Abstract
Translational control plays a crucial role during gametogenesis in organisms as different as worms and mammals. Mouse knockout models have highlighted the essential function of many RNA-binding proteins during spermatogenesis. Herein we have investigated the expression and function during mammalian male meiosis of Sam68, an RNA-binding protein implicated in several aspects of RNA metabolism. Sam68 expression and localization within the cells is stage specific: it is expressed in the nucleus of spermatogonia, it disappears at the onset of meiosis (leptotene/zygotene stages), and it accumulates again in the nucleus of pachytene spermatocytes and round spermatids. During the meiotic divisions, Sam68 translocates to the cytoplasm where it is found associated with the polysomes. Translocation correlates with serine/threonine phosphorylation and it is blocked by inhibitors of the mitogen activated protein kinases ERK1/2 and of the maturation promoting factor cyclinB-cdc2 complex. Both kinases associate with Sam68 in pachytene spermatocytes and phosphorylate the regulatory regions upstream and downstream of the Sam68 RNA-binding motif. Molecular cloning of the mRNAs associated with Sam68 in mouse spermatocytes reveals a subset of genes that might be posttranscriptionally regulated by this RNA-binding protein during spermatogenesis. We also demonstrate that Sam68 shuttles between the nucleus and the cytoplasm in secondary spermatocytes, suggesting that it may promote translation of specific RNA targets during the meiotic divisions.
INTRODUCTION
Spermatogenesis is a remarkable example of cellular differentiation in eukaryotic organisms. After a series of mitotic divisions that amplify the number of spermatogonia, germ cells enter the meiotic phase and four haploid spermatids are produced from each primary spermatocyte. The prophase of the first meiotic division lasts several days in the mouse testis (Handel and Eppig, 1998) and allows primary spermatocytes to undergo homologous recombination and to accumulate mRNAs and proteins that will be needed later on during the spermatogenic differentiation (Kleene, 1996). At the onset of spermiogenesis, transcription and translation become temporally uncoupled because elongating spermatids are transcriptionally incompetent. This is due to the profound remodelling of the chromatin, operated through the substitution of nucleosomal histones with the protamines, which renders the nucleus more compact and unable to transcribe new messengers (Sassone-Corsi, 2002). Nevertheless, de novo protein synthesis is ensured by the mRNAs that elongating spermatids inherit from primary spermatocytes and round spermatids and that are translated several days after their synthesis (Braun, 2000).
The pattern of translational activity during spermatogenesis varies according to the cellular differentiation of germ cells (Kleene, 1996). Similarly to proliferating somatic cells, mitotic spermatogonia readily utilize the transcribed mRNAs (Cataldo et al., 1999). By contrast, although the transcriptional activity of primary spermatocytes and round spermatids is copious, only a very small percentage (5–10%) of these mRNAs are actively translated and most of them are accumulated as ribonucleoproteins (RNPs; Biggiogera et al., 1990). Interestingly, it was shown that in rat primary spermatocytes the levels of poly(A) mRNAs is greater in the nucleus than in the cytoplasm (Morales and Hecht, 1994, Morales and Hecht, 1994), suggesting that nuclear RNPs also participate to mRNA storage and/or export processes.
The fundamental role played by RNPs during gametogenesis is suggested by their high levels of expression in germ cells (Venables and Eperon, 1999). Accordingly, knockout mouse models for RNA-binding proteins expressed in different phases of gametogenesis, like MSY2, Tenr, Miwi, Mili, and Dazl (Deng and Lin, 2002; Saunders et al., 2003; Kuramochi-Miyagawa et al., 2004; Connolly et al., 2005; Yang et al., 2005, Yang et al., 2005), display a sterile phenotype. Remarkably, gametogenesis is under a tight translational control of gene expression by RNA-binding proteins also in lower organisms like Caenorhabditis elegans and Drosophila melanogaster (Kuersten and Goodwin, 2003).
An emerging class of proteins involved in RNA homeostasis is represented by the STAR (signal transduction and activation of RNA metabolism) family (Vernet and Artzt, 1997). STAR proteins are conserved across eukaryotes and may represent a direct link between signal transduction pathways and RNA metabolism. They bind RNA through a GSG (Grp33/Sam68/GLD-1) domain, which contains a single KH domain (hnRNP K homology domain) flanked by conserved N- and C-terminal sequences named QUA1 and QUA2 domains required for homodimerization and specificity in RNA binding (Vernet and Artzt, 1997; Lukong and Richard, 2003). Interestingly, GLD-1, a STAR protein of C. elegans, is a temporal regulator of sexual identity and it is necessary for meiotic prophase progression of germ cells (Francis et al., 1995; Jan et al., 1999; Goodwin and Ellis, 2002). Accumulation of GLD-1 is required to switch from proliferation to meiosis during gametogenesis (Marin and Evans, 2003; Hansen et al., 2004). In females, GLD1 interacts with several mRNAs during the meiotic prophase, suppressing their translation and allowing the differentiation of germ cells into oocytes. At the end of the meiotic prophase, GLD1 expression ceases and the repressed target mRNAs begin to be translated (Lee and Schedl, 2001). Null mutations in this gene cause oocytes to exit meiosis and to enter mitotic divisions, which ultimately results in neoplastic transformation and gonadal germ cell tumors (Francis et al., 1995).
Two close homologues of GLD-1 are highly expressed in testis: Sam68 (Src-associated in mitosis, 68 kDa) and SLM-2 (Sam68-like molecule 2; Venables et al., 1999). However, their precise pattern of expression and function are still largely unknown. Sam68 contains five proline-rich sequences that represent docking sites for several SH3 and WW domain-containing proteins (reviewed by Lukong and Richard, 2003). It also contains a tyrosine-rich C-terminal tail that is phosphorylated by Src-kinases in mitosis (Fumagalli et al., 1994; Taylor and Shalloway, 1994) and a nuclear localization signal embedded in this region (Wu et al., 1999). Sam68 has been proposed to participate to many aspects of RNA metabolism, such as mRNA splicing (Hartmann et al., 1999; Matter et al., 2002), export (Li et al., 2002a), and cytoplasmic utilization of viral RNA (Coyle et al., 2003). Given its localization in the nucleus, the effect of Sam68 on cytoplasmic utilization of RNAs remains particularly elusive. Moreover, most of the results were obtained by protein or target overexpression. Thus, the cellular mRNAs regulated by Sam68 are still unknown, leaving open questions on the physiological function of this STAR protein. A possible role of Sam68 in cell cycle progression was suggested by the observation that ablation of its gene from chicken fibroblasts caused a delay of the G2/M phase (Li et al., 2002b). This result indicates that Sam68 is required during the late phases of mitotic divisions.
In the present work, we have investigated the expression and the function of Sam68 during mouse spermatogenesis. We found that the protein is down-regulated at the onset of meiosis and it accumulates again during the pachytene stage and until spermiogenesis begins. Interestingly, Sam68 is localized in the cytoplasm during the meiotic divisions and it associates with the polysomes engaged in active translation. Translocation of Sam68 from the nucleus to the polysomes correlates with its phosphorylation by ERK1/2 and MPF. Moreover, direct cloning experiments reveals that Sam68 binds mRNAs encoded by genes required for the spermatogenetic program. Thus, our studies suggest a novel role for Sam68 as a mRNA carrier that shuttles from the nucleus to the cytoplasm and may facilitate translation during the meiotic divisions.
MATERIALS AND METHODS
Immunohistochemistry
Tissues were fixed in Bouin's solution for 2 h. After fixation, samples were dehydrated, incubated in xylene, embedded in paraffin, and sectioned using standard histological protocols. Immunohistochemical staining was performed on 4-μm-thick sections. Briefly, tissues slides were deparaffinized with xylene and than rehydrated through three changes of alcohol. After washing, endogenous peroxidase activity was blocked by incubation with 0.3% hydrogen peroxide in distilled water for 10 min. Antigen retrieval was carried out by incubating tissue sections in boiling 1 mM ethilenediaminetetraacetic acid (EDTA, pH 8.0) for 30 min. The slides were then rinsed in phosphate buffered solution (phosphate-buffered saline [PBS]) with 0.1% Tween 20 (PBS-T) and subsequently, incubated with a polyclonal rabbit anti-Sam68 (Santa Cruz Biotechnology, Santa Cruz, CA; SC-133 1:400 dilution) for 1 h at room temperature. After washing, antibody detection was accomplished using a biotin-streptavidin horseradish peroxides detection kit (DAKO, Carpinteria, CA; EnVision/HRP) according to manufacturer's instructions.
Cell Isolation, Culture, and Treatments
Testes from 23- to 28-d-old CD1 mice (Charles River Italia, Calco, Italy) were used to obtain pachytene spermatocytes, secondary spermatocytes, and round spermatids by elutriation technique as described previously (Sette et al., 1999). Spermatogonia were obtained from mice 8-d-old as described previously (Rossi et al., 1993). After elutriation, pachytene spermatocytes were cultured in minimal essential medium, supplemented with 0.5% bovine serum albumin (BSA), 1 mM sodium pyruvate, 2 mM sodium lactate, at a density of 106 cells/ml at 32°C in a humidified atmosphere containing 95% air and 5% CO2. After 6 h, cells were treated with U0126 10 –30 μM overnight or roscovitine 50 –100 μM for 45 min before the addition of 0.5 μM okadaic acid (OA; Calbiochem, San Diego, CA) or equal volumes of the solvent dimethyl sulfoxide (DMSO). Cultures were continued for an additional 6 h to induce metaphase I entry (Wiltshire et al., 1995). At the end of the incubation, cells were collected by centrifugation and immediately used or frozen at –80°C.
