSSKII+::1.8-kb SspI pFR18, 4.8 kb, Apr Ems | This study |
| pFBYC050E | pFBYC050::1.1-kb BamHI pGhost9:ISS1, 7.72 kb, Apr Emr | This study |
| pFBYC50E | pFBYC050E with 2.5-kb PvuII fragment deleted, Emr | This study |
| pFBYC065 | pFBYC050E with 312-kb ExoIIIc fragment deleted, Apr Emr | This study |
| pFBYC060K | pFBYC050E with 482-kb KpnI-KpnI fragment deleted, Apr Emr | This study |
| pFBYC062S | pFBYC050E with 1.4-kb SalI-SspI fragment deleted, Apr Emr | This study |
| pFBYC068S | pFBYC050E with 1.9-kb SalI-AflIII fragment deleted, Apr Emr | This study |
| pFBYC051E | pFBYC051::1.1-kb BamHI pGhost9:ISS1, 7.72 kb, Apr Emr | This study |
| pFBYC06 | pRW5.6 with HindIII deleted | 4 |
| pFBYC069 | pFBYC051E::669-bp P59, dvnA fragment from pFBYC06 | This study |
| pFBYC070 | pFBYC069::485-bp dvnA::mesY, mesI PCR product | This study |
CNCM, Collection Nationale de Cultures de Microorganisms, France.
DSMZ, Deutsche Sammlung von Mikroorganismen and Zellkulturen, Germany.
ExoIII, exonuclease III.
L. mesenteroides subsp. mesenteroides Y110, which originated from goats' milk, was isolated and grown in MRS (Difco) broth or agar (1.2%, wt/vol) at 30°C. It contains five plasmids (6). The smallest, pTXL1 (2.6 kb) (6), was inserted into the unique SalI site of pBSSKII+ to yield pFBYC050. General genetic techniques used were as previously described (25, 28). Escherichia coli competent cells were prepared and transformed according to the method of Hanahan (16). The resulting transformant strains were propagated at 37°C in LB (28) broth or on agar (1.5%, wt/vol) containing ampicillin at a final concentration of 100 μg ml−1. The sequences of pTXL1 were determined on both strands with an Auto-read sequencing kit (Pharmacia) and with appropriate primers by using an automated laser fluorescence DNA sequencer (Pharmacia). Sequence analyses were performed with the Genetics Computer Group sequence analysis software package (University of Wisconsin). The G+C content in pTXL1 is 34%, in accordance with the G+C percentage of Leuconostoc subsp. chromosomes and plasmids previously described (10, 12). Surprisingly, sequence analysis revealed no similarities with genes encoding known proteins involved in replication, such as the replication initiation protein required for RCR plasmids. Generally, small cryptic plasmids replicate by an RCR mechanism and their sequence bears a gene encoding their replicase (19). No sequence resembling a double-strand origin or a single-strand origin involved in the conversion of single-strand DNA (ssDNA) intermediates into a double-stranded DNA plasmid molecule was identified in the pTXL1 nucleotidic sequence.
