Abstract
Oligonucleotide probes and colony hybridization (CH) were applied to enumerate organisms of the genus Legionella in cooling tower water. The CH counts indicated almost the same results as CFU counts in cultivated samples derived from the water. It was concluded that it is possible to substitute the CH procedure for the conventional one.
Legionella is the etiologic agent of Legionnaires' disease. The combination of isolation on selective medium and identification by DNA probe is useful in attempting to detect proliferating Legionella organisms (1, 2). Here we report two kinds of oligonucleotide probes, one targeting 16S rRNA and the other targeting a new target region, namely, an antigen common to the genus Legionella. Subsequently, we applied these probes to enumerate viable legionellae in various cooling tower water samples by using colony hybridization (CH). As a result of this investigation, we successfully identified and detected pinpoint Legionella colonies by using each of the probes.
Design and labeling of specific oligonucleotide probes for Legionella.
The probes LEG225 (6), CA/G L253, CA/G L301 (3, 4, 8), and L5S-1 (5), together with their sequences and target positions, are listed in Table 1. The probe L5S-1 was used as a positive control.
TABLE 1.
Probe sequences and target positions
| Probe | Sequence | Target position |
|---|---|---|
| LEG225 | 5′-AAgATTAgCCTgCgTCCgAT-3′ | 16S rRNA |
| CA/G L253 | 5′-ggTgATggTACTACTACTgC-3′ | Common antigen |
| CA/G L301 | 5′-gAAggTCACAAAgCAgTTgC-3′ | Common antigen |
| L5S (16) | 5′-CTCgAACTCAgAAgTCAAACA TTTCCgCgCCAATgATAgTgTgA ggCTTC-3′ | 5S rRNA |
The synthesized oligonucleotides were labeled with a digoxigenin oligonucleotide tailing kit (Boehringer Mannheim, Mannheim, Germany), in accordance with the manual.
Bacterial strains.
The bacterial strains used in this study, along with their sources, are listed in Tables 2 and 3. Legionella strains were obtained from the stock strains of the Department of Microbiology, Gifu University School of Medicine. The reference strains were supplied by the Department of Bacteriology, Juntendo University.
TABLE 2.
Legionella strains used for probe specificity examinations and dot blot hybridization results
| Legionella organism | Origina | Reaction with probe at indicated concn [pmol/ml]b
|
||||
|---|---|---|---|---|---|---|
| LEG225 [0.3] | CA/G L253 [1] | CA/G L301 [1] | CA/G L253 and CA/G L301 [1] | L5S-1 [0.3] | ||
| L. jordanis | GIFU 3193T | ++ | + | + | ++ | ++ |
| L. pneumophila | GIFU 9134T | ++ | ++ | ++ | ++ | ++ |
| L. bozemanae | GIFU 9140T | ++ | + | ++ | ++ | ++ |
| L. micdadei | GIFU 9141T | ++ | ++ | ++ | ++ | ++ |
| L. gormanii | GIFU 9142T | ++ | + | + | ++ | ++ |
| L. longbeachae | GIFU 9245T | ++ | + | ++ | ++ | ++ |
| L. pneumophila subsp. fraseri | GIFU 9246T | ++ | ++ | ++ | ++ | ++ |
| L. dumoffii | GIFU 9247T | ++ | + | ± | + | ++ |
| L. oakridgensis | GIFU 10061T | ++ | + | ± | ++ | ++ |
| L. wadsworthii | GIFU 10062T | ++ | ++ | − | ++ | ++ |
| L. feeleii | GIFU 10063T | ++ | + | + | ++ | ++ |
| L. sainthelensi | GIFU 10392T | ++ | + | − | ++ | ++ |
| L. hackeliae | GIFU 10740T | ++ | + | ± | + | ++ |