Flow Cytometry
Cells were fixed in 1% paraformaldehyde for 30 min, washed in PBS, and incubated for 16 h with 70% ethanol. After washing, cells are stained with RNAseA for 30 min at 37°C and then with propidium iodide (10 μg/ml) for additional 30 min at 37°C in the dark. Stained cells were analyzed on a FACSCalibur Flow Cytometer (Becton Dickinson, San Jose, Ca).
RNA Extraction and Northern Blot Analysis
RNA was purified from freshly isolated testes by using cold Trizol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Purified total RNAs (30 μg) were separated on 1.2% agarose gels and blotted onto Nylon membrane (Millipore, Bedford, MA). The Sam68 cDNA probe for Northern blot hybridization was prepared by RT-PCR amplification of mouse testis, by using specific oligonucleotide primers designed to span from the C-ter to the 3′ untranslated region (UTR) of the mouse Sam68 sequence (GI:):agg aat tcg gag cca tca cca gag gt (forward) and agg tcg acg aat tcc gtt ttt ttt taa ctc aac t (reverse). Sam68 and actin cDNAs were labeled by random priming with [α-32P]dNTPs and membranes were hybridized using the QuickHyb kit (Stratagene, La Jolla, CA) according to manufacturer's instructions. After washes, membranes were exposed overnight at –80°C with intensifier screens for autoradiography.
Immunoprecipitation Assay
Isolated spermatocytes were resuspended in lysis buffer (100 mM NaCl, 10 mM MgCl2, 30 mM Tris-HCl, pH 7.5, 1 mM dithiothreitol [DTT], protease inhibitor cocktail), supplemented with 1% Triton-X-100 (for Sam68) or 0.1% Nonidet P40 (for cyclin B1 and ERKs) and kept on ice for 10 min. Soluble extracts were separated by centrifugation at 10,000 × g for 10 min. Cell extracts (300 –500 μg of total proteins) were precleared for 1 h on a mixture of Protein A- and Protein G-Sepharose beads (Sigma-Aldrich, St. Louis, MO) and collected as supernatant after centrifugation for 1 min at 1000 × g. Supernatants were incubated with 1 μg of anti-Sam68 (Santa Cruz, SC-333), anti-Cyclin B1 (Santa Cruz, SC-245), or a mixture of anti-ERK1 and anti-ERK2 antibodies (Santa Cruz, SC-093, SC-154) for 2 h at 4°C under constant shaking. Protein A-/Protein G-Sepharose beads were preadsorbed with 0.05% BSA before the incubation with the immunocomplexes for 1 additional hour. Hence, beads were washed three times with lysis buffer and absorbed proteins were eluted in SDS-sample buffer for Western blot analysis.
Immunokinase Assays
Immunocomplexes bound to Sepharose beads obtained from immunoprecipitation of cell extracts were rinsed twice with either kinase buffer (50 mM HEPES, pH 7.5, 5 mM β-glycerophosphate, 5 mM MnCl2, 5 mM NaF, 0.1 mM sodium orthovanadate, 1 mM DTT, protease inhibitor cocktail). Kinase reactions were carried out in 50 l for 20 min at 30°C in kinase buffer supplemented with 10 μM [γ-32P]ATP (0.2 Ci/l), and the appropriate GST-Sam68 fusion protein as substrate (2 μg). Reactions were stopped either by adding SDS-sample buffer and analyzed by SDS-PAGE and autoradiography.
[32P]Orthophosphate Metabolic Labeling
Isolated spermatocytes were cultured as described above. After 12 h, the medium was replaced with phosphate-free minimal essential medium and carrier-free [32P]orthophosphate (0.3 mCi/ml), and spermatocytes were incubated for an additional 2 h. Hence, cells were treated with or without 0.5 μM OA for 6 h. At the end of the incubation, cells were washed three times with PBS, and cells were resuspended in lysis buffer supplemented with 1% Triton X-100 and incubated for 15 min on ice. After centrifugation, supernatants were preadsorbed to Protein A-Sepharose beads for 1 h to reduce the nonspecific binding and then immunoprecipitated for 2 h at 4°C with anti-Sam68 or anti-IgGs and new Protein A-Sepharose beads in the presence of 0.1% BSA. After three washes, the immunoprecipitates were eluted in sample buffer. Samples were separated on a 10% SDS-PAGE, the gel was dried, and radioactivity was analyzed by autoradiography.
Polysome-RNPs Fractionation by Sucrose Gradients
Isolated spermatocytes were homogenized in lysis buffer (100 mM NaCl, 10 mM MgCl2, 30 mM Tris-HCl [pH 7.5], 1 mM DTT, 30 U/ml RNasin) supplemented with 1% Triton X-100. After 5 min of incubation on ice, the lysates were centrifuged for 10 min at 12,000 × g at 4°C. The supernatants were loaded on a 15–50% (wt/vol) sucrose gradients and sedimented by centrifugation for 110 min at 37,000 rpm in a Beckman SW41 rotor (Fullerton, CA). Each gradient was collected in 10 fractions, proteins were precipitated from each fraction as previously reported (Zalfa et al., 2003) and analyzed by Western blot.
Plasmid Construction
The N-terminal (1–95), GSG (96-275), and C-terminal (276–443) portions of human Sam68 were amplified by PCR from pCDNA4-GFP-Sam68 (a generous gift from Julian Venables (University of Newcastle, UK; see Paronetto et al., 2003 for details) with the following oligonucleotides: agg aat tca tgc agc gcc ggg acg atc ctg cc and agg tcg act cag acc gag gct gtg gcc ga for GST-Sam681–95, agg aat tca aga tgg agc cgg aga ata ag and agg tcg aca ggt act ccg ttc aag tag g for GST-Sam6896–277, agg aat tcg gag tac ctg aac cct ct, and agg tcg act taa taa cgt cca tat gga tgc tc for GST-Sam68276–443. Amplified cDNAs were inserted into the pGEX-4T1 vector in frame with the GST sequence to generate the fusion proteins.
Expression and Purification of GST Fusion Proteins
Plasmids (pGEX)-containing GST fusion proteins were transformed into theE. coli BL21 strain, grown at 30°C in LB medium to an OD600 = 0.6 before induction with 0.5 mM isopropyl-β-thiogalactopyranoside (IPTG, Sigma-Aldrich) for 3 h. GST fusion proteins were purified from bacterial lysates on glutathione-agarose (Sigma-Aldrich) as previously described (Sette et al., 1998) and analyzed by SDS-PAGE and Coomassie blue staining to test purity and integrity.
PolyU/PolyA-Sepharose Pulldown Assay
Isolated spermatocytes cultured as described above and then harvested in lysis buffer (100 mM NaCl, 10 mM MgCl2, 30 mM Tris-HCl [pH 7.5], 1 mM DTT, 30 U/ml RNasin) supplemented with 1% Triton X-100. After 5 min of incubation on ice, and the lysates were centrifuged for 10 min at 12,000 × g at 4°C. Cell extracts, 100 μg, were precleared with Sepharose beads (Sigma) and then incubated for 90 min with poly-U-or poly-A-Sepharose beads (Amersham, Piscataway, NJ). After three washes with lysis buffer, beads were eluted in sample buffer for Western blot analysis.
Western blot Analysis
Cell extracts or immunoprecipitated proteins were diluted in SDS sample buffer and boiled for 5 min. Proteins were separated on 10% SDS-PAGE gels and transferred to polyvinylidene fluoride Immobilon-P membranes (Millipore) using a semidry blotting apparatus (Bio-Rad, Richmond, CA). Membranes were saturated with 5% nonfat dry milk in PBS containing 0.1% Tween 20 for 1 h at room temperature,, and incubated with the following primary antibody (1:1000 dilution) overnight at 4°C: rabbit anti-β-actin (Sigma-Aldrich); anti-mouse anti-cyclin-B1, rabbit anti-Sam68, mouse anti-Sam68 (SC-91), rabbit anti-ERK2, anti-phosphotyrosine (PY20), anti-phosphoserine (Chemicon, Temecula, CA), anti-phosphothreonine, MPM2 (1:500, Upstate Biotechnology, Lake Placid, NY) or rabbit anti-Staufen (generous gift of Dr. Juan Ortin), rabbit anti-S6 (Cell Signaling, Beverly, MA) and rabbit anti-eIF4E (Cell Signaling). Primary antibodies were all from Santa Cruz Biotechnology, unless specified otherwise. Secondary anti-mouse or anti-rabbit IgGs conjugated to horseradish peroxidase (Amersham) were incubated with the membranes for 1 h at room temperature at a 1:10000 dilution in PBS containing 0.1% Tween 20. Immunostained bands were detected by chemiluminescent method (Santa Cruz Biotechnology).
Immunofluorescence Analysis
Secondary spermatocytes were fixed for 10 min in 4% paraformaldehyde and processed for immunofluorescence analysis using the rabbit anti-Sam68 antibody (1:500) as previously reported for other antigens (Sette et al., 1999; Di Agostino et al., 2002).