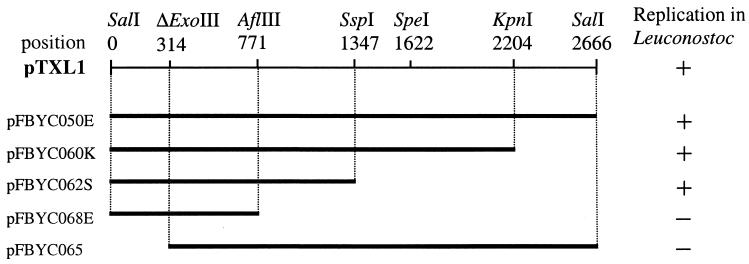
To locate the region required for pTXL1 replication, the erythromycin cassette originating from pGhost9:ISS1 was inserted into pFBYC050 (Fig. 1). Plasmid pFBC050E was therefore constructed by insertion of the 1.1-kb BamHI cassette into the unique BamHI site in the multiple cloning site of pFBYC050. Erythromycin-resistant transformants of E. coli were selected on brain heart infusion agar plates (Difco) containing 150 μg of erythromycin ml−1. Erythromycin-resistant transformants of Leuconostoc subsp. strains were grown at 30°C in MRS (Difco) broth or agar (1.2%, wt/vol) containing erythromycin (5 μg ml−1). As this construct was able to transform Leuconostoc mesenteroides subsp. dextranicum DSM20484 to Emr, a series of subclones was constructed to further locate the minimal region, which allowed replication (Fig. 1). We found that the minimal replicon required for the replication of pTXL1 resides within the 1.3-kb SalI-SspI fragment (Fig. 1). Within this minimum replicon three palindromic structures designated IRI, IRII, and IRIII were identified (Fig. 2). IRI, -II, and -III could form hairpin loops, with calculated changes in the free energy of formation (ΔGo) of −15, −22, and −10 kcal mol−1, respectively. Three direct repeats, DRI, DRII, and DRIII, were identified (Fig. 1). IRI and DRI may be required for plasmid replication because the construct pFBYC065, from which these repeats were deleted (Fig. 1), was not able to replicate in Leuconostoc. Sequence analysis revealed two small open reading frames, ORF1 and ORF2, extending from positions 1001 to 1339 and from positions 883 to 644, respectively. By scanning data banks for sequences deduced from ORF1 (80 amino acids) and ORF2 (113 amino acids), we did not find homology to any known protein. A deletion of ORF1 and ORF2 in the construct pFBYC068E abolished plasmid replication in Leuconostoc, suggesting that both ORFs may be involved in plasmid replication.
FIG. 1.
Identification of the minimal replicon from pTXL1. The restriction map of pTXL1 is shown with the positions of the restriction sites. Partial enzymatic deletion by exonuclease III is denoted by ΔExoIII with the size of the deletion.
FIG. 2.
Nucleotide sequences of the pTXL1 minimum replicon-linearized sequence and the deduced amino acid sequences for putative ORF1 and ORF2. The arrows with solid lines indicate inverted repeats (IRI, IRII, and IRIII), and the arrows with dashed lines indicate direct repeats (DRI, DRII, and DRIII).
Generally, small LAB plasmids use RCR. RCR implicates the formation of an ssDNA intermediate of the plasmid (29). Therefore, the ability of pTXL1 to generate ssDNA was examined as described previously (5). Leuconostoc cells were grown in MRS medium. Rifampin was added at 100 μg ml−1 in order to inhibit RNA polymerase (22). Whole-cell lysates were prepared as described previously (25, 26). S1 nuclease (Gibco-BRL) treatment was performed according to the method of Noirot-Gros et al. (26). ssDNA was detected by nonalkaline Southern blot hybridization. DNA transfer from agarose gel to a Hybond-N+ nylon membrane (Amersham) was adapted from the work of Sambrook et al. (28). The DNA denaturation step was omitted. DNA probes consisted of purified DNA restriction fragments that were specific for each plasmid and labeled by use of a random priming kit (Gibco-BRL) and [α-32P]dCTP (Amersham). Hybridization of the transferred DNA with the labeled probes was performed according to the method of te Riele et al. (29). Nonspecific hybridizations were avoided by washing the membrane at 62°C in 1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate, after which the membrane was exposed to an autoradiography film.