| L. jamestowniensis | GIFU 10741T | ++ | ++ | ++ | ++ | ++ |
| L. cherrii | GIFU 10742T | ++ | ++ | + | ++ | ++ |
| L. rubrilucens | GIFU 10743T | ++ | − | − | + | ++ |
| L. maceachernii | GIFU 10745T | ++ | + | ± | + | ++ |
| L. spiritensis | GIFU 11199T | ++ | − | ± | + | ++ |
| L. israelensis | GIFU 11367T | ± | − | + | ++ | ++ |
| L. parisiensis | GIFU 11745T | ++ | ± | + | ++ | ++ |
| L. santicrucis | GIFU 11746T | ++ | ± | − | + | ++ |
| L. steigerwaltii | GIFU 11747T | ++ | ++ | ++ | ++ | ++ |
| L. erythra | GIFU 11748T | ++ | ± | − | + | ++ |
| L. birminghamensis | GIFU 11749T | + | ++ | ++ | ++ | ++ |
| L. anisa | GIFU 12075T | ++ | ± | ± | ++ | ++ |
| L. cincinnatiensis | GIFU 12201T | ++ | ++ | ± | ++ | ++ |
| L. quinlivaniic | GIFU 12648T | |||||
| L. moravica | GIFU 12649T | ± | ++ | ± | ++ | ++ |
| L. brunensis | GIFU 12655T | ++ | − | ++ | ++ | ++ |
| L. tucsonensis | GIFU 12656T | ++ | ++ | ++ | ++ | ++ |
| L. adelaidensis | GIFU 13562T | ++ | + | ++ | ++ | ++ |
| L. fairfieldensis | GIFU 13563T | ++ | + | ± | ++ | ++ |
| L. gratiana | GIFU 13564T | ++ | ± | ± | ++ | ++ |
| L. lansingensis | GIFU 13565T | ++ | ++ | ± | ++ | ++ |
| L. shakespearei | GIFU 13566T | ± | ++ | ++ | ++ | ++ |
| L. pneumophila subsp. pascullei | GIFU 13567T | ++ | ++ | ++ | ++ | ++ |
| Total hybridization rate for Legionella spp. [%] | 91 | 74 | 57 | 100 | 100 | |
GIFU, Gifu University School of Medicine.
++, Strong hybridization signal; +, moderately strong hybridization signal; ±, weak hybridization signal; −, no hybridization signal.
DNA extraction failed.
TABLE 3.
Reference strains used for probe specificity examinations and dot blot hybridization results
| Organism | Origina | Reaction with probe at indicated concn [pmol/ml]b
|
||||
|---|---|---|---|---|---|---|
| LEG225 [0.3] | CA/G L253 [1] | CA/G L301 [1] | CA/G L253 and CA/G L301 [1] | L5S-1 [0.3] | ||
| Enterobacter cloacae | ATCC 23355T | − | − | − | ± | ++ |
| Escherichia coli | ATCC 25922T | − | − | − | ± | ++ |
| Klebsiella pneumoniae | ATCC 13883T | − | ++ | ± | ++ | ++ |
| Proteus vulgaris | ATCC 13315T | − | − | ± | ± | ++ |
| Pseudomonas aeruginosa | ATCC 27853T | − | − | − | ± | ++ |
| Salmonella enterica serovar Typhimurium | ATCC 14028T | − | − | − | ± | ++ |
| Serratia marcescens | ATCC 8100T | ± | − | − | ± | ++ |
| Staphylococcus aureus | ATCC 12600T | − | ± | − | ± | ++ |
| Total hybridization rate [%] | 0 | 12.5 | 0 | 12.5 | 100 | |
ATCC, American Type Culture Collection.
++, Strong hybridization signal; +, moderately strong hybridization signal; ±, weak hybridization signal; −, no hybridization signal.
Cell lysis for DNA extraction and purification.
Legionella strains were cultivated at 37°C for 3 to 5 days on a 0.1% (wt/vol) α-ketoglutarate (BCYEα) agar plate (Eiken Chemical Co., Tokyo, Japan). About 108 to 109 cells were supplied in 533 μl of 10 mM Tris-HCl-1 mM EDTA buffer (pH 8.0). Sixty μl of 10% sodium dodecyl sulfate (SDS) (final concentration, 1%) and 6 μl of 10-mg/ml proteinase K solution (final concentration, 100 μg/ml) were added. After the solutions were mixed, they were incubated for 1 h at 37°C.