Identification of Target mRNAs for Sam68
Primary spermatocytes 40 × 106 cells) were homogenized in lysis buffer (10 mM HEPES, pH 7.4, 150 mM NaCl, 10 mM MgCl2, 15 mM EGTA, 1 mM DTT, 100 U/ml Rnasin, 0.5% Triton X-100). After addition of BSA (0.05%) and yeast tRNA (0.1 μg/ml), soluble extracts (6 mg) were cleared for 60 min with Protein A-Sepharose beads (50 μl) and for 2 h with 10 μg rabbit preimmune IgGs preadsorbed to Protein A-Sepharose beads (50 μl). At the end of the incubation, precleared extracts were divided in two and incubated for additional 3 h with either 10 μg rabbit preimmune IgGs or 10 μg rabbit anti-Sam68 IgGs preadsorbed to Protein A-Sepharose beads (50 μl). At the end of the incubation, beads were washed three times with lysis buffer in the presence of (RNAse-free)-DNAse (Roche, Indianapolis, IN), wash twice with lysis buffer and then incubated with 50 μg of proteinase K (Roche). RNA was extracted by standard procedure and used for cloning into the SMART cDNA Library (Clontech) according to manufacturer's instructions or for RT-PCR analysis with specific oligonucleotides (see Supplementary Figure S4). The presence of cloned inserts was verified by PCR using the plasmid as template and the M13 oligonucleotides annealing to the regions flanking the Multiple Cloning Site of the pDNR-LIB plasmid. The identity of the insert was determined by direct sequencing.
GST Pulldown Assay of the mRNA-Sam68 Interactions
GST and GST-Sam68 were purified from E. coli as described above. Purified proteins (2 μg) were equilibrated for 1 h in 50 mM TrisCl, pH 7.4, 100 mM KCl, 2 mM MgCl2, 1 mM EDTA, 1 mM DTT, 100 U/ml Rnasin, 0.2% Nonidet P40 supplemented with BSA (0.05%) and yeast tRNA (0.1 μg/ml). Then, purified total RNA from primary spermatocytes (20 μg) was added to the beads and incubated for 2 h at 4°C under constant shaking. Washes of the beads and RNA extraction and RT-PCR were performed as described in the paragraph above.
RESULTS
Expression and Subcellular Localization of Sam68 during Mouse Spermatogenesis
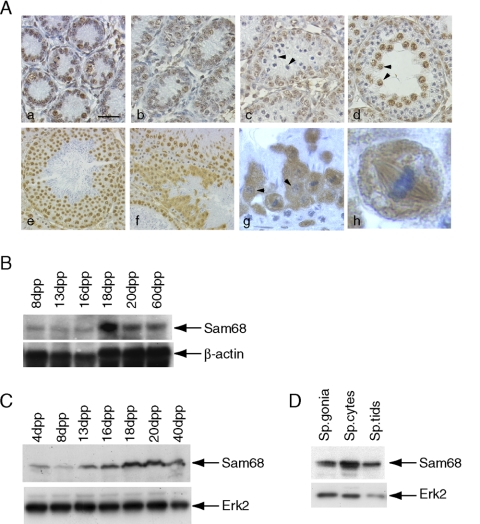
The STAR protein Sam68 is highly expressed in the adult testis (Venables et al., 1999). To investigate more accurately the pattern of expression of Sam68 during postnatal testis development, we performed an immunohistochemical analysis. Sam68 is expressed in the nuclei of both Sertoli cells and spermatogonia of 4-d post partum (dpp) and 8-dpp testes (Figure 1A, a and b). Sam68 is absent in leptotene and zygotene spermatocytes at the onset of meiosis in 12 dpp testis (Figure 1Ac; arrowheads indicate zygotene spermatocytes), whereas it accumulates again when germ cells reach the pachytene stage in 19 dpp testis (Figure 1Ad; arrowheads indicate pachytene spermatocytes). Interestingly, Sam68 displays a stage-specific expression and localization also in the adult testis (Figure 1A, e and f), with highest levels in the testicular stages approaching and following the meiotic divisions (stages XI and XII). In stage XII tubules, Sam68 is localized in the cytoplasm of metaphase I spermatocytes, in which the protein appears associated with the meiotic spindle (Figure 1A, g and h), and of secondary spermatocytes, in which the nucleus has formed but Sam68 remains mainly localized in the cytoplasm (Figure 1Ag; arrowheads indicate secondary spermatocytes). Sam68 relocalizes in the nucleus of stage I round spermatids and its expression levels begin to decrease at the onset of spermiogenesis (Figure 1Ae). The protein is completely absent in elongated spermatids (Figure 1A, e and f) and mature spermatozoa (unpublished data). Higher magnification images are available in Supplementary Figure S1. Immunofluorescence analysis of germ cells isolated by elutriation confirmed the results of the immunohistochemistry (Supplementary Figure S2).
Figure 1.
Expression of Sam68 in mouse postnatal testis. (A) Immunohistochemistry of testis at different developmental stages: in 4 dpp testis (a) and in 8 dpp testis (b), Sam68 is expressed in both the somatic Sertoli cells and the spermatogonia, the only germ cells present at this stage; in 12-dpp testis (c) Sam68 is still expressed by mitotic spermatogonia but it is absent in the first meiotic cells that appear in the seminiferous tubule, the leptotene and zygotene (indicated by arrowheads) spermatocytes; in 19-dpp testis, Sam68 accumulates again in large pachytene spermatocytes (indicated by arrowheads) approaching the meiotic divisions. In adult testis, Sam68 is most abundant in stage XI (e) and stage XII (f) tubules: it shows nuclear localization in pachytene spermatocytes and round spermatids (e) and cytoplasmic localization in metaphase and secondary spermatocytes (f and g). At metaphase, Sam68 appears localized on the meiotic spindle (g, and inset in h). Scale bar, (a– d) 30 μm, (e and f) 45 μm, (g) 15 μm, and (h) 3 μm. (B) Northern blot analysis of Sam68 mRNA in developing testis. Total RNA, 30 μg, extracted from mouse testis at different ages after birth were separated on 1.5% agarose gels, transferred onto nylon membranes, and probed with either labeled Sam68 (top panel) or actin (bottom panel) cDNAs. (C) Western blot analysis of Sam68 protein in developing testis. of cell extracts, 10 μg, from mouse testis at different ages after birth were analyzed by Western blot with a mouse monoclonal anti-Sam68 antibody (top panel). To determine that equal proteins were loaded, the blot was also probed with anti-Erk2 antibody (bottom panel). (D) Western blot analysis of Sam68 in isolated germ cells using the same antibodies as in C shows that Sam68 is most abundant in pachytene spermatocytes.
Northern blot analysis of total RNA obtained from testes at different ages showed a progressive increase of Sam68 mRNA in prepuberal mice, with a peak of expression at 18 dpp (Figure 1B), when late pachytene spermatocytes are the most abundant population of germ cells in the seminiferous tubules and the meiotic divisions begin (Bellve et al., 1977).
Next, we examined the expression of Sam68 in testicular and germ cell extracts by Western blot analysis. By using a monoclonal antibody, we observed a progressive increase of Sam68 expression during postnatal testicular development with a peak around age 18 –20 dpp (Figure 1C, top panel), which paralleled the expression of Sam68 mRNA. A similar result was obtained by using a polyclonal antibody for Sam68 (unpublished data). When the expression of Sam68 was analyzed in isolated germ cells, we observed that the protein is expressed at highest levels in pachytene spermatocytes and it is reduced in spermatogonia and round spermatids (Figure 1D). Hence, the expression of Sam68 peaks when meiotic germ cells approach the cell divisions.
Sam68 Translocates to the Cytoplasm and Associates with Polysomes during the Meiotic Divisions
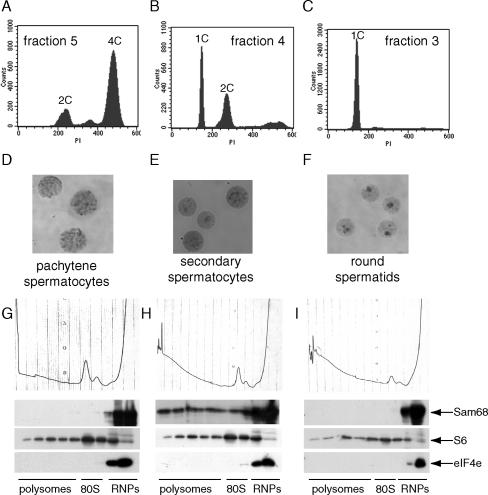
To investigate the subcellular localization of Sam68 in more detail, we isolated enriched populations of germ cells at different stages of differentiation. By using the elutriation procedure to separate cells from 24- to 28-dpp testes, we obtained fractions in which the population consisted of ∼80% primary spermatocytes, as determined by the 4C DNA content (FACS analysis in Figure 2A), or 95% haploid spermatids with a 1C DNA content (Figure 2C). An intermediate fraction with 70% haploid cells (1C) and 30% diploid cells with a 2C DNA content (Figure 2B) was also obtained. Air-dried nuclear preparations of these fractions showed that 4C cells were mainly pachytene spermatocytes (Figure 2D), whereas haploid cells were round spermatids (Figure 2F). Interestingly, the intermediate fraction contained round spermatids and secondary spermatocytes (Figure 2E), which show cytoplasmic staining of Sam68 (Figure 1Ag; Supplementary Figures S1 and S2).
Figure 2.