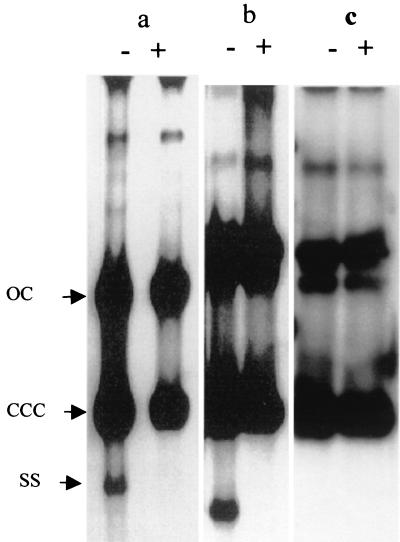
As a control, we used strains of L. mesenteroides subsp. dextranicum DSM20484 containing pFBYC18E (Fig. 3a) or pGhost9:ISS1 (Fig. 3b), two well-described RCR plasmids (5, 23). In addition to hybridizing to the open circular forms and covalently closed circular forms of the plasmids, the probes specific for pFBYC18E (Fig. 3a) and pGhost9:ISS1 (Fig. 3b) hybridized to a faint fast-migrating band suspected of being an ssDNA intermediate. Their sensitivity to S1 nuclease demonstrated their single-stranded nature. Similarly, whole-cell DNA from L. mesenteroides subsp. dextranicum DSM20484 containing pFBYC50E was tested with pTXL1 as a probe (Fig. 3c). The hybridization failed to detect any single-stranded intermediate of pTXL1. This result strongly suggests that pTXL1 does not belong to the RCR plasmid family. This indirect proof, added to the absence of any ORF encoding a replicase as well as the lack of single-strand and double-strand origin sequences, is consistent with a mode of replication differing from the rolling-circle mechanism.
FIG. 3.
Detection of single-stranded intermediates of pFBYC18E (a), pGhost9:ISS1 (b), and the pTXL1 derivative (c) in L. mesenteroides subsp. dextranicum DSM20484. A nonalkaline Southern blot hybridization analysis was performed with radiolabeled pFBYC18E (a), pGhost9:ISS1 (b), and pTXL1 (c) probes. Whole-cell DNA was prepared from MRS cultures containing rifampin. DNA preparations treated with endonuclease S1 are indicated by a +, and untreated preparations are indicated by a −. OC, open circular DNA; CCC, covalently closed circular DNA; SS, ssDNA.
RCR plasmids are less stable when they contain foreign DNA (20), which limits their use as cloning vectors. To investigate the stability of pTXL1 as a cloning vector, single colonies of Leuconostoc transformants carrying various endogenous plasmids were used to inoculate MRS broth without erythromycin. The cultures were maintained in mid-log phase and serially transferred until approximately 100 generations were obtained. Plasmid stability was then estimated by comparing the numbers of CFU on selective (5 μg of erythromycin ml−1) and nonselective (without antibiotic) agar plates. Results presented in Table 2 show that, in contrast to what occurs with pGhost9:ISS1 or pFR18 derivatives, the segregational stability of pTXL1 derivatives was not affected by a large insertion of foreign DNA. In addition, Southern blot analysis of plasmid DNA prepared from erythromycin-resistant colonies cultivated for 100 generations without selection pressure revealed no structural change in the plasmid (data not shown).
TABLE 2.
Stability of pTXL1 derivatives during extended growth under nonselective conditions
| Plasmid | Replicon | % of DSM20484 clones containing the plasmid after 100 generationsa | Reference |
|---|---|---|---|
| pGhost9:ISS1 | pWV01 | 76 | 23 |
| pFBYC050E | pTXL1 | 100 | This study |
| pFBYC50E | pTXL1 | 100 | This study |
| pFBYC018E | pFR18 | 3 | 5 |
| pFBYC18E | pFR18 | 100 | 5 |
Percentages are averages of results from duplicate experiments and were determined as percentages of erythromycin-resistant colonies.