Reference microorganisms except Staphylococcus aureus were cultured in Luria-Bertani broth overnight and harvested. About 108 to 109 cells were suspended in 2 ml of 10 mM Tris-HCl-1 mM EDTA buffer (pH 8.0). One hundred μl of 10% SDS (final concentration, 0.5%) and 20 μl of 10-mg/ml proteinase K solution (final concentration, 100 μg/ml) were added. After the solutions were mixed, the mixture was incubated for 1 h at 37°C. Then 353 μl of 5 M NaCl and 282 μl of cetyltrimethylammonium bromide-NaCl were added, and the solution was incubated for 10 min at 65°C. Staphylococcus aureus was cultured overnight in Luria-Bertani broth and harvested. The cells were washed twice in 2 ml of 10 mM Tris-HCl-10 mM EDTA buffer (pH 8.0). The pellet was suspended in 2 ml of 10 mM Tris-HCl-10 mM EDTA buffer (pH 8.0) and then mixed sufficiently with 40 μl of achromopeptidase (50,000 U/ml in 10 mM NaCl). The solution was incubated at 55°C until the cells were lysed.
The subsequent DNA extraction and purification procedures were performed as described previously (7).
Detection of legionella colonies.
The cultivation and identification of legionella colonies from the cooling tower water were performed as described previously (1). The replicated agar plates were reincubated for another 2 to 3 days at 37°C until the colonies grew to a visible size.
Testing of oligonucleotide probe specificity.
One hundred ng of nucleic acid was blotted onto a 0.22-μm-pore-size Biodyne A nylon membrane (Poll Europe Ltd.). Subsequent dot blot hybridization was performed under the standard 5× SSC procedure (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) in a digoxigenin system (Boehringer Mannheim).
CH.
The concentrated cooling tower water was cultured on a WYOα agar plate at 37°C for 2 to 5 days. Colonies were transferred from plates to 0.22-μm-pore-size (82-mm-diameter) Biodyne A nylon membranes (Poll Europe) by using procedures described previously (7). After replication, the membranes were placed colony-side-up on the paper (3MM; Whatman) and were saturated with 5% Triton X-100 in 10 mM Tris-HCl-1 mM EDTA (pH 8.0) for 1 h at 60°C. The membranes were transferred onto the paper (3MM); supplemented with 0.1 mg of proteinase K per ml in 10 mM Tris-HCl (pH 8.0) containing 10 mM NaCl, 1 mM sodium citrate, and 1.5% SDS; and incubated for 1 h at 60°C to lyse the cells. Subsequently, denaturing and neutralization of the DNA were performed as described previously (7). In order to remove the protein, the DNA fixed in the membranes was incubated overnight at 37°C with gentle agitation in a solution including 0.1 mg of proteinase K per ml, 10 mM Tris-HCl (pH 8.0), 150 mM NaCl, and 0.5% SDS. The membranes were washed twice for 5 min at room temperature with about 50 ml of 6× SSC per 100 cm2.
Hybridization was carried out by using the same procedure described for dot blot hybridization, except that 10% blocking reagent was used.
Specificity of the oligonucleotide probes.
Probe LEG225 hybridized to 32 of the tested strains of Legionella spp. (91%) (Table 2). No false-positive hybridization occurred with the reference strain (Table 3). Probes CA/G L253 and CA/G L301 could hybridize 26 strains (74%) and 18 strains (54%) of the Legionella species tested, respectively. A mixed probe consisting of probes CA/G L253 and CA/G L301 together could hybridize 100% of the Legionella species tested (Table 2), but false-positive hybridization occurred with Klebsiella pneumoniae (Table 3).