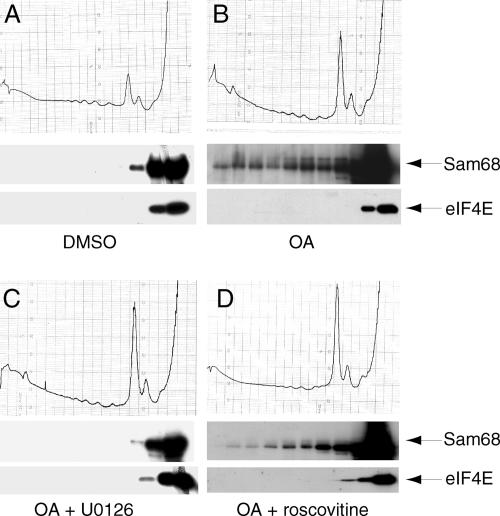
Sam68 cosediments with the polysomes in secondary spermatocytes. Germ cells isolated from mouse testis by elutriation were analyzed for DNA content by FACS. Cells from fraction 5 (A), 4 (B), or 3 (C) were fixed in 1% paraformaldehyde and stained with propidium iodide for the analysis. Nuclei of cells obtained from the same fractions were processed for cytological analysis and stained with Giemsa (D–F). Fractionation on sucrose gradients of cell extracts obtained from fraction 5 (G), or 4 (H), or 3 (I): absorbance profiles at 254 nm show the distribution of soluble RNP, single ribosome and polysomes (top panels); Western blot analyses of each fraction from the gradients indicate the distribution of Sam68 (second row panels), of the ribosomal S6 protein (third row panels), used as standard of distribution of the ribosome (80S) and the polysomes, and of initiation factor eIF4E (bottom panels), used as standard of distribution of smaller RNPs.
Sam68 is mainly nuclear but it has been reported to affect cytoplasmic utilization of viral RNA (Coyle et al., 2003). To test whether translocation of Sam68 to the cytoplasm during the meiotic divisions is associated with a function of this RNA-binding protein in mRNA translation, we performed fractionations of cell extracts by sucrose gradients (Zalfa et al., 2003). As shown by the 254-nm absorbance profiles (Figure 2, G–I), this sedimentation allows separation of the RNP complexes from the entire ribosome (80S) and from polysomes engaged in translation. Western blot analysis of each single fraction of the gradients revealed that Sam68 associates only with the RNPs in primary spermatocytes and haploid round spermatids (Figure 2, G and I), when the protein resides in the nucleus. However, in secondary spermatocytes, in which the protein is localized in the cytoplasm, a substantial portion of Sam68 is associated with the polysomes (Figure 2H). The quality of the fractionation procedures was confirmed by the absorbance profiles as well as by the distribution of the ribosomal protein S6 (in fractions containing the small ribosomal subunit, the 80S and the polysomes) and of the initiation factor eIF4E (exclusively in the RNP fractions; Figure 2, G–I).
Phosphorylation of Sam68 Correlates with Cytoplasmic Localization during the Meiotic Divisions
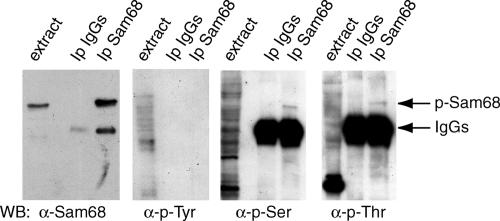
Nucleo-cytoplasmic shuttling of proteins is often regulated by phosphorylation events. Because Sam68 is phosphorylated by several kinases (Lukong and Richard, 2003), we set out to investigate if translocation of Sam68 to the cytoplasm and its association with polysomes during the meiotic divisions required phosphorylation. Sam68 was immunoprecipitated from the population enriched in secondary spermatocytes and analyzed by Western blot with different anti-phospho-residue antibodies. Figure 3 shows that Sam68 is phosphorylated in ser/thr residues, but not in tyrosines, indicating that the protein is posttranslationally modified during the meiotic divisions.
Figure 3.
Sam68 is phosphorylated in serine and threonine in secondary spermatocytes. Cells from fraction 4 of the elutriation were homogenized in the presence of 1% Triton X-100 and soluble extracts were precleared on protein A-Sepharose beads before using them for immunoprecipitation with either preimmune IgGs or rabbit anti-Sam68 IgGs preadsorbed to protein A-Sepharose beads. Cell extracts and immunoprecipitated proteins were analyzed by Western blot using rabbit anti-Sam68 (first panel), mouse anti-phospho-tyrosine (second panel), rabbit anti-phosphothreonine (third panel), or rabbit anti-phosphoserine (forth panel) antibodies. The bands corresponding to Sam68 and the rabbit IgGs (not detected by the anti-mouse IgGs used in the second panel) are pointed by arrows on the right side of the figure.
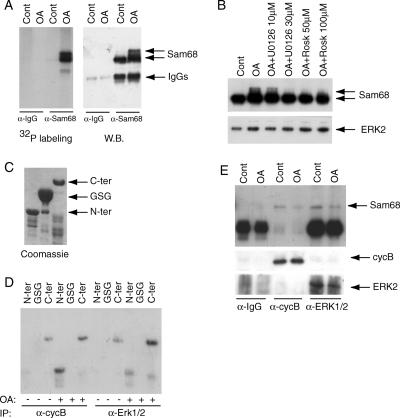
Treatment of pachytene spermatocytes with the phosphatase inhibitor OA in culture induces their progression into metaphase I (MI), characterized by normal condensation of bivalent chromosomes and synaptonemal complex breakdown (Wiltshire et al., 1995). To determine if Sam68 is phosphorylated when pachytene spermatocytes enter the meiotic divisions, we performed an in vivo metabolic labeling with 32P-orthophosphate. Immunoprecipitation of Sam68 from labeled spermatocytes followed by autoradiography demonstrated that the protein is strongly phosphorylated upon OA-induced entry into MI (∼50% under these conditions; Di Agostino et al., 2002). Moreover, phosphorylation caused a distinctive shift in the electrophoretic mobility of the protein (Figure 4A).
Figure 4.
Sam68 is phosphorylated upon entry into metaphase of primary spermatocytes. (A) Pachytene spermatocytes were cultured in phosphate-free minimal essential medium and carrier-free [32P]orthophosphate (0.3 mCi/ml), for 2 h,, and then treated with 0.5 μM OA or DMSO for 6 h. At the end of the incubation, cell extracts were immunoprecipitated with rabbit preimmune or anti-Sam68 IgGs. Autoradiography (left panel) reveals that Sam68 is strongly phosphorylated upon OA-induced entry into MI, but not in pachytene spermatocyte (control) and phosphorylation caused an electrophoretic mobility shift of the protein as revealed by Western blot analysis (right panel). No phosphorylated bands were precipitated by preimmune IgGs. (B) Pachytene spermatocytes were pretreated with selective inhibitors of MPF (roscovitine) or of MAPK pathway (U0126) at different concentration before the incubation with OA. Equal amounts of proteins were loaded on the SDS PAGE as revealed by the Erk2 antibody (bottom panel). (C) Coomassie blue staining of a SDS-PAGE with the purified GST-fusion proteins used for the kinase assay. (D) MPF and MAPKs were isolated by immunoprecipitation with anti-cyclin B1 or anti-ERK1/2 antibodies from pachytene and MI spermatocytes. Chimeric GST-Sam68(N-ter), GST-Sam68-(GSG), and GST-Sam68-(C-ter) were used as substrates for MPF and MAPKs in an immunokinase assays in vitro. Autoradiography reveals that MPF from MI spermatocytes phosphorylated both the N- and C-terminal regions of Sam68 whereas ERK1/2 mainly phosphorylated the C-terminal region. Neither kinase phosphorylated the GSG RNA-binding domain in this assay. (E) Western blot analysis of the IgG, cyclin B1 and Erk1/2 immunoprecipitates from the kinase assay (D) probed with anti-Sam68 antibody: Sam68 binds both cyclin B1 and Erk1/2, but this interaction is inhibited by OA stimulation.
It has been described that OA-induced meiotic progression requires activation of the maturation promoting factor (MPF; Wiltshire et al., 1995), a complex of cyclin-dependent kinase cdc2 and the regulatory subunit cyclin B1, and of the extracellular signal-regulated protein kinases (ERKs; Sette et al., 1999), also known as the mitogen-activated protein kinases (MAPKs). To determine if MPF or ERKs were responsible for the phosphorylation of Sam68, pachytene spermatocytes were pretreated with selective inhibitors of these kinases before the incubation with OA. Using the electrophoretic mobility shift to monitor phosphorylation of Sam68, it was observed that both the MAPK-pathway inhibitor U0126 and the MPF inhibitor roscovitine inhibited phosphorylation (Figure 4B), indicating that both kinases contribute to the phosphorylation of Sam68 during the meiotic progression. Measurements of MAPK and MPF activity in immunoprecipitates from these cell extracts confirmed that they were selectively inhibited by these inhibitors (Supplementary Figure S3).