Taking into account the overall properties of the pTXL1 replicon described above, it appeared to be a good alternative for use in developing Leuconostoc and other LAB cloning vectors. Indeed, the pTXL1 replicon was shown to be able to replicate in various Leuconostoc species (cremoris LC; mesenteroides subsp. mesenteroides Y110, Y105, and FR52; and mesenteroides subsp. dextranicum DSM20484), Pediococcus acidilactici P120, and Lactobacillus sakei 23K but not in E. coli (data not shown). Preparation of electrocompetent LAB cells and electroporation were as described previously (27). P. acidilactici P120 was grown in MRS (Difco) broth or agar (1.2%, wt/vol) at 30°C. MRS medium supplemented with 1% glucose was used for Lactobacillus sakei 23K propagation at 30°C. Interestingly, the transformation frequencies observed with the pTXL1 derivatives (pFBYC050E and pFBYC50E) were similar to those observed with the control RCR plasmid pGhost9:ISS1. In Leuconostoc strains, the transformation frequencies per microgram quantities of pTXL1 derivatives or pGhost9/ISS1 were about 105 transformants in L. mesenteroides subsp. dextranicum DSM20484 and about 101 in L. mesenteroides subsp. mesenteroides Y110. In the Lactobacillus sakei 23K and P. acidilactici P120 strains, the transformation frequencies were 105 for both plasmids. In addition, this replicon was compatible with the pWV01 replicon that served as a basis for most genetic tools devoted to LAB. Consequently, pTXL1 was used to design a narrow-host-range food-grade vector.
We decided to transform an industrial L. cremoris strain aiming at the heterologous production of the anti-Listeria bacteriocin mesentericin Y105, originally produced by L. mesenteroides subsp. mesenteroides Y110.
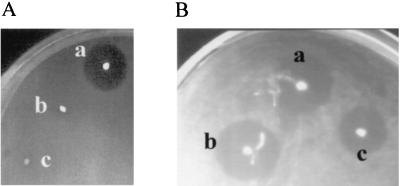
A 669-bp SmaI-HindIII restriction fragment containing the constitutive lactococcal p59 promoter, a ribosome-binding site, and part of the divergicin A signal peptide gene (4) was cloned into pFBYC051 (Table 1) that had been restricted with SmaI and HindIII enzymes. This construct, named pFBYC069, was restricted with HindIII and ligated with a 518-bp HindIII restriction fragment containing the mesentericin Y105 structural gene (mesY) and its immunity gene (mesI). The resulting plasmid, pFBYC070, was introduced into the L. mesenteroides subsp. dextranicum DSM20484 strain devoid of anti-Listeria activity and sensitive to mesentericin Y105 and into the industrial L. cremoris LC strain devoid of anti-Listeria activity. Production of mesentericin Y105 by recombinant L. mesenteroides subsp. dextranicum DSM20484 and recombinant L. cremoris LC were examined by a rapid purification method (4) and characterized by mass spectroscopy analysis (data not shown). Bacteriocin activity was assayed against Listeria ivanovii BUG 497 as described previously (1). Figure 4 shows that recombinant L. mesenteroides subsp. dextranicum DSM20484 and L. cremoris LC producing mesentericin Y105 displayed bactericidal activity against Listeria, unlike nonrecombinant Leuconostoc strains.
FIG. 4.
Bacteriocin activity assays against Listeria ivanovii BUG 497 overlaying Leuconostoc strains. (A) L. mesenteroides subsp. mesenteroides Y105 (a); nonrecombinant L. mesenteroides subsp. dextranicum DSM20484 (b), and nonrecombinant L. cremoris LC (c); (B) L. mesenteroides subsp. mesenteroides Y105; (a) Leuconostoc mesenteroides subsp. dextranicum DSM20484 containing pFBYC070 (b), and Leuconostoc cremoris LC containing pFBYC070 (c).
These results proved that the L. mesenteroides subsp. dextranicum DSM20484 strain and the industrial strain L. cremoris LC were able to express the bacteriocin from the pTXL1 derivative vector at a level similar to that of the natural producer.