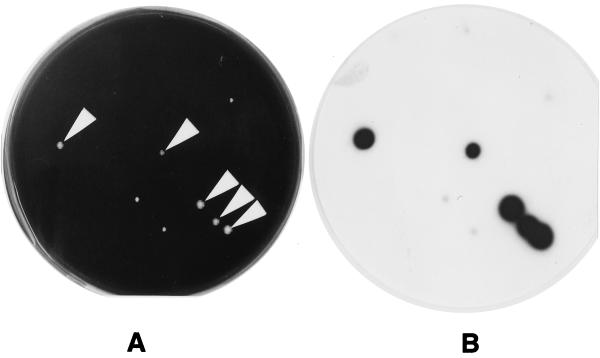
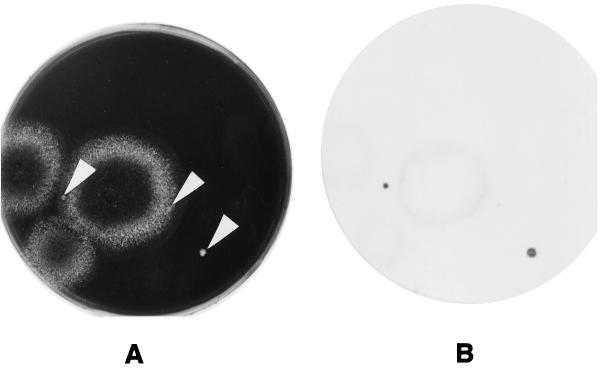
Other bacteria and fungi which grew on the WYOα agar plates frequently disturbed the growth of legionellae and of the screened legionella colonies. However, as shown in Fig. 1, the signals from Legionella colonies clearly discriminated among these other microorganisms. A Legionella colony formed within a fungus colony was also detected (Fig. 2).
FIG. 1.
Cultivated colonies of Legionella spp. from cooling tower water samples and CH results. The sample water was concentrated and mixed with 0.2 M KCl-HCl buffer (pH 2.2) for 4 min at 25°C. Then, 0.1 ml of mixture was streaked on WYOα agar. The plate was incubated at 37°C. (A) The plate after 4 days of incubation, just before the membrane was laid on the culture. The white arrows indicate Legionella colonies. (B) Signals of X-ray film developed after the plate shown in panel A was exposed to a membrane which was hybridized with a mixed probe consisting of probes CA/G L253 and CA/G L301.
FIG. 2.
Cultivated colonies of Legionella spp. from cooling tower water samples and CH results. The sample water was concentrated and mixed with 0.2 M KCl-HCl buffer (pH 2.2) for 4 min at 25°C. Then, 0.1 ml of mixture was streaked on WYOα agar. The plate was incubated at 37°C. (A) The plate after 4 days of incubation, just before the membrane was laid on the culture. The white arrows indicate Legionella colonies. (B) Signals of X-ray film developed after the plate shown in panel A was exposed to a membrane which was hybridized with a mixed probe consisting of probes CA/G L253 and CA/G L301.
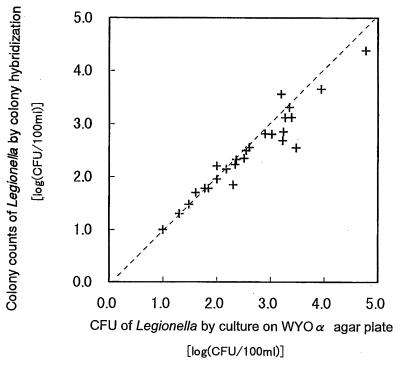
The results of CH counts of organisms from cooling tower water by using probe LEG225 and the results of CFU counts by cultivation revealed a close relationship, with a regression coefficient of 0.96 (Fig. 3).
FIG. 3.
Regression analysis of results of Legionella colony counts from samples of cooling tower water by CH with probe LEG225 against results of CFU counts by cultivation on WYOα agar plates. Regression coefficient, 0.96.