ERKs and MPF Isolated from Mouse Spermatocytes Directly Phosphorylate Sam68
To determine if the phosphorylation of Sam68 during the meiotic progression was due to the direct action of MPF and MAPKs, the kinases were isolated by immunoprecipitation with anti-cyclin B1 or anti-ERK1/2 antibodies from pachytene and MI spermatocytes. The GSG RNA-binding domain (aa 96 –275) of Sam68 is flanked by an N-terminal (aa 1–95) and C-terminal (aa 276–443) regions that are known to participate to protein-protein interactions and posttranslational modifications (Lukong and Richard, 2003). Chimeric GST-Sam68(N-ter), GST-Sam68(GSG), and GST-Sam68(C-ter) were produced in E. coli and purified proteins (Figure 4C) were used as substrates for immunokinase assays in vitro. As shown in Figure 4D, MPF from MI spermatocytes (treated with OA) phosphorylated both the N- and C-terminal regions of Sam68, whereas ERK1/2 mainly phosphorylated the C-terminal region. Neither kinase phosphorylated the GSG RNA-binding domain in this assay, whereas both kinases showed reduced or minimal activity when they were isolated from pachytene spermatocytes (Figure 4D). Beads from control immunoprecipitation using preimmune IgGs were inactive in this assay (unpublished data). Interestingly, coimmunoprecipitation experiments indicated that Sam68 was specifically associated with both cyclin B1 and ERKs in pachytene spermatocytes and that the interaction was weakened upon MI entry (Figure 4E). These results suggest that Sam68 may dissociate from the complexes with MPF and ERKs upon phosphorylation.
Phosphorylation of Sam68 Triggers Its Translocation to the Polysomes
Sam68 is phosphorylated by ERKs and MPF, and it cosediments with the polysomes during the meiotic divisions. To functionally correlate these events, we studied the localization of Sam68 on polysomal gradients during the OA-induced meiotic progression in cultured spermatocytes. Cell extracts from pachytene or OA-induced MI spermatocytes were fractionated on sucrose gradients, and each fraction was analyzed by Western blot. Similarly to what observed in secondary spermatocytes isolated directly from testis, Sam68 translocates from the RNP fractions to those containing the polysomes upon OA-induced meiotic progression (Figure 5, A and B).
Figure 5.
Sam68 translocates to the polysomes during the G2/M transition of primary spermatocytes. Pachytene spermatocytes were treated for 4 h with either DMSO (control) (A) or 0.5 μM OA alone (B) or after pretreatment with 30 μM U0126 (C) or 100 μM roscovitine (D). Absorbance profiles are shown in the top panels, whereas Western blot analyses of each fraction are shown below. Middle panels show the distribution of Sam68, and bottom panels show the distribution of eIF4E.
To determine if phosphorylation was required for the translocation of Sam68, pachytene spermatocytes were preincubated with either MPF or MAPK inhibitors before induction with OA. Sucrose gradient fractionations indicated that inhibition of ERKs by U0126 blocked the translocation of Sam68 to the “heavy” polysomes, and the protein remained stalled either on the single ribosome or with the RNPs (Figure 5C). Inhibition of MPF by roscovitine also partially blocked OA-induced translocation of Sam68 to the polysomes fractions (Figure 5D). These results indicate that phosphorylation is required for translocation of Sam68 to the fractions containing polysomes in cultured spermatocytes.
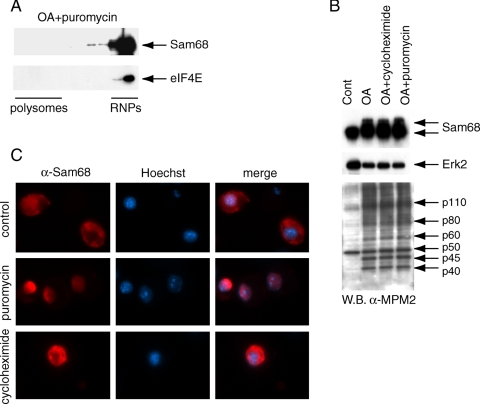
The shift of Sam68 to the polysomal fractions during the meiotic divisions may indicate its association with the translational machinery or cosedimentation due to association with another multimolecular complex. To distinguish between these events, we treated pachytene spermatocytes with OA in the presence of puromycin, which blocks the elongation step of translation and causes premature disassembling of polysomes from the mRNAs (Davis et al., 1974). Interestingly, under these conditions, OA was unable to induce the translocation of Sam68 (Figure 6A), even though it did not block phosphorylation of Sam68 nor the general pattern of OA-induced protein phosphorylation (Figure 6B). Remarkably, we found that treatment of secondary spermatocytes with puromycin caused the return of Sam68 in the nucleus (Figure 6C), whereas treatment with cycloheximide, which freezes the translational machinery without disassembling the polysomes (Stefani et al., 2004), did not alter cytoplasmic localization of the protein in these cells (Figure 6C). These data indicate that cytoplasmic localization of Sam68 requires its association with the translational machinery.
Figure 6.
Translocation of Sam68 to the cytoplasm requires assembled polysomes. (A) Pachytene spermatocytes were incubated for 4 h with 0.5 μM OA in the presence of 100 μg/ml puromycin. Cell extracts were fractionated on sucrose gradients as in Figure 5 and each fraction of the gradient was analyzed in Western blot by using the anti-Sam68 (top panel) or anti-eIF4E (bottom panel) antibody. (B) Pachytene spermatocytes were incubated for 4 h with 0.5 μM OA in the presence or absence of 1 mM of puromycin or of 100 μg/ml cycloheximide. Cell extracts were analyzed in Western blot by using the anti-Sam68 (top panel), anti-ERK2 (middle panel), or the MPM2 (bottom panel) antibody, which recognizes mitotic-type phosphorylations. (C) Immunofluorescence analysis of Sam68 (rabbit polyclonal antibody, 1:500) in secondary spermatocytes cultured for 4 h in the presence of methanol (control) or of 1 mM of puromycin or of 100 μg/ml cycloheximide.
Phosphorylation by ERKs and MPF Does Not Impair the Binding of Sam68 to Synthetic RNA
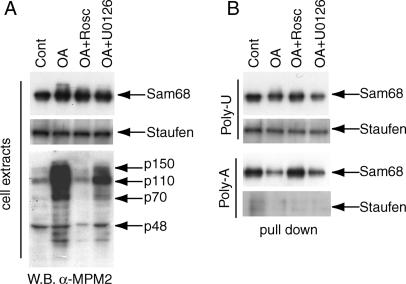
Tyrosine phosphorylation by Src-like kinases decreases the affinity of Sam68 for synthetic poly-U oligonucleotides (Wang et al., 1995). To test whether the meiotic ser/thr phosphorylation affected the RNA-binding affinity of Sam68 by similar assays, we performed pulldown experiments using poly-U- and poly-A-Sepharose beads. Pachytene spermatocytes were cultured for 6 h with OA in the presence or absence of either roscovitine or U0126. Cell extracts were incubated with either poly-U- or poly-A-Sepharose beads and bound proteins were analyzed by Western blot. Sam68 from pachytene spermatocyte extracts efficiently binds to poly-U and poly-A beads and treatment with OA only slightly decreased the affinity of Sam68 for the synthetic RNAs. The kinase inhibitors U0126 did not significantly affect the interaction, whereas roscovitine partially reverted the effect of OA (Figure 7). When the same samples were probed for Staufen, another RNA-binding protein expressed in spermatocytes (Saunders et al., 2000), we found that OA did not modulate its electrophoretic mobility nor its binding to poly-U-agarose beads, indicating that the modulation of the affinity of Sam68 was somewhat specific. Staufen did not bind to poly-A-agarose beads.
Figure 7.
Binding of Sam68 to synthetic RNA is not prevented by ERK- and MPF-mediated phosphorylation. Isolated pachytene spermatocytes were incubated for 4 h in the absence (control) or presence of 0.5 μM OA. Some of the cells were also pretreated with either 30 μM U0126 or 50 μM roscovitine to prevent activation of ERKs or MPF, respectively. (A) A portion of the cell extracts was used for Western blot analysis of Sam68 (top panel), Staufen (middle panel), or the MPM2 antibody (bottom panel). (B) Cell extracts from A were incubated with poly-U- or poly-A-agarose beads and proteins bound to synthetic RNA were analyzed by Western blot using the anti-Sam68 (first and third panels) or anti-Staufen antibody (second and fourth panel).
These results suggest that ser/thr phosphorylation of Sam68 modulates, rather than impairs, its ability to bind RNA.