We described here a new Leuconostoc plasmid, the first small non-RCR-type Leuconostoc plasmid identified to date. Its food-grade origin combined with its small size, high stability, and narrow host range makes pTXL1 a food-grade vector of interest. Derivatives of pTXL1 should be useful for constructing Leuconostoc starter strains with improved fermentative, flavor-enhancing, and bacteriocin-producing abilities.
Nucleotide sequence accession number.
The EMBL accession number for the complete nucleotide sequence of pTXL1 linearized at its unique SalI site reported in this paper is AJ272077.
Acknowledgments
This work was achieved as part of the program BIOAVENIR (contract 780227), supported by Rhône-Poulenc with the participation of the Ministère de la Recherche et de l'Espace and the Ministère de l'Industrie et du Commerce Extérieur.
We are grateful to Monique Zagorec for her contribution to the transfer of plasmids in Lactobacillus sakei. We thank Laurent Jannière for helpful discussion on single-stranded DNA detection.
REFERENCES
- 1.Ahn, C., and M. E. Stiles. 1990. Plasmid-associated bacteriocin production by a strain of Carnobacterium piscicola from meat. Appl. Environ. Microbiol. 56:2503-2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bates, E. E., and H. J. Gilbert. 1989. Characterization of a cryptic plasmid from Lactobacillus plantarum. Gene 85:253-258. [DOI] [PubMed] [Google Scholar]
- 3.Berthier, F., M. Zagorec, M. C. Champomier-vergès, and S. D. Ehrlich. 1996. Efficient transformation of Lactobacillus sake by electroporation. Microbiology 142:1273-1279. [DOI] [PubMed] [Google Scholar]
- 4.Biet, F., J. M. Berjeaud, R. W. Worobo, Y. Cenatiempo, and C. Fremaux. 1998. Heterologous expression of the bacteriocin mesentericin Y105 using the dedicated transport system and the general secretion pathway. Microbiology 144:2845-2854. [DOI] [PubMed] [Google Scholar]
- 5.Biet, F., Y. Cenatiempo, and C. Fremaux. 1999. Characterization of pFR18, a small cryptic plasmid from Leuconostoc mesenteroides subsp. mesenteroides FR52, and its use as a food grade vector. FEMS Microbiol. Lett. 179:375-383. [DOI] [PubMed] [Google Scholar]
- 6.Biet, F., Y. Cenatiempo, and C. Fremaux. May 1999. Non RCR Leuconostoc plasmid capable of being transferred into lactic acid bacteria; use as cloning and expressing tool. French patent WO9924591.
- 7.Brito, L., G. Vieira, M. A. Santos, and H. Paveia. 1996. Nucleotide sequence analysis of pOg32, a cryptic plasmid from Leuconostoc oenos. Plasmid 36:49-54. [DOI] [PubMed] [Google Scholar]
- 8.Chopin, A., M. C. Chopin, A. Moillo-Batt, and P. Langella. 1984. Two plasmid-determined restriction and modification systems in Streptococcus lactis. Plasmid 11:260-263. [DOI] [PubMed] [Google Scholar]
- 9.Clewell, D. B., Y. Yagi, G. M. Dunny, and S. K. Schultz. 1974. Characterization of three plasmid deoxyribonucleic acid molecules in a strain of Streptococcus faecalis: identification of a plasmid determining erythromycin resistance. J. Bacteriol. 117:283-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coffey, A., A. Harrington, K. Kearney, C. Daly, and G. Fitzgerald. 1994. Nucleotide sequence and structural organization of the small, broad-host-range plasmid pCI411 from Leuconostoc lactis 533. Microbiology 140:2263-2269. [DOI] [PubMed] [Google Scholar]
- 11.Fremaux, C., M. Aigle, and A. Lonvaud-Funel. 1993. Sequence analysis of Leuconostoc oenos DNA: organization of pLo13, a cryptic plasmid. Plasmid 30:212-223. [DOI] [PubMed] [Google Scholar]
- 12.Garvie, E. I., V. Zezula, and V. A. Hill. 1974. Guanine plus cytosine content of the deoxyribonucleic acids of the leuconostocs and some heterofermentative lactobacilli. Int. J. Syst. Bacteriol. 24:248-251. [Google Scholar]
- 13.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gruss, A., R. Dabert, S. Sozhamannan, and S. D. Ehrlich. 1990. Replication of ssDNA plasmids is sensitive to DNA content. In R. P. Novick (ed.), Molecular biology of the staphylococci. VCH Publishers, New York, N.Y.