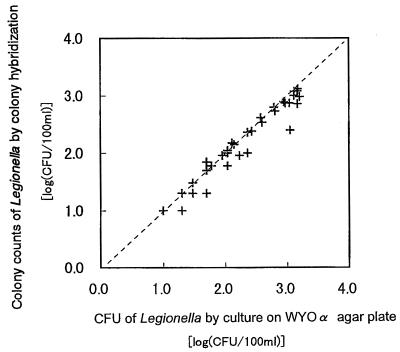
In the case of the mixed probe consisting of probes CA/G L253 and CA/G L301 together, the results of CH counts were generally in agreement with those of CFU counts by plate culture; the regression coefficient was 0.98 (Fig. 4). The results of CH counts were slightly lower than those yielded by CFU counts of WYOα agar plate cultures. This tendency became remarkable in the samples that contained more than 100 CFU per 100 ml. This phenomenon is considered to result from the creation of large spots on the membrane by individual adjacent colonies because the colonies were aggregated after the process of colony transfer and lysing. But this phenomenon did not significantly affect the result of Legionella enumeration. The CH method has excellent features, including the ability to simultaneously perform enumeration of colonies and identification of every colony on a single plate.
FIG. 4.
Regression analysis of results of Legionella colony counts from samples of cooling tower water by CH with a mixed probe consisting of probes CA/G L253 and CA/G L301 against results of CFU counts by cultivation on WYOα agar plates. Regression coefficient, 0.98.
Grimont et al. (2) reported that pinpoint Legionella pneumophila colonies were detectable by the use of a DNA probe labeled with a radioactive isotope after 4 days of incubation on BCYEα agar. In our study, we demonstrated the ability to enumerate Legionella colonies by DNA probe without radioactive-isotope labeling after the samples were cultured for 3 days.
Acknowledgments
We thank Hirohisa Tasiro and Takao Numakura for stimulating discussions. We are grateful to Eiko Yabuuchi and Katsunori Furuhata for their invaluable suggestions.
REFERENCES
- 1.Furuhata, K., W. Satoh, M. Wakasa, S. Shimahata, T. Itoh, and M. Fukuyama. 1999. Colony hybridization method for rapid detection of Legionella spp. biocontrol. Science 4:89-92. [Google Scholar]
- 2.Grimont, P. A. D., F. Grimont, N. Desplaces, and P. Tchen. 1985. DNA probe specific for Legionella pneumophila. J. Clin. Microbiol. 21:431-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hemmingsen, S. M., C. Woolford, S. M. van der Vies, K. Tilly, D. T. Dennis, C. P. Georgopoulos, R. W. Hendrix, and R. J. Ellis. 1988. Homologous plant and bacterial proteins chaperone oligomeric protein assembly. Nature 333:330-334. [DOI] [PubMed] [Google Scholar]
- 4.Hindersson, P., N. Hoiby, and J. Bangsborg. 1991. Sequence analysis of the Legionella micdadei groELS operon. FEMS Microbiol. Lett. 77:31-38. [DOI] [PubMed] [Google Scholar]
- 5.Mahbubaini, M. H., A. K. Bej, R. Miller, L. Haff, J. DiCesare, and R. M. Atlas. 1990. Detection of Legionella with polymerase chain reaction and gene probe methods. Mol. Cell. Probes 4:175-187. [DOI] [PubMed] [Google Scholar]
- 6.Miyamoto, H., H. Yamamoto, K. Arima, J. Fujii, K. Maruta, K. Izu, T. Shiomori, and S.-I. Yoshida. 1997. Development of a new seminested PCR method for detection of Legionella species and its application to surveillance of legionellae in hospital cooling tower water. Appl. Environ. Microbiol. 63:2489-2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 8.Sampson, J. S., S. P. O'Connor, B. P. Holloway, B. B. Plikaytis, G. M. Carlone, and L. W. Mayer. 1990. Nucleotide sequence of htpB, the Legionella pneumophila gene encoding the 58-kilodalton (kDa) common antigen, formerly designated the 60-kDa common antigen. Infect. Immun. 58:3154-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]