Identification of the mRNAs That Are Targets of Sam68 in Primary Spermatocytes
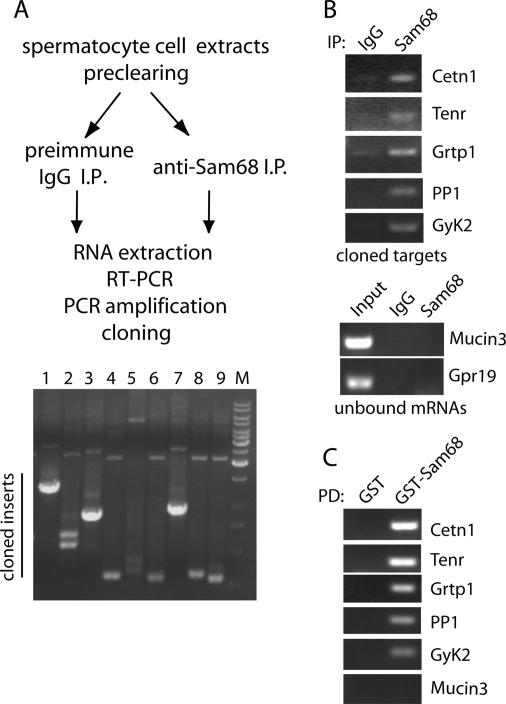
To identify the cellular targets of Sam68 in primary spermatocytes, the protein was immunoprecipitated with the anti-Sam68 antibody from cell extracts prepared under conditions that preserve RNA and RNPs integrity (Zalfa et al., 2003). RNA was extracted from the immunoprecipitates, subcloned into a plasmid, and transformed in E. coli (Figure 8A). Direct sequencing of the clones obtained revealed five potential target mRNAs of Sam68 (Table 1). Interestingly, two of these targets encode proteins that play a crucial role in mouse spermatogenesis: the RNA-binding protein TENR and the protein phosphatase PP1-gamma (Varmuza et al., 1999; Connolly et al., 2005). A third target, Centrin-1, is a testis-specific isoform of Centrin, a centrosomal protein which is required for formation of motile apparatus of spermatids in Marsilea vestita and whose expression is regulated at the translational level during gametogenesis (Klink and Wolniak, 2001). The potential role played by the other two targets (Grtp1 and GyK2) during spermatogenesis is currently unknown. To confirm the interaction of these targets with Sam68, we performed a similar immunoprecipitation experiment followed by RT-PCR using specific oligonucleotides for the mRNAs identified. All targets were amplified from the RNAs extracted from anti-Sam68 immunoprecipitates but not from the control immunoprecipitates (Figure 8B). On the contrary, other mRNAs highly expressed in primary spermatocytes like mucin3 and Gpr19 (Rossi et al., 2004), were not amplified by either immunoprecipitated samples (Figure 8B). To test whether interaction with the targets was direct, we used purified GST-Sam68 and total RNA purified primary spermatocytes in pulldown experiments coupled to RT-PCR analysis. As shown in Figure 8C, all targets efficiently bound to GST-Sam68 but not to GST alone, indicating that no additional RNA-binding proteins were required for interaction of Sam68 with its targets in vitro.
Figure 8.
Identification of target mRNAs for Sam68 from mouse primary spermatocytes. (A) Schematic representation of the experimental procedure used (see Materials and Methods for details). The panel at the bottom shows a representative analysis by PCR of the clones obtained. Clones containing an insert larger than 500 base pairs (for example, lanes 1, 3, and 7 in the figure) were submitted to sequence analysis for identification. Out of 30 clones analyzed, we identified five potential targets of Sam68 (Table 1); the remaining clones contained self-ligated vector. (B) Coimmunoprecipitation of Sam68 with target mRNAs. Spermatocyte cell extracts were immunoprecipitated with either control IgGs or anti-Sam68 IgGs, RNA was extracted and used for RT-PCR (20 cycles) with oligonucleotides specific for Centrin 1 (Cetn1), Tenr, Grtp1, PP1, and glycerol kinase 2 (GyK2). As control we used oligonucleotides for mRNAs highly expressed in spermatocytes (mucin3 and Gpr19) but not found in the screen for Sam68 targets. The panels show agarose gels with the amplified products stained with ethidium bromide. (C) Pulldown assay of the Sam68 target mRNAs. Total spermatocyte RNA, 20 μg, was incubated with 2 μg of either GST or GST-Sam68 preadsorbed to glutathione-agarose beads. Bound RNAs were extracted and analyzed by RT-PCR as described in B.
Table 1.
Description of the mRNAs bound to Sam68 in mouse primary spermatocytes
| Target mRNA | Description | Role in spermatogenesis |
|---|---|---|
| Centrin 1 (Ctn1) NM_007593 | Centrosome component; Ca2+ binding protein of 172 aa. | Not reported in mammals. Centrin is required for the formation of motile apparatus of spermatids in Marsilea vestita (Klink and Wolniak, 2001). |
| Testis nuclear RNA-binding protein (Tenr) NM_009350 | RNA-binding protein expressed only in male germ cells from mid-pachytene to elongating spermatids. | Required for normal spermiogenesis in mice. Tenr—/— sperm show decreased motility and malformed heads (Connolly et al., 2005). |
| GH-regulated TBC protein 1 (Grtp1) AF329833 | Putative Rab-like GTPase activator of 258 aa, novel gene up-regulated by growth hormone. | Its expression is most abundant in testis, it increases postpuberally. Function unknown (Lu et al., 2001). |
| Protein phosphatase type 1 (PP1, gamma isoform) AK152625 | Serine-threonine phosphatase involved in several cellular functions (i.e., cell cycle regulation, apoptosis) | PP1-gamma—/— male mice are infertile (meiotic defects, impaired spermiogenesis; Varmuza et al., 1999). |
| Glycerol kinase 2 (GyK2) BC061147 | Involved in carbohydrate and lipid metabolism. Encoded by an autosomal retrotrasposon gene, testis specific expression. (Pan et al., 1999) | Unknown. |
DISCUSSION
Sam68 is a RNA-binding protein reported to act at several steps in RNA metabolism, such as splicing, export, and cytoplasmic utilization of mRNAs (Lukong and Richard, 2003). However, its precise cellular function and putative mRNA targets are still obscure. Because of the nuclear localization of Sam68, its potential role/s in the regulation of translation remains the most elusive. Herein, we have demonstrated that Sam68 translocates to the cytoplasm in a regulated manner during the meiotic divisions. Sam68 remains in the cytoplasm of secondary spermatocytes and it is found associated with the polysomes engaged in translation. This association can be recapitulated by triggering meiotic progression of cultured pachytene spermatocytes and we have demonstrated that it requires phosphorylation of Sam68 by MPF and ERKs. Notably, all the experiments in this work were performed with endogenous Sam68 and interacting kinases from primary cultures of mouse spermatocytes, and did not involve overexpression of proteins. Our results suggest for the first time that phosphorylation regulates a cell cycle-dependent translocation of Sam68 from the nucleus to the cytoplasm where it associates with the translational machinery.
The association of Sam68 with the polysomes in primary spermatocytes induced to enter the first metaphase and in secondary spermatocytes was observed by the cosedimentation after sucrose gradient fractionation of cell extracts. The possibility exists that upon disassembly of the nuclear envelope, Sam68 forms a multimolecular complex that cosediments with the polysomes but it is distinct from it. However, we have demonstrated that inhibition of polysome formation by puromycin prevents the translocation of Sam68 to the heavy fractions of the gradients without affecting its phosphorylation. Moreover, the localization of Sam68 in the cytoplasm of secondary spermatocytes was also impaired by the treatment with puromycin, and the protein returned into the nucleus. These results strongly suggest that localization of Sam68 in the cytoplasm during the meiotic divisions requires the association of the protein with active polysomes. Hence, the cytoplasmic localization of Sam68 in secondary spermatocytes might be due to its sequestration on the translational machinery rather than to active nuclear export. It is possible that Sam68 allows the accumulation of mRNAs for factors required to complete the second meiotic division of secondary spermatocytes, which is very rapid and does not have canonical G1 and S phases. These mRNAs would be recruited to the translational machinery as a consequence of Sam68 phosphorylation by MPF and ERKs at metaphase of the first meiotic division. In line with this hypothesis, we found that Sam68 binds the mRNA for PP1-gamma, a protein required for normal meiotic divisions and spermiogenesis, as demonstrated by the knockout mouse model (Varmuza et al., 1999).
Our demonstration that Sam68 is capable of shuttling between nuclear RNPs and polysomes may help explain some of the effects attributed to Sam68 on the cytoplasmic utilization of target mRNAs. Although the data in the literature have been obtained with infected viral mRNAs (McBride et al., 1996; Coyle et al., 2003), they may also apply to cellular targets that are physiologically regulated by Sam68. Relocalization of Sam68 to the cytoplasm was also obtained by infection of HeLa cells with poliovirus (McBride et al., 1996), and it was shown that Sam68 interacted with the viral RNA-dependent RNA polymerase in the cytoplasm. The mechanism by which the RNA polymerase attracts Sam68 in the cytoplasm was not addressed in that study; however, in line with our results it is possible to hypothesize that it sequesters Sam68 during its cell cycle-dependent journey to the polysomes.
Sam68, like other STAR proteins, has been proposed as a link between signal transduction pathways and the regulation of RNA metabolism. Through five proline-rich sequences located upstream and downstream of the GSG RNA-binding domain, Sam68 can interact with signaling molecules containing SH3 or WW domains (Lukong and Richard, 2003). The in vivo function of Sam68 is still unknown but genetic disruption of its gene in chicken fibroblasts resulted in defective growth due to delayed G2/M progression (Li et al., 2002b). Interestingly, a recent RNAi screen for genes required for HeLa cells G2/M progression has uncovered the role of several RNA-binding proteins in mitotic spindle assembly (Kittler et al., 2004). Although Sam68 was not found in this screen, our observation that it associates with the meiotic spindle at metaphase suggests participation of this RNA-binding protein to RNA-mediated spindle functions. Herein we identify the first cellular mRNAs bound by endogenous Sam68. Interestingly, two of the five targets identified encode for proteins involved in spindle assembly during the G2/M transition: the centrosomal protein Centrin 1 and the protein phosphatase PP1-gamma (Tournebize et al., 1997). It will be interesting to determine if these mRNAs, or the corresponding somatic isoforms, are target of Sam68 also in mitotic cells and if their expression is regulated posttranscriptionally by this RNA-binding protein.