- 15.Gruss, A., and S. D. Ehrlich. 1989. The family of highly interrelated single-stranded deoxyribonucleic acid plasmids. Microbiol. Rev. 53:231-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmid DNA. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 17.Hayes, F., P. Vos, G. F. Fitzgerald, W. M. de Vos, and C. Daly. 1991. Molecular organization of the minimal replicon of novel, narrow-host-range, lactococcal plasmid pCI305. Plasmid 25:16-26. [DOI] [PubMed] [Google Scholar]
- 18.Hechard, Y., B. Derijard, F. Letellier, and Y. Cenatiempo. 1992. Characterization and purification of mesentericin Y105, an anti-Listeria bacteriocin from Leuconostoc mesenteroides. J. Gen. Microbiol. 138:2725-2731. [DOI] [PubMed] [Google Scholar]
- 19.Jannière, L., A. Gruss, and S. D. Ehrlich. 1993. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics, p. 625-644. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Plasmids. American Society for Microbiology, Washington, D.C.
- 20.Kiewiet, R., S. Bron, K. de Jonge, G. Venema, and J. F. Seegers. 1993. Theta replication of the lactococcal plasmid pWVO2. Mol. Microbiol. 10:319-327. [PubMed] [Google Scholar]
- 21.Kiewiet, R., J. Kok, J. F. Seegers, G. Venema, and S. Bron. 1993. The mode of replication is a major factor in segregational plasmid instability in Lactococcus lactis. Appl. Environ. Microbiol. 48:726-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leenhouts, K. J., B. Tolner, S. Bron, J. Kok, G. Venema, and J. F. Seegers. 1991. Nucleotide sequence and characterization of the broad-host-range lactococcal plasmid pWVO1. Plasmid 26:55-66. [DOI] [PubMed] [Google Scholar]
- 23.Maguin, E., H. Prevost, S. D. Ehrlich, and A. Gruss. 1996. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J. Bacteriol. 178:931-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mathieu, F., I. S. Suwandhi, N. Rekhif, J. B. Milliere, and G. Lefebvre. 1993. Mesenterocin 52, a bacteriocin produced by Leuconostoc mesenteroides ssp. mesenteroides FR 52. J. Appl. Bacteriol. 74:372-379. [DOI] [PubMed] [Google Scholar]
- 25.Muriana, P. M., and T. R. Klaenhammer. 1987. Conjugal transfer of plasmid-encoded determinants for bacteriocin production and immunity in Lactobacillus acidophilus 88. Appl. Environ. Microbiol. 53:553-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noirot-Gros, M. F., V. Bidnenko, and S. D. Ehrlich. 1994. Active site of the replication protein of the rolling circle plasmid pC194. EMBO J. 13:4412-4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raya, R. R., C. Fremaux, G. L. De Antoni, and T. R. Klaenhammer. 1992. Site-specific integration of the temperate bacteriophage φadh into the Lactobacillus gasseri chromosome and molecular characterization of the phage (attP) and bacterial (attB) attachment sites. J. Bacteriol. 174:5584-5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 29.te Riele, H., B. Michel, and S. D. Ehrlich. 1986. Single-stranded plasmid DNA in Bacillus subtilis and Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 83:2541-2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zuniga, M., I. Pardo, and S. Ferrer. 1996. Nucleotide sequence of plasmid p4028, a cryptic plasmid from Leuconostoc oenos. Plasmid 36:67-74. [DOI] [PubMed] [Google Scholar]