It has been demonstrated that Sam68 is regulated at the posttranslational level by cell cycle-dependent phosphorylation in serine, threonine, and tyrosine residues. Phosphorylation of Sam68 is strongly increased upon entry into mitosis and this protein was initially identified as the main substrate of the tyrosine kinase Src in mitosis (Fumagalli et al., 1994; Taylor and Shalloway, 1994). Tyrosine phosphorylation by Src and Fyn decreased the affinity of Sam68 for bound poly-U RNA oligonucleotides (Wang et al., 1995). Moreover, overexpression of constitutively active Fyn facilitates accumulation of Sam68 in the nucleus of transfected cells (Paronetto et al., 2003). Metabolic labeling of HeLa and NIH3T3 cells with 32P-orthophosphate highlighted a cell cycle-dependent phosphorylation of Sam68 also in serine and threonine residues. During the G1 and S phases of the cycle, Sam68 is preferentially phosphorylated in serine residues by unknown kinases, whereas during the G2/M progression it is strongly phosphorylated in threonine residues (Resnick et al., 1997). The main kinase responsible for mitotic threonine phosphorylation was MPF, although some residual activity could be ascribed to other unknown kinases. The metabolic labeling of mouse spermatocytes reported here shows that Sam68 is strongly phosphorylated also during the meiotic G2/M transition. Furthermore, MPF appears to be one of the responsible kinases in primary spermatocytes; in addition, we show that ERKs are also required for meiotic phosphorylation of Sam68 in male germ cells. Our results indicate that phosphorylation by both kinases is required for the translocation of Sam68 from the RNPs to the polysomes during meiotic divisions.
It was previously reported that mitotic phosphorylation of Sam68 by MPF did not alter its ability to bind synthetic RNA (Resnick et al., 1997). On the other hand, we have observed that a mild but significant phosphorylation-dependent change in RNA affinity occurs during cell cycle progression of primary spermatocytes and it is blocked by inhibition of MPF. To highlight this slight decrease in binding to poly-U or poly-A beads after meiotic phosphorylation, we needed to use small amount of total cell extracts (∼100 μg). This observation suggests that, unlike tyrosine phosphorylation, serine/threonine phosphorylation might tune, rather than impair, the ability of Sam68 to interact with RNA. This modulation could possibly loosen the interaction with target RNAs and favor the access to other RNA-binding proteins. A hint in this direction comes from the demonstration that phosphorylation of Sam68 by ERKs in response to signaling pathways modulates alternative splicing of a transfected minigene (Matter et al., 2002). Similarly to our results with synthetic RNA, it was shown that ERKs did not inhibit binding to the RNA target used, but it rendered a hidden donor site accessible for splicing. Because phosphorylation of Sam68 by ERKs and MPF is required for its interaction with the polysomes in meiosis, we suggest that posttranslational modification of Sam68 may affect translational efficiency of target mRNAs in addition to alternative splicing. Although phosphorylation of Sam68 by ERKs in mitosis has not yet been reported, our results indicate that it could occur under conditions in which these kinases are activated at the G2/M transition in somatic cells, such us after irradiation (Dent et al., 2003).
Mammalian spermatogenesis is under the control of several RNA-binding proteins that participate to posttranscriptional control of gene expression. Some of these proteins are localized to the cytoplasm, like Miwi, Mili, and MSY2 (Deng and Lin, 2002; Kuramochi-Miyagawa et al., 2004; Yang et al., 2005, Yang et al., 2005) and others are nuclear, like Tenr (Connolly et al., 2005), indicating that regulation of mRNA metabolism at several steps is crucial for the differentiation of male germ cells. Remarkably, Tenr is one of the cellular mRNA target of Sam68 in primary spermatocytes, suggesting that Sam68 may regulate its expression posttranscriptionally. It is important to mention that in lower organisms like C. elegans and D. melanogaster gametogenesis is regulated by several RNA-binding proteins that control each other at the translational level to ensure the correct progression of the developmental program (Kuersten and Goodwin, 2003). It will be of interest to determine if Sam68 is part of a similar program of translational control of gene expression in mammalian gametogenesis.
Sam68 accumulates in pachytene spermatocytes and round spermatids, cells displaying very high transcriptional activity. This observation suggests that one of the functions of Sam68 in meiosis is to allow the storage of mRNAs when the genome is suitable for high levels of transcription and they are accumulated for the later differentiation stages of spermatogenesis. Sam68 is also expressed by mitotic spermatogonia, but it is down-regulated at the onset of meiosis and it is completely absent in zygotene spermatocytes. Because Sam68 expression is tightly regulated during spermatogenesis, we suggest that this RNA-binding protein is a new candidate that plays a crucial role during this differentiation process.
Supplementary Material
Acknowledgments
We thank Drs. Alessia Di Florio and Enrica Bianchi for preparation of the pGEX-4T1-Sam68 and pGEM-Sam68UTR vectors, Susanna Dolci for preparation of mouse spermatogonia, Julian Venables for the GFP-Sam68 vector, Juan Ortin for the Staufen antibody, Federica Capolunghi and Rita Carsetti for help with FACS analysis, and Pellegrino Rossi for helpful discussion throughout this study. The work was supported by grants from Telethon (GGP04118), AIRC, and Ministry of Education (PRIN 2004).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–06–0548) on October 12, 2005.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Bellve, A. R., Cavicchia, J. C., Millette, C. F., O'Brien, D. A., Bhatnagar, Y. M., and Dym, M. (1977). Spermatogenic cells of the prepuberal mouse. Isolation and morphological characterization. J. Cell Biol. 74, 68–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggiogera, M., Fakan, S., Leser, G., Martin, T. E., and Gordon, J. (1990). Immunoelectron microscopic visualization of ribonucleoprotein in the chromatoid body of mouse spermatids. Reprod. Dev. 26, 150 –158. [DOI] [PubMed] [Google Scholar]
- Braun, R. E. (2000). Temporal control of protein synthesis during spermatogenesis. Int. J. Androl. 23(Suppl 2), 92–94. [DOI] [PubMed] [Google Scholar]
- Cataldo, L., Mastrangelo, M. A., and Kleene, K. C. (1999). A quantitative sucrose gradient analysis of the translational activity of 18 mRNA species in testis from adult mice. Mol. Hum. Reprod. 5, 206 –213. [DOI] [PubMed] [Google Scholar]
- Connolly, C. M., Dearth, A. T., and Braun, R. E. (2005). Disruption of murine Tenr results in teratospermia and male infertility. Dev. Biol. 278, 13–21. [DOI] [PubMed] [Google Scholar]
- Coyle, J. H., Guzik, B. W., Bor, Y. C., Jin, L., Eisner-Smerage, L., Taylor, S. J., Rekosh, D., and Hammarskjold, M. L. (2003). Sam68 enhances the cytoplasmic utilization of intron-containing RNA and is functionally regulated by the nuclear kinase Sik/BRK. Mol. Cell. Biol. 23, 92–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, B. D., Tai, P-C., and Wallacep, B. J. (1974). Complex interactions of antibiotics with the ribosome. In: Ribosomes, A.T.M. Nomura, P. Lengyel, eds., Cold Spring Harbor, NY: Cold Spring Harbor Laboratory, 771–789.
- Deng, W., and Lin, H. (2002). miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev. Cell 2, 819–830. [DOI] [PubMed] [Google Scholar]
- Dent, P., Yacoub, A., Fisher, P. B., Hagan, M. B., and Grant, S. (2003). MAPK pathways in radiation responses. Oncogene 22, 5885–5896. [DOI] [PubMed] [Google Scholar]
- Di Agostino, S., Rossi, P., Geremia, R., and Sette, C. (2002). The MAPK pathway triggers activation of Nek2 during chromosome condensation in mouse spermatocytes. Development 129, 1715–1727. [DOI] [PubMed] [Google Scholar]
- Francis, R., Barton, M. K., Kimble, J., and Schedl, T. (1995). gld-1, a tumor suppressor gene required for oocyte development in Caenorhabditis elegans. Genetics. 139, 579–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli, S., Totty, N. F., Hsuan, J. J., and Courtneidge, S. A. (1994). A target for Src in mitosis. Nature 368, 871– 874. [DOI] [PubMed] [Google Scholar]
- Goodwin, E. B., and Ellis, R. E. (2002). Turning clustering loops: sex determination in Caenorhabditis elegans. Curr. Biol. 12, R111–R120. [DOI] [PubMed] [Google Scholar]
- Handel, M. A., and Eppig, J. J. (1998). Sexual dimorphism in the regulation of mammalian meiosis. Curr. Top. Dev. Biol. 37, 333–358. [DOI] [PubMed] [Google Scholar]
- Hansen, D., Wilson-Berry, L., Dang, T., and Schedl, T. (2004). Control of the proliferation versus meiotic development decision in the C. elegans germline through regulation of GLD-1 protein accumulation. Development 131, 93–104. [DOI] [PubMed] [Google Scholar]
- Hartmann, A. M., Nayler, O., Schwaiger, F. W., Obermeier, A., and Stamm, S. (1999). The interaction and colocalization of Sam68 with the splicing-associated factor YT521-B in nuclear dots is regulated by the Src family kinase p59(fyn). Mol. Biol. Cell 10, 3909 –3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan, E., Motzny, C. K., Graves, L. E., and Goodwin, E. B. (1999). The STAR protein GLD-1 is a translational regulator of sexual identity in Caenorhabditis elegans. EMBO J. 18, 258 –269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittler, R. et al. (2004). An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature. 432, 1036 –1040. [DOI] [PubMed] [Google Scholar]
- Kleene, K. C. (1996). Patterns of translational in the mammalian testis. Reprod. Dev. 43, 268 –281. [DOI] [PubMed] [Google Scholar]
- Klink, V. P., and Wolniak, S. M. (2001). Centrin is necessary for the formation of the motile apparatus in spermatids of Marsilea. Mol. Biol. Cell 12, 761–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales, C. R., and Hecht, N. B. (1994). Poly(A)+ ribonucleic acid s are enriched in spermatocyte nuclei but not in chromatoid bodies in the rat testis. Biol. Reprod. 50, 309 –319. [DOI] [PubMed] [Google Scholar]
- Kuersten, S., and Goodwin, E. B. (2003). The power of the 3′ UTR: translational control and development. Nat. Rev. Genet. 4, 626–637. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa et al. (2004). Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development 131, 839–849. [DOI] [PubMed] [Google Scholar]
- Lee, M. H., and Schedl, T. (2001). Identification of in vivo mRNA targets of GLD-1, a maxi-KH motif containing protein required for C. elegans germ cell development. Genes Dev. 15, 2408 –2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Liu, Y., Kim, B. O., and He, J. J. (2002a). Direct participation of Sam68, the 68-kilodalton Src-associated protein in mitosis, in the CRM1-mediated Rev nuclear export pathway. J Virol. 76, 8374–8382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q. H., Haga, I., Shimizu, T., Itoh, M., Kurosaki, T., and Fujisawa, J. (2002b). Retardation of the G2-M phase progression on gene disruption of RNA binding protein Sam68 in the DT40 cell line. FEBS Lett. 525, 145–150. [DOI] [PubMed] [Google Scholar]
- Lu, C., Kasik, J. Stephan, D. A., Yang, S., Sperling, M. A., Menon, R. K. (2001). Grtp1, a novel gene regulated by growth hormone. Endocrinology 142, 4568–4571. [DOI] [PubMed] [Google Scholar]
- Lukong, K. E., and Richard, S. (2003). Sam68, the KH domain-containing superSTAR. Biochim. Biophys. Acta 1653, 73– 86. [DOI] [PubMed] [Google Scholar]
- Marin, V. A., and Evans, T. (2003). Translational repression of a C. elegans Notch mRNA by the STAR/KH domain protein GLD-1. Development 130, 2623–2632. [DOI] [PubMed] [Google Scholar]
- Matter, N., Herrlich, P., and Konig, H. (2002). Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature 420, 691– 695. [DOI] [PubMed] [Google Scholar]
- McBride, A. E., Schlegel, A., and Kirkegaard, K. (1996). Human protein Sam68 relocalization and interaction with poliovirus RNA polymerase in infected cells. Proc. Natl. Acad. Sci. USA 93, 2296 –2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales, C. R., and Hecht, N. B. (1994). Poly(A)+ ribonucleic acids are enriched in spermatocyte nuclei but not in chromatoid bodies in the rat testis. Biol. Reprod. 50, 309 –319. [DOI] [PubMed] [Google Scholar]
- Pan, Y., Decker, W. K., Huq, A. H., and Craigen, W. J. (1999). Retrotransposition of glycerol kinase-related genes from the X chromosome to autosomes: functional and evolutionary aspects. Genomics 59, 282–290. [DOI] [PubMed] [Google Scholar]
- Paronetto, M. P., Venables, J. P., Elliott, D. J., Geremia, R., Rossi, P., and Sette, C. (2003). Tr-kit promotes the formation of a multimolecular complex composed by Fyn, PLCgamma1 and Sam68. Oncogene 22, 8707– 8715. [DOI] [PubMed] [Google Scholar]
- Resnick, R. J., Taylor, S. J., Lin, Q., and Shalloway, D. (1997). Phosphorylation of the Src substrate Sam68 by Cdc2 during mitosis. Oncogene 15, 1247–1253. [DOI] [PubMed] [Google Scholar]
- Rossi, P., Dolci, S., Albanesi, C., Grimaldi, P., and Geremia, R. (1993). Direct evidence that the mouse sex-determining gene Sry is expressed in the somatic cells of male fetal gonads and in the germ cell line in the adult testis. Reprod. Dev. 34, 369 –373. [DOI] [PubMed] [Google Scholar]
- Rossi, P., et al. (2004). Analysis of the gene expression profile of mouse male meiotic germ cells. Gene Expr. Patterns 4, 267–281. [DOI] [PubMed] [Google Scholar]
- Sassone-Corsi, P. (2002). Unique chromatin remodeling and transcriptional regulation in spermatogenesis. Science 296, 2176 –2178. [DOI] [PubMed] [Google Scholar]
- Saunders, P.T.K., Pathirana, S., Maguire, S. M., Doyle, M., Wood, T., and Bownes, M. (2000). Mouse staufen genes are expressed in germ cells during oogenesis and spermatogenesis. Mol. Hum. Reprod. 11, 983–991. [DOI] [PubMed] [Google Scholar]
- Saunders, P. T., Turner, J. M., Ruggiu, M., Taggart, M., Burgoyne, P. S., Elliott, D., and Cooke, H. J. (2003). Absence of mDazl produces a final block on germ cell development at meiosis. Reproduction 126, 589 –597. [DOI] [PubMed] [Google Scholar]
- Sette, C., Bevilacqua, A., Geremia, R., and Rossi, P. (1998). Involvement of phospholipase Cgamma1 in mouse egg activation induced by a truncated form of the C-kit tyrosine kinase present in spermatozoa. J. Cell Biol. 142, 1063–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette, C., Barchi, M., Bianchini, A., Conti, M., Rossi, P., and Geremia, R. (1999). Activation of the mitogen-activated protein kinase ERK1 during meiotic progression of mouse pachytene spermatocytes. J. Biol. Chem. 274, 33571– 33579. [DOI] [PubMed] [Google Scholar]
- Stefani, G., Fraser, C. E., Darnell, J. C., and Darnell, R. B. (2004). Fragile X mental retardation protein is associated with translating polyribosomes in neuronal cells. J. Neurosci. 24, 7272–7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, S. J., and Shalloway, D. (1994). An RNA-binding protein associated with Src through its SH2 and SH3 domains in mitosis. Nature 368, 867– 871. [DOI] [PubMed] [Google Scholar]
- Tournebize, R., Andersen, S. S., Verde, F., Doree, M., Karsenti, E., and Hyman, A. A. (1997). Distinct roles of PP1 and PP2A-like phosphatases in control of microtubule dynamics during mitosis. EMBO J. 16, 5537–5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varmuza, S., Jurisicova, A., Okano, K., Hudson, J., Boekeleide, K., and Skipp, E. B. (1999). Spermiogenesis is impaired in mice bearing a targeted mutation in the protein phosphatase 1cgamma gene. Dev. Biol. 205, 98–110. [DOI] [PubMed] [Google Scholar]
- Venables, J. P., and Eperon, I. (1999). The roles of RNA-binding proteins in spermatogenesis and male infertility. Curr. Opin. Genet. Dev. 9, 346 –354. [DOI] [PubMed] [Google Scholar]
- Venables, J. P., Vernet, C., Chew, S. L., Elliott, D. J., Cowmeadow, R. B., Wu, J., Cooke, H. J., Artzt, K., and Eperon, I. C. (1999). T-STAR/ETOILE: a novel relative of SAM68 that interacts with an RNA-binding protein implicated in spermatogenesis. Hum. Mol. Genet. 8, 959 –969. [DOI] [PubMed] [Google Scholar]
- Vernet, C., and Artzt, K. (1997). STAR, a gene family involved in signal transduction and activation of RNA. Trends Genet. 13, 479–484. [DOI] [PubMed] [Google Scholar]
- Yang, J., Medvedev, S., Yu, J., Tang, L. C., Agno, J. E., Matzuk, M. M., Schultz, R. M., and Hecht, N. B. (2005). Absence of the DNA-/RNA-binding protein MSY2 results in male and female infertility. Proc. Natl. Acad. Sci. USA. 102, 5755–5760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. L., Richard, S., and Shaw, A. S. (1995). P62 association with RNA is regulated by tyrosine phosphorylation. J. Biol. Chem. 270, 2010 –2013. [DOI] [PubMed] [Google Scholar]
- Wiltshire, T., Park, C., Caldwell, K. A., and Handel, M. A. (1995). Induced premature G2/M transition in pachytene spermatocytes includes events unique to meiosis. Dev. Biol. 169, 557–567. [DOI] [PubMed] [Google Scholar]
- Wu, J., Zhou, L., Tonissen, K., Tee, R., and Artzt, K. (1999). The quaking I-5 protein (QKI-5) has a novel nuclear localization signal and shuttles between the nucleus and the cytoplasm. J. Biol. Chem. 274, 29202–29210. [DOI] [PubMed] [Google Scholar]
- Yang, J., Medvedev, S., Yu, J., Tang, L. C., Agno, J. E., Matzuk, M. M., Schultz, R. M., and Hecht, N. B. (2005). Absence of the DNA-/RNA-binding protein MSY2 results in male and female infertility. Proc. Natl. Acad. Sci. USA 102, 5755–5760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalfa, F., Giorgi, M., Primerano, B., Moro, A., Di Penta, A., Reis, S., Oostra, B., and Bagni, C. (2003). The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell 112, 317–327. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.